Abstract
With the proliferating studies of human cancers by single-cell RNA sequencing technique (scRNA-seq), cellular heterogeneity, immune landscape and pathogenesis within diverse cancers have been uncovered successively. The exponential explosion of massive cancer scRNA-seq datasets in the past decade are calling for a burning demand to be integrated and processed for essential investigations in tumor microenvironment of various cancer types. To fill this gap, we developed a database of Cancer Single-cell Expression Map (CancerSCEM, https://ngdc.cncb.ac.cn/cancerscem), particularly focusing on a variety of human cancers. To date, CancerSCE version 1.0 consists of 208 cancer samples across 28 studies and 20 human cancer types. A series of uniformly and multiscale analyses for each sample were performed, including accurate cell type annotation, functional gene expressions, cell interaction network, survival analysis and etc. Plus, we visualized CancerSCEM as a user-friendly web interface for users to browse, search, online analyze and download all the metadata as well as analytical results. More importantly and unprecedentedly, the newly-constructed comprehensive online analyzing platform in CancerSCEM integrates seven analyze functions, where investigators can interactively perform cancer scRNA-seq analyses. In all, CancerSCEM paves an informative and practical way to facilitate human cancer studies, and also provides insights into clinical therapy assessments.
INTRODUCTION
Since Tang et al. first reported the completely unbiased approach of whole-transcriptome mRNA sequencing at single cell resolution in 2009 (1), this field has developed rapidly over the past decade. Thanks to the studies of cellular transcriptome heterogeneity, scRNA-seq is being widely applied to answer many essential questions in developmental biology, neurosciences, oncology, and immunology (2,3). Particularly in oncology, scRNA-seq becomes indispensable means for studies in tumor microenvironment (4), heterogeneity (5), pathogenesis (6), metastasis and invasion (7) and treatment and diagnosis of diverse tumors (8). For instance, Zhang et al. depicted sequentially the dynamic immune landscape of T cells in liver cancer, non-small-cell lung cancer and hepatocellular carcinoma and also revealed novel biomarkers associated with the mal-prognosis (9–11). Maynard et al. studied metastatic lung cancer across 30 patients before and during targeted therapy, and found cancer cells that survived from treatments expressed an alveolar-regenerative cell signature, suggesting of therapy-induced primitive cell-state transition (12). Zhang et al. performed scRNA-seq analyses on immune and stromal populations from colorectal cancer patients, identifying specific macrophage and conventional dendritic cell subsets as key mediators of cellular cross-talk in the specific tumor microenvironment (TME) (13). These researches have greatly improved our understanding of human cancer progression and also undoubtedly facilitated clinical diagnoses and treatments.
However, to date there are only several single cell databases available to the public, despite of daily eruptive scRNA-seq datasets. Comprehensive databases, such as Single Cell Portal, PanglaoDB (14), Single Cell Expression Atlas (15), Human Cell Atlas Data Portal (16) and scRNASeqDB (17), catalogue a large number of single-cell expression datasets for both healthy and diseased tissues, majority of which are from human and mouse samples. These databases, in general, merely contain elementary analyses of cell clustering and differential gene expression profiling. Other disease- or cancer-specific databases like CancerSEA (18) and TISCH (19) may provide some extra valuable annotations and are widely used in tumor single-cell studies. Although CancerSEA is the first database released in 2018 to decode distinct functional of cancer cells at single-cell resolution, back then, it combined 49 cancer-related scRNA-seq datasets and 128 518 single cells. Meanwhile, it solely focused on 14 functional states of 41 900 cancer cells instead of considering immune or stromal cells in TME. As for TISCH, it integrated single-cell transcriptomic profiles of nearly 2 million cells from 76 tumor datasets across 27 cancer types, and provided detailed cell type identifications, gene expression comparison between datasets and cell types, and gene set enrichment analysis as well. Nonetheless, all the downloaded datasets in TISCH were in fixed expression matrices and had no uniform methods for reads quality control and gene expression quantification. These issues might possibly lead to analysis bias or reduce comparability across diverse databases. In sum, these integration of cross-platform single-cell datasets, accurate cell type identifications and comprehensive online analyzing platforms are still somehow insufficient, and leaving a huge challenge to human cancer studies.
Here, we present CancerSCEM (Cancer Single-cell Expression Map), a public database of collecting, analyzing, visualizing scRNA-seq data for human cancer samples. Database version 1.0 collected public scRNA-seq datasets involving 638 341 high-quality single cells from 208 samples across 20 types of human cancers. Multiscale data analyses for TME profiling and functional gene annotation were performed with in-house pipeline, and a comprehensive online analyzing platform was equipped in CancerSCEM. In short, we expect CancerSCEM will significantly reinforce scRNA-seq databases for human cancers studies.
DATA COLLECTION AND PROCESSING
Cancer single cell RNA-seq data collection and expression quantification
CancerSCEM was established from hundreds of cancer related scRNA-seq datasets from GEO (https://www.ncbi.nlm.nih.gov/geo/) (20), ArrayExpress (https://www.ebi.ac.uk/arrayexpress/) (21), Single Cell Expression Atlas – EBI (https://www.ebi.ac.uk/gxa/sc/home) (15), GSA (https://ngdc.cncb.ac.cn/gsa/) (22) and ZENODO (https://zenodo.org/record/3937811). Briefly, a total of 208 samples across 20 human cancer types and 5 construction protocols (10X Genomics, Smart-seq2, Drop-seq, Seq-Well and Microwell) were collected and processed. Raw sequencing datasets and expression matrices consist of 82.69% and 17.31%, respectively (Supplementary Table S1). For data from 10X Genomics platform, we consistently used Cell Ranger v5.0 (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome) (23) with its built-in reference data refdata-gex-GRCh38-2020-A.tar.gz to construct UMI (unique molecular identifier) count matrix for each individual sample. For each non-10X Genomics dataset, a standard strict reads quality control was initially performed by Fastp v0.20.0 (https://github.com/OpenGene/fastp) (24) and Trimmomatic v0.36 (http://www.usadellab.org/cms/?page=trimmomatic) (25). zUMIs v2.9.4f (https://github.com/sdparekh/zUMIs) was then adopted for high-quality reads alignment and gene expression quantification (26).
Cell quality control, unsupervised clustering and ‘three-step’ cell-type annotation
After expression matrix construction, DoubletFinder v2.2.0 (https://github.com/comodr/doubletfinder) was applied to doublets removal (7% per 10 000 cells) for all datasets (27), and R package Seurat v3.2.3 (https://satijalab.org/seurat/) was utilized to perform a series of analyses: cell quality control (200 ≤ nfeatures ≤ 5000 and MT < 10%), PCA (Principal Component Analysis) dimension reduction, tSNE (t-distributed Stochastic Neighbor Embedding) and UMAP (Uniform Manifold Approximation and Projection) clustering with personalized principal component numbers and clustering resolutions and etc. (28). Crucially, based on the collection and curation of biomarker genes for different cell types from Cell Marker database (29), cell-type annotation in CancerSCEM was achieved by a combined ‘three-step’ strategy: (i) scCancer v2.2.0 (https://github.com/HealthVivo/scCancer) (30) and CopyKAT v1.0.4 (https://github.com/navinlabcode/copykat) (31) were used for the copy number variation assessment for 10X Genomics and other sequencing datasets, respectively. A group of marker genes, such as EPCAM, KRT8, KRT18, KRT19 and EGFR in glioblastoma cells that represent cancer cells or cancer stem cells, were investigated in parallel. Cells with significantly abnormal CNV levels and high expression levels of above marker genes were defined as malignant cells, (ii) Manual annotations were subsequently performed based on the expression of dozens of canonical markers for common cell types like T cells (e.g.CD3D, CD3E), B cells (e.g. MS4A1, BANK1), Macrophages/Monocytes (e.g. CD68, CD14), Mast cells (e.g. SLC18A2, ASIC4), Endothelial cells (e.g. VWF, PECAM1), Fibroblasts (e.g. FAP, NECTIN1), Oligodendrocytes (e.g. OLIG1, PLP1) and Astrocytes (e.g. SLC1A3, GFAP) and etc. A complete list of marker genes used in our database is accessible on the Documents webpage. Only when a considerable number of marker genes have significantly specific expression in the target cell cluster (P-value < 0.01), their corresponding cell types can be determined, (iii) SingleR has been among the top tools in cell type annotation analysis in recent years (32,33), here we chose SingleR v1.4.1 (https://github.com/dviraran/SingleR) for immune cell subtype recognition (34). We further classified T cells and B cells into subtypes in CancerSCEM. In samples of Mixed phenotype Acute Leukemia and Acute Myeloid Leukemia, multiple cell types except malignant cells were identified due to the complications of cell type annotation in liquid tumor. Finally, a total of 33 cell types including immune cell subtypes were identified, and the FindMarkers function in Seurat was used to perform differential gene expression analysis for each specific cell type (28).
In-depth TME profiling at single-cell resolution and TCGA bulk RNA-seq data integration
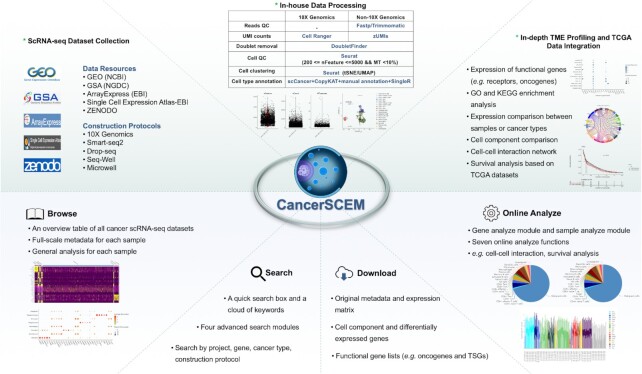
In addition to the conventional single cell analyses mentioned above, a series of special analyses were performed as well. First, thousands of functional receptor-ligand gene-pairs were curated from multiple data resources including CelltalkDB (35), SingleCellSingalR (36), Cellinker (37), Cell–Cell Interaction Database (38), and another review article (39). Oncogenes and tumor suppressor genes (TSGs) were collected from Cancer Gene Census (CGC) (40), OncoKB (41), Network of Cancer Genes (NCG) (42), TSGene (43), IntOGene (44) and another cancer gene clinical care study (45). We further screened out those genes supported by at least three data resources and exhibited their original expression patterns in our database. Second, we collected bulk RNA-seq data across 13 cancer projects from TCGA (46) and profiled the expression patterns for these target gene in different cancer types at tissue level. These profiles serve as references to the single-cell-level expression studies. Third, GO and KEGG enrichment analyses on differentially expressed genes were performed for each specific cell type using R package clusterProfiler v3.14.3 (http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html) (47–49). Gene expression correlation was calculated by R package Hmisc v4.4.2 (https://cran.r-project.org/web/packages/Hmisc/index.html). Cell component comparison was assessed by both chi-square test and Wilcoxon signed rank test. Cell–cell interaction networks were constructed by CellphoneDB (https://www.cellphonedb.org/) (50). Survival analysis was performed by R package survival v3.2.7 (https://cran.r-project.org/web/packages/survival/index.html) and survminer v0.4.9 (https://github.com/kassambara/survminer). Static figures in the database were created by ggplot2 v3.3.3 (https://github.com/tidyverse/ggplot2) (51), and an overview of the data processing workflow is shown in Figure 1.
Figure 1.
An overview of data processing (above) and four functional modules (below) equipped in CancerSCEM. Cancer scRNA-seq data were mainly collected from five data resources. Beside conventional analysis, several advanced analyses such as cell interaction network construction and survival analysis were also performed. To support visualization and exploration, a user-friendly web interface for CancerSCEM was developed where users can browse, search, online analyze and download all the metadata and analytical results of interest.
DATABASE IMPLEMENTATION
CancerSCEM was constructed by standard database development techniques. Thymeleaf (a Java template engine), HTML5, CSS, AJAX, JQuery and Bootstrap were used for rendering and interactive operations of front-end pages. Spring Boot was used as the basic architecture of the back-end system. MySQL was served as a container for data storage and Mybatis as an accesser to the container. Echarts, Highcharts, svg3dtagcloud.js and plotly.js were adopted for building interactive graphs. Bootstrap Table was used to construct data tables, and finally, bioinformatics analyses were implemented by in-house R scripts. To better comply with CancerSCEM infrastructure, we advise following browsers: Google Chrome (v56.0 and up), Opera (v53.0 and up), Safari (v11.1 and up) or Firefox (v64.0 and up).
DATABASE CONTENTS AND FEATURES
Cancer scRNA-seq dataset summary and additional statistics
CancerSCEM was originally designed to extensively collect all public single-cell RNA-seq datasets of diverse human cancer types, and investigate the immune profiles and gene expression dynamics in the specific TME. Database version 1.0 was released online on 12 June 2021, including full-scale metadata and multi-level analytical results of a total of 208 scRNA-seq datasets (samples) across 28 projects and 20 human cancer types. After reads and cell quality control, 638 341 high-quality cells were reserved. The datasets collected from GEO (NCBI), GSA (NGDC), ArrayExpress (EBI), Single Cell Expression Atlas – EBI and ZENODO accounted for 139 (66.83%), 24 (11.54%), 23 (11.06%), 16 (7.69%) and 6 (2.88%), respectively. Cancer types involved in the database and the number of samples within each cancer type were listed in Supplementary Table S1. Among these samples, 125 (60.1%), 39 (18.75%), 36 (17.31%), 4 (1.92%) and 4(1.92%) datasets were produced by 10X Genomics, Microwell, Seq-Well, Smart-seq2 and Drop-seq techniques, respectively. Cell counts in each sample ranged from 105 in LUSC to 28 764 in GBM with the median of 2 270, were partially associated with the sequencing techniques. The average UMI counts and detected gene counts of each sample across 208 samples ranged correspondingly from 558 to 927 701 and from 358 to 3 971. Statistically, there are 33 cell types were finalized in CancerSCEM. A total number of 36 601 genes including 36 widely-used immune checkpoint molecules, 3 853 receptor-ligand pairs, 488 oncogenes and 523 TSGs were associated with gene expression analyses, and 13 TCGA cancer projects covering 5 554 donors (TCGA-BLCA, TCGA-LUAD, TCGA-COAD and etc.) were adopted in CancerSCEM for tissue-level gene expression profiling and online survival analysis.
A browse interface for retrieving cancer scRNA-seq datasets
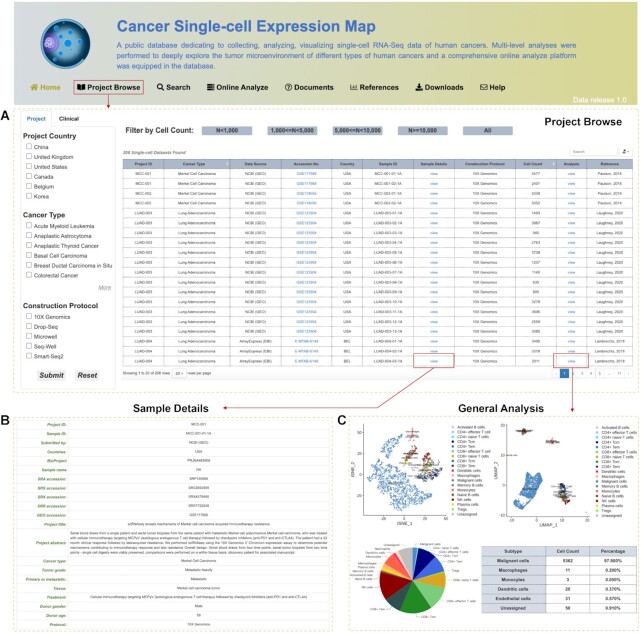
Multiscale data analyses for 208 cancer samples were processed at single-cell resolution, and users can browse, search, online analyze and download all the metadata and analytical results of interest. An overview and interactive table on the Project Browse page present all collected cancer scRNA-seq projects, with information ranging from the unique newly-assigned project ID, cancer type, sample ID, sample details, cell count to the data construction protocol (Figure 2A). The Sample Details and Analysis columns in the table additionally provide hyperlinks to the detailed information of the tumor sample (Figure 2B, e.g. accession number, donor age, tumor grade and clinical treatments) and the comprehensive analytical results for each dataset, respectively. In detail, there are three main modules on the Analysis page, consisting of Data Statistics and tSNE/UMAP Visualization, Tumor Microenvironment and Functional Genes' Expression (Figure 2C). The cell-type annotation, cell component (including malignant cells, immune cells and stromal cells) and their corresponding functional enrichment analyses on differentially expressed genes show the landscape of particular TME. The changes in the composition of different immune cells and stromal cells can confine or promote tumor growth and progression depending on the context according to previous studies (52,53). The list of significantly positively expressed genes in different cell types exhibites the information of essential biomarkers for single-cell studies. Moreover, the expression patterns of these collected high-confidence functional genes (60 receptors, 42 ligands, 205 oncogenes and 228 TSGs) will undoubtedly improve our understanding of their potential impacts on diverse tumors.
Figure 2.
Demonstration of browse interfaces in CancerSCEM. (A) An overview table of all collected cancer scRNA-seq projects and samples. (B) An extended page presenting the full-scale metadata of each cancer sample. (C) A general analysis page including comprehensive analytical results related to the cancer TME and functional gene expression dynamics.
User-friendly searching modules for data inquiry
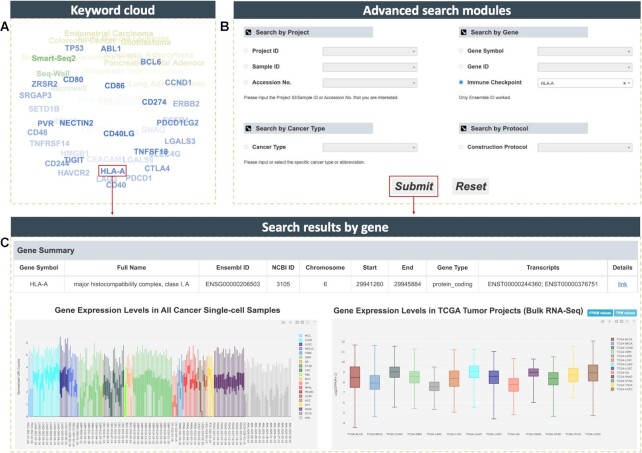
To better access datasets or genes of interest, CancerSCEM provides several inquiry methods: (i) A quick search box and a keyword cloud (Figure 3A) are presented on the home page, and both equipped for real-time querying by specifying cancer type, gene or data protocol; (ii) Four advanced search modules on the search page (Figure 3B): in term of projects, users can specify a project/sample ID or an accession number, or select a particular cancer type or construction protocol, an overview of the target projects or datasets as well as comprehensive analytical results will be obtained (Figure 2); in terms of genes, by searching a gene symbol or gene ID users can quickly view gene summary and expression distributions at both single-cell and bulk RNA-seq levels (Figure 3C). Currently, the immune checkpoint molecules such as CTLA4 and PDCD1 were widely-used as targets for immunotherapies in clinical practice (54,55). However, it is still unclear which treatment to different cancer patients will respond more effectively, and thus, this question drives further investigations into optimal co-inhibitory receptors for each cancer type (56,57). To solve this issue, 36 widely-used immune checkpoint molecules such as PDCD1, TIGIT and LAG3 were listed in both keyword cloud and the Search by Gene module, researchers and clinicians thereby have a direct access to their expression profiles across different cancer types, and quickly narrow down the best targets for further researches or immunotherapies.
Figure 3.
Illustration of data querying in CancerSCEM. (A) Keyword cloud is shown on the home page including a majority of cancer types, data sequencing techniques, and several biological or clinical gene symbols. (B) Four advanced searching modules queried by project, gene, cancer type or protocol. (C) Returned results including gene summary, gene expression profiles across both single cell and bulk RNA-seq datasets.
Comprehensive multiple-dimensional online data exploration
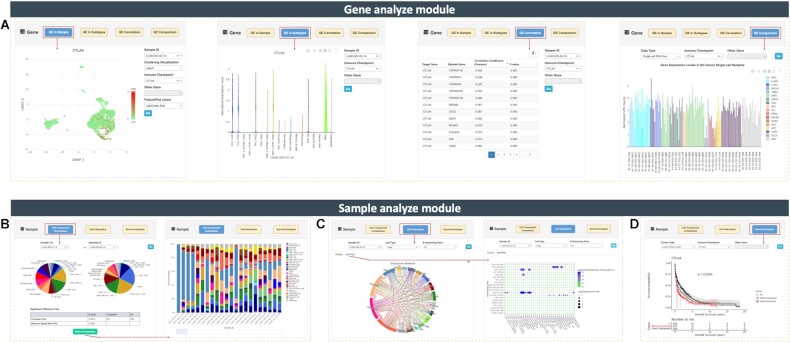
Gene analyzing module. It contains four functions: (i) Gene Expression (GE) in Sample—whole expression profiling of the target gene in specified cancer single-cell sample in tSNE or UMAP manner; (ii) GE in Subtypes—gene expressions in different cell types or subtypes in the sample; (iii) GE Correlation—pearson correlation calculation between any two genes in the specific sample ranked by P-values and (iv) GE Comparison—gene expression comparison across different scRNA-seq or TCGA bulk RNA-seq datasets (Figure 4A). With the addition of these functions, users can find out cell types that contribute the highest expressions of a given gene, and further uncover their underlying biological roles during tumor genesis and development. Users can also compare the expression of a given gene (especially for 36 immune checkpoint molecules) across all different cancer types at both single-cell or tissue levels, and it is much easier to identify novel predictive biomarkers (e.g. HMGB1 significantly highly expressed in mixed-phenotype acute leukemia).
Figure 4.
Screen captures of seven online analyzing functions equipped in CancerSCEM. (A) Four analyze functions equipped in Gene Analyze module. (B) Cell component comparison and overall cell component across 208 samples. (C) Cell–cell interaction networks with quantifying expression intensities of receptor-ligand pairs. (D) Survival analysis based on TCGA bulk RNA-seq data and clinical survival data.
Cell component comparison in sample analyzing module
Cell component and proportion, consisting of malignant/immune/stromal cells, can be compared between any two single-cell samples using chi-square test and Wilcoxon signed rank test (Figure 4B, left panel). Users can evaluate the global immunity of different samples or different cancer types according to the immune cell proportion. Diverse composition of immune and stromal cells may impact tumor growth and progression on different ways (52). For example, the more CD8 + effector T cells, the higher cytotoxic activities the tissue harvests, thus might damage some specific foreign antigens or malignant tumor cells (58). By clicking on View All Samples button, the cell component for 208 samples will pop up (Figure 4B, right panel).
Cell interaction network construction across different cell types
Increasing cell interaction evidence suggests that cell–cell interaction plays critical roles in driving cancer progression (13,59). Therefore, a function of cell-cell interaction network construction was integrated in Sample Analyze module. By using the expressions of receptor–ligand pairs, the biological relevance across different cell types can be quantified within the dynamic tumor ecosystem. Users need to specify a sample and a cell type, by which, an interaction network circos image (http://circos.ca/) will be generated with a positive integral on each connection. These connections represent the communication intensity between two cell types (Figure 4C, left panel). For example, the majority of the samples are harboring endothelial cells which, in general, actively and widely interact with other cell types (e.g. macrophages, dendritic cells and fibroblasts). A maximum of 50 pairs can be displayed with interacting intensities and calculated P-values (Figure 4C, right panel).
Survival analysis in sample analyzing module
Based on the bulk RNA-seq data and clinical survival data collected from TCGA, the module of online survival analysis allows users, especially clinicians, rapidly grasp the correlation between the expression levels of a given gene and clinical prognosis of the specified cancer type. This may also aid in fishing out useful candidate genes for constructing prognosis prediction panel in clinical practice, and ultimately serve in the targeting therapy in future. Here, we provide a case study to demonstrate the excellent performance of this module. By applying survival analysis, we profiled a high expression of CTLA4. This gene is significantly associated with worse prognosis of lung adenocarcinoma with P-value < 0.0001 (Figure 4D). CTLA4 ia also well-known as a immune checkpoint molecule and a cytotoxic T lymphocyte associated protein coding gene. The protein of CTLA4 is known as CD152 which is one of the leukocyte differentiation antigens. CTLA4 binding and molecule B7 liganding can effectively induce the T cell non-responsiveness, resulting in the negative regulation of immune response (60). We further double checked the expression profiles of CTLA4 in Gene Analyze module and, not surprisingly, CTLA4 significantly highly expressed in some T cell subtypes especially in regulatory T cells (Tregs) of LUAD samples (Figure 4A), evidently identical with previous studies (61). Taken together, this online anlysis using both single-cell and bulk RNA sequencing data proves our platform is a powerful tool for cancer scRNA-Seq data exploration in a neoteric interactive mode.
Data retrieving and maintenance
Users can perform customized analyses of their choice by retrieving a varity of information: original metadata with raw sequencing dataset sources, normalized gene expression matrices, cell component for each single-cell datasets and differential expressed gene list for each specific cell type. The complete lists of biological and clinical functional molecules (receptors, ligands, oncogenes and TSGs) and cell interaction analytical results are available as well.
DISCUSSION AND FUTURE DIRECTIONS
The rapid growth of cancer single cell sequencing datasets rises various challenges for scRNA-seq data studies. Data integration into a comprehensive platform across different experimentations, different samples, even different species is one of these challenges. Besides, inter-experimental comparisons and visual presentations are problematic as well. Several large-scale studies upon human cancers have depicted the dynamic immune landscapes and revealed novel biomarkers associated with the poor prognosis in clinical trials (9–11). Some studies obtained the inner primitive cell-state transition mechanisms (12) and identified key mediators of cellular cross-talk in the specific TME (13). To better and easier compare diverse human cancer types, a public open access and cancer-specific single-cell expression database for diverse types of human cancers, CancerSCEM, was developed. In comparison with existing single cell expression databases, CancerSCEM mainly features: (i) the relatively consistant data processing and the tailored ‘three-step’ strategy for cell type annotation, combining multiple-level information from numerous tools and manual annotations, significantly improve the accuracy of cell type identification; (ii) it has the competency of hosting more abundant analyses such as gene expression of functional molecules, cell component and diversity analysis, cell–cell interactions and survival analysis; (iii) it integrates of an unprecedented online analyzing functions (two modules and seven functions) and facilitates cancer single-cell data explorations in multiple levels and (iv) it offers the availability of expression profiles for all curated immune checkpoint molecules.
By comprehensively profiling the expression of immune checkpoint molecules across 208 single-cell samples, we observed that HMGB1, LGALS3 and CD48 have expressed in almost all cancer types with significantly higher expressions in average. This suggests they have the universal roles in clinical cancer immunotherapies. While in contrast, the expression of several widely-used checkpoints (such as LAG3, PDCD1 and TIGIT) exhibited dynamic variation characteristics across different samples or cancer types. This might demonstrate the inconsistent responses of patients in immunotherapy by using these molecules (Supplementary Figure S1). Taken all, CancerSCEM presents a functionally complete, human cancer-centered, multi-level analyzed and broadly applicable scRNA-seq resource. We believe that CancerSCEM will facilitate our understanding of cancer TMEs and tumorigenesis mechanisms at single-cell resolution, and also will provide valuable information for cancer preventions and clinical researches.
To date, CancerSCEM has already catalogued 208 scRNA-seq datasets across 20 human cancer types. With numerous new cancer single cell projects being carried out, more scRNA-seq datasets are being conducted in CancerSCEM. This will cover more cancer types, such as esophageal carcinoma and uveal melanoma (62,63). Moreover, other sequencing data types like spatial transcriptome data, chromatin accessibility data, DNA methylation data, histone modifications data and chromosome conformation data at single-cell resolution are also expected to be integrated for better understanding the TME profiles and regulatory mechanisms of diverse cancers (64). In addition, we will be constructing an effective method for cell type annotation of liquid tumors. In the long run, a batch of novel practical tools will be integrated in our database to facilitate network modeling, malignant cell categorizing, clinical immunotherapy response prediction and cross-experimental or cross-sample dataset integration (65,66), which will undoubtedly improve graphical user interface experience. In a word, as one of most important database resources in National Genomics Data Center, CancerSCEM will be continuously interpretating more datasets, developing more handy analytical modules and ultimately, supporting global human cancer researches, clinical applications and beyond.
DATA AVAILABILITY
CancerSCEM is a database of single-cell expression map across various human cancers (https://ngdc.cncb.ac.cn/cancerscem).
Supplementary Material
ACKNOWLEDGEMENTS
We thank a number of users for reporting bugs and providing suggestions. The results of bulk expression profiling and survival analysis published here are based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga.
Contributor Information
Jingyao Zeng, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Yadong Zhang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Yunfei Shang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jialin Mai, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Shuo Shi, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Mingming Lu, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Congfan Bu, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Zhewen Zhang, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Zaichao Zhang, Department of Biology, The University of Western Ontario, London, Ontario N6A 5B7, Canada.
Yang Li, Beijing Tongren Eye Center, Beijing key Laboratory of Intraocular Tumor Diagnosis and Treatment, Beijing Tongren Hospital, Capital Medical University, Beijing 100730, China.
Zhenglin Du, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China.
Jingfa Xiao, National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38030400]; National Natural Science Foundation of China [31970634 and 31771465]; National Key Research Program of China [2016YFB0201702, 2020YFA0907001]; CAS Key Technology Talent Program (to Z.D.); Specialized Research Assistant Program of the Chinese Academy of Sciences [202044]; China Postdoctoral Science Foundation [2021M693109]. Funding for open access charge: National Natural Science Foundation of China [31970634].
Conflict of interest statement. None declared.
REFERENCES
- 1. Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau J., Tuch B.B., Siddiqui A.et al.. mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods. 2009; 6:377–382. [DOI] [PubMed] [Google Scholar]
- 2. Li L., Xiong F., Wang Y., Zhang S., Gong Z., Li X., He Y., Shi L., Wang F., Liao Q.et al.. What are the applications of single-cell RNA sequencing in cancer research: a systematic review. J. Exp. Clin. Cancer Res. 2021; 40:163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhang Y., Wang D., Peng M., Tang L., Ouyang J., Xiong F., Guo C., Tang Y., Zhou Y., Liao Q.et al.. Single-cell RNA sequencing in cancer research. J. Exp. Clin. Cancer Res. 2021; 40:81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ren X., Zhang L., Zhang Y., Li Z., Siemers N., Zhang Z.. Insights gained from single-cell analysis of immune cells in the tumor microenvironment. Annu. Rev. Immunol. 2021; 39:583–609. [DOI] [PubMed] [Google Scholar]
- 5. Yuan J., Levitin H.M., Frattini V., Bush E.C., Boyett D.M., Samanamud J., Ceccarelli M., Dovas A., Zanazzi G., Canoll P.et al.. Single-cell transcriptome analysis of lineage diversity in high-grade glioma. Genome Med. 2018; 10:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fendler A., Bauer D., Busch J., Jung K., Wulf-Goldenberg A., Kunz S., Song K., Myszczyszyn A., Elezkurtaj S., Erguen B.et al.. Inhibiting WNT and NOTCH in renal cancer stem cells and the implications for human patients. Nat. Commun. 2020; 11:929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geng S., Wang J., Zhang X., Zhang J.J., Wu F., Pang Y., Zhong Y., Wang J., Wang W., Lyu X.et al.. Single-cell RNA sequencing reveals chemokine self-feeding of myeloma cells promotes extramedullary metastasis. FEBS Lett. 2020; 594:452–465. [DOI] [PubMed] [Google Scholar]
- 8. Zhang J., Guan M., Wang Q., Zhang J., Zhou T., Sun X.. Single-cell transcriptome-based multilayer network biomarker for predicting prognosis and therapeutic response of gliomas. Brief. Bioinform. 2020; 21:1080–1097. [DOI] [PubMed] [Google Scholar]
- 9. Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q.et al.. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017; 169:1342–1356. [DOI] [PubMed] [Google Scholar]
- 10. Guo X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R.et al.. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018; 24:978–985. [DOI] [PubMed] [Google Scholar]
- 11. Zhang Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D.et al.. Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. 2019; 179:829–845. [DOI] [PubMed] [Google Scholar]
- 12. Maynard A., McCoach C.E., Rotow J.K., Harris L., Haderk F., Kerr D.L., Yu E.A., Schenk E.L., Tan W., Zee A.et al.. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 2020; 182:1232–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., He Y., Wang L., Zhang Q., Kim A.et al.. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020; 181:442–459. [DOI] [PubMed] [Google Scholar]
- 14. Franzen O., Gan L.M., Bjorkegren J.L.M.. PanglaoDB: a web server for exploration of mouse and human single-cell RNA sequencing data. Database (Oxford). 2019; 2019:baz046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Papatheodorou I., Moreno P., Manning J., Fuentes A.M., George N., Fexova S., Fonseca N.A., Fullgrabe A., Green M., Huang N.et al.. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020; 48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lindeboom R.G.H., Regev A., Teichmann S.A.. Towards a human cell atlas: taking notes from the past. Trends Genet. 2021; 37:625–630. [DOI] [PubMed] [Google Scholar]
- 17. Cao Y., Zhu J., Jia P., Zhao Z.. scRNASeqDB: a database for RNA-seq based gene expression profiles in human single cells. Genes. 2017; 8:368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z.et al.. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; 47:D900–D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sun D., Wang J., Han Y., Dong X., Ge J., Zheng R., Shi X., Wang B., Li Z., Ren P.et al.. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021; 49:D1420–D1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets - update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Athar A., Fullgrabe A., George N., Iqbal H., Huerta L., Ali A., Snow C., Fonseca N.A., Petryszak R., Papatheodorou I.et al.. ArrayExpress update - from bulk to single-cell expression data. Nucleic Acids Res. 2019; 47:D711–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y.et al.. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021; 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng G.X., Terry J.M., Belgrader P., Ryvkin P., Bent Z.W., Wilson R., Ziraldo S.B., Wheeler T.D., McDermott G.P., Zhu J.et al.. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen S., Zhou Y., Chen Y., Gu J.. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parekh S., Ziegenhain C., Vieth B., Enard W., Hellmann I.. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. GigaScience. 2018; 7:giy059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mcginnis C.S., Murrow L.M., Gartner Z.J.. DoubletFinder: doublet detection in single-cell RNA sequencing data using artificial nearest neighbors. Cell Syst. 2019; 8:329–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hao Y., Hao S., Andersen-Nissen E., Mauck W.M. 3rd, Zheng S., Butler A., Lee M.J., Wilk A.J., Darby C., Zager M.et al.. Integrated analysis of multimodal single-cell data. Cell. 2021; 184:3573–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M.et al.. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019; 47:D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo W., Wang D., Wang S., Shan Y., Liu C., Gu J.. scCancer: a package for automated processing of single-cell RNA-seq data in cancer. Brief. Bioinform. 2021; 22:bbaa127. [DOI] [PubMed] [Google Scholar]
- 31. Gao R., Bai S., Henderson Y.C., Lin Y., Schalck A., Yan Y., Kumar T., Hu M., Sei E., Davis A.et al.. Delineating copy number and clonal substructure in human tumors from single-cell transcriptomes. Nat. Biotechnol. 2021; 39:599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bhattacharya A., Hamilton A.M., Troester M.A., Love M.I.. DeCompress: tissue compartment deconvolution of targeted mRNA expression panels using compressed sensing. Nucleic Acid Res. 2021; 49:e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Amezquita R.A., Lun A.T.L., Becht E., Carey V.J., Carpp L.N., Geistlinger L., Marini F., Rue-Albrecht K., Risso D., Soneson C.et al.. Orchestrating single-cell analysis with Bioconductor. Nat. Methods. 2020; 17:137–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R.et al.. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019; 20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shao X., Liao J., Li C., Lu X., Fan X.. CellTalkDB: a manually curated database of ligand-receptor interactions in humans and mice. Brief. Bioinform. 2021; 22:bbaa269. [DOI] [PubMed] [Google Scholar]
- 36. Cabello-Aguilar S., Alame M., Kon-Sun-Tack F., Fau C., Lacroix M., Colinge J.. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 2020; 48:e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Y., Liu T., Wang J., Zou B., Li L., Yao L., Chen K., Ning L., Wu B., Zhao X.et al.. Cellinker: a platform of ligand-receptor interactions for intercellular communication analysis. Bioinformatics. 2021; 37:2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ximerakis M., Lipnick S.L., Innes B.T., Simmons S.K., Adiconis X., Dionne D., Mayweather B.A., Nguyen L., Niziolek Z., Ozek C.et al.. Single-cell transcriptomic profiling of the aging mouse brain. Nat. Neurosci. 2019; 22:1696–1708. [DOI] [PubMed] [Google Scholar]
- 39. Armingol E., Officer A., Harismendy O., Lewis N.E.. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 2021; 22:71–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sondka Z., Bamford S., Cole C.G., Ward S.A., Dunham I., Forbes S.A.. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018; 18:696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chakravarty D., Gao J., Phillips S.M., Kundra R., Zhang H., Wang J., Rudolph J.E., Yaeger R., Soumerai T., Nissan M.H.et al.. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Repana D., Nulsen J., Dressler L., Bortolomeazzi M., Venkata S.K., Tourna A., Yakovleva A., Palmieri T., Ciccarelli F.D.. The Network of Cancer Genes (NCG): a comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019; 20:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhao M., Pora K., Ramkrishna M., Zhao J., Zhao Z.. TSGene 2.0: an updated literature-based knowledgebase for tumor suppressor genes. Nucleic Acids Res. 2016; 44:D1023–D1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martinez-Jimenez F., Muinos F., Sentis I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C., Mularoni L., Pich O., Bonet J., Kranas H.et al.. A compendium of mutational cancer driver genes. Nat. Rev. Cancer. 2020; 20:555–572. [DOI] [PubMed] [Google Scholar]
- 45. Okur V., Chung W.K.. The impact of hereditary cancer gene panels on clinical care and lessons learned. Cold Spring Harb. Mol. Case Stud. 2017; 3:a002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tomczak K., Czerwinska P., Wiznerowicz M.. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp. Oncol. 2015; 19:A68–A77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yu G., Wang L.G., Han Y., He Q.Y.. ClusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gene Onotology Consortium. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R.. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020; 15:1484–1506. [DOI] [PubMed] [Google Scholar]
- 51. Klaus and Galensa. ggplot2: elegant graphics for data (2nd ed.). Comput. Rev. 2017; 58:457–458. [Google Scholar]
- 52. Rozenblatt-Rosen O., Regev A., Oberdoerffer P., Nawy T., Hupalowska A., Rood J.E., Ashenberg O., Cerami E., Coffey R.J., Demir E.. The human tumor atlas network: charting tumor transitions across space and time at single-cell resolution. Cell. 2020; 181:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Atiya H., Frisbie L., Pressimone C., Coffman L.. Mesenchymal stem cells in the tumor microenvironment. Adv. Exp. Med. Biol. 2020; 1234:31–42. [DOI] [PubMed] [Google Scholar]
- 54. Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S.et al.. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014; 371:2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boussiotis V.A. Molecular and biochemical aspects of the PD-1 checkpoint pathway. N. Engl. J. Med. 2016; 375:1767–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Anderson A.C., Joller N., Kuchroo V.K.. Lag-3, Tim-3, and TIGIT: co-inhibitory receptors with specialized functions in immune regulation. Immunity. 2016; 44:989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Eskiocak U., Guzman W., Wolf B., Cummings C., Milling L., Wu H.J., Ophir M., Lambden C., Bakhru P., Gilmore D.C.. Differentiated agonistic antibody targeting CD137 eradicates large tumors without hepatotoxicity. JCI Insight. 2020; 5:e133647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Farhood B., Najafi M., Mortezaee K.. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: a review. J. Cell. Physiol. 2019; 234:8509–8521. [DOI] [PubMed] [Google Scholar]
- 59. Zhang M., Yang H., Wan L., Wang Z., Wang H., Ge C., Liu Y., Hao Y., Zhang D., Shi G.et al.. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 2020; 73:1118–1130. [DOI] [PubMed] [Google Scholar]
- 60. Bassi R., Fornoni A., Doria A., Fiorina P.. CTLA4-Ig in B7-1-positive diabetic and non-diabetic kidney disease. Diabetologia. 2016; 59:21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ji D., Song C., Li Y., Xia J., Wu Y., Jia J., Cui X., Yu S., Gu J.. Combination of radiotherapy and suppression of Tregs enhances abscopal antitumor effect and inhibits metastasis in rectal cancer. J. Immunother. Cancer. 2020; 8:e000826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yang L., Zhang X., Hou Q., Huang M., Zhang H., Jiang Z., Yue J., Wu S.. Single-cell RNA-seq of esophageal squamous cell carcinoma cell line with fractionated irradiation reveals radioresistant gene expression patterns. BMC Genomics. 2019; 20:611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Durante M.A., Rodriguez D.A., Kurtenbach S., Kuznetsov J.N., Sanchez M.I., Decatur C.L., Snyder H., Feun L.G., Livingstone A.S., Harbour J.W.. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020; 11:496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stuart T., Satija R.. Integrative single-cell analysis. Nat. Rev. Genet. 2019; 20:257–272. [DOI] [PubMed] [Google Scholar]
- 65. Saadatpour A., Lai S., Guo G., Yuan G.C.. Single-cell analysis in cancer genomics. Trends Genet. 2015; 31:576–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Suva M.L., Tirosh I.. Single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol. Cell. 2019; 75:7–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
CancerSCEM is a database of single-cell expression map across various human cancers (https://ngdc.cncb.ac.cn/cancerscem).