Abstract
As a means to aid in the investigation of viral infection mechanisms and identification of more effective antivirus targets, the availability of a source which continually collects and updates information on the virus and host ncRNA-associated interaction resources is essential. Here, we update the ViRBase database to version 3.0 (http://www.virbase.org/ or http://www.rna-society.org/virbase/). This update represents a major revision: (i) the total number of interaction entries is now greater than 820,000, an approximately 70-fold increment, involving 116 virus and 36 host organisms, (ii) it supplements and provides more details on RNA annotations (including RNA editing, RNA localization and RNA modification), ncRNA SNP and ncRNA-drug related information and (iii) it provides two additional tools for predicting binding sites (IntaRNA and PRIdictor), a visual plug-in to display interactions and a website which is optimized for more practical and user-friendly operation. Overall, ViRBase v3.0 provides a more comprehensive resource for virus and host ncRNA-associated interactions enabling researchers a more effective means for investigation of viral infections.
INTRODUCTION
Increasing evidence has accrued indicating that non-coding RNAs (ncRNAs) play critical roles in the process of viral infection (1–4). For example, viral ncRNAs can control both viral and cellular gene expressions to facilitate completion of the viral life cycle (2,5), and some cellular ncRNAs can affect viral replication and even directly target viral genomes (2,6–9). Thus, a continuously updated comprehensive database on virus and host ncRNA-associated interaction resources is essential for the progress of virology.
The first version of ViRBase, which was released in 2015 (10), contained only ∼12 000 experimentally validated interactions involving about 60 viruses and 20 hosts. Recently, with increasing attention to virus research and rapid advancements in sequencing technology, there has been an explosive increase in the characterization of experimentally validated and computationally predicted virus and host ncRNA-associated interactions (11,12). Especially, it has been reported that ncRNAs may serve as potential therapeutic targets for COVID-19 (1,12). Therefore, it is clear that collection of ncRNA-associated interaction information as well as supplying more annotation data and prediction tools will substantially contribute to viral study.
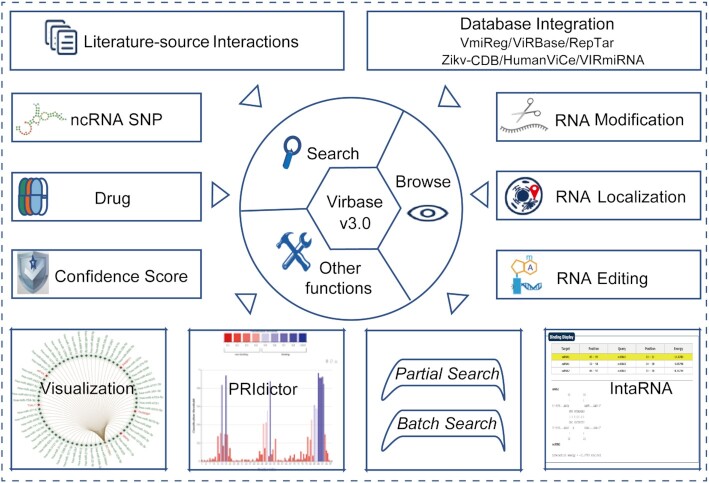
Motivated by aforesaid opinion, here we make an update of ViRBase to version 3.0 (http://www.virbase.org/ or http://www.rna-society.org/virbase/). This update offers a number of notable advantages: (i) it includes an integration of new data on virus and host ncRNA-associated interactions from the literature and five additional databases, (ii) it adds more detailed information on RNA annotations (including RNA editing, RNA localization and RNA modification) as well as ncRNA SNP and ncRNA-drug related information and (iii) it also provides two additional tools for predicting binding sites (IntaRNA (13) and PRIdictor), a visual plug-in to display interactions and new functions for partial and batch searches. This ViRBase v3.0 contains >820 000 interaction entries with detailed annotations and adds several new useful tools. Accordingly, it now serves as a more comprehensive platform which allows users to query, visualize, analyze and download virus and host ncRNA-associated interactions (Figure 1).
Figure 1.
Overview of ViRBase v3.0.
MATERIALS AND METHODS
Data collection and organization
ViRBase v3.0 integrates experimentally validated and computationally predicted virus and host ncRNA-associated interactions from the literature as well as five other databases. PubMed literature (mainly from 2016 to 2021) were reviewed with examples of keyword combinations being ‘RNA AND virus’ and ‘miRNA AND virus’ as well as related combinations. More than 148 000 virus and host ncRNA-associated interaction entries were identified and added to our database. ViRBase v3.0 also integrated experimentally validated virus and host miRNA-associated interactions from VIRmiRNA (14) and VmiReg (15) databases and computationally predicted interactions from HumanViCe (16), RepTar (17), VmiReg (15) and Zikv-CDB (18) databases. As a result, ViRBase v3.0 contains a total of >820 000 virus and host ncRNA-associated interaction entries, including 116 viruses (involving 36 virus families, Supplementary Table S1) and 36 host organisms.
RNA annotations, ncRNA SNP information and ncRNA drug-related information were added to the ViRBase v3.0. Detailed information on RNA editing sites was obtained from DARNED (19), Lncediting (20) and RADAR (21), RNA subcellular localization from RNALocate v2.0 (22), RNA modification sites from RMBase v2.0 (23), ncRNA SNP information from LincSNP v3.0 (24) and miRNASNP-v3.0 (25) and ncRNA-drug related information from ncDR (26), NoncoRNA (27), NRDTD (28) and RNAInter (29). Transcript and protein sequences from Refseq (30) and miRBase (31) databases were added to ViRBase v3.0 as a means to represent target sites predicted by miRanda (32), RIsearch (33) or PRIdictor (34). miRanda and RIsearch were used as tools for predicting RNA-RNA interactions, while PRIdictor was used to predict RNA-Protein interactions. In addition, information on experimentally verified RNA-binding sites in proteins was incorporated within this version, as documented in PDB (35), RBPDB (36) and RsiteDB (37).
MiRNA symbols were obtained from the miRBase (31) and other RNA and protein symbols were collected according to the NCBI Gene or Ensembl database (38), while NCBI aliases were also provided. Virus names were standardized according to the NCBI taxonomy database (39) and information on virus strains was added according to that as contained in the literature (Supplementary Table S2). Entrez ID, miRBase accession, PubChem Compound CID and their external links were provided as means to help users access associated information from external resources.
Confidence score
In ViRBase v3.0, virus and host ncRNA-associated interactions were derived from different evidence resources, such as data from experimental evidences or computational predictions. Similar to the miRTarBase and RNAInter databases (29,40), these data were divided into strong experimental evidence (e.g. RNA immunoprecipitation and luciferase reporter assay results), weak experimental evidence (e.g. RNA-seq) and computational prediction evidence. To assess the credibility of interactions with multiple evidence types, each interaction entry was assigned a confidence score (S) based on the following formula:
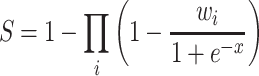 |
where i is the evidence type, either strong (Ss) or weak (Sw) experimental evidence, while Sp is the computational prediction method and x is the number of evidence resources. The weight factors Ws, Ww and Wp were set to 1, 0.65 and 0.25, respectively. If x = 0, the weight factor (Wi) was set to 0. Only well-supported ncRNA-associated interactions could achieve a score approaching 1 (scores ranged from 0 and 1), suggesting that this approach served as an effective tool for filtering relatively reliable virus and host ncRNA-associated interactions.
RESULTS
ViRBase v3.0 statistics
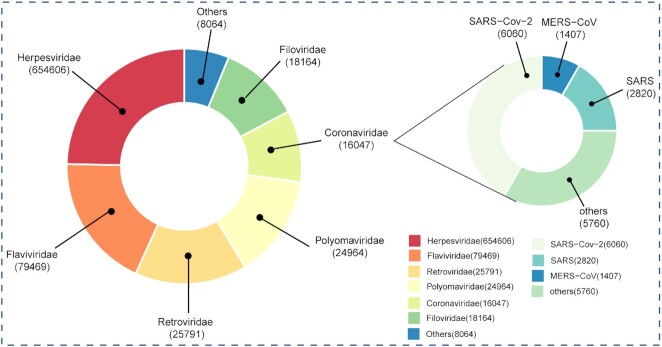
Currently, ViRBase v3.0 documents 151 196 experimentally validated and 675 909 computationally predicted virus and host ncRNA-associated interactions, which consists of 248 virus–virus, 10 643 host–virus, 109 398 host–host and 706 816 virus–host interactions (Figure 2A). Based on RNA types, these interactions involve 819 276 miRNA–RNA, 11 221 lncRNA–RNA, 2915 other ncRNA–RNA, 1454 circRNA–RNA and 258 ncRNA–protein (Figure 2B). Similarly, based on virus families, there are 654 606 Herpesviridae, 79 469 Flaviviridea, 25 791 Retroviridae, 24 964 Polyomaviridae, 18 164 Filovirdae, 16 047 Coronaviridae and 8064 other interactions (Figure 3). Notably, of the 16 047 Coronaviridae interactions, 6060, 2820, 1407 and 5760 are associated with the SARS-Cov-2, SARS, MERS-Cov and other coronaviruses, respectively (Figure 3).
Figure 2.
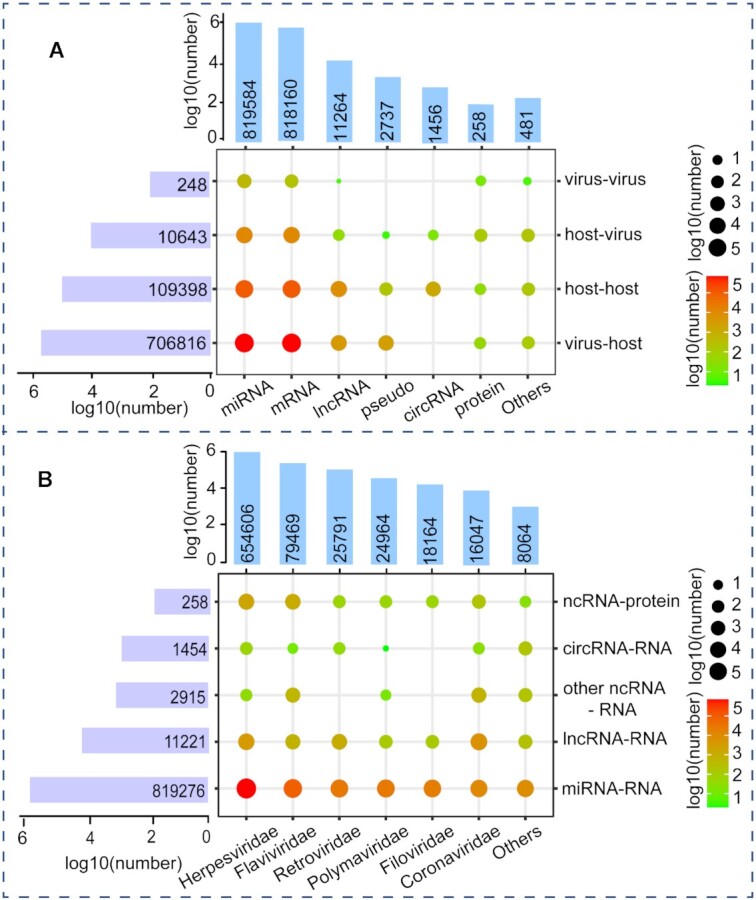
Distribution of interaction types, including virus and host interactions (A) and ncRNA-associated interactions (B).
Figure 3.
Statistics on ViRBase v3.0, numbers of virus and host ncRNA-associated interactions in all virus families and coronavridae.
Now, this ViRBase v3.0 contains >56 000 non-redundant RNAs and 13 RNA types. Among the ncRNAs, miRNA, lncRNA, circRNA, snoRNA, snRNA, misc_RNA, nsRNA, scaRNA, shRNA and pseudo are included. Compared to previous version, the number of viruses increases from 60 to 116, while the number of host organisms enhances from 20 to 36.
Data feature and database usage
In addition to basic information, supporting evidence, confidence scores and references, ViRbase v3.0 supplies detailed information on a number of factors, such as target regions, RNA annotations (including RNA editing, RNA localization and modification), ncRNA SNP information as well as ncRNA-drug related information. The data of target regions offer information on binding regions between interactors. RNA annotations provide editing (or modification) positions, editing (or modification) types as well as subcellular localizations. ncRNA SNP information provides information on SNP sites and base changes in miRNA or lncRNA sequences. ncRNA-drug related information contains drug names, compound ID and four types of ncRNA-drug associations (drug–target, sensitivity, resistant and interactions).
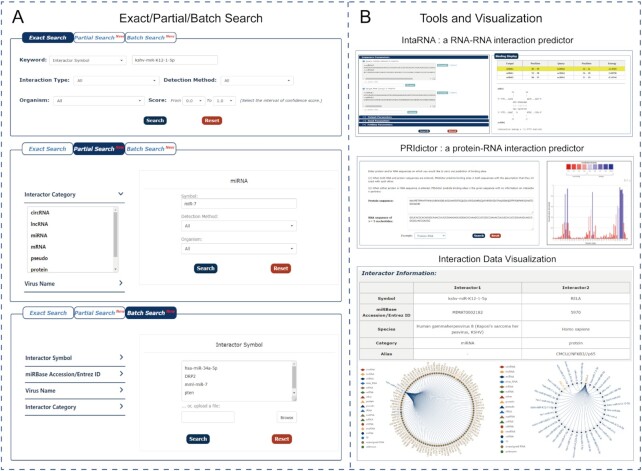
ViRbase v3.0 also enables a more user-friendly web interface. For example, to search efficiently, ViRBase v3.0 adds new functions for partial and batch searches. Partial search allows to directly search interactions using non-standardized or unidentified interactor or virus names, while batch search allows to input a list of official symbols/IDs, virus names or upload files of text format to obtain virus and host ncRNA-associated interactions. In this way, users can perform an ‘Exact Search’ to obtain search results, a ‘Partial Search’ to search for interested interactors or viruses, or a ‘Batch Search’ to customize their query content in batch (as shown in Figure 4A). Moreover, with the addition of a ‘filter’ function, users can further search the interactions with a specific characteristic, after performing the Exact/Partial/Batch Search. And, all search results can be directly downloaded at any step. ViRBase v3.0 also updates the download option in the ‘Browse’ page, where users can browse and download virus and host ncRNA-associated interaction data by interaction type, detection method or organism. In this way, it provides ‘Download’ as well as ‘API’ services for users to flexibly retrieve all virus and host ncRNA-associated interaction information. Furthermore, ViRbase v3.0 offers several tools to meet diverse needs encountered by users. For example, two prediction tools (IntaRNA and PRIdictor) are added. IntaRNA accurately predicts interactions between two RNA molecules, while PRIdictor reliably predicts protein-RNA interactions (Figure 4B), and a visual plug-in is provided to display these interactions. Clicking on any edge of the network can redirect users to the page of the corresponding interaction entry (Figure 4B).
Figure 4.
New search functions and tools. (A) Snapshot of exact, partial and batch searches as described in the search options. (B) Presentation of serval tools in ViRbase v3.0 including IntaRNA, PRIdictor and a visual plug-in.
CONCLUSION AND FUTURE DIRECTIONS
ViRBase v3.0 provides a more comprehensive platform for accessing virus and host ncRNA-associated interaction repositories as achieved with the addition of increased coverage, annotation and tools. It incorporates more data, over 820 000 virus and host ncRNA-associated interaction entries, a 70-fold increment. Moreover, it provides detailed annotations from sequences to binding sites and various external links, as well as supplying two new tools for predicting RNA-associated binding sites and a visual plug-in to display interactions. In this way, ViRBase v3.0 offers the opportunity to stimulate and enhance current virology research. With the current and growing interest in virus research combined with the emergence of new sequencing technologies, experimental methods and prediction algorithms, the resources and information available for accessing virus and host ncRNA-associated interactions will continue at an exponential rate. Therefore, while our current ViRBase v3.0, serves to fulfil this goal, we plan to continuously expand and improve our ViRBase as a means of providing an uninterrupted, reliable and comprehensive updated resource that will be available for the entire community.
Supplementary Material
Contributor Information
Jun Cheng, Affiliated Foshan Maternity and Child Healthcare Hospital, Southern Medical University (Foshan Maternity & Child Healthcare Hospital), Foshan 528000, China.
Yunqing Lin, Center for Cell Lineage and Atlas (CCLA), Bioland Laboratory (Guangzhou Regenerative Medicine and Health Guangdong Laboratory), Guangzhou 510005, China.
Linfu Xu, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Kechen Chen, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Qi Li, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Kaixin Xu, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Lin Ning, Dermatology Hospital, Southern Medical University, Guangzhou 510091, China.
Juanjuan Kang, Affiliated Foshan Maternity and Child Healthcare Hospital, Southern Medical University (Foshan Maternity & Child Healthcare Hospital), Foshan 528000, China.
Tianyu Cui, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Yan Huang, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Xiaoyang Zhao, State Key Laboratory of Organ Failure Research, Department of Developmental Biology, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China.
Dong Wang, Department of Bioinformatics, School of Basic Medical Sciences, Southern Medical University, Guangzhou 510515, China; Dermatology Hospital, Southern Medical University, Guangzhou 510091, China.
Yanhui Li, Institute of Cardiovascular Sciences and Key Laboratory of Molecular Cardiovascular Sciences, Ministry of Education, Peking University Health Science Center, Beijing, PR China.
Xi Su, Affiliated Foshan Maternity and Child Healthcare Hospital, Southern Medical University (Foshan Maternity & Child Healthcare Hospital), Foshan 528000, China.
Bin Yang, Dermatology Hospital, Southern Medical University, Guangzhou 510091, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key Research and Development Project of China [2017YFA0105001, 2019YFA0801800]; National Natural Science Foundation of China [82073418, 82070109, 81770104, 62002153]; Guangdong Basic and Applied Basic Research Foundation [2019A1515010784, 2019A1515110701]; China Postdoctoral Science Foundation [2020M682623, 2020M682785]; Foshan Medicine Dengfeng Project [2019–2021]. Funding for open access charge: Postdoctoral research funding of Affiliated Foshan Maternity and Child Healthcare Hospital.
Conflict of interest statement. None declared.
REFERENCES
- 1. Henzinger H., Barth D.A., Klec C., Pichler M.. Non-coding RNAs and SARS-related coronaviruses. Viruses. 2020; 12:1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Skalsky R.L., Cullen B.R.. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010; 64:123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mocarski E.S., Upton J.W., Kaiser W.J.. Viral infection and the evolution of caspase 8-regulated apoptotic and necrotic death pathways. Nat. Rev. Immunol. 2011; 12:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ghosh Z., Mallick B., Chakrabarti J.. Cellular versus viral microRNAs in host-virus interaction. Nucleic Acids Res. 2009; 37:1035–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Klase Z., Houzet L., Jeang K.T.. MicroRNAs and HIV-1: complex interactions. J. Biol. Chem. 2012; 287:40884–40890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boliar S., Russell D.G.. Lnc(ing)RNAs to the “shock and kill” strategy for HIV-1 cure. Mol Ther Nucleic Acids. 2021; 23:1272–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Haque M.M., Murale D.P., Lee J.S.. Role of microRNA and Oxidative Stress in Influenza A Virus Pathogenesis. Int. J. Mol. Sci. 2020; 21:8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang P., Xu J., Wang Y., Cao X.. An interferon-independent lncRNA promotes viral replication by modulating cellular metabolism. Science. 2017; 358:1051–1055. [DOI] [PubMed] [Google Scholar]
- 9. Chen L., Zhou Y., Li H.. LncRNA, miRNA and lncRNA-miRNA interaction in viral infection. Virus Res. 2018; 257:25–32. [DOI] [PubMed] [Google Scholar]
- 10. Li Y., Wang C., Miao Z., Bi X., Wu D., Jin N., Wang L., Wu H., Qian K., Li C.et al.. ViRBase: a resource for virus-host ncRNA-associated interactions. Nucleic Acids Res. 2015; 43:D578–D582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huang J.T., Chen J.N., Gong L.P., Bi Y.H., Liang J., Zhou L., He D., Shao C.K.. Identification of virus-encoded circular RNA. Virology. 2019; 529:144–151. [DOI] [PubMed] [Google Scholar]
- 12. Alam T., Lipovich L.. miRCOVID-19: potential targets of human miRNAs in SARS-CoV-2 for RNA-based drug discovery. Noncoding RNA. 2021; 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mann M., Wright P.R., Backofen R.. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017; 45:W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Qureshi A., Thakur N., Monga I., Thakur A., Kumar M.. VIRmiRNA: a comprehensive resource for experimentally validated viral miRNAs and their targets. Database (Oxford). 2014; 2014:bau103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shao T., Zhao Z., Wu A., Bai J., Li Y., Chen H., Jiang C., Wang Y., Li S., Wang L.et al.. Functional dissection of virus-human crosstalk mediated by miRNAs based on the VmiReg database. Mol. Biosyst. 2015; 11:1319–1328. [DOI] [PubMed] [Google Scholar]
- 16. Ghosal S., Das S., Sen R., Chakrabarti J.. HumanViCe: host ceRNA network in virus infected cells in human. Front Genet. 2014; 5:249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Elefant N., Berger A., Shein H., Hofree M., Margalit H., Altuvia Y.. RepTar: a database of predicted cellular targets of host and viral miRNAs. Nucleic Acids Res. 2011; 39:D188–D194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pylro V.S., Oliveira F.S., Morais D.K., Cuadros-Orellana S., Pais F.S., Medeiros J.D., Geraldo J.A., Gilbert J., Volpini A.C., Fernandes G.R.. ZIKV - CDB: a collaborative database to guide research linking SncRNAs and ZIKA virus disease symptoms. PLoS Negl Trop Dis. 2016; 10:e0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kiran A., Baranov P.V.. DARNED: a DAtabase of RNa EDiting in humans. Bioinformatics. 2010; 26:1772–1776. [DOI] [PubMed] [Google Scholar]
- 20. Gong J., Liu C., Liu W., Xiang Y., Diao L., Guo A.Y., Han L.. LNCediting: a database for functional effects of RNA editing in lncRNAs. Nucleic Acids Res. 2017; 45:D79–D84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ramaswami G., Li J.B.. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2014; 42:D109–D113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cui T., Dou Y., Tan P., Ni Z., Liu T., Wang D., Huang Y., Cai K., Zhao X., Xu D.et al.. RNALocate v2.0: an updated resource for RNA subcellular localization with increased coverage and annotation. Nucleic Acids Res. 2021; 10.1093/nar/gkab825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xuan J.J., Sun W.J., Lin P.H., Zhou K.R., Liu S., Zheng L.L., Qu L.H., Yang J.H.. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018; 46:D327–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gao Y., Li X., Shang S., Guo S., Wang P., Sun D., Gan J., Sun J., Zhang Y., Wang J.et al.. LincSNP 3.0: an updated database for linking functional variants to human long non-coding RNAs, circular RNAs and their regulatory elements. Nucleic Acids Res. 2021; 49:D1244–D1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu C.J., Fu X., Xia M., Zhang Q., Gu Z., Guo A.Y.. miRNASNP-v3: a comprehensive database for SNPs and disease-related variations in miRNAs and miRNA targets. Nucleic Acids Res. 2021; 49:D1276–D1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dai E., Yang F., Wang J., Zhou X., Song Q., An W., Wang L., Jiang W.. ncDR: a comprehensive resource of non-coding RNAs involved in drug resistance. Bioinformatics. 2017; 33:4010–4011. [DOI] [PubMed] [Google Scholar]
- 27. Li L., Wu P., Wang Z., Meng X., Zha C., Li Z., Qi T., Zhang Y., Han B., Li S.et al.. NoncoRNA: a database of experimentally supported non-coding RNAs and drug targets in cancer. J. Hematol. Oncol. 2020; 13:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen X., Sun Y.Z., Zhang D.H., Li J.Q., Yan G.Y., An J.Y., You Z.H.. NRDTD: a database for clinically or experimentally supported non-coding RNAs and drug targets associations. Database (Oxford). 2017; 2017:bax057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kang J., Tang Q., He J., Li L., Yang N., Yu S., Wang M., Zhang Y., Lin J., Cui T.et al.. RNAInter v4.0: RNA interactome repository with redefined confidence scoring system and improved accessibility. Nucleic Acids Res. 2021; 10.1093/nar/gkab997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Haft D.H., DiCuccio M., Badretdin A., Brover V., Chetvernin V., O’Neill K., Li W., Chitsaz F., Derbyshire M.K., Gonzales N.R.et al.. RefSeq: an update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018; 46:D851–D860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kozomara A., Birgaoanu M., Griffiths-Jones S.. miRBase: from microRNA sequences to function. Nucleic Acids Res. 2019; 47:D155–D162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S.. Human MicroRNA targets. PLoS Biol. 2004; 2:e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wenzel A., Akbasli E., Gorodkin J.. RIsearch: fast RNA-RNA interaction search using a simplified nearest-neighbor energy model. Bioinformatics. 2012; 28:2738–2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tuvshinjargal N., Lee W., Park B., Han K.. PRIdictor: protein-RNA Interaction predictor. Biosystems. 2016; 139:17–22. [DOI] [PubMed] [Google Scholar]
- 35. Burley S.K., Berman H.M., Bhikadiya C., Bi C., Chen L., Di Costanzo L., Christie C., Dalenberg K., Duarte J.M., Dutta S.et al.. RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. Nucleic Acids Res. 2019; 47:D464–D474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cook K.B., Kazan H., Zuberi K., Morris Q., Hughes T.R.. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2011; 39:D301–D308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shulman-Peleg A., Nussinov R., Wolfson H.J.. RsiteDB: a database of protein binding pockets that interact with RNA nucleotide bases. Nucleic Acids Res. 2009; 37:D369–D373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cunningham F., Achuthan P., Akanni W., Allen J., Amode M.R., Armean I.M., Bennett R., Bhai J., Billis K., Boddu S.et al.. Ensembl 2019. Nucleic Acids Res. 2019; 47:D745–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Huang H.Y., Lin Y.C., Li J., Huang K.Y., Shrestha S., Hong H.C., Tang Y., Chen Y.G., Jin C.N., Yu Y.et al.. miRTarBase 2020: updates to the experimentally validated microRNA-target interaction database. Nucleic Acids Res. 2020; 48:D148–D154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.