Abstract
Viral infectious diseases are a devastating and continuing threat to human and animal health. Receptor binding is the key step for viral entry into host cells. Therefore, recognizing viral receptors is fundamental for understanding the potential tissue tropism or host range of these pathogens. The rapid advancement of single-cell RNA sequencing (scRNA-seq) technology has paved the way for studying the expression of viral receptors in different tissues of animal species at single-cell resolution, resulting in huge scRNA-seq datasets. However, effectively integrating or sharing these datasets among the research community is challenging, especially for laboratory scientists. In this study, we manually curated up-to-date datasets generated in animal scRNA-seq studies, analyzed them using a unified processing pipeline, and comprehensively annotated 107 viral receptors in 142 viruses and obtained accurate expression signatures in 2 100 962 cells from 47 animal species. Thus, the VThunter database provides a user-friendly interface for the research community to explore the expression signatures of viral receptors. VThunter offers an informative and convenient resource for scientists to better understand the interactions between viral receptors and animal viruses and to assess viral pathogenesis and transmission in species. Database URL: https://db.cngb.org/VThunter/.
INTRODUCTION
The COVID-19 pandemic has caused huge loss of human life, economic recession, and social disruption worldwide, underscoring the destructive impact of infectious diseases on human health and global security. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can infect various animals, including bats, pangolins, cats, dogs, ferrets and minks (1–6). With the occurrence of anthropogenic transmission of SARS-CoV-2 to animals, the potential host range of this virus continues to raise concerns within the scientific community. Moreover, the emergence of SARS-CoV-2 also highlights the need for more rapid host range identification upon the emergence of novel pathogens.fun
In recent decades, tremendous achievements have been made to characterize the host range and tissue tropism of viruses using traditional methods like epidemiological investigations or animal infection experiments. While these approaches are essential to elucidate bona fide viral infection in animals, it is impossible to carry out large-scale screening on the versatile species that might be susceptible to this pathogen, due to the limited availability of virus/animal/experimental resources. Recent advances in scRNA-seq technology has opened up new ways to identify all cell types in various tissues and/or organs and profile gene expression landscapes at single cell resolution, which holds tremendous potential to predict the potential cell types, tissues and organs targeted by viruses based on the expression profiles of their receptors in all cell types. For example, we conducted scRNA-seq for 11 representative species in pets, livestock, poultry, and wildlife to build the expression pattern profiles of all cell types to screen the potential target cell types and hosts of SARS-CoV-2 in a previous study (7), where cats were found to be highly susceptible to SARS-CoV-2, in accordance with serological and experimental findings by other researchers (3,6,8). Because cellular receptors play a critical role in the cell entry process of virus, identifying the tissue tropism is the first step towards understanding the pathogenesis and transmission of viruses in different hosts, thus laying the foundation for the prevention and control of putative outbreaks (9). Predicting host and tissue tropism based on comprehensive gene expression patterns at single-cell resolution is promising but presents challenges for laboratory biologists and experimentalists as the huge amount of data obtained from scRNA-seq studies can be daunting for those with limited backgrounds in bioinformatics.
Several databases have been developed to make available the rapidly increasing volume of scRNA-seq data. Raw data and expression matrix datasets produced in scRNA-seq studies can be submitted to several freely available primary archives, such as ArrayExpress (10), and Gene Expression Omnibus (GEO) (11) for academic publication. In addition, several value-added databases have been developed based on manual curation and comprehensive integration of numerous datasets produced in scRNA-seq studies, such as CancerSEA (12), CellMarker (13), SC2disease (14) and TISCH (15), which are mainly produced for researches on human disease and cancers. Currently, multidimensional integrating analysis between viral receptor information and all publicly available gene expression patterns profiled by scRNA-Seq to determine the host tropism of animal viruses is in urgent need. Unfortunately, there is no comprehensive database available for bench scientists and researchers to conveniently obtain viral receptor expression information on the tissue/organ specific cell types of the various animal species.
To fill this gap, we collected and manually curated 285 up-to-date scRNA-seq datasets, which included 2 100 962 cells from 47 animal species. We analyzed the datasets using a unified processing pipeline, integrated them with expert-curated receptor information of 142 viruses, and obtained accurate expression signatures of the viral receptors in 47 animal species. Information on viral receptor expression signatures is fundamental for understanding the molecular mechanisms underlying host infection by viruses. Thus, we also developed a comprehensive and user-friendly database, named VThunter, to ensure that the curated data were publicly available and could be easily utilized. In short, VThunter is a-value-added database with transformative information to facilitate study of the cross-species transmission mechanisms of animal viruses.
DATA COLLECTION AND DATABASE CONTENT
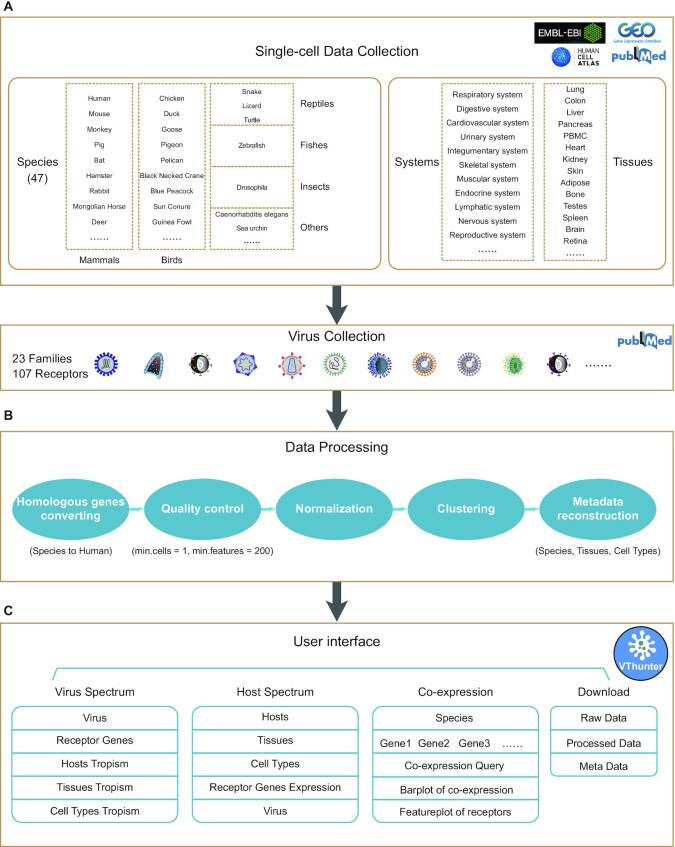
In total, 285 animal scRNA-seq datasets generated from 2 100 962 cells in 47 animal species were collected and used to predict the cell types targeted by viruses (Figure 1A, Supplementary Data 1 and Supplementary Data 2). The list of these 285 scRNA-Seq datasets is available on the database ‘Download’ page, and includes detailed metadata for each dataset, such as data source, time, technology, species name, sample tissue, treatment, cell number and URL for related literature. scRNA-seq datasets were retrieved based on literature search and downloaded from multiple scRNAseq data repositories including Gene Expression Omnibus (NCBI/GEO) (11), Human Cell Atlas Portal (HCA) (16), Single Cell Expression Atlas (EMBL-EBI/SCEA) (17) and Mouse Cell Atlas (MCA) (18). The information of receptor information of 142 animal viruses were obtained from the Viral Receptor database (19) and UniProt (20) (Supplementary Data 3).
Figure 1.
Overview of data collection, data processing and functional modules of VThunter database. (A) Diagram of species and tissues of collected scRNA-Seq datasets. (B) Steps of unified pipeline to process the curated scRNA-Seq datasets. (C) Overview of functional modules and user interfaces.
All the literatures of the studies generating the above scRNA-seq datasets were manually confirmed by a group of experienced researchers and all the scRNA-seq dataset were processed with a unified analyzing pipeline (Figure 1B). Briefly, they are processed with steps composed of both utilities packaged in Seurat v3.0 and in-house scripts according to previously study (21,22). Firstly, we conduct the quality control by filtering out cells with expressed genes <200 and genes those expressed in <1 cell for each dataset. Then, function of ‘NormalizedData’ in Seurat v3.0 were used to normalize the sparse single cell gene expression matrix. The highly variable genes were identified using the function ‘FindVariableGenes’ and the top 2000 highly variable genes were used for dimensionality reduction using principal component analysis (PCA). Based on the PCA elbow plot, the top 20 PCs were selected and used for clustering. Based on the transcriptomic profiles resulted from the scRNA-seq datasets, the expression patterns of virus receptor genes in various cell types were investigated. In total, the expression signature of 107 viral receptors in all obtained cell types of various tissues from 47 animal species were generated. The 107 viral receptors could be recognized and potentially infected by 142 viruses from 23 families.
DATABASE CONSTRUCTION AND USER INTERFACE
VThunter could be publicly and freely accessed through web browser by bench researchers worldwide. The web application of VThunter was implemented on a high-performance Linux server with open-source software. VThunter was equipped with a real-time search engine. VThunter's web interface allows users to intuitively browse and exactly query the expression signature of viral receptors at single-cell resolution. Figure 1C shows the schematic workflow and main functional modules of this database. The navigation menu contains seven icons including ‘Home’, ‘Virus Spectrum’, ‘Host Spectrum’, ‘Demo’, ‘Co-expression’, ‘Download’ and ‘Help’, which could lead users to the functional interfaces. On the ‘Home’ page, there are four main elements in addition to the header and navigation menu, including search forms for virus receptors or virus target genes, galleries for representative viral and animal species, and statistics related to data resources maintained by VThunter (Figure 2). If users are interested with searching the host spectrum of certain virus, they could query the virus in the search box or select it in the virus gallery to enter a virus page with comprehensive information of viral receptor and host tropism including target genes and target species with the expression profiles of the target genes in tissues and cell types. If a researcher only wants to fucus on a specific animal species, they could select the species of interest in the animal species gallery and enter an animal species page where receptor expression profiles of all viruses that may potentially infect that animal species will be present.
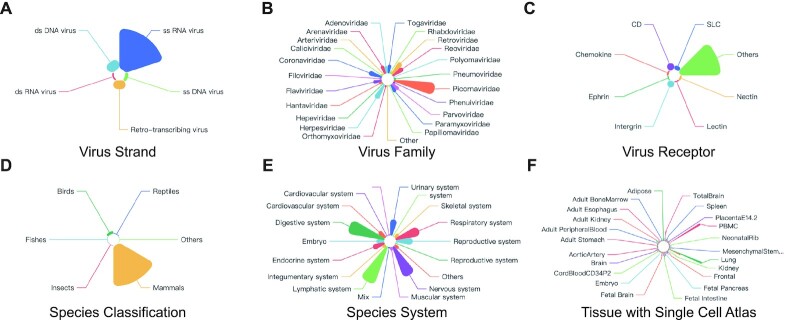
Figure 2.
Statistics of viruses of receptors and animal species with scRNA-Seq datasets in this database. Pie chart showing the percentage of viruses of curated receptors according to their strand type (A), viruses of curated receptors according to their family type (B), according to their receptors (C), curated scRNA-Seq datasets according to species classification (D), curated scRNA-Seq datasets according to species system (E), curated scRNA-Seq datasets according to organs (F).
On the ‘Virus Spectrum’ page, users could browse all the viruses with comprehensive receptor and host tropism information collected in this database. The ‘Check Details’ button under a virus icon will lead users to the virus page (Figure 3). Similarly, users could select a certain animal species in the ‘Host Spectrum’page and click the ‘Check Details’ button under the species icon to enter an animal species page (Figure 4). On the ‘Demo’ page, users can quickly view the content and format of data retrievable from VThunter. In the ‘Download’ page, links of all the raw data and resultant files maintained in this database are provided for interested researchers to conduct further analysis to meet their personalized needs. In the ‘Help’ page, a graphical operation guide is prepared for new users to get used to query relevant information easily, which will help them fully use the resource in VThunter. Besides, the ‘Co-expression’ module is also implemented for further inspect the co-expression genes of the certain viral receptors based on the comprehensive scRNA-seq expression profiles in VThunter (Figure 5).
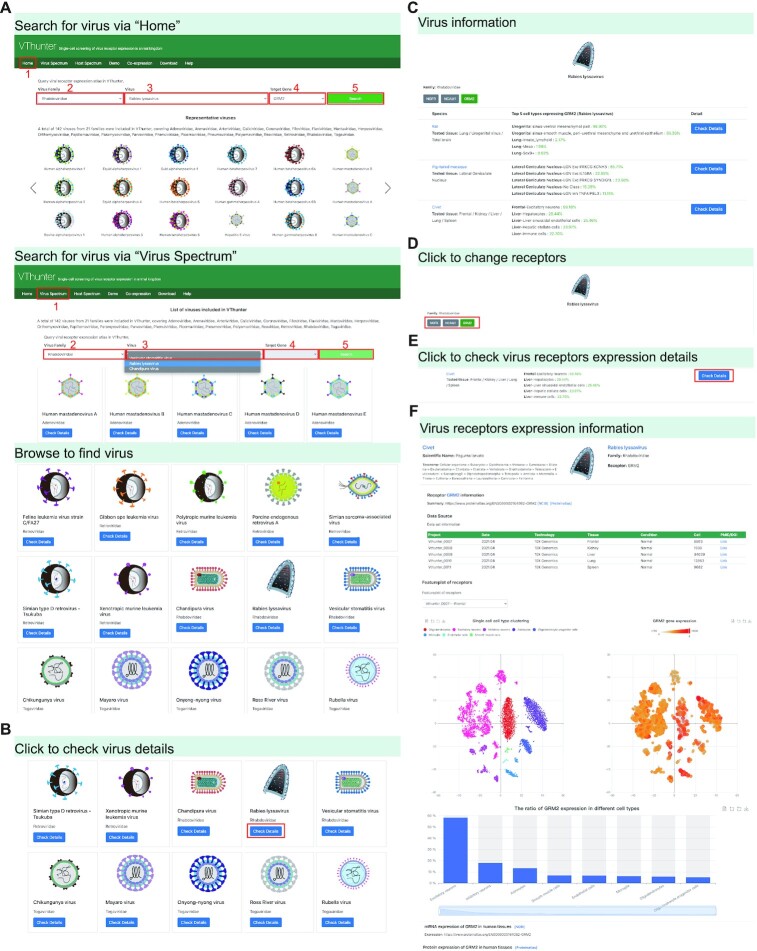
Figure 3.
Demonstration of Virus Spectrum module. (A) Searching for virus via ‘Home’ page or ‘Virus spectrum’. (B) Operation to obtain detail information in virus page. (C) Screenshot of virus information page. (D) Operation to view different receptor information of each virus in virus page. (E, F) Operation and screenshot of checking scRNA-seq expression profile of viral receptors.
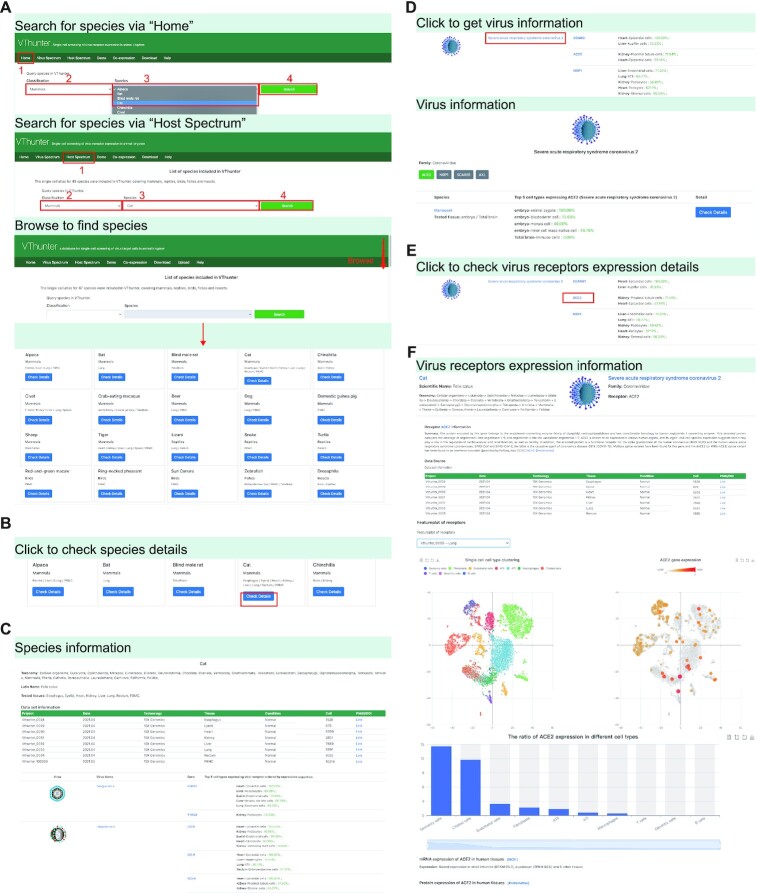
Figure 4.
Demonstration of Host Spectrum module. (A) Screenshot of searching animal species via ‘Home’ page or ‘Host spectrum’ page. (B) Operation to check detail information of certain species. (C) Screenshot of species information page. (D) Operation to check information of potential viruses in species page. (E, F) Operation and screenshot of checking scRNA-seq expression profile of virus receptors in species page.
Figure 5.
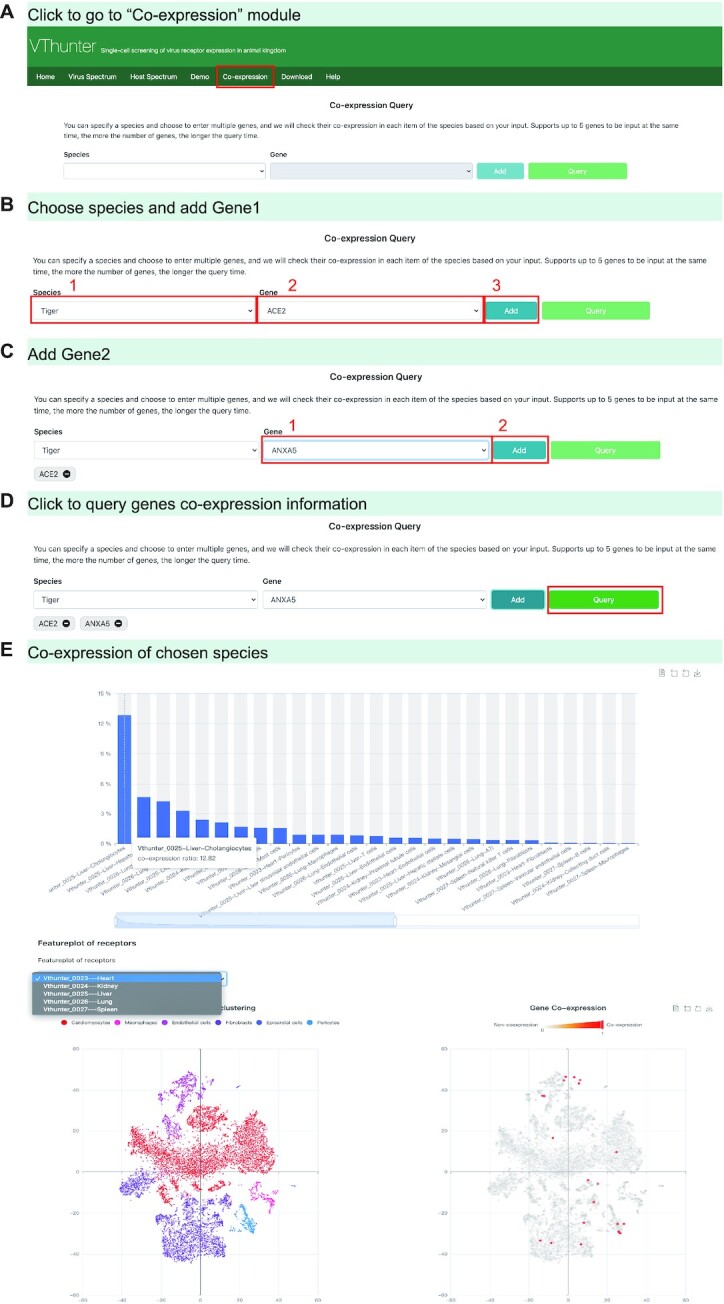
Demonstration of gene co-expression module. (A–D) Operations to enter ‘Co-expression’ page and perform co-expression query. (E) Screenshot of co-expression information of given genes.
APPLICATION CASE
VThunter is a comprehensive database designed for virological study, where users can search for viruses of interest to obtain information on host tropism, including target tissues and cell types in certain animal species, and to investigate viruses potentially infecting an animal species.
If a researcher wonders what animal species may be targeted by Rabies lyssavirus, they could conduct the following steps to obtain the relevant information as shown in Figure 3: (i) Select ‘Rhabdoviridae’ in virus family option list → select ‘Rabies lyssavirus’ in virus option list → select ‘GRM2’ → click ‘Search’. (ii) (optional) Find the Rabies lyssavirus in the representative virus gallery or in the ‘Virus Spectrum’ page, then click the ‘Check Details’ button under the virus icon. (iii) Users will be guided to the virus tropism page. In this page, animals with expression record of GRM2 will be displayed. If we click on the ‘Check Details’ button on the right of a specific animal, such as civet, the taxonomy lineage information for civet and the literature-based general information about Rabies lyssavirus and its infecting receptor are provided. All the scRNA-seq datasets collected in this database and relevant metadata including tissue type, animal health status, experimental details will be given in a data source form. After choosing a certain dataset, taking ‘Vthunter_007’ for example, an overall cell type cluster figure will be displayed on the left and the gene expression of gene GRM2 in each cell types will be displayed on the right. Furthermore, a boxplot showing the expression level of gene GRM2 in different tissues of civet will also be provided.
In addition to querying host tropism for a certain virus, we may also want to know which viruses can affect the health of a certain animal species. Here, we take cat as the animal of interest (Figure 4). Briefly, our search process involves three steps: (i) click on the cat specie icon in the representative galley in the homepage, or, enter the ‘Host Spectrum’ page → select ‘Mammals’ in the classification option list and select ‘Cat’ in the species option list → click ‘Search’ → enter the cat page (ii) in this page, you could have an overview of what viruses might attack cat, what genes be targeted as the receptor and what tissues of cat have the expression of the target genes. (iii) After clicking the link of target gene ‘ACE2’ as the receptor of SARS-CoV-2, users will be led to the page showing the details of scRNA-seq studies conducted on cat and the expression levels of ACE2 in each cell type and different tissues.
The above simple search steps highlight the user-friendly and highly interactive interface of VThunter, which can help users explore the expression signatures of viral receptors. VThunter provides the expression signatures of viral receptors in all cell types of 47 animal species at single-cell resolution to help clarify the interactions between host cells and viral surface proteins. In addition, it also provides quick download access to all raw data and resultant files maintained in the database to meet personalized needs. These features support VThunter as a reliable and useful database for the study of the cross-species transmission mechanisms of animal viruses.
SUMMARY AND FUTURE PERSPECTIVES
With the rapid accumulation of scRNA-Seq data from more and more species, it is time to fully archive and apply these resources in virological study, especially during the emergence of a novel animal virus. We believe host range assessment based on archived cellular receptor profiles could serve as an effective surrogate to narrow down the suspected host list and guide experimental designs for bench scientists, given that viral entry is the single step of infection and transmission in complete viral life cycle. Here, we have presented VThunter to reach this goal, where the expression signature of 107 viral receptors utilized by 142 viruses in various cell types of the tissues from 47 animal species is freely accessible. However, the expression level of a viral receptor can sometimes be low, but the virus may still be capable of infecting various tissues or cells (23). Therefore, researchers are advised to keep these limitations in mind and use scRNA-seq data wisely.
Further extension will be conducted in the following aspects. First, feedbacks and suggestions from users will be addressed timely to improve the performance and scientific value of the database. Second, more comprehensive scRNA-seq datasets produced in the future studies and latest achievement of viral receptor will be collected at regular period and incorporated into this database in time. Third, other multi-omics datasets like proteomics, metabolism related to the animal virus infection and transmission are also expected to be manually curated and integrated into this database in the future. Fourth, as emerging study and validation of viral infection in various species is released, the information will be constantly curated by experts and integrated with relevant data in this database.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Lei Chen and Dr Yiquan Wu for constructive suggestion and feedback from users.
Contributor Information
Dongsheng Chen, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Cong Tan, BGI-Shenzhen, Shenzhen 518083, China.
Peiwen Ding, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Lihua Luo, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Jiacheng Zhu, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Xiaosen Jiang, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Zhihua Ou, BGI-Shenzhen, Shenzhen 518083, China; Shenzhen Key Laboratory of Unknown Pathogen Identification, BGI-Shenzhen, Shenzhen 518083, China.
Xiangning Ding, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Tianming Lan, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yixin Zhu, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yi Jia, BGI-Shenzhen, Shenzhen 518083, China.
Yanan Wei, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Runchu Li, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Qiuyu Qin, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Chengcheng Sun, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Wandong Zhao, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Zhiyuan Lv, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Haoyu Wang, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Wendi Wu, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Yuting Yuan, BGI-Shenzhen, Shenzhen 518083, China; Department of Physiology, School of Basic Medical Sciences, Binzhou Medical University, Yantai 264003, China.
Mingyi Pu, BGI-Shenzhen, Shenzhen 518083, China; School of Basic Medicine, Qingdao University, Qingdao 266071, China.
Yuejiao Li, BGI-Shenzhen, Shenzhen 518083, China.
Yanan Zhang, BGI-Shenzhen, Shenzhen 518083, China; Tsinghua-Berkeley Shenzhen Institute (TBSI), Tsinghua University, Shenzhen, 518055, China.
Ashley Chang, BGI-Shenzhen, Shenzhen 518083, China.
Guoji Guo, Center for Stem Cell and Regenerative Medicine, Zhejiang University School of Medicine, Hangzhou 310058, China.
Yong Bai, BGI-Shenzhen, Shenzhen 518083, China.
Xin Jin, BGI-Shenzhen, Shenzhen 518083, China; School of Medicine, South China University of Technology, Guangzhou 510006, Guangdong, China; Guangdong Provincial Key Laboratory of Human Disease Genomics, Shenzhen Key Laboratory of Genomics, BGI-Shenzhen, Shenzhen 518083, China.
Huan Liu, BGI-Shenzhen, Shenzhen 518083, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Self-funding.
Conflict of interest statement. None declared.
REFERENCES
- 1. Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L.. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020; 579:270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lam T.T., Jia N., Zhang Y.W., Shum M.H., Jiang J.F., Zhu H.C., Tong Y.G., Shi Y.X., Ni X.B., Liao Y.S.et al.. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature. 2020; 583:282–285. [DOI] [PubMed] [Google Scholar]
- 3. Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z.. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020; 368:1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Munnink B.B.O., Sikkema R.S., Nieuwenhuijse D.F., Molenaar R.J., Munger E., Molenkamp R., Van Der Spek A., Tolsma P., Rietveld A., Brouwer M.. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science. 2021; 371:172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sit T.H.C., Brackman C.J., Ip S.M., Tam K.W.S., Law P.Y.T., To E.M.W., Yu V.Y.T., Sims L.D., Tsang D.N.C., Chu D.K.W.et al.. Infection of dogs with SARS-CoV-2. Nature. 2020; 586:776–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang Q., Zhang H., Huang K., Yang Y., Hui X., Gao J., He X., Li C., Gong W., Zhang Y.et al.. SARS-CoV-2 neutralizing serum antibodies in cats: a serological investigation. 2020; bioRxiv doi:03 April 2020, preprint: not peer reviewed 10.1101/2020.04.01.021196. [DOI]
- 7. Chen D., Sun J., Zhu J., Ding X., Lan T., Zhu L., Xiang R., Ding P., Wang H., Wang X.. Single-cell screening of SARS-CoV-2 target cells in pets, livestock, poultry and wildlife. 2020; bioRxiv doi:14 June 2020, preprint: not peer reviewed 10.1101/2020.06.13.149690. [DOI] [Google Scholar]
- 8. Halfmann P.J., Hatta M., Chiba S., Maemura T., Fan S., Takeda M., Kinoshita N., Hattori S.I., Sakai-Tagawa Y., Iwatsuki-Horimoto K.et al.. Transmission of SARS-CoV-2 in domestic cats. N. Engl. J. Med. 2020; 383:592–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maginnis M.S. Virus–receptor interactions: the key to cellular invasion. J. Mol. Biol. 2018; 430:2590–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Athar A., Füllgrabe A., George N., Iqbal H., Huerta L., Ali A., Snow C., Fonseca N.A., Petryszak R., Papatheodorou I.. ArrayExpress update–from bulk to single-cell expression data. Nucleic Acids Res. 2019; 47:D711–D715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2012; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yuan H., Yan M., Zhang G., Liu W., Deng C., Liao G., Xu L., Luo T., Yan H., Long Z.et al.. CancerSEA: a cancer single-cell state atlas. Nucleic Acids Res. 2019; 47:D900–D908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang X., Lan Y., Xu J., Quan F., Zhao E., Deng C., Luo T., Xu L., Liao G., Yan M.et al.. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019; 47:D721–D728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhao T., Lyu S., Lu G., Juan L., Zeng X., Wei Z., Hao J., Peng J.. SC2disease: a manually curated database of single-cell transcriptome for human diseases. Nucleic Acids Res. 2021; 49:D1413–D1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun D., Wang J., Han Y., Dong X., Ge J., Zheng R., Shi X., Wang B., Li Z., Ren P.et al.. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021; 49:D1420–D1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rozenblatt-Rosen O., Stubbington M.J., Regev A., Teichmann S.A.. The Human Cell Atlas: from vision to reality. Nature News. 2017; 550:451. [DOI] [PubMed] [Google Scholar]
- 17. Papatheodorou I., Moreno P., Manning J., Fuentes A.M.-P., George N., Fexova S., Fonseca N.A., Füllgrabe A., Green M., Huang N.. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020; 48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Han X., Wang R., Zhou Y., Fei L., Sun H., Lai S., Saadatpour A., Zhou Z., Chen H., Ye F.. Mapping the mouse cell atlas by microwell-seq. Cell. 2018; 172:1091–1107. [DOI] [PubMed] [Google Scholar]
- 19. Zhang Z., Zhu Z., Chen W., Cai Z., Xu B., Tan Z., Wu A., Ge X., Guo X., Tan Z.. Cell membrane proteins with high N-glycosylation, high expression and multiple interaction partners are preferred by mammalian viruses as receptors. Bioinformatics. 2019; 35:723–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. The UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang L., Zhu J., Wang H., Xia J., Liu P., Chen F., Jiang H., Miao Q., Wu W., Zhang L.et al.. A high-resolution cell atlas of the domestic pig lung and an online platform for exploring lung single-cell data. J. Genet. Genomics. 2021; 48:411–425. [DOI] [PubMed] [Google Scholar]
- 22. Zhu J., Chen F., Luo L., Wu W., Dai J., Zhong J., Lin X., Chai C., Ding P., Liang L.et al.. Single-cell atlas of domestic pig cerebral cortex and hypothalamus. Sci. Bull. 2021; 66:1448–1461. [DOI] [PubMed] [Google Scholar]
- 23. Sungnak W., Huang N., Bécavin C., Berg M., Queen R., Litvinukova M., Talavera-López C., Maatz H., Reichart D., Sampaziotis F.. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020; 26:681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.