Abstract
The National Genomics Data Center (NGDC), part of the China National Center for Bioinformation (CNCB), provides a family of database resources to support global research in both academia and industry. With the explosively accumulated multi-omics data at ever-faster rates, CNCB-NGDC is constantly scaling up and updating its core database resources through big data archive, curation, integration and analysis. In the past year, efforts have been made to synthesize the growing data and knowledge, particularly in single-cell omics and precision medicine research, and a series of resources have been newly developed, updated and enhanced. Moreover, CNCB-NGDC has continued to daily update SARS-CoV-2 genome sequences, variants, haplotypes and literature. Particularly, OpenLB, an open library of bioscience, has been established by providing easy and open access to a substantial number of abstract texts from PubMed, bioRxiv and medRxiv. In addition, Database Commons is significantly updated by cataloguing a full list of global databases, and BLAST tools are newly deployed to provide online sequence search services. All these resources along with their services are publicly accessible at https://ngdc.cncb.ac.cn.
INTRODUCTION
The National Genomics Data Center (NGDC), part of the China National Center for Bioinformation (CNCB), was officially founded in 2019. Since then, CNCB-NGDC is constructed by joint efforts and collaborations from three institutions of Chinese Academy of Sciences, namely, Beijing Institute of Genomics, Institute of Biophysics and Shanghai Institute of Nutrition and Health as well as several partners (https://ngdc.cncb.ac.cn/partners). In the past several years, an increasing number of large-scale high-throughput sequencing projects have been carried out in biomedical research worldwide, resulting in vast amounts of multi-omics data that are continually generated at ever-growing rates and scales. Therefore, CNCB-NGDC is devoted to empowering accelerated progresses in life and health sciences by providing open access to a suite of database resources through big data archive, curation, integration and analysis (1–5).
Nowadays, rapid advances in single-cell sequencing technologies have opened a new era for biomedical research, paving the way to delineate cellular composition diversity and elucidate complex mechanisms of organ development and diseases at single-cell resolution (6,7). In addition, large-scale cohort-based precision medicine studies have identified new biomarkers and drug targets, greatly promoting the development of more effective means for disease diagnosis, molecular subtyping and medical treatment (8). To synthesize such growing data and knowledge, CNCB-NGDC has made considerable efforts in the past year by developing new resources and updating relevant resources. Particularly, due to the coronavirus disease (COVID-19) pandemic that is still a global health threat to our human being, CNCB-NGDC has continued to put enormous efforts in daily update of SARS-CoV-2 genome sequences, variants, haplotypes and literature (https://ngdc.cncb.ac.cn/ncov) (9,10). Moreover, Database Commons is significantly updated to provide open access to a full list of worldwide biological databases, and BLAST tools are newly deployed to support online sequence search services. Here, we provide a brief overview of new developments and recent updates in CNCB-NGDC and present its core resources and services (Figure 1). All these resources and their derived services are publicly available in the home page of CNCB-NGDC at https://ngdc.cncb.ac.cn.
Figure 1.
Core database resources of CNCB-NGDC in terms of database categories. A full list of data resources, which contains links to each resource, is available at https://ngdc.cncb.ac.cn/databases.
NEW DEVELOPMENTS
CancerSCEM
CancerSCEM (https://ngdc.cncb.ac.cn/cancerscem, detailed in (11) in this issue) is an open access database of cancer single-cell expression map. In the current version, it integrates a total of 638 341 high-quality cells from 208 samples across 20 human cancer types. CancerSCEM provides comprehensive metadata and multi-scale analyzed results, including cell type annotation, cell component statistics, gene expression profile (curated receptor–ligand gene pairs, oncogenes and tumor suppressor genes), cell–cell interaction network and survival analysis. Most importantly, equipped with the newly constructed comprehensive online analysis platform, CancerSCEM allows users to perform cancer scRNA-seq data exploration in a real-time and interactive mode.
CeDR Atlas
CeDR Atlas (https://ngdc.cncb.ac.cn/cedr, detailed in (12) in this issue) is a knowledge base reporting computational inference of cellular drug response for hundreds of cell types from various tissues. By collecting the fast-growing single-cell transcriptome profiles generated by multiple international consortiums and other available labeled datasets, tissue and cell type specific drug response analysis was conducted to provide direct references for cellular drug response profiles, including not only disease cell types but also normal cell types. Currently, CeDR Atlas maintains the results of 582 single-cell data objects for human, mouse and cell lines. Specifically, it hosts 188 157 significant cell type-drug associations for human, 42 660 for mouse and 10 299 for cell lines.
Cell Taxonomy
Cell Taxonomy (https://ngdc.cncb.ac.cn/celltaxonomy) is a curated repository of cell types and cell markers covering a wide range of species, tissues and conditions. Based on manual curation of 3402 publications, it presents a standardized and well-structured taxonomy for 2650 cell types and collects 25 087 associated cell markers in 157 conditions and 296 tissues across 21 species. In addition, Cell Taxonomy incorporates 564 single-cell RNA-seq datasets and provides multifaceted characterization for cell types and cell markers by enrichment analysis, cellular component similarity estimation and quality assessment of cell markers and cell clusters. Taken together, Cell Taxonomy is of great utility for cell type characterization and accurate selection of cell markers and reference datasets, functioning as a fundamental reference cellular resource for a wide range of single-cell research.
CompoDynamics
CompoDynamics (https://ngdc.cncb.ac.cn/compodynamics, detailed in (13) in this issue) is a comprehensive database of sequence compositions of coding sequences (CDSs) and genomes for a wide range of species. CompoDynamics characterizes rich sequence compositions (nucleotide content, codon usage and amino acid usage) and derived molecular features (coding potential, physicochemical property and phase separation) for 118 689 747 high-quality CDSs and 34 562 genomes across 24 995 species. In addition, multiple tools are provided to enable comparative analyses of sequence compositions and features across different species and gene groups. Collectively, CompoDynamics bears great potential to help us reveal sequence composition dynamics across genes and genomes, providing a fundamental resource for a broad spectrum of biological studies.
OMIX
The Open Archive for Miscellaneous Data (OMIX; https://ngdc.cncb.ac.cn/omix), a new member of the GSA family, aims to meet users’ needs for archiving miscellaneous data that are unsuitable for storing in GSA/GSA-Human. It allows different data types (e.g. microarray and genotype), accepts various omics data (e.g. lipidome, metabolome and proteome) and houses analyzed results and related research data (e.g. clinical information, demographic data and questionnaire). OMIX features straightforward submission interfaces and offers open-access and controlled-access data management strategies. As of September 2021, OMIX has archived 269 data submissions with 13.3 Terabytes (TB), among which 115 have controlled access.
OpenLB
The Open Library of Bioscience (OpenLB; https://ngdc.cncb.ac.cn/openlb) provides easy and open access to a large number of biological literatures. In the current version, it contains ∼33 million abstract texts from PubMed (14), bioRxiv and medRxiv. OpenLB provides both simple keyword query and advanced search functionalities, in order to help users search publications in a convenient and customized manner. In addition, OpenLB aims to provide seamless links with CNCB-NGDC database resources, associating scientific literature with omics data and curated information if available so that users can easily find both publications and their related data/information. Ongoing efforts of OpenLB include the integration of more literature types, deployment of named entity recognition tool and development of manuscript submission service.
RDBSB
The Registry and Database of Bioparts for Synthetic Biology (RDBSB; https://www.biosino.org/rdbsb) is a finely curated resource for catalytic bioparts, incorporating comprehensive information of biopart sequence and functions (including catalytic processes, qualitative and quantitative parameters and biopart expression). RDBSB collects 366 045 catalytic bioparts, and 72 180 of them are manually curated with experimental evidence from literature mining. In addition, RDBSB collects relevant experimental conditions, such as pH, temperature and chassis, etc., which are crucial for pathway design in a given chassis.
Regeneration Roadmap
Regeneration Roadmap (https://ngdc.cncb.ac.cn/regeneration, detailed in (15) in this issue) is a comprehensive database collecting and standardizing experimental data generated in regeneration research. In the current version, Regeneration Roadmap systematically and comprehensively collects regenerative information over 1.96 million data entries across 10 species and 34 tissues, including regeneration-related genes, bulk and single-cell transcriptomics, epigenomics and pharmacogenomics data. In this database, users can easily explore regulatory and expression changes of regeneration-associated genes in different species or tissues. Together, Regeneration Roadmap provides the research community with a long awaited and valuable data resource featuring convenient computing and visualization tools.
RECENT UPDATES
BioProject and BioSample
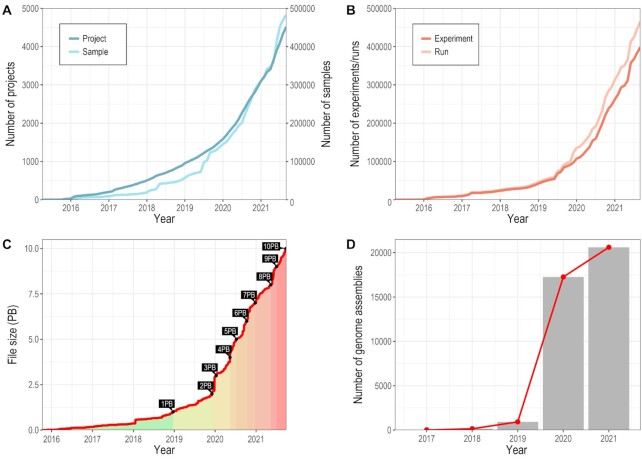
BioProject (https://ngdc.cncb.ac.cn/bioproject) and BioSample (https://ngdc.cncb.ac.cn/biosample) are two public repositories of biological research projects and samples, respectively. They collect descriptive metadata on biological projects and samples investigated in experiments and provide centralized accesses to all public projects and samples as well as cross links to their related data resources. BioProject organizes a huge volume of projects, involving multi-omics sequencing efforts, genome-wide association studies and variation analyses. BioSample supports a wide scope of sample types, including human, plant, animal, microbe, virus, pathogen and metagenome. Up to September 2021, there are a total of 4514 biological projects and 482 577 samples submitted by 2538 users from 514 organizations (Figure 2A), presenting a rapid increase by comparison with 2288 projects and 176 288 samples in August 2020.
Figure 2.
Statistics of data submissions. (A) Data statistics of BioProject and BioSample. (B) Data statistics of Experiments and Runs in GSA. (C) Timeline of data growth in GSA. (D) Statistics of genome assemblies in GWH. All statistics are frequently updated and publicly available at https://ngdc.cncb.ac.cn/bioproject, https://ngdc.cncb.ac.cn/biosample and https://ngdc.cncb.ac.cn/gsa and https://ngdc.cncb.ac.cn/gwh.
GSA and GSA-Human
The Genome Sequence Archive (GSA; https://ngdc.cncb.ac.cn/gsa) (16,17) is a public data repository for archiving raw sequence reads. GSA accepts worldwide data submissions, performs data curation and quality control, and provides free open access to all publicly available data without restrictions. In addition, GSA for Human (GSA-Human; https://ngdc.cncb.ac.cn/gsa-human) (17), serving as an important partner database of GSA, features controlled-access and security services for human genetics-related data and accepts data submissions of various studies, including disease, cohort, cell line, clinical pathogen and human-associated metagenome. As of September 2021, GSA together with GSA-Human has reached a milestone of over 10 PB of raw sequencing data archived as well as 398 322 experiments and 465 245 runs (Figure 2B and C), showing the doubled volume by comparison with the previous release last August (∼4.6 PB). In particular, GSA-Human has accommodated 5.6 PB of raw sequence data since its inception in 2018, demonstrating that human genetic data are growing at an unprecedented rate and scale.
Genome Warehouse
The Genome Warehouse (GWH; https://ngdc.cncb.ac.cn/gwh) is a public repository of genome-scale data for a wide range of species (18). By September 2021, GWH has housed a total of 20 606 submitted genome assemblies covering 1,251 species (Figure 2D), presenting a doubled increase in contrast to the previous release (9337 assemblies in 2020). Among them, 9886 genome assemblies have been publicly released and reported in 97 articles of 47 journals. Particularly, GWH has received the submission of 1660 SARS-CoV-2 genome assemblies, which were further integrated into the 2019 novel coronavirus resources (2019nCoVR) (9,10). Moreover, compared with the previous release, GWH has been significantly upgraded by providing sequence alignment service via BLAST (19) and supplying encrypted links for reviewing unpublic data. Collectively, GWH serves as an important resource for genome assembly data to support genomic research throughout the world.
Gene Expression Nebulas
The Gene Expression Nebulas (GEN; https://ngdc.cncb.ac.cn/gen) is a data portal integrating transcriptomic profiles at both bulk and single-cell levels in various conditions across multiple species (detailed in (20) in this issue). In the current version, GEN houses a collection of transcriptomic profiles of 323 datasets covering 50 500 samples and 15 540 169 cells across 30 species involving 17 animals, 10 plants, 2 protists, and 1 fungus, grown from 191 studies covering 23 073 experiments and 410 149 cells derived from human and 4 plants. In particular, GEN integrates a full range of transcriptomic profiles on gene expression, RNA editing and alternative splicing for 10 bulk datasets. Moreover, it accommodates value-added gene annotations based on differential expression analysis across diverse experimental conditions and cell clusters. Of note, curated annotation information of plant RNA editing factors from Plant Editosome Database (PED) (21), editome-disease associations from Editome-Disease Knowledgebase (EDK) (22) and RT-qPCR reference genes from Internal Control Genes (ICG) (23) are also interconnected to expand the scope of knowledge for corresponding genes.
MethBank
The Methylation Bank (MethBank; https://ngdc.cncb.ac.cn/methbank) (24,25) is a comprehensive database of DNA methylation data. The current version of MethBank incorporates 855 single-base resolution methylomes (SRMs), 93 936 775 methylation profiles of genes, 6 945 524 methylated CpG Islands and 304 884 differentially methylated promoters based on whole-genome bisulfite sequencing data, exhibiting significant updates relative to the previous version in August 2020 (394 SRMs, 19 701 343 methylation profiles, 1 258 420 methylated CpG Islands and 304 884 differentially methylated promoters). Based on 4577 450K DNA methylation samples from normal peripheral blood, MethBank also offers 692 methylation sites closely associated with age, 2335 sites with constant methylation levels across different ages, 53 211 age-specific differentially methylated cytosines and 1899 age-specific differentially methylated regions.
scMethBank
The single-cell methylation bank (scMethBank; https://ngdc.cncb.ac.cn/methbank/scm) is a public data portal that integrates a comprehensive collection of single-cell DNA methylation data (detailed in (26) in this issue). In the past year, scMethBank has rapidly grown from 3166 samples in August 2020 to 8328 samples currently, involving 29 cell types and 67 619 genes with curated metadata in human and mouse. Based on uniformed data processing, it presents whole-genome DNA methylation profiles at single-nucleotide resolution in various biological contexts and developmental stages. Accordingly, user-friendly web interfaces for data search, download, visualization and online tools for downstream analysis are implemented in scMethBank.
LncRNAWiki
LncRNAWiki (https://ngdc.cncb.ac.cn/lncrnawiki) is a wiki-based database for community-curation of human long non-coding RNAs (lncRNAs) (27,28). The current version of LncRNAWiki 2.0 is significantly updated by (i) providing a new curation model with more informative and essential annotation items, (ii) developing a new web system based on MySQL/Java (instead of MediaWiki) that is capable of organizing all contents in a structured manner, (iii) improving the community-annotation submission functionality and providing more user-friendly web interfaces and (iv) equipping with online tools for ID conversion and functional prediction. Consequently, LncRNAWiki 2.0 incorporates 2512 lncRNAs and their annotations compared to 2056 featured lncRNAs in LncRNAWiki 1.0 in 2020, thus providing an up-to-date picture of experimentally validated and functionally annotated lncRNAs in human.
piRBase
The updated version of piRBase v3.0 (http://bigdata.ibp.ac.cn/piRBase) (29) is a comprehensive database of piRNA sequences. In current release of piRBase, the number of non-redundant piRNA sequence increases from 173 million in last August to 181 million, and the species reaches 44 compared to 21 in August 2020. In view of the huge amount of piRNAs, it provides users with gold standard piRNA sequence sets. In order to further expand the research on piRNA function, potential information of splicing-junction piRNA and piRNA variants is also included in piRBase, offering an alternative explanation for possible mechanism of piRNAs. In addition, it integrates piRNA-related information on a variety of diseases, like cancers, cardiovascular diseases, stroke and Alzheimer. Also, piRBase presents regulatory network of piRNAs in a visualized manner and provides the expression of piRNAs in different tissues and cell lines.
EWAS Open Platform
The EWAS Open Platform (https://ngdc.cncb.ac.cn/ewas) is an open platform for epigenome-wide association studies (EWAS), including EWAS Atlas, EWAS Data Hub and EWAS Toolkit (detailed in (30) in this issue). As an EWAS knowledgebase, EWAS Atlas (https://ngdc.cncb.ac.cn/ewas/atlas) has grown from 577 267 associations in August 2020 to 617 018 associations curated from 910 publications, covering 618 traits and 3382 cohorts in September 2021 (31). As a data portal of EWAS Open Platform, EWAS Data Hub (https://ngdc.cncb.ac.cn/ewas/datahub) integrates 115 852 samples (in contrast to 95 783 samples in August 2020) of standardized DNA methylation array data (450K and EPIC/850k) (32) and the corresponding metadata involving 925 tissues/cells and 528 diseases (33). EWAS Toolkit (https://ngdc.cncb.ac.cn/ewas/toolkit) is newly developed to provide downstream analysis and network visualization, such as trait enrichment, genomic location enrichment, GO and KEGG enrichment, chromatin state and histone modification enrichment, tissue methylation, expression regulation, motif enrichment and EWAS knowledge graph.
GWAS Atlas
GWAS Atlas (https://ngdc.cncb.ac.cn/gwas) (34) is a curated resource of genome-wide variant-trait associations in plants and animals. In contrast to 78 950 associations in August 2020, the current version of GWAS Atlas has archived a total of 96 141 associations across seven cultivated plants and five domesticated animals, manually curated from 1350 studies in 367 publications. As a result, a total of 23 880 genes and 862 traits were annotated and presented based on a set of ontologies. Together, GWAS Atlas provides high-quality curated GWAS associations for plants and animals, and accordingly serves as a valuable resource for genetic research of important traits and breeding application.
BrainBase
BrainBase (https://ngdc.cncb.ac.cn/brainbase, detailed in (35) in this issue) is a curated knowledgebase for brain diseases with the aim to provide a whole picture of brain diseases and associated genes. Compared to the previous version that contains 4248 associations and 3996 genes in August 2020, the current version houses 7175 disease-gene associations, spanning a total of 123 brain diseases and linking with 5662 genes. It also integrates 16 591 drug–target interactions covering 2118 drugs/chemicals and 623 genes, and presents specific genes in light of expression specificity in brain tissue/regions/cerebrospinal fluid/cells. In addition, BrainBase incorporates multi-omics datasets to identify glioma featured genes with potential clinical significance.
dbDEMC
The database of Differentially Expressed MicroRNAs in human Cancers (dbDEMC, https://www.biosino.org/dbDEMC) is an integrated database for storing and annotating potential cancer-related microRNAs (miRNAs), retrieved by analyzing large numbers of miRNA expression profiling studies. Compared with the previous version (2224 differentially expressed miRNAs [DEMs] in 36 cancer types from 209 expression profiling data sets), dbDEMC version 3.0 integrates more data entries, containing 3268 DEMs in 40 cancer types curated from 807 experiments in human, mouse and rat. It is also updated by enhancing the visualization functionalities for expression heatmap, regulatory network, gene ontology, KEGG pathway map and miRNA expression boxplot. In addition, dbDEMC incorporates experimentally validated targets for the DEMs. Therefore, dbDEMC will play an important role in characterizing molecular functions and regulatory mechanisms of DEMs in human cancers.
SARS-CoV-2 Resources
The 2019 Novel Coronavirus Resources (2019nCoVR; https://bigd.big.ac.cn/ncov) (36,37) contains a comprehensive collection of all publicly available SARS-CoV-2 genome sequences with quality evaluation and value-added manual annotations. Consequently, it houses a global landscape of genomic variants and haplotypes, visualizes the spatiotemporal change for each variant and constructs haplotype network maps for the course of the outbreak. More importantly, it provides the hierarchical epidemiological lineage browser to easily capture the leading edge of pandemic transmission (38). Besides, 2019nCoVR offers a set of online tools for SARS-CoV-2 genome assembly and annotation, variant identification and effect annotation, genome tracing and haplotype construction as well as a full collection of literatures on COVID-19 (9). Notably, all SARS-CoV-2 genome sequences, variants, haplotypes and literatures are updated daily since January 2020. Meantime, a patient-centric resource named integrative CT images and clinical features for COVID-19 (iCTCF) is developed to archive chest CT images and 130 types of clinical features as well as laboratory-confirmed SARS-CoV-2 clinical status, providing a useful tool for improving diagnosis and treatment of COVID-19 patients (39).
iDog
iDog (https://ngdc.cncb.ac.cn/idog) is an integrated omics data resource for domestic dog (Canis lupus familiaris) and wild canids (40). In the current version, iDog is updated by integrating 27 ancient dog samples with 6 544 496 unique SNPs and including 26 cell clusters with 105 057 single cells for dog brain tissue. As a result, a total of 71 050 194 unique SNPs in 722 samples, 481 breeds, 806 diseases and 1170 genotype-to-phenotype pairs from 1192 experiments and 62 high-quality RNA-seq projects are integrated, dramatically increasing from 42 871 184 SNPs and 594 genotype-to-phenotype pairs in August 2020. Additionally, iDog provides an online classification tool used to predict the dog breed by using deep learning method. As a data resource of the Dog 10K Genomes Project (http://dog10k.big.ac.cn), with these functions and data, iDog provides freely browse, search and download services for worldwide users.
iSheep
iSheep (https://ngdc.cncb.ac.cn/isheep) (41) is the most comprehensive genetic database specific for the Ovis species. It provides an integrated resource for sheep comprising of 82 689 498 genetic variants from 2778 samples and a wealth of information on genotype and phenotype association. In the past year, 1418 world’s breed information entries are newly curated from 19 public databases, and new online tools are implemented for comparing the SNPs between two or more individual genomes and visualizing the genomic locations of variants. Additionally, iSheep also provides the reference and annotation resources of other 10 species.
SorGSD
The Sorghum Genome Science Database (SorGSD, previously named as Sorghum Genome SNP Database; https://ngdc.cncb.ac.cn/sorgsd) (42) is updated by expanding to 289 sorghum lines including 33 825 236 SNPs and 5 722 385 small INDELs compared with 48 sorghum lines in August 2020. It also added phenotypic data and panicle pictures of critical accessions. Currently, SorGSD also implements new tools including ID Conversion, Homologue Search, Blast and Genome Browser for online data analysis and provides general information related to sorghum research, such as 44 online sorghum resources and 162 literature references. Collectively, SorGSD contains large-scale genomic variations and phenotypic information and thus serves as a critical resource for the global sorghum researchers.
Database Commons
Database Commons (https://ngdc.cncb.ac.cn/databasecommons) is a catalogue of worldwide biological databases, aiming to provide open access to a full list of global databases and their descriptive metadata manually curated from their associated publications. Currently, with the efforts of 53 curators, it catalogues a total of 5455 databases involving 8133 publications and 2095 organizations throughout the world, showing a growth by comparison with the previous version (5064 databases, 7595 publications and 1944 organizations) in August 2020. Based on this, Database Commons provides a global landscape of publicly available databases, allowing users to access and browse databases by customized filters and yielding a series of informative statistics in terms of country, institution, database category, year, citation, etc.
NGDC Education
NGDC Education (https://ngdc.cncb.ac.cn/education) is an open education resource that provides a series of educational materials. This past year, two courses, viz., Bioinformatics and Genomics Data Analysis, were newly added by the courtesy of Prof. Yu Xue from Huazhong University of Science and Technology and Prof. Cheng Li from Peking University, respectively. In addition, biographies of the late Profs. Xiaocheng Gu of Peking University and Bailin Hao of Fudan University were added. Early in the 1990s, Prof. Gu established the Center for Bioinformatics in Peking University to provide bioinformatics resources and services for domestic and international users. Prof. Hao made great contributions to the bioinformatics research, particularly his CVTree algorithm for bacterial genome classification (43,44) and advocate of establishing the CNCB since the 1990s. Their personal profiles, articles, and videos (if available) can be found at NGDC Education. In addition, in coordination with the Global Biodiversity and Health Big Data (BHBD) Alliance, we promote open sharing of educational materials as well as multi-omics data throughout the world.
Tools
Users’ needs of sequence search and comparison are growing with the expansion of various database resources in CNCB-NGDC. BLAST tools (https://ngdc.cncb.ac.cn/blast) are newly deployed, providing online services of different sequence alignment types developed by National Center for Biotechnology Information (NCBI) (45) with featured databases, for instance, GWH transcripts, LncBook human lncRNA sequences, 10K protist species genomes and SARS-CoV-2 genome sequences. In particular, to support worldwide studies on SARS-CoV-2, a series of genomic analysis tools on coronavirus are also established (https://ngdc.cncb.ac.cn/ncov/online/tools) (37), which cover sequencing quality control, de novo assembly and variant calling, haplotype network construction, genome tracing and lineage identification. Besides, computational identification of long non-coding RNAs (https://ngdc.cncb.ac.cn/lgc) (46) and EWAS Toolkit for functional enrichment and network visualization (https://ngdc.cncb.ac.cn/ewas/toolkit) (47) are also presented. And BIG Search, a distributed and scalable search engine, has been updated by including standardized data indexes from all resources in CNCB-NGDC, 39 partner resources (see details at https://ngdc.cncb.ac.cn/partners) as well as European Bioinformatics Institute (EBI) resources based on EBI Search RESTful API (48), NCBI resources powered by NCBI Entrez (49) and the AlphaFold Protein Structure Database (50).
CONCLUDING REMARKS
This year, several core resources of CNCB-NGDC have been listed as recommended repositories (e.g. nucleic acid sequences and genetic variations) by major publishers such as Cell Press, Elsevier and Springer Nature, greatly accelerating the rapid deposition and public sharing of biomedical big data at a global scale. Additionally, we keep paying efforts to build close collaborations with INSDC (International Nucleotide Sequence Database Collaboration) (51), as testified by the open sharing of SARS-CoV-2 genome data with NCBI. Importantly, 2019nCoVR has been significantly updated by frequent data integration and web interface improvement. Meanwhile, to deal with the explosive growth of multi-omics data, CNCB-NGDC provides a suite of database resources, which are newly developed and frequently updated, to accept worldwide data submissions and provide value-added annotations and curated knowledge. Ongoing efforts include, but not limited to, optimization and automation of data submission, curation and analysis procedures, infrastructure upgrade for big data storage and transfer, and development of new tools and pipelines to support worldwide genetic and genomic research. As one of the major global centers, CNCB-NGDC will continue to expand and offer a series of data resources and services to benefit a wide range of research in life and health sciences.
DATA AVAILABILITY
All the resources can be accessed at https://ngdc.cncb.ac.cn.
ACKNOWLEDGEMENTS
We thank our users for submitting data, sending suggestions, reporting bugs and getting involved in community curation. CNCB-NGDC is indebted to its funders, including the Ministry of Science and Technology and the Ministry of Finance of the People’s Republic of China as well as Chinese Academy of Sciences.
APPENDIX
Corresponding author: Yongbiao Xue1,2,3,*
Co-corresponding authors: Yiming Bao1,2,3,*, Zhang Zhang1,2,3,*, Wenming Zhao1,2,3,*, Jingfa Xiao1,2,3,*, Shunmin He3,4,*, Guoqing Zhang3,5,*, Yixue Li3,5,*, Guoping Zhao3,5,6,7,*, Runsheng Chen4,8,*
CNCB-NGDC MEMBERS (Arranged by project role and then by contribution except for Team Leader (TL), as indicated)
CancerSCEM: Jingyao Zeng1,2,#, Yadong Zhang1,2#, Yunfei Shang1,2,3,#, Jialin Mai1,2,3, Shuo Shi1,2,3, Mingming Lu1,2,3, Congfan Bu1,2, Zhewen Zhang1,2,3, Zhenglin Du1,2, Jingfa Xiao1,2,3,* (TL)
CeDR Atlas: Yinying Wang2,9,#, Hongen Kang1,2,3,9,#, Tianyi Xu1,2, Lili Hao1,2, Yiming Bao1,2,3,*, Peilin Jia1,2,9,# (TL)
Cell Taxonomy: Shuai Jiang1,2,#, Qiheng Qian1,2,3,#, Tongtong Zhu1,2,3,#, Yunfei Shang1,2,3, Wenting Zong1,2,3, Tong Jin1,2,3, Yuansheng Zhang1,2,3, Dong Zou1,2, Yiming Bao1,2,3, Jingfa Xiao1,2,3,* (TL), Zhang Zhang1,2,3,* (TL)
CompoDynamics: Shuai Jiang1,2,#, Qiang Du1,2,3,#, Changrui Feng1,2,3,#, Lina Ma1,2,# (TL)
OMIX: Sisi Zhang1,2,#, Anke Wang1,2,#, Lili Dong1,2, Yanqing Wang1,2,# (TL)
OpenLB: Dong Zou1,2,#, Zhang Zhang1,2,3,*
RDBSB: Wan Liu5,#, Xing Yan10,#, Yunchao Ling5,#, Guoping Zhao5,10, Zhihua Zhou10, Guoqing Zhang5,*
Regeneration Roadmap: Wang Kang2,3,9,11,#, Tong Jin1,2,3,#, Tao Zhang1,2,3,#, Shuai Ma3,11,12,13,#, Haoteng Yan14,15,#, Zunpeng Liu3,12,13,16, Zejun Ji12,13,16, Yusheng Cai11,12,13, Si Wang14,15, Moshi Song3,11,12,13, Jie Ren2,3,9,12, Qi Zhou3,12,13,16, Jing Qu3,12,13,16,#, Weiqi Zhang2,3,9,12,#, Yiming Bao1,2,3,*, Guanghui Liu3,11,12,13,14,15,#
BioProject & BioSample & GSA & BIG Submission & infrastruction: Xu Chen1,2,#, Tingting Chen1,2,#, Sisi Zhang1,2,#, Yanling Sun1,2,#, Caixia Yu1,2, Bixia Tang1,2, Junwei Zhu1,2, Lili Dong1,2, Shuang Zhai1,2, Yubin Sun1,2, Qiancheng Chen1,2, Xiaoyu Yang1,2, Xin Zhang1,2, Zhengqi Sang1,2, Yonggang Wang1,2, Yilin Zhao1,2, Huanxin Chen1,2, Li Lan1,2, Yanqing Wang1,2,* (TL), Wenming Zhao1,2,3,* (TL)
Genome Warehouse: Yingke Ma1,2,#, Yaokai Jia1,2,#, Xinchang Zheng1,2,#, Meili Chen1,2,# (TL)
Gene Expression Nebulas: Yuansheng Zhang1,2,3,#, Dong Zou1,2,#, Tongtong Zhu1,2,3,#, Tianyi Xu1,2,#, Ming Chen1,2,3,#, Guangyi Niu1,2,3, Wenting Zong1,2,3, Rong Pan1,2,3, Wei Jing1,2,3, Jian Sang1,2,3,†, Chang Liu1,2,3, Yujia Xiong17, Yubin Sun1,2, Shuang Zhai1,2, Huanxin Chen1,2, Wenming Zhao1,2,3, Jingfa Xiao1,2,3, Yiming Bao1,2,3, Lili Hao1,2,# (TL)
MethBank: Mochen Zhang1,2,3,#, Guoliang Wang1,2,3,#, Dong Zou1,2,#, Lizhi Yi1,2,3,#, Wei Zhao1,2,3,#, Wenting Zong1,2,3, Song Wu1,2,3, Zhuang Xiong1,2,3, Rujiao Li1,2,#(TL)
scMethBank: Wenting Zong1,2,3,#, Hongen Kang1,2,3,#, Zhuang Xiong1,2,3, Yingke Ma1,2, Tong Jin1,2,3, Zheng Gong1,2,3, Lizhi Yi1,2,3, Mochen Zhang1,2,3, Song Wu1,2,3, Guoliang Wang1,2,3, Rujiao Li1,2,# (TL)
LncRNAWiki: Lin Liu1,2,3,#, Zhao Li1,2,3,#, Chang Liu1,2,3,#, Dong Zou1,2, Qianpeng Li1,2,3, Changrui Feng1,2,3, Wei Jing1,2,3, Sicheng Luo1,2,3,18, Lina Ma1,2,# (TL)
piRBase: Jiajia Wang3,4,#, Yirong Shi3,4,#, Honghong Zhou4,#, Peng Zhang4, Tingrui Song4, Yanyan Li4, Shunmin He3,4,* (TL)
EWAS Open Platform: Zhuang Xiong1,2,3,#, Fei Yang1,2,3,#, Mengwei Li1,2,#, Wei Zhao1,2,3, Guoliang Wang1,2,3, Zhaohua Li1,2,3, Yingke Ma1,2, Dong Zou1,2, Wenting Zong1,2,3, Hongen Kang1,2,3, Yaokai Jia1,2, Xinchang Zheng1,2, Rujiao Li1,2,#(TL)
GWAS Atlas: Dongmei Tian1,2,#, Xiaonan Liu1,2,3,#, Cuiping Li1,2, Xufei Teng1,2,3, Shuhui Song1,2,3,# (TL)
BrainBase: Lin Liu1,2,3,#, Yang Zhang1,2,3,#, Guangyi Niu1,2,3,#, Qianpeng Li1,2,3, Zhao Li1,2,3, Tongtong Zhu1,2,3, Changrui Feng1,2,3, Xiaonan Liu1,2,3, Yuansheng Zhang1,2,3, Tianyi Xu1,2, Ruru Chen1,2,3,18, Xufei Teng1,2,3, Rongqin Zhang1,2,3, Dong Zou1,2, Lina Ma1,2,# (TL)
dbDEMC: Feng Xu19,#, Yifan Wang5,#, Yunchao Ling5, Chenfen Zhou5, Haizhou Wang19, Andrew E. Teschendorff5, Yungang He19,#, Guoqing Zhang5,*, Zhen Yang19,#
2019nCoVR: Shuhui Song1,2,3,#, Lina Ma1,2,#, Dong Zou1,2,#, Dongmei Tian1,2,#, Cuiping Li1,2,#, Junwei Zhu1,2,#, Lun Li1,2,#, Na Li1,2,#, Zheng Gong1,2,3, Meili Chen1,2, Anke Wang1,2, Yingke Ma1,2, Xufei Teng1,2,3, Ying Cui1,2,3, Guangya Duan1,2,3, Mochen Zhang1,2,3,20, Tong Jin1,2,3, Gangao Wu1,2,3,Tianhao Huang1,2,3, Enhui Jin1,2,3, Wei Zhao1,2,3, Hailong Kang1,2,3, Zhonghuang Wang1,2,3, Zhenglin Du1,2, Yadong Zhang1,2, Rujiao Li1,2, Jingyao Zeng1,2, Lili Hao1,2, Shuai Jiang1,2, Hua Chen2,9, Mingkun Li2,9, Jingfa Xiao1,2,3, Zhang Zhang1,2,3,* (TL), Wenming Zhao1,2,3,* (TL), Yongbiao Xue1,2,3,* (TL), Yiming Bao1,2,3,* (TL)
iCTCF: Wanshan Ning21,#, Yu Xue21,#
iDog: Bixia Tang1,2,#, Yanhu Liu16,22,#, Yanling Sun1,2,#, Guangya Duan1,2,3,#, Ying Cui1,2,3,#, Qijun Zhou16,22, Lili Dong1,2, Enhui Jin1,2,3, Xingyan Liu3,23, Longlong Zhang3,22, Bingyu Mao3,22, Shihua Zhang3,23, Yaping Zhang3,16,22, Guodong Wang3,16,22,# (TL), Wenming Zhao1,2,3,* (TL)
iSheep: Zhonghuang Wang1,2,3,#, Qianghui Zhu3,16,#, Xin Li16, Junwei Zhu1,2, Dongmei Tian1,2, Hailong Kang1,2,3, Cuiping Li1,2, Sisi Zhang1,2, Shuhui Song1,2,3, Menghua Li (TL)16,24, Wenming Zhao1,2,3,* (TL)
SorGSD: Yuanming Liu3,25,#, Zhonghuang Wang1,2,3,#, Hong Luo25, Junwei Zhu1,2, Xiaoyuan Wu25, Dongmei Tian1,2, Cuiping Li1,2, Wenming Zhao1,2,3,* (TL), Haichun Jing3,25,26,# (TL)
SSO: Junwei Zhu1,2,# (TL), Bixia Tang1,2,#
Database Commons: Dong Zou1,2,#, Lin Liu1,2,3,#, Yitong Pan1,2,3, Chang Liu1,2,3, Ming Chen1,2,3, Xiaonan Liu1,2,3, Yuansheng Zhang1,2,3, Zhao Li1,2,3, Changrui Feng1,2,3, Qiang Du1,2,3, Ruru Chen1,2,3,18, Tongtong Zhu1,2,3, Lina Ma1,2,# (TL)
NGDC Education: Dong Zou1,2,#, Shuai Jiang1,2, Zhang Zhang1,2,3,* (TL)
Coronavirus analysis platform: Zheng Gong1,2,3,#, Junwei Zhu1,2,#, Cuiping Li1,2,#, Shuai Jiang1,2,#, Lina Ma1,2, Bixia Tang1,2, Dong Zou1,2, Meili Chen1,2, Yubin Sun1,2, Leisheng Shi2,3,9, Shuhui Song1,2,3, Zhang Zhang1,2,3, Mingkun Li2,9, Jingfa Xiao1,2,3, Yongbiao Xue1,2,3, Yiming Bao1,2,3, Zhenglin Du1,2,3,#, Wenming Zhao1,2,3,*
LGC: Zhao Li1,2,3, Qiang Du1,2,3, Shuai Jiang1,2, Lina Ma1,2,#, Zhang Zhang1,2,3,*
EWAS Toolkit: Zhuang Xiong1,2,3,#, Mengwei Li1,2,#, Dong Zou1,2, Wenting Zong1,2,3, Rujiao Li1,2,#(TL)
BLAST: Meili Chen1,2,#, Zhenglin Du1,2,#, Wenming Zhao1,2,3,*, Yiming Bao1,2,3,*, Yingke Ma1,2,# (TL)
BHBD: Xin Zhang1,2, Li Lan1,2, Yongbiao Xue 1,2,3,*, Yiming Bao1,2,3,*
Writing Group: Shuai Jiang1,2,#, Changrui Feng1,2,3,#, Wenming Zhao1,2,3,*, Jingfa Xiao1,2,3,*, Yiming Bao1,2,3,*, Zhang Zhang1,2,3,*
CNCB-NGDC PARTNERS (Listed in alphabetical order by database names)
BBCancer: Zhixiang Zuo27, Jian Ren27
CancerSEA: Xinxin Zhang28, Yun Xiao28, Xia Li28
CellMarker: Xinxin Zhang28, Yun Xiao28, Xia Li28
CGDB: Dan Liu21, Chi Zhang21, Yu Xue21
CGGA: Zheng Zhao17, Tao Jiang17
circAtlas: Wanying Wu29, Fangqing Zhao29
CirFunBase: Xianwen Meng30, Ming Chen30,
dbPSP & THANATOS: Di Peng21, Yu Xue21
DEG & DoriC: Hao Luo31,32,33, Feng Gao31,32,33
DrLLPS: Wanshan Ning21, Yu Xue21
EPSD & WERAM: Shaofeng Lin21, Yu Xue21
EVAtlas: Chuijie Liu21, Anyuan Guo21
GenTree: Hao Yuan3,34, Tianhan Su3,34, Yong E. Zhang3,34,35
HCL: Yincong Zhou30, Ming Chen30, Guoji Guo36
iEKPD: Shanshan Fu21, Xiaodan Tan21, Yu Xue21
iUUCD: Weizhi Zhang21, Yu Xue21
LeukemiaDB: Mei Luo21, Anyuan Guo21
lnCAR: Yubin Xie27, Jian Ren27
MCA: Yincong Zhou30, Ming Chen30, Guoji Guo36
MiCroKiTS: Chenwei Wang21, Yu Xue21
msRepDB: Xingyu Liao37,38, Xin Gao37, Jianxin Wang38
PEA: Guiyan Xie21, Anyuan Guo21
PceRBase: Chunhui Yuan30, Ming Chen30
PlantRegMap: Feng Tian39, Dechang Yang39, Ge Gao39
PLMD: Dachao Tang21, Yu Xue21
PncStres: Wenyi Wu30, Ming Chen30
PTMD: Yujie Gou21, Cheng Han21, Yu Xue21, Qinghua Cui40,41
RhesusBase: Xiangshang Li42, Chuan-Yun Li42
RMVar: XiaoTong Luo27, Jian Ren27
SEECancer: Xinxin Zhang28, Yun Xiao28, Xia Li28
* To whom correspondence should be addressed. Tel: +86 10 84097261; Email: ybxue@big.ac.cn
Correspondence may also be addressed to Yiming Bao. Tel: +86 10 84097858; Email: baoym@big.ac.cn
Correspondence may also be addressed to Zhang Zhang. Tel: +86 10 84097261; Email: zhangzhang@big.ac.cn
Correspondence may also be addressed to Wenming Zhao. Tel: +86 10 84097636; Email: zhaowm@big.ac.cn
Correspondence may also be addressed to Jingfa Xiao. Tel: +86 10 84097443; Email: xiaojingfa@big.ac.cn
Correspondence may also be addressed to Shunmin He. Tel: +86 10 64807279; Email: heshunmin@ibp.ac.cn
Correspondence may also be addressed to Guoqing Zhang. Tel: 13524783378; Email: gqzhang@picb.ac.cn
Correspondence may also be addressed to Yixue Li. Tel: +86 21 54920086; Email: yxli@sibs.ac.cn
Correspondence may also be addressed to Guoping Zhao. Tel: +86 21 54924000; Email: gpzhao@sibs.ac.cn
Correspondence may also be addressed to Runsheng Chen. Tel: +86 10 64888543; Email: crs@ibp.ac.cn
#The authors wish it to be known that, in their opinion, these authors should be regarded as Joint First Authors.
1National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
2China National Center for Bioinformation, Beijing 100101, China
3University of Chinese Academy of Sciences, Beijing 100049, China
4National Genomics Data Center & Key Laboratory of RNA Biology, Center for Big Data Research in Health, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China
5National Genomics Data Center & Bio-Med Big Data Center, Key Laboratory of Computational Biology, CAS-MPG Partner Institute for Computational Biology, Shanghai Institute of Nutrition and Health, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Xuhui, Shanghai 200031, China
6CAS-Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, 300 Fenglin Road, Xuhui, Shanghai 200032, China
7Center for Quantitative Synthetic Biology, Institute of Synthetic Biology, Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences, Shenzhen 518000, China
8Guangdong Geneway Decoding Bio-Tech Co. Ltd, Foshan, 528316, China
9CAS Key Laboratory of Genomic and Precision Medicine, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China
10CAS Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China
11State Key Laboratory of Membrane Biology, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China
12Institute for Stem cell and Regeneration, CAS, Beijing 100101, China
13Beijing Institute for Stem Cell and Regenerative Medicine, Beijing 100101, China
14Advanced Innovation Center for Human Brain Protection, and National Clinical Research Center for Geriatric Disorders, Xuanwu Hospital Capital Medical University, Beijing 100053, China
15Aging Translational Medicine Center, Xuanwu Hospital, Capital Medical University, Beijing 100053, China
16Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, 650223, China
17Beijing Neurosurgical Institute, Capital Medical University, Beijing 100069, China
18Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China
19Shanghai Key Laboratory of Medical Epigenetics, the International Co-laboratory of Medical Epigenetics and Metabolism, Ministry of Science and Technology, Institutes of Biomedical Sciences, Fudan University, Shanghai 200032, China
20School of Future Technology, University of Chinese Academy of Sciences, Beijing 100049, China
21Key Laboratory of Molecular Biophysics of Ministry of Education, Hubei Bioinformatics and Molecular Imaging Key Laboratory, Center for Artificial Intelligence Biology, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan 430074, Hubei, China
22State Key Laboratory of Genetic Resources and Evolution, Kunming Institute of Zoology, Chinese Academy of Sciences, Kunming 650223, China
23NCMIS, CEMS, RCSDS, Academy of Mathematics and Systems Science, Chinese Academy of Sciences, Beijing, 100190, China
24College of Animal Science and Technology, China Agricultural University, Beijing 100193, China
25Key Laboratory of Plant Resources, Institute of Botany, Chinese Academy of Sciences, Beijing, 100093, China
26Engineering Laboratory for Grass-Based Livestock Husbandry, Chinese Academy of Sciences, Beijing, 100093, China
27State Key Laboratory of Oncology in South China, Cancer Center, Collaborative Innovation Center for Cancer Medicine, School of Life Sciences, Sun Yat-sen University, Guangzhou 510060, China
28College of Bioinformatics Science and Technology, Harbin Medical University, Harbin, Heilongjiang 150081, China
29Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing 100101, China
30Department of Bioinformatics, College of Life Sciences, The First Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou 310058, China
31Department of Physics, School of Science, Tianjin University, Tianjin 300072, China
32Frontiers Science Center for Synthetic Biology and Key Laboratory of Systems Bioengineering (Ministry of Education), Tianjin University, Tianjin 300072, China
33SynBio Research Platform, Collaborative Innovation Center of Chemical Science and Engineering (Tianjin), Tianjin 300072, China
34Key Laboratory of Zoological Systematics and Evolution and State Key Laboratory of Integrated Management of Pest Insects and Rodents, Institute of Zoology, Chinese Academy of Sciences, Beijing 100101, China
35CAS Center for Excellence in Animal Evolution and Genetics, Chinese Academy of Sciences, Kunming, Yunnan 650223, China
36Center for Stem Cell and Regenerative Medicine, Zhejiang University School of Medicine, Hangzhou 310000, China
37Computational Bioscience Research Center (CBRC), Computer, Electrical and Mathematical Sciences and Engineering Division, King Abdullah University of Science and Technology (KAUST), Thuwal 23955, Saudi Arabia
38Hunan Provincial Key Lab on Bioinformatics, School of Computer Science and Engineering, Central South University, Changsha 410083, China
39Biomedical Pioneering Innovation Center (BIOPIC), Beijing Advanced Innovation Center for Genomics (ICG), Center for Bioinformatics (CBI), and State Key Laboratory of Protein and Plant Gene Research at School of Life Sciences, Peking University, Beijing 100871, China
40Department of Biomedical Informatics, School of Basic Medical Sciences, MOE Key Lab of Cardiovascular Sciences, Center for Noncoding RNA Medicine, Peking University, Beijing 100190, China
41Center of Bioinformatics, Key Laboratory for NeuroInformation of Ministry of Education, School of Life Science and Technology, University of Electronic Science and Technology of China, Chengdu, Sichuan 610054, China
42Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Institute of Molecular Medicine, College of Future Technology, Peking University, Beijing 100190, China
Contributor Information
CNCB-NGDC Members and Partners:
Yongbiao Xue, Yiming Bao, Zhang Zhang, Wenming Zhao, Jingfa Xiao, Shunmin He, Guoqing Zhang, Yixue Li, Guoping Zhao, Runsheng Chen, Jingyao Zeng, Yadong Zhang, Yunfei Shang, Jialin Mai, Shuo Shi, Mingming Lu, Congfan Bu, Zhewen Zhang, Zhenglin Du, Jingfa Xiao, Yinying Wang, Hongen Kang, Tianyi Xu, Lili Hao, Yiming Bao, Peilin Jia, Shuai Jiang, Qiheng Qian, Tongtong Zhu, Yunfei Shang, Wenting Zong, Tong Jin, Yuansheng Zhang, Dong Zou, Yiming Bao, Jingfa Xiao, Zhang Zhang, Shuai Jiang, Qiang Du, Changrui Feng, Lina Ma, Sisi Zhang, Anke Wang, Lili Dong, Yanqing Wang, Dong Zou, Zhang Zhang, Wan Liu, Xing Yan, Yunchao Ling, Guoping Zhao, Zhihua Zhou, Guoqing Zhang, Wang Kang, Tong Jin, Tao Zhang, Shuai Ma, Haoteng Yan, Zunpeng Liu, Zejun Ji, Yusheng Cai, Si Wang, Moshi Song, Jie Ren, Qi Zhou, Jing Qu, Weiqi Zhang, Yiming Bao, Guanghui Liu, Xu Chen, Tingting Chen, Sisi Zhang, Yanling Sun, Caixia Yu, Bixia Tang, Junwei Zhu, Lili Dong, Shuang Zhai, Yubin Sun, Qiancheng Chen, Xiaoyu Yang, Xin Zhang, Zhengqi Sang, Yonggang Wang, Yilin Zhao, Huanxin Chen, Li Lan, Yanqing Wang, Wenming Zhao, Yingke Ma, Yaokai Jia, Xinchang Zheng, Meili Chen, Yuansheng Zhang, Dong Zou, Tongtong Zhu, Tianyi Xu, Ming Chen, Guangyi Niu, Wenting Zong, Rong Pan, Wei Jing, Jian Sang, Chang Liu, Yujia Xiong, Yubin Sun, Shuang Zhai, Huanxin Chen, Wenming Zhao, Jingfa Xiao, Yiming Bao, Lili Hao, Mochen Zhang, Guoliang Wang, Dong Zou, Lizhi Yi, Wei Zhao, Wenting Zong, Song Wu, Zhuang Xiong, Rujiao Li, Wenting Zong, Hongen Kang, Zhuang Xiong, Yingke Ma, Tong Jin, Zheng Gong, Lizhi Yi, Mochen Zhang, Song Wu, Guoliang Wang, Rujiao Li, Lin Liu, Zhao Li, Chang Liu, Dong Zou, Qianpeng Li, Changrui Feng, Wei Jing, Sicheng Luo, Lina Ma, Jiajia Wang, Yirong Shi, Honghong Zhou, Peng Zhang, Tingrui Song, Yanyan Li, Shunmin He, Zhuang Xiong, Fei Yang, Mengwei Li, Wei Zhao, Guoliang Wang, Zhaohua Li, Yingke Ma, Dong Zou, Wenting Zong, Hongen Kang, Yaokai Jia, Xinchang Zheng, Rujiao Li, Dongmei Tian, Xiaonan Liu, Cuiping Li, Xufei Teng, Shuhui Song, Lin Liu, Yang Zhang, Guangyi Niu, Qianpeng Li, Zhao Li, Tongtong Zhu, Changrui Feng, Xiaonan Liu, Yuansheng Zhang, Tianyi Xu, Ruru Chen, Xufei Teng, Rongqin Zhang, Dong Zou, Lina Ma, Feng Xu, Yifan Wang, Yunchao Ling, Chenfen Zhou, Haizhou Wang, Andrew E Teschendorff, Yungang He, Guoqing Zhang, Zhen Yang, Shuhui Song, Lina Ma, Dong Zou, Dongmei Tian, Cuiping Li, Junwei Zhu, Lun Li, Na Li, Zheng Gong, Meili Chen, Anke Wang, Yingke Ma, Xufei Teng, Ying Cui, Guangya Duan, Mochen Zhang, Tong Jin, Gangao Wu, Tianhao Huang, Enhui Jin, Wei Zhao, Hailong Kang, Zhonghuang Wang, Zhenglin Du, Yadong Zhang, Rujiao Li, Jingyao Zeng, Lili Hao, Shuai Jiang, Hua Chen, Mingkun Li, Jingfa Xiao, Zhang Zhang, Wenming Zhao, Yongbiao Xue, Yiming Bao, Wanshan Ning, Yu Xue, Bixia Tang, Yanhu Liu, Yanling Sun, Guangya Duan, Ying Cui, Qijun Zhou, Lili Dong, Enhui Jin, Xingyan Liu, Longlong Zhang, Bingyu Mao, Shihua Zhang, Yaping Zhang, Guodong Wang, Wenming Zhao, Zhonghuang Wang, Qianghui Zhu, Xin Li, Junwei Zhu, Dongmei Tian, Hailong Kang, Cuiping Li, Sisi Zhang, Shuhui Song, Menghua Li, Wenming Zhao, Yuanming Liu, Zhonghuang Wang, Hong Luo, Junwei Zhu, Xiaoyuan Wu, Dongmei Tian, Cuiping Li, Wenming Zhao, Haichun Jing, Junwei Zhu, Bixia Tang, Dong Zou, Lin Liu, Yitong Pan, Chang Liu, Ming Chen, Xiaonan Liu, Yuansheng Zhang, Zhao Li, Changrui Feng, Qiang Du, Ruru Chen, Tongtong Zhu, Lina Ma, Dong Zou, Shuai Jiang, Zhang Zhang, Zheng Gong, Junwei Zhu, Cuiping Li, Shuai Jiang, Lina Ma, Bixia Tang, Dong Zou, Meili Chen, Yubin Sun, Leisheng Shi, Shuhui Song, Zhang Zhang, Mingkun Li, Jingfa Xiao, Yongbiao Xue, Yiming Bao, Zhenglin Du, Wenming Zhao, Zhao Li, Qiang Du, Shuai Jiang, Lina Ma, Zhang Zhang, Zhuang Xiong, Mengwei Li, Dong Zou, Wenting Zong, Rujiao Li, Meili Chen, Zhenglin Du, Wenming Zhao, Yiming Bao, Yingke Ma, Xin Zhang, Li Lan, Yongbiao Xue, Yiming Bao, Shuai Jiang, Changrui Feng, Wenming Zhao, Jingfa Xiao, Yiming Bao, Zhang Zhang, Zhixiang Zuo, Jian Ren, Xinxin Zhang, Yun Xiao, Xia Li, Xinxin Zhang, Yun Xiao, Xia Li, Dan Liu, Chi Zhang, Yu Xue, Zheng Zhao, Tao Jiang, Wanying Wu, Fangqing Zhao, Xianwen Meng, Ming Chen, Di Peng, Yu Xue, Hao Luo, Feng Gao, Wanshan Ning, Yu Xue, Shaofeng Lin, Yu Xue, Chuijie Liu, Anyuan Guo, Hao Yuan, Tianhan Su, Yong E Zhang, Yincong Zhou, Ming Chen, Guoji Guo, Shanshan Fu, Xiaodan Tan, Yu Xue, Weizhi Zhang, Yu Xue, Mei Luo, Anyuan Guo, Yubin Xie, Jian Ren, Yincong Zhou, Ming Chen, Guoji Guo, Chenwei Wang, Yu Xue, Xingyu Liao, Xin Gao, Jianxin Wang, Guiyan Xie, Anyuan Guo, Chunhui Yuan, Ming Chen, Feng Tian, Dechang Yang, Ge Gao, Dachao Tang, Yu Xue, Wenyi Wu, Ming Chen, Yujie Gou, Cheng Han, Yu Xue, Qinghua Cui, Xiangshang Li, Chuan-Yun Li, XiaoTong Luo, Jian Ren, Xinxin Zhang, Yun Xiao, and Xia Li
FUNDING
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38030200, XDB38050300, XDB38030400, XDA19050302, XDA19090116, XDA24040201, XDB38030100, XDA12030100, XDB38040300]; National Key Research and Development Program of China [2019YFA0801801, 2018YFA0801405, 2018YFD1000505, 2018YFC2000100, 2018YFC1406902, 2018YFC0910400, 2018YFC0310602, 2018YFA0903700, 2018YFA0900704, 2017YFC1201200, 2017YFC0908405, 2017YFC0908404, 2017YFC0908403, 2017YFC0907505, 2017YFC0907503, 2017YFC0907502, 2016YFE0206600, 2016YFC0906403, 2016YFC0903003, 2016YFC0901904, 2016YFC0901903, 2016YFC0901702, 2016YFC0901604, 2016YFC0901603, 2016YFB0201702, 2016YFA0501704, 2021YFC0863300, 2016YFC0902500, 2018YFA0900700]; National Natural Science Foundation of China [91731303, 81670462, 31970565, 31871328, 31871294, 31701117, 31970647, 21621004, 31801104, 31771465, 31771410, 31771388, 31671360, 81701567, 31571358, 31525014, 1470330, 31961130380, 31711530221, 31771477, 31571366, 31822030, 31801113, 31801154, 31771458, 91940303, 91940306, 31661143031, 31730110, 31871281, 31970634, 31930021, 32025009, 31970633, 32100520, 62002388, 61772557, 81761168038, 82161148009]; International Partnership Program of the Chinese Academy of Sciences [153F11KYSB20160008, 153D31KYSB20170121]; 13th Five-year Informatization Plan of Chinese Academy of Sciences [XXH13505-05]; Genomics Data Center Construction of Chinese Academy of Sciences [WX145XQ07-04]; Fundamental Research Funds for the Central Universities [2019kfyRCPY043]; UK Royal Society-Newton Advanced Fellowship [NAF\R1\191094]; Key Program of the Chinese Academy of Sciences [KJZD-EW-L14]; Key Research Program of Frontier Sciences of the Chinese Academy of Sciences [QYZDJ-SSW-SYS009]; Key Technology Talent Program of the Chinese Academy of Sciences; 100 Talent Program of the Chinese Academy of Sciences; K.C. Wong Education Foundation; Youth Innovation Promotion Association of the Chinese Academy of Sciences [2019104, 2018134, 2017141]; National Key R&D Program [SQ2017YFSF090210]; China Postdoctoral Science Foundation [2019M652623, 2018M632830]; China 863 Program [2015AA020108]; Open Biodiversity and Health Big Data Program of IUBS; Professional Association of the Alliance of International Science Organizations [ANSO-PA-2020–07]; Funds for Basic Resources Investigation Research of the Ministry of Science and Technology [2018FY10080002]; Special Project on National Science and Technology Basic Resources Investigation [2019FY100102]; CAS Pioneer 100-Talent program; Key Research Program of the Chinese Academy of Sciences [KFZD-SW-219–5]; Zhang jiang special project of national innovation demonstration zone [ZJ2018-ZD-013]; Science and Technology Service Network Initiative of Chinese Academy of Sciences; Hunan Provincial Science and Technology Program [2018wk4001]; 111 Project [B18059]; King Abdullah University of Science and Technology (KAUST) [FCC/1/1976–18-01, FCC/1/1976–23-01, FCC/1/1976–25-01, FCC/1/1976–26-01, REI/1/0018–01-01, REI/1/4216–01-01, REI/1/4437–01-01, REI/1/4473–01-01, URF/1/4352–01-01, URF/1/4379–01-01, REI/1/4742–01-01, URF/1/4098–01-01]; Chinese Academy of Sciences [KFJ-BRP-017–79]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38030200].
Conflict of interest statement. None declared.
REFERENCES
- 1. CNCB-NGDC Members and Partners. Database resources of the national genomics data center, china national center for bioinformation in 2021. Nucleic Acids Res. 2021; 49:D18–D28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. National Genomics Data Center Members and Partners Database resources of the national genomics data center in 2020. Nucleic Acids Res. 2020; 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. BIG Data Center Members Database resources of the big data center in 2019. Nucleic Acids Res. 2019; 47:D8–D14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. BIG Data Center Members Database resources of the big data center in 2018. Nucleic Acids Res. 2018; 46:D14–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. BIG Data Center Members The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang Y., Navin N.E.. Advances and applications of single-cell sequencing technologies. Mol. Cell. 2015; 58:598–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Potter S.S. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018; 14:479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bilkey G.A., Burns B.L., Coles E.P., Mahede T., Baynam G., Nowak K.J.. Optimizing precision medicine for public health. Front Public Health. 2019; 7:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao W.M., Song S.H., Chen M.L., Zou D., Ma L.N., Ma Y.K., Li R.J., Hao L.L., Li C.P., Tian D.M.et al.. The 2019 novel coronavirus resource. Yi Chuan. 2020; 42:212–221. [DOI] [PubMed] [Google Scholar]
- 10. Song S., Ma L., Zou D., Tian D., Li C., Zhu J., Chen M., Wang A., Ma Y., Li M.et al.. The global landscape of SARS-CoV-2 genomes, variants, and haplotypes in 2019nCoVR. Genomics Proteomics Bioinformatics. 2021; 18:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zeng J., Zhang Y., Shang Y., Mai J., Shi S., Lu M., Bu C., Zhang Z., Zhang Z., Li Y.et al.. CancerSCEM: A database of single-cell expression map across various human cancers. Nucleic Acids Res. 2021; 10.1093/nar/gkab905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang Y., Kang H., Xu T., Hao L., Bao Y., Jia P.. CeDR Atlas: a knowledgebase of cellular drug response. Nucleic Acids Res. 2021; 10.1093/nar/gkab972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang S., Du Q., Feng C., Ma L., Zhang Z.. CompoDynamics: a comprehensive database for characterizing sequence composition dynamics. Nucleic Acids Res. 2021; 10.1093/nar/gkab979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fiorini N., Lipman D.J., Lu Z.. Towards PubMed 2.0. Elife. 2017; 6:e28801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kang W., Jin T., Zhang T., Ma S., Yan H., Liu Z., Ji Z., Cai Y., Wang S., Song M.et al.. Regeneration Roadmap: database resources for regenerative biology. Nucleic Acids Res. 2022; 10.1093/nar/gkab870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Y., Song F., Zhu J., Zhang S., Yang Y., Chen T., Tang B., Dong L., Ding N., Zhang Q.et al.. GSA: genome sequence archive. Genomics Proteom. Bioinform. 2017; 15:14–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., Dong L., Zhang Z., Yu C., Sun Y.et al.. The genome sequence archive family: toward explosive data growth and diverse data types. Genomics Proteom. Bioinform. 2021; 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen M., Ma Y., Wu S., Zheng X., Kang H., Sang J., Xu X., Hao L., Li Z., Gong Z.et al.. Genome warehouse: a public repository housing genome-scale data. Genomics Proteom. Bioinform. 2021; 10.1016/j.gpb.2021.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J.. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997; 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang Y., Zou D., Zhu T., Xu T., Chen M., Niu G., Zong W., Pan R., Jing W., Sang J.et al.. Gene Expression Nebulas (GEN): a comprehensive data portal integrating transcriptomic profiles across multiple species at both bulk and single-cell levels. Nucleic Acids Res. 2022; 10.1093/nar/gkab878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li M., Xia L., Zhang Y., Niu G., Li M., Wang P., Zhang Y., Sang J., Zou D., Hu S.et al.. Plant editosome database: a curated database of RNA editosome in plants. Nucleic Acids Res. 2019; 47:D170–D174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niu G., Zou D., Li M., Zhang Y., Sang J., Xia L., Li M., Liu L., Cao J., Zhang Y.et al.. Editome Disease Knowledgebase (EDK): a curated knowledgebase of editome-disease associations in human. Nucleic Acids Res. 2019; 47:D78–D83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sang J., Wang Z., Li M., Cao J., Niu G., Xia L., Zou D., Wang F., Xu X., Han X.et al.. ICG: a wiki-driven knowledgebase of internal control genes for RT-qPCR normalization. Nucleic Acids Res. 2018; 46:D121–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li R., Liang F., Li M., Zou D., Sun S., Zhao Y., Zhao W., Bao Y., Xiao J., Zhang Z.. MethBank 3.0: a database of DNA methylomes across a variety of species. Nucleic Acids Res. 2018; 46:D288–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zou D., Sun S., Li R., Liu J., Zhang J., Zhang Z.. MethBank: a database integrating next-generation sequencing single-base-resolution DNA methylation programming data. Nucleic Acids Res. 2015; 43:D54–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zong W., Kang H., Xiong Z., Ma Y., Jin T., Gong Z., Yi L., Zhang M., Wu S., Wang G.et al.. scMethBank: a database for single-cell whole genome DNA methylation maps. Nucleic Acids Res. 2022; 10.1093/nar/gkab833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ma L.N., Li A., Zou D., Xu X.J., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu L., Li Z., Liu C., Zou D., Li Q., Feng C., Jing W., Luo S., Zhang Z., Ma L.. LncRNAWiki 2.0: a knowledgebase of human long non-coding RNAs with enhanced curation model and database system. Nucleic Acids Res. 2021; 10.1093/nar/gkab998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang J., Zhang P., Lu Y., Li Y., Zheng Y., Kan Y., Chen R., He S.. piRBase: a comprehensive database of piRNA sequences. Nucleic Acids Res. 2019; 47:D175–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Xiong Z., Yang F., Li M., Ma Y., Zhao W., Wang G., Li Z., Zheng X., Zou D., Zong W.et al.. EWAS Open Platform: integrated data, knowledge and toolkit for epigenome-wide association study. Nucleic Acids Res. 2021; 10.1093/nar/gkab972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li M., Zou D., Li Z., Gao R., Sang J., Zhang Y., Li R., Xia L., Zhang T., Niu G.et al.. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019; 47:D983–D988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiong Z., Li M., Ma Y., Li R., Bao Y.. GMQN: A reference-based method for correcting batch effects as well as probes bias in HumanMethylation BeadChip. 2021; bioRxiv doi:07 September 2021, preprint: not peer reviewed 10.1101/2021.09.06.459116. [DOI] [PMC free article] [PubMed]
- 33. Xiong Z., Li M., Yang F., Ma Y., Sang J., Li R., Li Z., Zhang Z., Bao Y.. EWAS Data Hub: a resource of DNA methylation array data and metadata. Nucleic Acids Res. 2020; 48:D890–D895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tian D., Wang P., Tang B., Teng X., Li C., Liu X., Zou D., Song S., Zhang Z.. GWAS Atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020; 48:D927–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu L., Zhang Y., Niu G., Li Q., Li Z., Zhu T., Feng C., Liu X., Zhang Y., Xu T.et al.. BrainBase: a curated knowledgebase for brain diseases. Nucleic Acids Res. 2022; 10.1093/nar/gkab987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song S., Ma L., Zou D., Tian D., Li C., Zhu J., Chen M., Wang A., Ma Y., Li M.et al.. The Global Landscape of SARS-CoV-2 Genomes, Variants, and Haplotypes in 2019nCoVR. Genomics Proteomics Bioinformatics. 2020; 18:749–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gong Z., Zhu J.W., Li C.P., Jiang S., Ma L.N., Tang B.X., Zou D., Chen M.L., Sun Y.B., Song S.H.et al.. An online coronavirus analysis platform from the National Genomics Data Center. Zool Res. 2020; 41:705–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rambaut A., Holmes E.C., O’Toole A., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G.. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 2020; 5:1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ning W., Lei S., Yang J., Cao Y., Jiang P., Yang Q., Zhang J., Wang X., Chen F., Geng Z.et al.. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020; 4:1197–1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tang B., Zhou Q., Dong L., Li W., Zhang X., Lan L., Zhai S., Xiao J., Zhang Z., Bao Y.et al.. iDog: an integrated resource for domestic dogs and wild canids. Nucleic Acids Res. 2019; 47:D793–D800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang Z.H., Zhu Q.H., Li X., Zhu J.W., Tian D.M., Zhang S.S., Kang H.L., Li C.P., Dong L.L., Zhao W.M.et al.. iSheep: an integrated resource for sheep genome, variant and phenotype. Front Genet. 2021; 12:714852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liu Y., Wang Z., Wu X., Zhu J., Luo H., Tian D., Li C., Luo J., Zhao W., Hao H.et al.. SorGSD: updating and expanding the sorghum genome science database with new contents and tools. Biotechnol. Biofuels. 2021; 14:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Qi J., Luo H., Hao B.. CVTree: a phylogenetic tree reconstruction tool based on whole genomes. Nucleic Acids Res. 2004; 32:W45–W47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu Z., Hao B.. CVTree update: a newly designed phylogenetic study platform using composition vectors and whole genomes. Nucleic Acids Res. 2009; 37:W174–W178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 46. Wang G., Yin H., Li B., Yu C., Wang F., Xu X., Cao J., Bao Y., Wang L., Abbasi A.A.et al.. Characterization and identification of long non-coding RNAs based on feature relationship. Bioinformatics. 2019; 35:2949–2956. [DOI] [PubMed] [Google Scholar]
- 47. Li M., Zou D., Li Z., Gao R., Sang J., Zhang Y., Li R., Xia L., Zhang T., Niu G.et al.. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019; 47:D983–D988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Madeira F., Park Y.M., Lee J., Buso N., Gur T., Madhusoodanan N., Basutkar P., Tivey A.R.N., Potter S.C., Finn R.D.et al.. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019; 47:W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gibney G., Baxevanis A.D.. Searching NCBI databases using entrez. Curr. Protoc. Hum. Genet. 2011; 10.1002/0471142905.hg0610s71. [DOI] [PubMed] [Google Scholar]
- 50. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arita M., Karsch-Mizrachi I., Cochrane G.. The international nucleotide sequence database collaboration. Nucleic Acids Res. 2021; 49:D121–D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the resources can be accessed at https://ngdc.cncb.ac.cn.