Abstract
Poly(A) polymerase (PAP) plays an essential role in polyadenylation of mRNA precursors, and it has long been thought that mammalian cells contain only a single PAP gene. We describe here the unexpected existence of a human PAP, which we call neo-PAP, encoded by a previously uncharacterized gene. cDNA was isolated from a tumor-derived cDNA library encoding an 82.8-kDa protein bearing 71% overall similarity to human PAP. Strikingly, the organization of the two PAP genes is nearly identical, indicating that they arose from a common ancestor. Neo-PAP and PAP were indistinguishable in in vitro assays of both specific and nonspecific polyadenylation and also endonucleolytic cleavage. Neo-PAP produced by transfection was exclusively nuclear, as demonstrated by immunofluorescence microscopy. However, notable sequence divergence between the C-terminal domains of neo-PAP and PAP suggested that the two enzymes might be differentially regulated. While PAP is phosphorylated throughout the cell cycle and hyperphosphorylated during M phase, neo-PAP did not show evidence of phosphorylation on Western blot analysis, which was unexpected in the context of a conserved cyclin recognition motif and multiple potential cyclin-dependent kinase (cdk) phosphorylation sites. Intriguingly, Northern blot analysis demonstrated that each PAP displayed distinct mRNA splice variants, and both PAP mRNAs were significantly overexpressed in human cancer cells compared to expression in normal or virally transformed cells. Neo-PAP may therefore be an important RNA processing enzyme that is regulated by a mechanism distinct from that utilized by PAP.
Posttranscriptional modification of pre-mRNA is a complex and tightly regulated process that is essential for the life of the cell. Polyadenylation of the 3′ end of mRNA, in the simplest view, is a two-step process involving cleavage of pre-mRNA and addition of the poly(A) tail. However, this event requires the coordinated interactions of at least a dozen different polypeptides (reviewed in references 4, 20, 30, and 41). One of these factors is poly(A) polymerase (PAP, also termed polynucleotide adenylyltransferase), a single-subunit enzyme that catalyzes 3′ poly(A) synthesis and also contributes to the endonucleolytic cleavage step. PAP gains site specificity by interacting with the multisubunit factors cleavage stimulation factor (CstF) and cleavage/polyadenylation specificity factor (CPSF), which recognize the 3′ G-U-rich element and the AAUAAA signal sequence in pre-mRNA, respectively. Successful polyadenylation of almost all eukaryotic mRNAs is required for the trafficking of mRNA from the nucleus to the cytoplasm (for an example, see reference 13), for enhancing the efficiency of translation (28), and for regulating mRNA degradation (39).
PAP is classified as a template-independent polymerase, a category formerly shared only by terminal deoxynucleotidyl transferase. The functional domains of PAP have been studied extensively (18, 24, 44), and the crystal structures of yeast and bovine PAP complexed with ATP have been recently solved (1, 19). There has been considerable evolutionary conservation of the amino acid sequence of the N-terminal catalytic domain, with extensive similarities from yeast to human. In vertebrates, the conserved structure of PAP also includes two bipartite nuclear localization signals (NLS) surrounding an S-T-rich C-terminal domain (CTD) and a 20-mer peptide at the extreme C terminus that interacts with RNA splicing factors (9, 35). The CTD of vertebrate PAPs is not conserved among different species to the same extent as the catalytic domain and, for example, is essentially lacking in yeast PAP (17). Hyperphosphorylation of the CTD by the cyclin-dependent kinase (cdk) p34cdc2-cyclin B represses PAP activity during M phase (5, 6), an event perceived as critical to efficient cell cycle progression, since expression of a PAP mutant unable to be fully phosphorylated significantly impedes cell proliferation in chicken DT40 cells (43). Multiple splice variants of vertebrate PAP mRNA have been described, some of which do not seem to be translated, would encode an inactive enzyme, and have been suggested to be products of autoregulation (23, 36, 42). The longest translated variant, PAP II, is thought to be the major form of the enzyme in most tissues. There are several other long-form mRNAs that encode functional enzymes, but the significance of this diversity is unknown.
Recently, a testis-specific PAP, called TPAP, was described and shown to be localized in the cytoplasm of mouse spermatocytes (15, 16). Interestingly, TPAP appears to be the product of a processed retroposon derived from an alternatively spliced form of PAP mRNA (42). Although the gene encoding TPAP was initially believed to be an inactive pseudogene (42), these more recent results suggest that it is active and that TPAP may function in cytoplasmic polyadenylation.
The present report describes our discovery and characterization of a form of PAP identified by molecular cloning from a human tumor cell cDNA library. Due to its identification and overexpression in human neoplasms, this molecule is designated neo-poly(A) polymerase (neo-PAP). Intriguingly, the intron-exon organization of the PAP and neo-PAP genes is almost identical, suggesting that they arose by gene duplication and subsequent recombination. We show that neo-PAP is indistinguishable in its biochemical functions in vitro from PAP. However, significant sequence dissimilarities in the CTD of neo-PAP compared to that of PAP as well as apparent differences in the phosphorylation of these two molecules suggest that each may be influenced by distinct regulatory controls.
MATERIALS AND METHODS
Sequence derivation and analysis.
Neo-PAP was first isolated as a 1.8-kb cDNA containing 0.5 kb of a partial 3′ coding sequence (CDS) and 1.3 kb of a 3′ untranslated region (UTR). This cDNA was cloned from an oligo(dT)-primed library derived from 1087-mel, a human malignant melanoma cell line. The cDNA was identified by the property that its protein product specifically stimulated CD4+ T lymphocytes from a patient with metastatic melanoma in experiments that are the subject of a separate report (S. Topalian et al., unpublished data). Using sequence-specific primers, two rounds of 5′ rapid amplification of cDNA ends (RACE) were performed on total RNA extracted from 1087-mel or the autologous Epstein-Barr virus-transformed B lymphocyte line 1087-EBV (Trizol reagent; GIBCO-BRL, Rockville, Md.) to isolate the 5′ CDS and 5′ UTR of neo-PAP according to the manufacturer's instructions (5′ RACE System; GIBCO-BRL). The entire cDNA sequence of neo-PAP, verified in multiple RACE clones derived from 1087-mel and 1087-EBV and in the original cDNA clone, contained 3,752 bp. To further validate this sequence, oligonucleotide PCR primers based on sequences derived from the 5′ RACE segment as well as from the original library clone were used to amplify cDNA clones containing the longest open reading frame of neo-PAP (2.2 kb) by performing reverse transcription (RT)-PCR on total RNA from 1087-mel or 1087-EBV. The forward PCR primer 5′-GGTTGGATGCCTCAGCCATAGTAAG-3′ terminated 125 bp upstream from the initiation codon, and the reverse primer 5′-GATTGCTTGTTCACTTAAGTGAGG-3′ ended 14 bp downstream of the stop codon. PCR was performed using a proofreading DNA polymerase (Vent DNA polymerase; New England Biolabs, Beverly, Mass.). PCR products were ligated into the pCR-Blunt II-TOPO vector (Zero Blunt TOPO PCR cloning kit; Invitrogen, Carlsbad, Calif.), and DNA sequencing was performed on seven individual cDNA clones, including six clones derived from 1087-mel and one from 1087-EBV, using the Big Dye Terminator Cycle Sequencing Kit (Perkin-Elmer/ABI). Sequences were determined with an ABI Prism 310 Genetic Analyzer (Perkin-Elmer). The complete neo-PAP cDNA sequence of 3.7 kb is shown in Fig. 1A. Database searches for nucleotide and deduced amino acid sequence similarities were performed with the BLAST program (http://www.ncbi.nlm.nih.gov/blast).
FIG. 1.
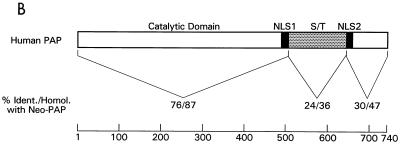
Nucleotide and amino acid sequences of neo-PAP. (A) cDNA and translated protein sequences of neo-PAP, with initiation and termination codons bolded and underlined. (B) Similarity between neo-PAP and human PAP II protein sequences. (C) Alignment of amino acid sequences of the C-terminal domains of neo-PAP and human PAP II. Asterisks indicate NLSs, nonconsensus cdk sites are in bold, consensus cdk sites are in bold and are underlined, plus signs indicate conserved amino acids, and minus signs indicate that no amino acid is present.
Plasmid constructs.
For expression of neo-PAP protein in Escherichia coli and subsequent purification, its CDS was cloned into the prokaryotic expression vector pET-14b, which encodes an N-terminal polyhistidine fusion tag for affinity purification (Novagen, Madison, Wis.). The neo-PAP CDS was amplified by PCR, using cDNA ligated into the pCR-Blunt II-TOPO vector (see above) as the template. The forward PCR primer 5′-CAGCTCGAGATGAAAGAGATGTCTGC-3′ and the reverse PCR primer 5′-TATCTCGAGTTACCGATTAAGGGTCAGTCG-3′ contained XhoI restriction sites (underlined) and translation initiation and termination codons (in bold). Complete DNA sequencing was performed on the neo-PAP insert after ligation into pET-14b. For expression and detection of neo-PAP in eukaryotic cells, the CDS was again amplified by PCR and cloned into the pEAK8 vector (Edge Biosystems, Gaithersburg, Md.). The forward primer 5′-CACCACGATATCCACCATGTACCCATACGATGTTCCAGATTACGCTATGAAAGAGATGTCTGC-3′ contained an EcoRV restriction site (underlined) and a 30-bp sequence encoding an N-terminal influenza virus hemagglutinin (HA) epitope tag for antibody-mediated detection (in italics) (3). The reverse PCR primer contained a NotI restriction site. Using a similar strategy, a cDNA encoding bovine PAP II with an N-terminal HA fusion tag was cloned into the HindIII and NotI sites of pEAK8. The bovine PAP II cDNA sequence corresponds to GenBank accession no. X61585 (36), and the translated protein is 98.5% identical to human PAP II.
Preparation of PAP I and neo-PAP proteins.
N-terminally His-tagged PAPs were expressed for 18 h at 15°C in 400 ml of Luria-Bertani buffer plus 200 mg of ampicillin/ml. E. coli BL21(DE3) cells were pelleted, resuspended in 12 ml of binding buffer (20 mM Tris-HCl [pH 7.4], 100 mM NaCl, 0.05% NP-40, 5 mM imidazole, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]), and sonicated. The supernatants were rocked for 2 h with 0.4 ml of Ni2+-nitriloacetic acid agarose (Qiagen Inc., Valencia, Calif.), washed with 20 column volumes of high-salt buffer (20 mM Tris-HCl [pH 7.4], 500 mM NaCl, 0.05% NP-40, 5 mM imidazole, 0.5 mM PMSF) and then washed with 5 column volumes of high-salt buffer containing 15 mM imidazole instead of 5 mM imidazole, and eluted with high-salt buffer containing 200 mM imidazole instead of 5 mM imidazole. Preparations were dialyzed against buffer D [20 mM HEPES (pH 7.9), 50 mM (NH4)2SO4, 20% glycerol, 0.2 mM EDTA, 0.5 mM dithiothreitol (DTT), 0.5 mM PMSF].
RNA substrates and polyadenylation and cleavage assays.
Plasmids pG3SVL-A and pG3L3-A, which contain the simian virus 40 (SV40) late and adenovirus-2 L3 polyadenylation sites (31), respectively, were digested with appropriate restriction enzymes and used as templates to synthesize 32P-labeled RNA substrates. Specific polyadenylation with the SV40 substrate was assayed in 12.5-μl reaction volumes containing 1 to 2 ng of labeled pre-RNA, 0.4 μl of purified CPSF fraction (21), 10 mM HEPES (pH 7.9), 0.5 mM MgCl2, 1 mM ATP, 25 mM (NH4)2SO4, 0.1 mM EDTA, 0.25 mM DTT, 0.25 mM PMSF, 2.5% (wt/vol) polyvinyl alcohol, 400 ng of tRNA, 0.16 U of RNasin (Promega, Madison, Wis.), 100 ng of bovine serum albumin (BSA), and the indicated amounts of recombinant PAPs. Specific cleavage with the L3 substrate was performed in 12.5-μl reaction volumes containing 1 to 2 ng of labeled pre-RNA, 3.7 μl of purified or partially purified proteins (CstF, CPSF, CFI, CFII), 30 ng of recombinant murine glutathione S-transferase-CTD, 9.6 mM HEPES (pH 7.9), 2 mM MgCl2, 1 mM dATP, 24 mM (NH4)2SO4, 0.12 mM EDTA, 0.24 mM DTT, 0.24 mM PMSF, 2.5% (wt/vol) polyvinyl alcohol, 250 ng of tRNA, 0.25 U of RNasin, 500 ng of BSA, and the indicated amounts of recombinant PAP (11). All reaction mixtures were incubated for 90 min at 30°C, and RNA products were isolated and fractionated on 5% polyacrylamide–8.3 M urea gels.
Incorporation of α-32P-labeled nucleotides and nonspecific polyadenylation assay.
Incorporation of α-32P-labeled nucleotides was done in 25 μl of buffer HMN (10 mM HEPES [pH 7.9], 1 mM MnCl2, 0.1% NP-40, 250 ng of BSA, 0.1 mM EDTA, 0.25 mM DTT, 0.25 mM PMSF, 0.5 mM ATP, 0.3 pmol of labeled nucleotides). tRNA (100 nM) as substrate and 50 nM PAPs were present in the reaction. Reaction mixtures were incubated at 37°C and terminated by application of the complete reaction mixture to a DE81 paper. The paper was washed with a solution containing 0.5 M Na2HPO4 (pH 7.0) and 70% ethanol and was counted in a scintillation counter. Nonspecific polyadenylation was also carried out as described for the specific polyadenylation assays, except that 1 mM MnCl2 replaced 0.5 mM MgCl2, purified CPSF was omitted, and reaction mixtures were incubated for 30 min at 30°C.
Immunofluorescent staining and confocal microscopy.
Exponentially growing HeLa cells (human cervical cancer; American Type Culture Collection, Manassas, Va.) were plated on glass coverslips in 24-well tissue culture plates and incubated overnight at 37°C and 5% CO2. The following day cells were transfected with 0.5 μg of the plasmid pEAK8/HA-neoPAP per well using Lipofectamine Plus (GIBCO-BRL). Twenty to 48 h later, cells were rinsed in phosphate-buffered saline (PBS), fixed in 4% paraformaldehyde–PBS, and permeabilized with 1% Triton X-100 (Sigma, St. Louis, Mo.) in 0.2% BSA–PBS. After being blocked with 20% goat serum at 37°C for 30 min and then rinsed in 0.2% BSA–PBS, samples were stained with a fluorscein isothiocyante-conjugated rat monoclonal antibody (MAb) (5 μg/ml) specific for the HA epitope YPYDVPDYA (clone 3F10; Roche Molecular Biochemicals, Indianapolis, Ind.) for 1 h at room temperature. Coverslips were rinsed with PBS and mounted onto microscope slides using GelMount (Biomeda Corp., Foster City, Calif.). Stained cells were examined on a Zeiss Axioplan microscope using the 100×/1.4 oil immersion objective (total magnification, ×1,400). Confocal images were generated on a Zeiss laser scanning microscope (LSM 510).
Western blots.
Subconfluent 293 cells (adenovirus-transformed human embryonic kidney epithelium; American Type Culture Collection) were transfected with the plasmid pEAK8/HA-neoPAP or pEAK8/HA-bovine PAP II using the Effectine reagent according to the manufacturer's instructions (Qiagen). Forty-eight hours later cells were harvested, washed, and lysed in radioimmunoprecipitation assay buffer (Boehringer Mannheim, Indianapolis, Ind.), containing detergents and the protease inhibitor PMSF, at a concentration of 2 × 107 cells/ml. Following centrifugation at 10,000 × g for 10 min, supernatants were collected and adjusted to pH 5.3. Samples were treated with potato acid phosphatase (Roche Biochemicals) at a final concentration of 5 U/ml at 37°C for 1 h and then were boiled for 3 min under nonreducing conditions and loaded into a 4 to 20% Tris-glycine acrylamide gel (Novex, San Diego, Calif.). Electrophoretically separated proteins were blotted onto a nitrocellulose membrane, which was incubated for 1 h with peroxidase-conjugated MAb (125 ng/ml) specific for HA (clone 3F10; Roche Biochemicals). Protein visualization was achieved with chemiluminescence (ECL detection system; Amersham Pharmacia Biotech, Piscataway, N.J.).
Northern blot analysis.
Total RNA was isolated from a variety of cultured cell lines and fresh peripheral blood lymphocytes (PBL) using the Trizol method (GIBCO-BRL) or the RNeasy Midi Kit (Qiagen). Cell lines initiated in our laboratory included the malignant melanoma from which neo-PAP was cloned (1087-mel) and its autologous transformed B-cell line (1087-EBV) and the prostate cancer cell lines 1532-CPTX, 1535-CPTX, and 1542-CPTX. Fresh cryopreserved PBL from patients 1087, 1532, and 1535 were autologous to the tumor cell lines mentioned above. The colon cancer cell lines CY13, LoVo, and SW480 were obtained from the American Type Culture Collection, as were 293 cells. Total RNA (10 μg per lane) was electrophoresed in a 1% agarose formaldehyde gel and transferred to a nylon membrane (Nytran; Schleicher & Schuell, Inc., Keene, N.H.). In addition, a Northern blot containing approximately 2 μg of poly(A)+ RNA isolated from 12 different fresh human tissues/lane was purchased from OriGene Technologies (Rockville, Md.). Hybridization of blots with radiolabeled oligonucleotide probes was performed at 68°C for 2 h according to the QuikHyb protocol (Stratagene, La Jolla, Calif.). After being washed with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% sodium dodecyl sulfate (SDS), blots were subjected to a high-stringency wash with 0.1× SSC–0.1% SDS at 60°C for 30 min, and then autoradiography was performed at −70°C. To synthesize oligonucleotide probes specific for neo-PAP or human PAP, RT-PCR was performed on total RNA from 1087-mel. The neo-PAP probe contained bp 27 to 325 (5′ to 3′) of the sequence shown in Fig. 1A, corresponding to a portion of 5′ UTR as well as 5′ CDS, and had no significant similarity to PAP. The PAP probe corresponded to bp 10 to 341 in the extreme 5′ coding region of human PAP, GenBank accession no. X76770 (32). Probes were radiolabeled by the random priming method (Lofstrand Labs, Gaithersburg, Md.). Blots were hybridized first with the probe for neo-PAP and then with β-actin, after which they were stripped and then hybridized with the probe for human PAP and then with β-actin again.
RT-PCR to assess normal tissue expression of neo-PAP.
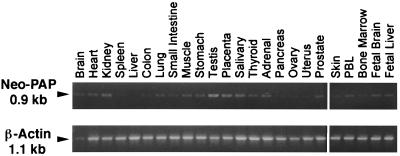
To further investigate the profile of neo-PAP mRNA expression in various normal adult and fetal human tissues, RT-PCR was performed using the Human Rapid-Scan Gene Expression Panel kit according to the manufacturer's instructions (OriGene Technologies, Rockville, Md.). This kit includes duplicate 96-well PCR plates containing first-strand cDNAs derived from 24 different human tissues, normalized to β-actin concentration and serially diluted over a 4-log range. PCR for neo-PAP was performed with a forward oligonucleotide primer corresponding to bases 1558 to 1576 (5′ to 3′) and a reverse primer corresponding to bases 2477 to 2464 (5′ to 3′) (see Fig. 1A), yielding a product of approximately 0.9 kb encoding the C terminus of neo-PAP. In the duplicate 96-well plate, PCR for β-actin was performed with primers covering the entire 1.1-kb coding region. For both neo-PAP and β-actin, 35 cycles of PCR were carried out at 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min. Approximately one third of the volume of each PCR was electrophoresed in a 0.8% agarose–Tris-borate-EDTA gel and stained with ethidium bromide.
Nucleotide sequence accession number.
The complete neo-PAP cDNA sequence was deposited in GenBank under accession number AF312211.
RESULTS
Sequence analysis.
A partial cDNA clone encoding the C terminus of neo-PAP was isolated from a melanoma-derived cDNA library. When expressed following transient transfection into genetically modified 293 cells according to procedures previously described (38), the protein product of this cDNA specifically stimulated melanoma-reactive autologous CD4+ T cells (Topalian et al., unpublished data). The complete cDNA sequence of neo-PAP was obtained using 5′ RACE. Neo-PAP cDNA contains 3,752 bp, with a 2,208-bp open reading frame predicted to encode a protein of 736 amino acids and having a molecular size of 82.8 kDa (Fig. 1A). Database queries for nucleotide similarities with molecules having known functions revealed the most significant similarity with human PAP (GenBank no. X76770), which showed regions of up to 86% nucleotide identity, followed by nonhuman PAPs with somewhat lower segmental identities. However, searching the expressed sequence tag and genome project databases revealed dozens of nucleotide entries with 95 to 100% identity to neo-PAP. Most of these sequences were derived from human placenta, fetus, or a wide variety of neoplasms, including leukemia, melanoma, and cancers originating from brain, colon, lung, stomach, endometrium, and pancreas. These findings suggested that rather than representing a previously unrecognized splice variant of PAP, neo-PAP might be transcribed from a gene distinct from the original PAP. This was confirmed by searching the human genome database (http://www.ncbi.nlm.nih.gov/genome/guide/) and finding that neo-PAP cDNA was nearly 100% identical in its entirety to sequences located on chromosome 2 (working draft NT 005399.1). In contrast, the original human PAP gene is located on chromosome 14 (40). Strikingly, the organization of the neo-PAP gene into 22 exons on chromosome 2 recapitulates the intron-exon structure defined for murine PAP II (42).
The deduced neo-PAP protein sequence of 736 amino acids was found, through protein database searching, to have an overall similarity of 71% to human PAP (Swissprot P51003). It also had approximately the same degree of similarity to nonhuman PAPs, which was expected due to the high degree of amino acid sequence conservation among the vertebrate PAPs. Neo-PAP was not significantly similar to other molecules with known functions and appears to be organized into functional domains that recapitulate those of the original PAP (Fig. 1B). An N-terminal region of almost 500 amino acids was 87% similar to the N-terminal catalytic domain of PAP, suggesting that neo-PAP might have a polymerase function. Also comparable to PAP, neo-PAP contained two bipartite NLSs (8) predicting nuclear localization of this protein. In the original PAP, the two NLSs surround an S-T-rich CTD containing multiple cdk phosphorylation sites that are critical for regulating polymerase function (5, 23). However, the sequence similarity between neo-PAP and PAP declined sharply in this region (to 36%), suggesting that these two molecules might be regulated differently. Figure 1C aligns the amino acid sequences of neo-PAP and PAP II, commencing at NLS1 and continuing through the C terminus. The original human PAP II contains seven cdk phosphorylation sites, including two consensus (T/SPXK/R) and five nonconsensus sites (T/SP), and it has been shown that full phosphorylation of all of these sites is required to repress enzymatic function during M phase (6). In comparison, neo-PAP contains nine cdk motifs, two of which are consensus sites. Thus, conservation of cdk sites by neo-PAP is significant, and even more so when considered in the context of the percent S+T in this region: neo-PAP contains only 23% S+T, compared to 34% for PAP. These findings suggest that regulation of neo-PAP function may occur through a phosphorylation mechanism despite striking sequence dissimilarities between neo-PAP and PAP in this region. Of note, the extreme C-terminal 20-mer peptide implicated in splicing regulation (35) is highly, although not perfectly, conserved between the two molecules.
Neo-PAP is biochemically indistinguishable from PAP.
In view of the significant degree of similarity between the amino acid sequences of the N-terminal region of neo-PAP and the catalytic domain of PAP, we next set out to determine whether neo-PAP functions like PAP in several in vitro functional assays. To this end, we first expressed a His-tagged derivative of neo-PAP in E. coli and purified the protein alongside an identically tagged version of bovine PAP I (for an example, see reference 5). Figure 2A displays a Coomassie-stained SDS gel of the two purified proteins. We then compared the activity of the two PAPs in so-called nonspecific poly(A) synthesis assays. Such assays measure the ability of PAP to catalyze primer-dependent poly(A) synthesis independent of both other polyadenylation factors and the sequence of the RNA primer, a property of the enzyme facilitated by the inclusion of Mn2+ instead of Mg2+ in reaction mixtures (for an example, see reference 23). Figure 2B displays a time course measuring the ability of each enzyme to incorporate [α-32P]ATP onto an unlabeled RNA primer. Both PAPs displayed equivalent activities and, as is expected for an authentic PAP, were unable to utilize GTP instead of ATP as a substrate. A related nonspecific assay utilizes a 32P-labeled RNA primer and unlabeled ATP as substrate and measures poly(A) synthesis by the change in size of the RNA primer. Figure 2C shows that increasing concentrations of each PAP resulted in comparable increases in the size of the primer. (The slight differences observed in size reflect the very sensitive nature of the assay and were not reproducible.)
FIG. 2.
Neo-PAP has nonspecific polyadenylation activity. (A) PAP I and neo-PAP proteins utilized in subsequent experiments were resolved on SDS–8% polyacrylamide gel electrophoresis and were Coomassie stained. Lanes 1 and 2, 1.2 μg of recombinant PAP I and neo-PAP expressed in and purified from E. coli. (B) Efficiencies of incorporation of α-32P-labeled nucleotides. The relative amounts of incorporated nucleotides [α-32P]ATP and [α-32P]GTP were measured. Assay conditions are described in Materials and Methods. (C) Increasing amounts (1, 2.5, 10, and 50 ng) of PAP I (lanes 2 through 5) and neo-PAP (lanes 7 through 10) were assayed in a nonspecific polyadenylation assay using a 172-nucleotide 32P-labeled RNA substrate. RNA products were resolved by denaturing polyacrylamide gel electrophoresis.
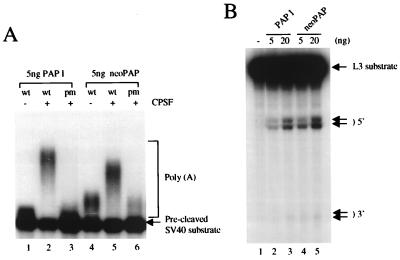
We next compared the two PAPs in a specific polyadenylation assay, which requires an AAUAAA-containing RNA primer and CPSF. Figure 3A displays the results of such an assay utilizing a 32P-labeled SV40 late pre-mRNA containing an intact AAUAAA (wild type) or a U-to-A mutation in the hexanucleotide (pm), purified CPSF, and 5 ng of either PAP I (lanes 1 to 3) or neo-PAP (lanes 4 to 6). Importantly, both PAPs displayed significant polyadenylation activity with the wild-type RNA that was reduced to background levels with the pm RNA. Note that low levels of poly(A) synthesis were detected with both PAPs in the absence of CPSF and that this activity was slightly higher with neo-PAP (lanes 1 and 4). This reflects nonspecific poly(A) synthesis analogous to that shown in Fig. 2. This low activity was also observed with the pm RNA but was reduced by the presence of CPSF (lanes 3 and 6), reflecting a well-established activity of CPSF, which is to inhibit nonspecific poly(A) synthesis under conditions that stimulate AAUAAA-dependent polyadenylation (for an example, see reference 27). But the important result is that neo-PAP, like PAP I, was highly active in AAUAAA- and CPSF-dependent polyadenylation. The length of the poly(A) synthesized by neo-PAP was slightly shorter than that synthesized by PAP I. The significance of this, if any, is not known.
FIG. 3.
Neo-PAP has specific polyadenylation and cleavage activity. (A) Recombinant PAP I, neo-PAP, or purified CPSF, alone or in the indicated combinations, was added to reaction mixtures containing either wild-type (wt; AAUAAA) or mutant (pm; AAAAAA) 32P-labeled pG3SVL-A pre-RNA. (B) Increasing amounts (5 and 20 ng) of PAP I (lanes 2 and 3) or neo-PAP (lanes 4 and 5) were assayed in a reconstitution cleavage assay (see Materials and Methods). Arrows indicate the positions of upstream (5′) and downstream (3′) cleavage products. In both panels A and B, RNA products were resolved by denaturing polyacrylamide gel electrophoresis.
PAP is also known to be required with most pre-mRNAs for the first step of polyadenylation, endonucleolytic cleavage (for examples, see reference 31). To determine whether neo-PAP is able to activate cleavage, we reconstituted 3′ cleavage reactions with purified CPSF, CstF, and neo-PAP or PAP I, plus partially purified CFI and CFII, using a 32P-labeled adenovirus L3 pre-mRNA (Fig. 3B). In the absence of PAP, cleavage was essentially undetectable (lane 1). But increasing concentrations of either PAP I (lanes 2 and 3) or neo-PAP (lanes 4 and 5) resulted in significant cleavage, and both enzymes displayed comparable activity. Note that cleavage was induced at two nearby sites, generating distinct 5′ and 3′ cleavage products. Significantly, this pattern, which has been observed previously in similar reconstitution assays (10), was identical with both PAPs. Taken together, our data indicate that the properties of bovine PAP I and human neo-PAP are essentially indistinguishable, suggesting that neo-PAP has the potential to function in pre-mRNA 3′ processing in the cell nucleus.
Subcellular localization of neo-PAP.
We next used immunofluorescence microscopy to examine directly the subcellular localization of neo-PAP. Specifically, the localization of an epitope-tagged neo-PAP following transient transfection of HeLa cells was determined. Twenty to 48 h after transfection with the plasmid pEAK/HA-neoPAP, cells were stained with a MAb specific for the N-terminal HA epitope tag. In a representative experiment shown in Fig. 4, the transiently expressed neo-PAP protein localized exclusively to the nuclei of HeLa cells. Similar results were obtained with transiently transfected COS-7 cells and stable transfectants of 293 cells (data not shown). These results are similar to those of previously published experiments with bovine PAP I and PAP II, which showed exclusively nuclear localization of those proteins when expressed by transient transfection (24).
FIG. 4.
Neo-PAP localizes exclusively to the nucleus. After a 20-h transfection, HeLa cells were stained with a fluorescein isothiocyanate-conjugated anti-HA MAb and examined using immunofluorescence microscopy. Magnification, ×1,400.
Differential phosphorylation of neo-PAP versus that of PAP II.
Previous work has demonstrated that hyperphosphorylation of classic PAP is an important mechanism by which its enzymatic activity is coordinately repressed during cell division (5). Western blots performed on extracts of human cells expressing PAP or cells transfected with plasmids encoding HA-tagged PAP have consistently demonstrated the apparent molecular size of PAP to be approximately 20 kDa greater than that predicted by the protein sequence. Site-directed mutagenesis or phosphatase treatment of cell extracts has shown that the low-mobility forms of classic PAP observed on Western blots reflect phosphorylation of multiple cdk sites in the CTD (6, 24, 32).
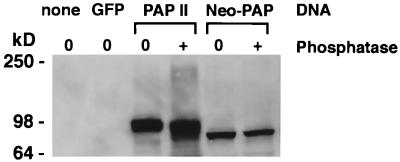
In light of the above findings, we next set out to investigate the phosphorylation state of neo-PAP by performing Western blots on extracts of 293 cells after a 48-h transfection with pEAK/HA-neoPAP or pEAK/HA-PAP II (Fig. 5). Unexpectedly, neo-PAP was detected as only a single protein band migrating at its predicted molecular size of 82.8 kDa. In contrast, bovine PAP II (predicted molecular size of 82.4 kDa, protein sequence 98.5% identical to human PAP II) presented as multiple bands in the range of 100 kDa. These results suggested that neo-PAP might not be phosphorylated under the same conditions that caused hyperphosphorylation of PAP. A single neo-PAP form migrating at its true molecular size was also observed in Western blots performed on extracts of stable transfectants of 293 cells and transiently transfected Epstein-Barr virus-transformed B-cell lines (data not shown), further suggesting that the predominant form of this enzyme might not be phosphorylated. To address this issue directly, extracts of transiently transfected 293 cells were treated with acid phosphatase prior to Western blotting. As shown in Fig. 5, the migration of neo-PAP was not influenced by phosphatase treatment, while the migration of PAP II shifted to a smear of higher-mobility species in the 80- to 100-kDa range, consistent with partial to complete dephosphorylation. Although apparent mobility shifts on Western blots are not conclusive evidence of phosphorylation states, these results suggest that neo-PAP and PAP are differentially phosphorylated and hence differentially regulated. This is especially interesting because both neo-PAP and PAP contain a conserved cyclin recognition motif (2) and multiple consensus and nonconsensus cdk phosphorylation sites that would predict similar, and not disparate, modes of regulation.
FIG. 5.
Neo-PAP and classic PAP appear to be differentially phosphorylated. Western blotting with an anti-HA epitope antibody was performed on extracts of 293 cells that were not transfected (“none”) or transfected with plasmids encoding green fluorescent protein (GFP), HA-PAP II, or HA-neoPAP. Some lysates (indicated with a plus sign) were treated with potato acid phosphatase prior to loading onto SDS–4 to 20% polyacrylamide gel electrophoresis. Cell equivalents were 1.6 × 105 per lane. Results are representative of three separate experiments.
Neo-PAP is overexpressed in human cancers.
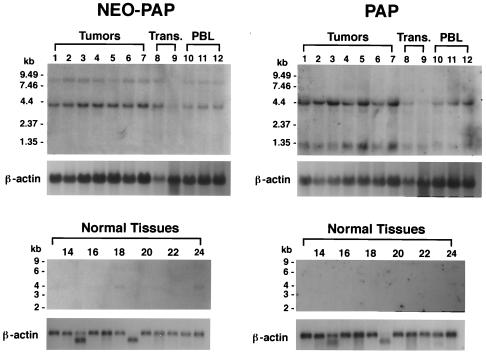
Having cloned and sequenced neo-PAP from a patient's melanoma cells and from virus-transformed B cells, we next set out to determine the profile of neo-PAP and PAP gene expression in other malignant, transformed, and normal human cells and tissues. Figure 6 shows Northern blots hybridized first with a probe specific for portions of the 5′ UTR and extreme 5′ CDS of neo-PAP and then stripped and hybridized with a probe specific for a comparable 5′ region in human PAP. Immediately evident are the different apparent splicing patterns of neo-PAP versus PAP mRNA. With PAP, we observed two dominant mRNA species of approximately 4.4 and 1.3 kb similar to those previously described for bovine PAP II and human PAP (36). In contrast, neo-PAP had one dominant mRNA species of approximately 4 kb (consistent with the cDNA sequence of 3.7 kb), but no smaller species were apparent. The significance of a faint 8-kb mRNA species observed for neo-PAP but not for PAP is unknown; this could represent an alternatively or incompletely processed mRNA. In a separate experiment (data not shown), a Northern blot containing total RNAs derived from various tumors and transformed cells was hybridized with the 5′ neo-PAP probe described above and then was stripped and hybridized again with a neo-PAP probe specific for the 3′ end of the CDS and entire 3′ UTR. Both probes revealed a dominant 4-kb mRNA species and minor band at 8 kb similar to the pattern shown in Fig. 6. These findings indicate that, unlike PAP, neo-PAP does not seem to generate splice variants.
FIG. 6.
Neo-PAP and PAP are both overexpressed in human cancers but have distinct splicing patterns. Northern blots containing 10 μg of total RNA/lane (upper right and left panels) or approximately 2 μg of poly(A)+ RNA/lane (lower right and left panels) were hybridized with a neo-PAP probe followed by β-actin and then stripped and reprobed with PAP, followed again with β-actin. Lane 1, 1087-mel; 2, 1532-CPTX; 3, 1535-CPTX; 4, 1542-CPTX; 5, CY13; 6, LoVo; 7, SW480; 8, 293 cells; 9, 1087-EBV; 10, 1087 PBL; 11, 1532 PBL; 12, 1535 PBL; 13, brain; 14, colon; 15, heart; 16, kidney; 17, liver; 18, lung; 19, muscle; 20, placenta; 21, small intestine; 22, spleen; 23, stomach; 24, testis. Blots probed with neo-PAP or PAP were exposed to film for 67 to 72 h or with β-actin for 2 to 2.5 h. Trans., transformed cells.
Despite apparent dissimilarities in RNA splicing, both neo-PAP and PAP were overexpressed by tumors compared to virally transformed or normal cells. The upper portion of Fig. 6 demonstrates, in one Northern blot, enhanced expression of both neo-PAP and PAP mRNAs in seven cancers, compared to that of two transformed cell lines and three fresh PBL specimens. The tumor specimens assayed in Fig. 6 included one melanoma (1087-mel, from which neo-PAP was originally cloned; lane 1), three prostate cancers (1532-CPTX, 1535-CPTX, and 1542-CPTX; lanes 2 to 4), and three colon cancers (CY13, LoVo, and SW480; lanes 5 to 7). In a separate experiment, RNAs from five other melanomas, two additional prostate cancers, and another colon cancer were assessed for neo-PAP expression by Northern blotting, and all were significantly positive (data not shown). The lower portion of Fig. 6 demonstrates that, among 12 different fresh normal human tissues, neo-PAP was weakly expressed and PAP could not be detected at all, even after prolonged film exposure. In this experiment, neo-PAP was expressed predominantly in testis (lane 24), but weaker signals were also seen in brain (lane 13) and lung (lane 18). In a repeat normal tissue Northern blot experiment, neo-PAP expression was observed only in testis (data not shown). Taken together, the data indicate that both PAP and neo-PAP mRNAs are expressed at low levels in normal tissues and overexpressed in tumors.
To further explore the normal tissue distribution of neo-PAP, RT-PCR was performed on 24 different adult and fetal human tissues. Figure 7 shows that, consistent with the Northern blot results, testicular expression was strongest, but most other tissues also showed weak expression of neo-PAP after 35 cycles of PCR. By visual assessment of RT-PCR products generated from cDNAs diluted over a 4-log range, the intensity of neo-PAP expression was estimated to be 1 to 2% of that of β-actin (not shown). Preferential neo-PAP expression in the testis, among normal tissues, is particularly interesting in view of the existence of the testis-specific PAP, TPAP, suggested to catalyze cytoplasmic polyadenylation of mRNAs in spermatocytes (GenBank number AF218840) (15, 16).
FIG. 7.
Expression of neo-PAP in testis and other normal tissues assessed by RT-PCR. Products of RT-PCR with primers specific for neo-PAP or β-actin were stained with ethidium bromide and were electrophoresed on a 0.8% agarose gel.
Mutational analysis of the CTD of neo-PAP.
Given the overexpression of PAP in cancer cells, we next wished to examine the possibility that the S-T-rich CTD of neo-PAP, bearing the signature of a regulatory domain, might harbor cancer-associated mutations. To this end we sequenced purified RT-PCR products spanning this region which were generated with the same PCR primers utilized for the experiment shown in Fig. 7. A total of 21 samples were analyzed, including those generated from the RNA preparations visualized in lanes 1 to 12 of the Northern blots shown in Fig. 6 as well as an additional 9 samples comprising five melanomas, two prostate cancers, one colon cancer, and one transformed human fibroblast line. Of note, 1087-mel, the tumor line from which neo-PAP was first isolated, was found to contain two neo-PAP alleles, one of which had a C-to-T mutation at position 2159 that appeared as two overlapping peaks on automated DNA sequencing. This mutation was confirmed by isolating and sequencing individual full-length neo-PAP cDNA clones from 1087-mel. Intriguingly, this caused a P-to-L missense mutation, destroying a consensus cdk phosphorylation site immediately preceding NLS2. It is conceivable that this mutation affects the growth of these cells (43). The C-to-T substitution in one allele in 1087-mel did not seem to represent a polymorphism, since it was not observed in 1087-EBV or 1087-PBL. However, aside from a silent point mutation discovered in 293 cells, no other mutations or polymorphisms were identified in these samples. Thus, it does not appear that the CTD of neo-PAP is exceedingly prone to mutation in cancers.
DISCUSSION
Processing of mRNA precursors, a critical step in gene expression, comprises an integrated series of reactions mediated by a large and complex set of factors (12). Poly(A) polymerase is a critical player in RNA processing: it not only catalyzes poly(A) synthesis and participates in endonucleolytic cleavage of pre-mRNA, but it also interacts with other proteins, including, for example, splicing factors that may help coordinate polyadenylation and splicing (35). The present report demonstrates the existence of a novel poly(A) polymerase, neo-PAP, which shares many of the characteristics of the originally described enzyme. Common to both PAPs are their protein domain organization, subcellular nuclear localization, and in vitro functions. Particularly intriguing, we show by Northern blotting that both PAPs are overexpressed in human tumors, a finding consistent with previous studies of PAP. Overexpression of PAP mRNA has been demonstrated in human carcinomas originating in breast, colon, ovary, and pancreas, compared to expression in normal tissue counterparts (22). Furthermore, the polyadenylation activity of crude or partially purified cell extracts has been shown to be significantly enhanced in acute leukemias compared to that in chronic leukemias and normal lymphocytes (33), and in aggressive forms compared to more indolent forms of breast cancer (29), suggesting that PAP levels might correlate with clinical prognosis. A role for PAP in sustaining activated or hyperproliferative cell states has also been postulated on the basis of experiments showing elevated polyadenylation activity or enhanced PAP mRNA in phytohemagglutinin-stimulated human lymphocytes (7) and factor VII-stimulated human fibroblasts (22), respectively. In retrospect, while previous investigations conducted with Northern blotting were specific for PAP, assays for enzymatic activity in cell extracts could have reflected expression of PAP, neo-PAP, or both.
The factors required for polyadenylation of nuclear mRNA precursors have been extensively studied in both mammals and yeast. Nearly all the factors have been cloned during the last decade, beginning with PAP in 1991 (17, 23, 36). Sequencing of bovine cDNAs immediately suggested the potential for diversity in PAP isoforms created by alternative splicing, and this was confirmed by more detailed analysis of the mouse gene and cDNAs (42). However, there was until very recently no evidence for a second PAP gene, or indeed any indications that more than a single gene existed for any of the known polyadenylation factors. Recent studies, though, have suggested that there may be related genes for many of the mammalian polyadenylation factors. A novel form of the RNA binding subunit of CstF, CstF-64, has been described and shown to be the product of a second CstF-64 gene (37). Intriguingly, similar to the functional retroposon TPAP gene mentioned above (15, 16), the novel CstF-64-related protein is expressed almost exclusively in the testis. The reasons for this are unclear. In the case of TPAP, which is largely cytoplasmic (apparently due to the absence of NLSs [15]), it could reflect a role in cytoplasmic polyadenylation known to occur in germ cells (25). The CstF-64 gene is localized on the X chromosome (37), so the CstF-64-related gene may function to ensure production of CstF-64 following X inactivation. It remains to be determined why neo-PAP appears to be preferentially expressed in testis. A recent analysis of the human genome suggested that related genes may exist for the majority of factors in the polyadenylation complex (34). Indeed, this analysis pointed to the possible existence of a third PAP-related gene in humans.
What might be the reasons for the existence of multiple PAP genes? Although our data did not uncover any significant differences in the biochemical properties of neo-PAP relative to those of PAP, it is conceivable that neo-PAP may have unique properties in vivo that allow it to modulate the efficiency of 3′-end formation or poly(A) tail length of specific genes. Alternatively, it might be functionally equivalent to PAP but exist to allow precise quantitative control of PAP levels in different tissues and/or cell growth states. It is known that PAP levels must be tightly regulated (43), and it may be that a second gene allows more precise control under a variety of conditions.
It is intriguing that neo-PAP went undiscovered for a decade after identification of PAP. Purification of PAP and cloning of PAP cDNAs were from bovine tissues (23, 36), and it is conceivable that neo-PAP is not present in bovine tissue or not expressed in the tissues analyzed. Alternatively, neo-PAP may behave poorly during extraction or purification. Whatever the reason, it now seems that similar or related proteins exist for many factors that function in gene expression in metazoans (34). This includes not only gene-specific regulatory factors but also many of the general factors that participate in transcription, splicing, and as mentioned above, polyadenylation. The existence of multiple forms of a polymerase, though, is unexpected, and is consistent with the idea that PAP plays an important role in gene control (for an example, see reference 43). Perhaps consistent with this possibility, it is noteworthy that key elements of the PAP regulatory region, such as the dual NLSs, putative cdk phosphorylation sites, and the very C-terminal splicing factor interaction domain, are well conserved. On the other hand, none of these motifs are 100% identical, and it is possible that future studies will reveal differences in the behavior of PAP and neo-PAP.
Despite their similarities, neo-PAP and PAP demonstrate unique properties. One difference is their mRNA splicing patterns: on Northern blots, neo-PAP displayed only the longest of the splice variants observed for PAP. Even more interesting, neo-PAP does not seem to be susceptible to the same regulatory controls as PAP. Although neo-PAP contains a conserved cyclin recognition motif (2) and multiple C-terminal cdk phosphorylation sites (5, 6), phosphorylation of neo-PAP could not be demonstrated in Western blots of transient or stable transfectants of proliferating human cells. Coupled with overexpression of neo-PAP message in cancers, these findings suggest that neo-PAP may represent an aberrantly regulated polymerase enzyme that supports rapid cell proliferation. The existence of such an enzyme was postulated by Jacob et al., who purified nuclear protein fractions with polyadenylation activity from rat hepatomas or normal rat liver and described their distinct properties (14, 26). However, the much smaller molecular size, signature amino acid composition, and hyperphosphorylated state of the rat hepatoma PAP are inconsistent with our characterization of neo-PAP. In future studies, it will be critical to analyze the expression of endogenous neo-PAP protein in normal tissues and cancer and to define gene-, tissue-, and cell cycle-specific regulation of PAP function in normal cells and how this is altered in tumor cells.
ACKNOWLEDGMENTS
We thank Paul Robbins for helpful ideas, Yong Li and Aaron Chen for DNA sequencing, Steven Rosenberg for advice and support, and Drew Pardoll for insights and critical review of the manuscript.
Work in the lab of J.L.M. was supported by NIH grant GM28983.
ADDENDUM
We have become aware of a recent GenBank entry by K. Perumal et al., accession number AY029162, the translated protein product of which is 99% identical to full-length neo-PAP.
REFERENCES
- 1.Bard J, Zhelkovsky A M, Helmling S, Earnest T N, Moore C L, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- 2.Bond G L, Prives C, Manley J L. Poly(A) polymerase phosphorylation is dependent on novel interactions with cyclins. Mol Cell Biol. 2000;20:5310–5320. doi: 10.1128/mcb.20.14.5310-5320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y-T, Holcomb C, Moore H-P H. Expression and localization of two low molecular weight GTP-binding proteins, Rab8 and Rab10, by epitope tag. Proc Natl Acad Sci USA. 1993;90:6508–6512. doi: 10.1073/pnas.90.14.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 5.Colgan D F, Murthy K G K, Prives C, Manley J L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- 6.Colgan D F, Murthy K G K, Zhao W, Prives C, Manley J L. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and non-consensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courtis N C, Trangas T T, Tsiapalis C M. Increase in the levels of activity of polyadenylic acid-metabolizing enzymes following phytohaemagglutinin stimulation of human lymphocytes. Mol Cell Biochem. 1987;75:33–42. doi: 10.1007/BF00231606. [DOI] [PubMed] [Google Scholar]
- 8.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 10.Hirose Y, Manley J L. Creatine phosphate, not ATP, is required for 3′ end cleavage of mammalian pre-mRNA in vitro. J Biol Chem. 1997;272:29636–29642. doi: 10.1074/jbc.272.47.29636. [DOI] [PubMed] [Google Scholar]
- 11.Hirose Y, Manley J L. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- 12.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 13.Huang Y, Carmichael G C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob S T, Terns M P, Maguire K A. Polyadenylate polymerases from normal and cancer cells and their potential role in messenger RNA processing: a review. Cancer Res. 1989;49:2827–2833. [PubMed] [Google Scholar]
- 15.Kashiwabara S, Zhuang T, Yamagata K, Noguchi J, Fukamizu A, Baba T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev Biol. 2000;228:106–115. doi: 10.1006/dbio.2000.9894. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y J, Lee Y, Chung J H. An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Lett. 2000;487:287–292. doi: 10.1016/s0014-5793(00)02367-x. [DOI] [PubMed] [Google Scholar]
- 17.Lingner J, Kellermann J, Keller W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae. Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- 18.Martin G, Keller W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and a catalytic domain, homologous to the family X polymerases and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- 19.Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 21.Murthy K G K, Manley J L. Characterization of the multisubunit cleavage polyadenylation specificity factor from calf thymus. J Biol Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 22.Pendurthi U R, Alok D, Rao L V M. Binding of factor VIIa to tissue factor induces alterations in gene expression in human fibroblast cells: up-regulation of poly(A) polymerase. Proc Natl Acad Sci USA. 1997;94:12598–12603. doi: 10.1073/pnas.94.23.12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raabe T, Bollum F J, Manley J L. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- 24.Raabe T, Murthy K G K, Manley J L. Poly(A) polymerase contains multiple functional domains. Mol Cell Biol. 1994;14:2946–2957. doi: 10.1128/mcb.14.5.2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richter J D. Cytoplasmic polyadenylation in development and beyond. Microbiol Mol Biol Rev. 1999;63:446–456. doi: 10.1128/mmbr.63.2.446-456.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rose K M, Jacob S T. Nuclear poly(A) polymerase from rat liver and hepatoma: comparison of properties, molecular weights and amino acid compositions. Eur J Biochem. 1976;67:11–21. doi: 10.1111/j.1432-1033.1976.tb10626.x. [DOI] [PubMed] [Google Scholar]
- 27.Ryner L C, Takagaki Y, Manley J L. Multiple forms of poly(A) polymerases purified from HeLa cells function in specific mRNA 3′-end formation. Mol Cell Biol. 1989;9:4229–4238. doi: 10.1128/mcb.9.10.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sachs A B, Sarnow P, Hentze M W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 29.Scorilas A, Talieri M, Ardavanis A, Courtis N, Dimitriadis E, Yotis J, Tsiapalis C M, Trangas T. Polyadenylate polymerase enzymatic activity in mammary tumor cytosols: a new independent prognostic marker in primary breast cancer. Cancer Res. 2000;60:5427–5433. [PubMed] [Google Scholar]
- 30.Shatkin A J, Manley J L. The ends of the affair: capping and polyadenylation. Nat Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 31.Takagaki Y, Ryner L C, Manley J L. Separation of characterization of a poly(A) polymerase and a cleavage/specificity factor required for pre-mRNA polyadenylation. Cell. 1988;52:731–742. doi: 10.1016/0092-8674(88)90411-4. [DOI] [PubMed] [Google Scholar]
- 32.Thuresson A-C, Astrom J, Astrom A, Gronvik K-O, Virtanen A. Multiple forms of poly(A) polymerases in human cells. Proc Natl Acad Sci USA. 1994;91:979–983. doi: 10.1073/pnas.91.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trangas T, Courtis N, Pangalis G A, Cosmides H V, Ioannides C, Papamichail M, Tsiapalis C M. Polyadenylic acid polymerase activity in normal and leukemic human leukocytes. Cancer Res. 1984;44:3691–3697. [PubMed] [Google Scholar]
- 34.Tuppler R, Perini G, Green M R. Expressing the human genome. Nature. 2001;409:832–833. doi: 10.1038/35057011. [DOI] [PubMed] [Google Scholar]
- 35.Vagner S, Vagner C, Mattaj I W. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- 36.Wahle E, Martin G, Schlitz E, Keller W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wallace A M, Dass B, Ravnik S E, Tonk V, Jenkins N A, Gilbert D J, Copeland N G, MacDonald C C. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc Natl Acad Sci USA. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang R-F, Wang X, Atwood A C, Topalian S L, Rosenberg S A. Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science. 1999;284:1351–1354. doi: 10.1126/science.284.5418.1351. [DOI] [PubMed] [Google Scholar]
- 39.Wilusz C J, Wormington M, Peltz S W. The cap-to-tail guide to mRNA turnover. Nat Rev Mol Cell Biol. 2001;2:237–246. doi: 10.1038/35067025. [DOI] [PubMed] [Google Scholar]
- 40.Yamauchi T, Sugimoto J, Hatakeyama T, Asakawa S, Shimizu N, Isobe M. Assignment of the human poly(A) polymerase (PAP) gene to chromosome 14q32.1-q32.2 and isolation of a polymorphic CA repeat sequence. J Hum Genet. 1999;44:253–255. doi: 10.1007/s100380050154. [DOI] [PubMed] [Google Scholar]
- 41.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao W, Manley J L. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol Cell Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao W, Manley J L. Deregulation of poly(A) polymerase interferes with cell growth. Mol Cell Biol. 1998;18:5010–5020. doi: 10.1128/mcb.18.9.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhelkovsky A M, Kessler M M, Moore C L. Structure-function relationships in the Saccharomyces cerevisiae poly(A) polymerase: identification of a novel RNA binding site and a domain that interacts with specificity factor(s) J Biol Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]