Abstract
Transcriptomic profiling is critical to uncovering functional elements from transcriptional and post-transcriptional aspects. Here, we present Gene Expression Nebulas (GEN, https://ngdc.cncb.ac.cn/gen/), an open-access data portal integrating transcriptomic profiles under various biological contexts. GEN features a curated collection of high-quality bulk and single-cell RNA sequencing datasets by using standardized data processing pipelines and a structured curation model. Currently, GEN houses a large number of gene expression profiles from 323 datasets (157 bulk and 166 single-cell), covering 50 500 samples and 15 540 169 cells across 30 species, which are further categorized into six biological contexts. Moreover, GEN integrates a full range of transcriptomic profiles on expression, RNA editing and alternative splicing for 10 bulk datasets, providing opportunities for users to conduct integrative analysis at both transcriptional and post-transcriptional levels. In addition, GEN provides abundant gene annotations based on value-added curation of transcriptomic profiles and delivers online services for data analysis and visualization. Collectively, GEN presents a comprehensive collection of transcriptomic profiles across multiple species, thus serving as a fundamental resource for better understanding genetic regulatory architecture and functional mechanisms from tissues to cells.
INTRODUCTION
Transcriptomic profiling, involving both transcriptional and post-transcriptional modifications or events at whole-genome level, is of great importance for uncovering functional elements across the three domains of life, including ‘Bacteria’, ‘Archaea’ and ‘Eukarya’ (1–3). High-throughput RNA sequencing (RNA-seq) (4), which can qualitatively and quantitatively capture any type of RNA, promises to help researchers characterize transcriptome comprehensively due to the capacities of whole-genome expression profiling (5–7), detection of novel RNA forms and variants (8–12) and genome reannotation (13,14). With the continuous developments of RNA-seq technology, it has become a routine and indispensable approach for systematically characterizing transcriptome across diverse developmental stages and physiological conditions (1,10,15–17). Of note, over the past years, transcriptomic studies have made the leap from bulk RNA-seq to single-cell RNA-seq (scRNA-seq), unveiling new insights into cell type classification and cellular heterogeneity exploration (18,19).
As RNA-seq has been widely used in a broad diversity of species worldwide, a huge amount of transcriptomic data has been generated at unprecedentedly exponential rates, accordingly posing great challenges in large-scale data aggregation and standardized processing. To facilitate more effective reuse, integration, and mining of those data, valuable efforts have been made to construct comprehensive or specialized database resources, such as Gene Expression Omnibus (GEO) (20), Expression Atlas (21), Human Cell Atlas (HCA) (22) and Genotype-Tissue Expression (GTEx) (23). Specifically, GEO, a widely used resource developed by NCBI (24), is devoted to archiving worldwide transcriptomic data (as well as other omics data), yet ignoring standardized data processing and structured metadata management. Expression Atlas in EBI (25), contains both bulk and single-cell expression profiles with unified processing, nevertheless lacking co/post-transcriptional events (e.g. RNA editing and splicing). HCA is specialized in human single-cell expression profiling, whereas GTEx focuses on human gene expression and regulation across tissues. To sum up, existing resources have two major shortcomings. First, none of them takes good account of transcriptomic profiles (e.g. expression, RNA editing, splicing, etc.). Second, they do not well curate and categorize experimental metadata under the framework of biological contexts. Given the large-scale data volumes and heterogeneous types of data and metadata, it is challenging to build a comprehensive database that integrates transcriptomic profiles at both bulk and single-cell levels, accompanying with standardized data processing, metadata curation, and online tools.
To address these challenges, here we present Gene Expression Nebulas (GEN, https://ngdc.cncb.ac.cn/gen/), an open-access data portal integrating transcriptomic profiles under various conditions across multiple species. It was originally established in 2016, along with the foundation of the National Genomics Data Center (NGDC; previously named as BIG Data Center) (26,27), China National Center for Bioinformation (CNCB). Since its inception, GEN, as one of the core resources in CNCB-NGDC, has been frequently updated by importing and processing datasets obtained from a variety of raw sequencing data archives. Unlike existing resources, GEN provides a curated collection of high-quality bulk and single-cell RNA-seq datasets with uniformed data processing and adopts a structured curation model to categorize diverse experimental conditions into different biological contexts. Accordingly, GEN features large-scale integration of diverse transcriptomic profiles and provides online tools for analysis and visualization of both bulk and single-cell RNA-seq data.
MATERIALS AND METHODS
Data collection
A number of high-throughput RNA-seq projects and their associated datasets were collected from several public raw sequencing databases, including Genome Sequence Archive (GSA, https://ngdc.cncb.ac.cn/gsa/) (28,29), Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) (30), European Nucleotide Archive (ENA, https://www.ebi.ac.uk/ena) (31) and DDBJ Sequence Read Archive (DRA, https://ddbj.nig.ac.jp/DRASearch/) (32). Only the datasets with median mapping rates ≥70% for bulk RNA-seq and ≥40% for scRNA-seq were kept for further processing. As a result, a total of 296 RNA-seq projects and 323 high-quality datasets were obtained.
Unified and standardized data processing
For bulk RNA-seq datasets, Fastp v0.20.0 (33) was used for trimming and filtering raw reads. And, HISAT2 v2.0.5 (34) was used for quick alignment to evaluate the data quality, and RseQC v2.6.4 (35) was implemented for inferring the strand specificity of the sequencing library. Then high-quality RNA-seq reads were aligned to the reference genome by STAR v2.7.1a (36). After that, quantification of gene/isoform assembly was performed by RSEM v1.3.1 (37) with default parameters. ‘Raw counts’, ‘FPKM’ (Fragments Per Kilobase of transcript per Million mapped fragments) and ‘TPM’ (Transcripts Per Million) values of each gene/isoform were calculated. For circular RNA (circRNA) expression analysis, the cleaned RNA-seq reads were mapped to the reference genome by BWA-MEM (38). Next, CIRCexplorer2 (39) and CIRI2 v2.0.6 (40) were used to identify circRNA candidates by recognizing the back-splicing junction (BSJ) reads (≥2) with default parameters.
Moreover, RNA editing sites were identified with the genome from GENCODE v33 (41) as reference. All known RNA editing sites were retrieved from REDIportal v2.0 (42) (http://srv00.recas.ba.infn.it/atlas/). Novel human RNA editing sites were detected by Parallel Strategy of REDItool 2.0 (43). To obtain more accurate novel editing sites, a filtration step was added for non-Alu regions using additional criteria as the non-Alu regions usually have sporadic editing sites. Meanwhile, pblat v1.0 (44) was used to discover the mismatched RNA-seq reads and multi-mapping reads, which were then trimmed to remove duplicate reads by using SAMtools v1.9 (45). Editing sites of both A-to-I and C-to-U were maintained for further analysis. RepeatMasker (http://www.repeatmasker.org) and SNP files used for annotating high-confidence novel RNA editing sites were both downloaded from UCSC (https://hgdownload.soe.ucsc.edu/downloads.html).
In addition, for alternative splicing analysis, high-quality RNA-seq reads were mapped to the reference genome by STAR. Then, detection of differentially spliced events was mainly executed with BAM files by rMATS v3.1.0 (46). The high-quality RNA-seq reads were mapped to the reference genome by STAR. Each ‘case’ group was compared to the ‘control’ group to identify differentially spliced events, and parameter of ‘–cstat 0.0001’ was used for 0.01% difference, to compute p-values and FDRs of splicing events with the absolute value of exon inclusion level (|Δψ|) > 0.01% cutoff.
For scRNA-seq datasets, notably, alignment approach was consistent with bulk RNA-seq datasets, while gene quantification tools varied with the data generated by different platforms/strategies to deal with cell barcodes and unique molecular identifiers (UMIs). Currently, pipelines for the three most commonly adopted scRNA-seq technologies were as follows (47–49): (i) for data generated by plate- or fluidigm-based protocol, such as Smart-seq2 (50) and SMARTer (Fluidigm C1) strategies, STAR v2.7.1a and RSEM v1.3.1 were used to align and calculate ‘raw counts’, ‘FPKM’ and ‘TPM’ values of each gene/isoform with the parameter ‘–single-cell-prior’; (ii) for data from droplet-based protocol including Drop-seq (51) and inDrop (52), dropEst v0.8.6 (53) was used to provide more accurate estimates of molecular counts in individual cells by barcode corrections, classification of cell quality, and diagnostic information about the droplet libraries; and (iii) specifically for data from 10× Genomics platform (54), CellRanger v3.1.0 (https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome) was implemented as a one-stop analysis pipeline for quality control, sample de-multiplexing, barcode processing and generation of feature-barcode matrices.
Collection of gene annotations
For all collected species, a wide range of gene functional annotations were extracted from Ensembl (55) and NCBI (24), roughly falling into basic information including genomic location and functional description, and associated terms or ontologies like Gene Ontology (GO) (56). Particularly, for Homo sapiens, housekeeping and tissue-specific genes were derived from GTEx (57), genes were annotated based on Disease Ontology (DO) (58) along with GO, and a gene structure visualization on the basis of Genome Browser (59) was provided. Furthermore, annotation information of editome-disease associations from Editome-Disease Knowledgebase (EDK, https://ngdc.cncb.ac.cn/edk) (60) and RT-qPCR reference genes from Internal Control Genes (ICG, http://icg.big.ac.cn) (61) were also included for corresponding genes, while external links to GTEx (https://www.gtexportal.org/home/) (23), REDIportal (http://srv00.recas.ba.infn.it/atlas/) (62) and GeneCard (https://www.genecards.org) (63) were added to each gene (if available).
Downstream analysis
A series of popular downstream analysis tools were implemented in GEN. For bulk RNA-seq data, four tools were included for different analysis purposes, namely, differential expression analysis with limma (64), weighted gene co-expression network analysis with WGCNA (65), functional enrichment analysis with clusterProfiler (66), and gene regulatory network inference with GENIE3 (67). For scRNA-seq data, Seurat (68) was integrated for the selection and filtration of cells based on quality-control metrics, data normalization and scaling, detection of high-variance genes, linear dimensional reduction (i.e. principal component analysis), graph-based clustering, visualization of cluster assignment and identification of cluster markers. Marker gene enrichment analysis was generated with Enrichr (69), and trajectory inference was performed with Monocle (70). Furthermore, SingleR (71) was employed to infer cell type identity of each cell independently by leveraging reference transcriptomic datasets of pure cell types. Here, reference datasets from Human Primary Cell Atlas (72), BLUEPRINT (73), and Human Immune Cell RNA-seq Data (74), Human Hematopoietic Cell RNA-seq Data (75) and DICE (Database of Immune Cell Expression, Expression quantitative trait loci (eQTLs) and Epigenomics) Project (76) were used for human cell type annotation, while those from Mouse RNA-seq Data (77) and Immunological Genome Project (ImmGen) (78) were used for mouse cell type annotation, separately.
Database implementation
GEN was implemented using Spring Boot (https://spring.io/projects/spring-boot; a framework easy to create stand-alone java applications) as the back-end framework. All data were stored and managed by using MySQL (https://dev.mysql.com; a free and popular relational database management system). To provide user-friendly and highly interactive web applications, web pages were constructed using HTML5 and rendered using JSP (https://jakarta.ee/specifications/pages/3.0/, Jakarta Server Pages, a template engine for web applications). Front-end interfaces were built using Semantic UI (https://semantic-ui.com; a development framework that helps create beautiful, responsive layouts HTML) and JQuery (https://jquery.com; a fast, small, and feature-rich JavaScript library). Furthermore, data visualization was built by HighCharts (https://www.highcharts.com; a JavaScript plug-in to create interactive charts), Plotly.js (https://plotly.com/javascript/; a high-level, declarative charting library) and DataTables (https://datatables.net; a plug-in for the jQuery JavaScript library to render HTML tables). Interactive visualization of scRNA-seq data was powered by Cerebro (79). Online tools were developed with Shiny (https://shiny.rstudio.com/, an R package to build interactive web applications).
DATABASE CONTENTS AND USAGE
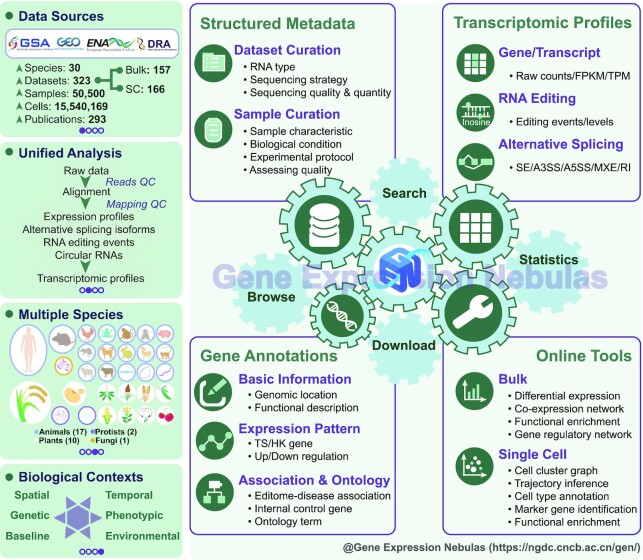
GEN features comprehensive integration, manual curation and standardized analysis of high-quality transcriptomic datasets at bulk and single-cell levels based on a structured curation model and uniformed data processing pipelines. More importantly, diverse experimental conditions of all incorporated datasets are categorized into more informative biological contexts. In the current version, GEN houses a collection of transcriptomic profiles of 323 datasets covering 50 500 samples and 15 540 169 cells across 30 species. For each dataset, a full range of transcriptomic profiles including gene expression, alternative RNA splicing and RNA editing (if applicable) are provided in GEN. Moreover, GEN accommodates value-added gene annotations based on differential expression analysis across diverse experimental conditions and cell clusters. Accordingly, GEN provides user-friendly web functionalities and applications for large-scale data query, retrieval, analysis and visualization (Figure 1).
Figure 1.
Database contents and features of Gene Expression Nebulas. Abbreviations used: SC, single-cell; TS, tissue-specific; HS, house-keeping; FPKM, fragments per kilobase of transcript per million mapped fragments; TPM, transcripts per million. SE: skipped exon; A3SS: alternative 3′ splice site; A5SS: alternative 5′ splice site; MXE: mutually exclusive exons; RI: retained intron.
Metadata curation and datasets
GEN adopts a structured curation model, incorporating manually curated items in light of dataset, profile (expression/splicing/editing), and sample: (i) datasets are annotated and categorized into six biological contexts of general interest, involving baseline, genetic (e.g. mutation, natural variation), phenotypic (e.g. disease, aging), environmental (e.g. abiotic stress, biotic stress), spatial (e.g. organism, tissue, cell type) and temporal (e.g. development, circadian, time series); (ii) Expression/splicing/editing profiles include the main steps and parameters of data processing together with reference genome and annotation details and (iii) samples contain a wealth of descriptive information, including basic information, sample characteristic, biological condition, experimental variable, experimental protocol, sequencing strategy and platform, quality assessment and data analysis procedure (reference genome, annotation file, software and parameter setting). All descriptive terms with controlled vocabularies are extracted and abstracted by manual curation of 293 published articles. In particular, diseases, tissues, and cell types are further linked to controlled terms from Disease Ontology (DO, https://disease-ontology.org) and BRENDA Tissue Ontology (BTO, http://www.ontobee.org/ontology/bto). More details about the curation model are publicly available at https://ngdc.cncb.ac.cn/gen/documentation.
Specifically, for each dataset, GEN provides a curated summary of metadata, covering species, tissue, healthy condition, RNA type, sample number, sequencing strategy, sequencing quality & quantity and experimental condition (https://ngdc.cncb.ac.cn/gen/browse/datasets, Figure 2A). To manage all collected datasets, GEN assigns an accession number prefixed with ‘GEND’ for each dataset. Moreover, since each dataset associates with specific sample(s) (prefixed with ‘GENS’), manual curation is conducted for all datasets by linking to controlled terms from DO and BTO via sample meta-information. As a result, all datasets incorporated in GEN cover 128 tissues and 46 cell types (originally curated from metadata provided by submitters). Based on these curated metadata, as a consequence, users can conveniently find the dataset(s) of interest. Structured metadata for all collected datasets is provided in a tabular form and also freely downloadable (https://ngdc.cncb.ac.cn/gen/download). Overall, bulk RNA-seq and scRNA-seq datasets involve 30 and 22 species, 89 and 64 tissues, respectively (Table 1). Regarding the specific biological contexts, GEN incorporates 153 baseline datasets, 323 spatial datasets, 83 temporal datasets, 58 environmental datasets, 55 genetic datasets and 148 phenotypic datasets, involving 84 diseases such as autism, cancer, diabetes, systemic lupus erythematosus (https://ngdc.cncb.ac.cn/gen/browse/datasets). Not surprisingly, Homo sapiens has the most abundant datasets, involving 192 datasets, 29 942 samples, 70 tissues and 84 diseases corresponding to 11 body systems (including cardiovascular, endocrine, gastrointestinal, hematopoietic, immune, integumentary, musculoskeletal, nervous, respiratory, reproductive and urinary system). More statistics of datasets and samples housed in GEN are summarized and publicly accessible on the statistics page (https://ngdc.cncb.ac.cn/gen/statistics).
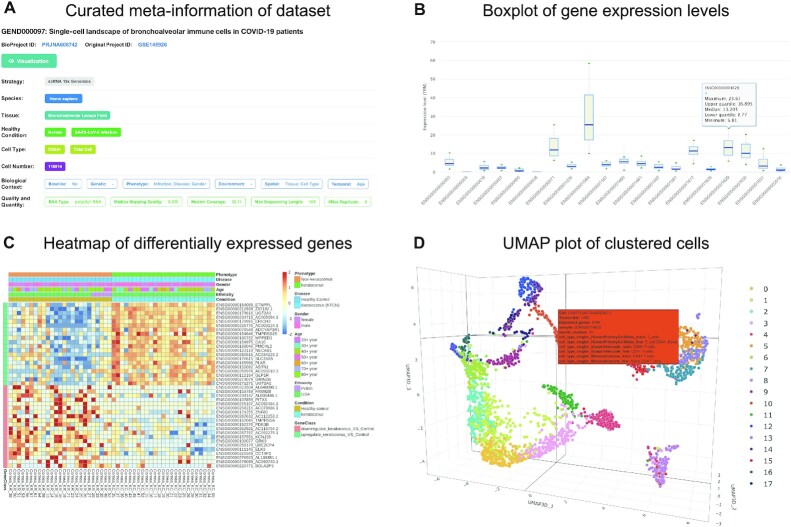
Figure 2.
Screenshots of database web interfaces. (A) Curated meta-information of dataset, including sequencing strategies, tissue, cell type, disease, biological context, quality and quantity and etc. (B) Boxplot of expression levels of multiple genes of interest across samples. (C) Heatmap of differentially expressed genes for bulk RNA-seq datasets. (D) Clustering results of single-cell RNA-seq dataset on a 3D UMAP plot where cells are color-coded by clusters.
Table 1.
Data statistics in Gene Expression Nebulas (as of August 2021)
| Kingdom | Species | #Datasets (bulk/single-cell) | #Samples | #Tissues | #Cells |
|---|---|---|---|---|---|
| Animalia | Homo sapiens | 192 (68/124) | 29 942 | 70 | 6 823 695 |
| Mus musculus | 11 (3/8) | 914 | 7 | 1 176 003 | |
| Drosophila melanogaster | 7 (1/6) | 14 800 | 4 | 3 837 235 | |
| Gallus gallus | 4 (1/3) | 329 | 7 | 42 129 | |
| Macaca mulatta | 4 (1/3) | 326 | 1 | 304 | |
| Rattus norvegicus | 4 (2/2) | 134 | 2 | 122 | |
| Capra hircus | 3 (1/2) | 86 | 3 | 59 | |
| Danio rerio | 3 (1/2) | 367 | 9 | 28 773 | |
| Bos taurus | 2 (1/1) | 142 | 3 | 100 | |
| Caenorhabditis elegans | 2 (1/1) | 12 | 2 | 130 713 | |
| Canis lupus familiaris | 2 (1/1) | 30 | 7 | 657 999 | |
| Macaca fascicularis | 2 (1/1) | 20 | 4 | 22 737 | |
| Ovis aries | 2 (1/1) | 21 | 8 | 11 380 | |
| Oryctolagus cuniculus | 2 (1/1) | 32 | 1 | 32 | |
| Schistosoma mansoni | 2 (1/1) | 15 | 2 | 55 930 | |
| Sus scrofa | 2 (1/1) | 32 | 1 | 32 | |
| Xenopus tropicalis | 2 (1/1) | 115 | 2 | 2 520 906 | |
| Plantae | Oryza sativa | 32 (31/1) | 1087 | 14 | 27 |
| Glycine max | 16 (16/0) | 499 | 8 | - | |
| Arabidopsis thaliana | 8 (5/3) | 242 | 7 | 220 188 | |
| Sorghum bicolor | 5 (5/0) | 462 | 7 | - | |
| Triticum aestivum | 3 (3/0) | 78 | 6 | - | |
| Glycine soja | 2 (2/0) | 34 | 6 | - | |
| Zea mays | 2 (2/0) | 480 | 1 | - | |
| Brassica napus | 1 (1/0) | 44 | 6 | - | |
| Gossypium hirsutum | 1 (1/0) | 14 | 1 | - | |
| Solanum lycopersicum | 1 (1/0) | 6 | 1 | - | |
| Protista | Plasmodium falciparum | 2 (1/1) | 208 | 0 | 180 |
| Dictyostelium discoideum | 2 (1/1) | 12 | 0 | 4988 | |
| Fungi | Saccharomyces cerevisiae | 2 (1/1) | 17 | 0 | 6637 |
| Total | 30 | 323 (157/166) | 50 500 | 128 | 15 540 169 |
Transcriptomic profiles at bulk and single-cell levels
GEN provides a full range of transcriptomic profiles characterizing both transcriptional and post-transcriptional regulations. For all collected datasets in GEN, expression profiles are quantified on both gene and transcript levels by three types of quantification methods, namely, raw read count number, FPKM and TPM. At the bulk level, GEN currently integrates gene expression profiles of 7412 samples from 157 datasets, involving 17 animals, 10 plants, 2 protists and 1 fungus, including Homo sapiens and model organisms such as Arabidopsis thaliana, Danio rerio, Drosophila melanogaster and Mus musculus (Table 1). Gene expression profiles can be visualized in heatmap/boxplot charts (Figure 2B). Moreover, GEN incorporates circRNA expression profiles of 456 samples from 10 human datasets. Based on the expression profiles, differentially expressed genes (DEGs) are identified between biological condition groups, which can be accessed in tabular form and visualized in heatmap charts (Figure 2C). In addition, GEN integrates a valuable collection of RNA editing events and alternative RNA splicing isoforms in 10 datasets with 574 human samples (involving 18 tissues and 16 diseases) as value-added profiles on co/post-transcriptional levels.
At the single-cell level, GEN provides high-quality expression profiles of 15 540 169 cells from 166 datasets covering 22 species (17 animals, 2 plants, 2 protists and 1 fungus), 64 tissues and 42 human diseases (Table 1). To reveal biological functions underlying expression profiles, further downstream analyses including cell clustering, identification of marker genes for each cluster and functional enrichment are performed. To facilitate easy access to cell clustering results for each dataset/sample, GEN is capable of visualizing the clustered cells using t-SNE and UMAP plots, which can be color-coded according to metadata information, cell clusters and inferred cell types (Figure 2D). Notably, in the current implementation, GEN presents cell type annotations for 121 datasets in H. sapiens and 7 datasets in M. Musculus since sufficient cell type annotation reference only exists for them (see details in Materials and Methods). In addition, marker genes for each cluster and gene enrichment analysis results can be browsed and downloaded.
Gene annotations and expression profiles
GEN provides an abundance of gene annotations for a total of 1 191 846 genes across 30 species. In addition to basic annotation (such as genomic location, biotype, functional description), GEN integrates value-added annotations derived from transcriptomic profiles, including quantitative (expression levels across different conditions) and qualitative (differential expression patterns between condition groups). For any specific gene(s), expression levels in a given dataset can be visualized by interactive heatmap and boxplot charts, and expression patterns from differential expression analysis (also applicable to the identification of marker genes for specific cell types) are annotated and incorporated in GEN. Moreover, GEN incorporates additional annotations for each gene, including editome-disease associations, internal control genes, and ontology terms (from GO, DO; see details in Materials and Methods). Consequently, GEN allows users to retrieve single or multiple genes by gene name/ID/symbol (https://ngdc.cncb.ac.cn/gen/browse/genes). Based on all collected annotations in GEN, users can conveniently find the genes of interest with specific annotations/profiles and investigate expression patterns across diverse biological conditions.
Online tools for data analysis and visualization
GEN is equipped with a series of online tools in aid of further downstream data analysis and visualization (see details in Materials and Methods). For bulk RNA-seq data, GEN offers online services for differential expression analysis, weighted gene co-expression network analysis (WGCNA), functional enrichment analysis and gene regulatory network inference. For scRNA-seq data, users can perform multiple analyses including quality control, data normalization, scaling and regression, dimensional reduction, graph-based clustering, and identification of marker genes for cell clusters (68). Furthermore, GEN is able to help users conduct gene enrichment analysis for cell markers, cell trajectory inference, and cell type annotation. Meanwhile, single-cell analysis results can be visualized by Cerebro (79), which allows interactive investigation and inspection of single-cell transcriptomic profiles incorporated in GEN. All these results can be downloaded in CSV and Excel formats and visualized images can be exported to PNG or PDF.
DISCUSSION AND FUTURE DEVELOPMENTS
GEN features systematic integration, manual curation and standardized data processing of 323 high-quality bulk and single-cell RNA-seq datasets across 30 species. It enables easy access to a comprehensive range of transcriptomic profiles, which are critical for unravelling both transcriptional and post-transcriptional regulatory mechanisms. Moreover, GEN provides abundant gene annotations based on value-added curation of transcriptomic profiles and delivers online services for bulk and single-cell data analysis and visualization.
Future directions of GEN include continuous integration and analysis of high-quality RNA-seq datasets with diverse sequencing strategies (e.g. miRNA-seq, single-cell spatial RNA-seq, nanopore long-read RNA-seq) across more species. Also, GEN will be frequently updated by enriching gene annotations based on manual curation of the ever-increasing transcriptomic profiles (13). Particularly, since the field of single-cell genomics is under rapid development, we will keep an eye on cutting-edge scRNA-seq analysis methods and make updates on GEN data processing pipelines accordingly. GEN will also provide online services to accept user-submitted expression profiles with quality control and manual curation. Furthermore, interconnections with external and internal database resources at multi-omics levels (e.g. variome (80), methylome (81) and interactome (82)) will be added and enhanced. Web tools for RNA editing profiling, alternative splicing detection and batch-effect correction across different technologies and conditions will be developed and/or implemented in GEN.
DATA AVAILABILITY
GEN is freely available online at https://ngdc.cncb.ac.cn/gen/ and does not require user to register.
ACKNOWLEDGEMENTS
We would like to thank Zhuojing Fan for her help on web interface design and a number of users for providing supports and reporting bugs. We also appreciate the anonymous reviewers for their valuable comments on this work.
Notes
Present address: Jian Sang, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
Contributor Information
Yuansheng Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dong Zou, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Tongtong Zhu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Tianyi Xu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Ming Chen, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Guangyi Niu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wenting Zong, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Rong Pan, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wei Jing, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jian Sang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Chang Liu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yujia Xiong, Beijing Neurosurgical Institute, Capital Medical University, Beijing 100069, China.
Yubin Sun, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Shuang Zhai, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Huanxin Chen, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Wenming Zhao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Jingfa Xiao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Yiming Bao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Lili Hao, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Zhang Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
FUNDING
National Key Research & Development Program of China [2017YFC0907502]; Special Investigation on Science and Technology Basic Resources of the MOST [2019FY100102]; Strategic Priority Research Program of the Chinese Academy of Sciences [XDA19050302, XDB38030200, XDB38030400]; The Youth Innovation Promotion Association of Chinese Academy of Science [2018134]; National Key Research & Development Program of China [2018YFC0309805]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008]; Genomics Data Center Construction of Chinese Academy of Sciences [WX145XQ07-04]; National Natural Science Foundation of China [32030021, 31871328]; Open Biodiversity and Health Big Data Programme of IUBS. Funding for open access charge: Special Investigation on Science and Technology Basic Resources.
Conflict of interest statement. None declared.
REFERENCES
- 1. Stubbington M.J.T., Rozenblatt-Rosen O., Regev A., Teichmann S.A.. Single-cell transcriptomics to explore the immune system in health and disease. Science. 2017; 358:58–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giacomello S., Salmen F., Terebieniec B.K., Vickovic S., Navarro J.F., Alexeyenko A., Reimegard J., McKee L.S., Mannapperuma C., Bulone V.et al.. Spatially resolved transcriptome profiling in model plant species. Nat. Plants. 2017; 3:17061. [DOI] [PubMed] [Google Scholar]
- 3. Bhadauria V., Popescu L., Zhao W.S., Peng Y.L.. Fungal transcriptomics. Microbiol. Res. 2007; 162:285–298. [DOI] [PubMed] [Google Scholar]
- 4. Wang Z., Gerstein M., Snyder M.. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009; 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao J., Spielmann M., Qiu X., Huang X., Ibrahim D.M., Hill A.J., Zhang F., Mundlos S., Christiansen L., Steemers F.J.et al.. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019; 566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Smirnov D.A., Zweitzig D.R., Foulk B.W., Miller M.C., Doyle G.V., Pienta K.J., Meropol N.J., Weiner L.M., Cohen S.J., Moreno J.G.et al.. Global gene expression profiling of circulating tumor cells. Cancer Res. 2005; 65:4993–4997. [DOI] [PubMed] [Google Scholar]
- 7. Schnable P.S., Hochholdinger F., Nakazono M.. Global expression profiling applied to plant development. Curr. Opin. Plant Biol. 2004; 7:50–56. [DOI] [PubMed] [Google Scholar]
- 8. Marioni J.C., Mason C.E., Mane S.M., Stephens M., Gilad Y.. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008; 18:1509–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alamancos G.P., Agirre E., Eyras E.. Methods to study splicing from high-throughput RNA sequencing data. Methods Mol. Biol. 2014; 1126:357–397. [DOI] [PubMed] [Google Scholar]
- 10. Stark R., Grzelak M., Hadfield J.. RNA sequencing: the teenage years. Nat. Rev. Genet. 2019; 20:631–656. [DOI] [PubMed] [Google Scholar]
- 11. Park E., Jiang Y., Hao L., Hui J., Xing Y.. Genetic variation and microRNA targeting of A-to-I RNA editing fine tune human tissue transcriptomes. Genome Biol. 2021; 22:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Z., Pan Z., Ying Y., Xie Z., Adhikari S., Phillips J., Carstens R.P., Black D.L., Wu Y., Xing Y.. Deep-learning augmented RNA-seq analysis of transcript splicing. Nat. Methods. 2019; 16:307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sang J., Zou D., Wang Z., Wang F., Zhang Y., Xia L., Li Z., Ma L., Li M., Xu B.et al.. IC4R 2.0: rice genome reannotation using massive RNA-Seq Data. Genom. Proteom. Bioinf. 2019; 18:161–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheng C.Y., Krishnakumar V., Chan A.P., Thibaud-Nissen F., Schobel S., Town C.D.. Araport11: a complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017; 89:789–804. [DOI] [PubMed] [Google Scholar]
- 15. Ray S., Satya P.. Next generation sequencing technologies for next generation plant breeding. Front. Plant Sci. 2014; 5:367–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodon J., Soria J.C., Berger R., Miller W.H., Rubin E., Kugel A., Tsimberidou A., Saintigny P., Ackerstein A., Brana I.et al.. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat. Med. 2019; 25:751–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang X., Kui L., Tang M., Li D., Wei K., Chen W., Miao J., Dong Y.. High-hroughput transcriptome profiling in drug and biomarker discovery. Front. Genet. 2020; 11:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong S., Zhang S., Fan X., Wu Q., Yan L., Dong J., Zhang H., Li L., Sun L., Pan N.et al.. A single-cell RNA-seq survey of the developmental landscape of the human prefrontal cortex. Nature. 2018; 555:524–528. [DOI] [PubMed] [Google Scholar]
- 19. Ren X., Wen W., Fan X., Hou W., Su B., Cai P., Li J., Liu Y., Tang F., Zhang F.et al.. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021; 184:1895–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Papatheodorou I., Moreno P., Manning J., Fuentes A.M.-P., George N., Fexova S., Fonseca N.A., Füllgrabe A., Green M., Huang N.et al.. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020; 48:D77–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Regev A., Teichmann S.A., Lander E.S., Amit I., Benoist C., Birney E., Bodenmiller B., Campbell P., Carninci P., Clatworthy M.et al.. The human cell atlas. Elife. 2017; 6:e27041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 2013; 45:580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D.et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. CNCB-NGDC Members and Partners. Database resources of the national genomics data center, china national center for bioinformation in 2021. Nucleic Acids Res. 2021; 49:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. BIG Data Center Members Database resources of the big data center in 2019. Nucleic. Acids. Res. 2019; 47:D8–D14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y.et al.. The genome sequence archive family: Toward explosive data growth and diverse data types. Genom. Proteom. Bioinform. 2021; 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B.. GSA: genome sequence archive. Genom. Proteom. Bioinform. 2017; 15:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kodama Y., Shumway M., Leinonen R.International Nucleotide Sequence Database, C. . The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Harrison P.W., Ahamed A., Aslam R., Alako B.T.F., Burgin J., Buso N., Courtot M., Fan J., Gupta D., Haseeb M.et al.. The european nucleotide archive in 2020. Nucleic Acids Res. 2021; 49:D82–D85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kodama Y., Mashima J., Kaminuma E., Gojobori T., Ogasawara O., Takagi T., Okubo K., Nakamura Y.. The DNA Data Bank of Japan launches a new resource, the DDBJ Omics Archive of functional genomics experiments. Nucleic Acids Res. 2012; 40:D38–D42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang L., Wang S., Li W.. RSeQC: quality control of RNA-seq experiments. Bioinformatics. 2012; 28:2184–2185. [DOI] [PubMed] [Google Scholar]
- 36. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013; arXiv doi:26 May 2013, preprint: not peer reviewedhttps://arxiv.org/abs/1303.3997.
- 39. Zhang X., Dong R., Zhang Y., Zhang J., Luo Z., Zhang J., Chen L., Yang L.. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016; 26:1277–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Gao Y., Zhang J., Zhao F.. Circular RNA identification based on multiple seed matching. Brief. Bioinform. 2018; 19:803–810. [DOI] [PubMed] [Google Scholar]
- 41. Frankish A., Diekhans M., Ferreira A.M., Johnson R., Jungreis I., Loveland J., Mudge J.M., Sisu C., Wright J., Armstrong J.et al.. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019; 47:D766–D773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Picardi E., D’Erchia A.M., Lo Giudice C., Pesole G.. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017; 45:D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Picardi E., Pesole G.. REDItools: high-throughput RNA editing detection made easy. Bioinformatics. 2013; 29:1813–1814. [DOI] [PubMed] [Google Scholar]
- 44. Wang M., Kong L.. pblat: a multithread blat algorithm speeding up aligning sequences to genomes. BMC Bioinformatics. 2019; 20:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Chen G., Ning B., Shi T.. Single-cell RNA-Seq technologies and related computational data analysis. Front Genet. 2019; 10:317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang X., Li T., Liu F., Chen Y., Yao J., Li Z., Huang Y., Wang J.. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-Seq systems. Mol. Cell. 2019; 73:130–142. [DOI] [PubMed] [Google Scholar]
- 49. Luecken M.D., Theis F.J.. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 2019; 15:e8746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Picelli S., Björklund Å.K., Faridani O.R., Sagasser S., Winberg G., Sandberg R.. Smart-seq2 for sensitive full-length transcriptome profiling in single cells. Nat. Methods. 2013; 10:1096–1098. [DOI] [PubMed] [Google Scholar]
- 51. Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M.et al.. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015; 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Klein A.M., Mazutis L., Akartuna I., Tallapragada N., Veres A., Li V., Peshkin L., Weitz D.A., Kirschner M.W.. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell. 2015; 161:1187–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Petukhov V., Guo J., Baryawno N., Severe N., Scadden D.T., Samsonova M.G., Kharchenko P.V.. dropEst: pipeline for accurate estimation of molecular counts in droplet-based single-cell RNA-seq experiments. Genome Biol. 2018; 19:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zheng G., Terry J., Belgrader P., Ryvkin P., Bent Z., Wilson R., Ziraldo S., Wheeler T., McDermott G., Zhu J.et al.. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017; 8:14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J.et al.. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019; 47:D330–D338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yanai I., Benjamin H., Shmoish M., Chalifa-Caspi V., Shklar M., Ophir R., Bar-Even A., Horn-Saban S., Safran M., Domany E.et al.. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005; 21:650–659. [DOI] [PubMed] [Google Scholar]
- 58. Schriml L.M., Arze C., Nadendla S., Chang Y.-W.W., Mazaitis M., Felix V., Feng G., Kibbe W.A.. Disease ontology: a backbone for disease semantic integration. Nucleic Acids Res. 2012; 40:D940–D946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L.et al.. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Niu G., Zou D., Li M., Zhang Y., Sang J., Xia L., Li M., Liu L., Cao J., Zhang Y.et al.. Editome Disease Knowledgebase (EDK): a curated knowledgebase of editome-disease associations in human. Nucleic Acids Res. 2019; 47:D78–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sang J., Wang Z., Li M., Cao J., Niu G., Xia L., Zou D., Wang F., Xu X., Han X.et al.. ICG: a wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 2018; 46:D121–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Picardi E., D’Erchia A.M., Lo Giudice C., Pesole G.. REDIportal: a comprehensive database of A-to-I RNA editing events in humans. Nucleic Acids Res. 2017; 45:D750–D757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Safran M., Dalah I., Alexander J., Rosen N., Iny Stein T., Shmoish M., Nativ N., Bahir I., Doniger T., Krug H.et al.. GeneCards Version 3: the human gene integrator. Database (Oxford). 2010; 2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K.. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015; 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Langfelder P., Horvath S.. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008; 9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L.et al.. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation. 2021; 2:100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huynh-Thu V.A., Irrthum A., Wehenkel L., Geurts P.. Inferring regulatory networks from expression data using tree-based methods. PLoS One. 2010; 5:e12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., Hao Y., Stoeckius M., Smibert P., Satija R.. Comprehensive integration of Single-Cell data. Cell. 2019; 177:1888–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A.et al.. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016; 44:W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014; 32:381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Aran D., Looney A.P., Liu L., Wu E., Fong V., Hsu A., Chak S., Naikawadi R.P., Wolters P.J., Abate A.R.et al.. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 2019; 20:163–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mabbott N.A., Baillie J.K., Brown H., Freeman T.C., Hume D.A.. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics. 2013; 14:632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Martens J.H.A., Stunnenberg H.G.. BLUEPRINT: mapping human blood cell epigenomes. Haematologica. 2013; 98:1487–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Monaco G., Lee B., Xu W., Mustafah S., Hwang Y.Y., Carre C., Burdin N., Visan L., Ceccarelli M., Poidinger M.et al.. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019; 26:1627–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Novershtern N., Subramanian A., Lawton L.N., Mak R.H., Haining W.N., McConkey M.E., Habib N., Yosef N., Chang C.Y., Shay T.et al.. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell. 2011; 144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Schmiedel B.J., Singh D., Madrigal A., Valdovino-Gonzalez A.G., White B.M., Zapardiel-Gonzalo J., Ha B., Altay G., Greenbaum J.A., McVicker G.et al.. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018; 175:1701–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Benayoun B.A., Pollina E.A., Singh P.P., Mahmoudi S., Harel I., Casey K.M., Dulken B.W., Kundaje A., Brunet A.. Remodeling of epigenome and transcriptome landscapes with aging in mice reveals widespread induction of inflammatory responses. Genome Res. 2019; 29:697–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heng T.S.P., Painter M.W.. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 2008; 9:1091–1094. [DOI] [PubMed] [Google Scholar]
- 79. Hillje R., Pelicci P.G., Luzi L.. Cerebro: interactive visualization of scRNA-seq data. Bioinformatics. 2020; 36:2311–2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li C., Tian D., Tang B., Liu X., Teng X., Zhao W., Zhang Z., Song S.. Genome Variation Map: a worldwide collection of genome variations across multiple species. Nucleic Acids Res. 2021; 49:D1186–D1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nawijn M., Carpaij O., Vieira Braga F., Berg M., Brouwer S., Kar G., Teichmann S., Van Den Berge M.. Novel cell types and altered cell states in asthma revealed by single-cell RNA sequencing of airway wall biopsies. Eur. Respir. J. 2018; 52:OA505. [Google Scholar]
- 82. Zhao Y., Wang J., Liang F., Liu Y., Wang Q., Zhang H., Jiang M., Zhang Z., Zhao W., Bao Y.et al.. NucMap: a database of genome-wide nucleosome positioning map across species. Nucleic Acids Res. 2019; 47:D163–D169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
GEN is freely available online at https://ngdc.cncb.ac.cn/gen/ and does not require user to register.