Abstract
The Sequence Read Archive (SRA, https://www.ncbi.nlm.nih.gov/sra/) stores raw sequencing data and alignment information to enhance reproducibility and facilitate new discoveries through data analysis. Here we note changes in storage designed to increase access and highlight analyses that augment metadata with taxonomic insight to help users select data. In addition, we present three unanticipated applications of taxonomic analysis.
INTRODUCTION
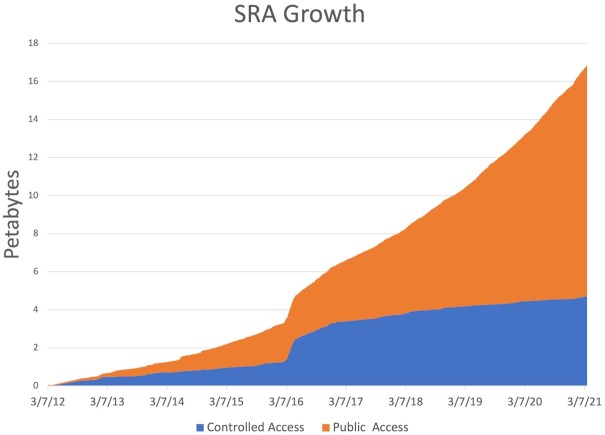
The Sequence Read Archive (SRA) for next-generation sequencing (NGS) data at the National Center for Biotechnology Information (NCBI) was established as part of the International Nucleotide Sequence Database Collaboration (INSDC) in 2009 (1). Experiencing ‘explosive’ growth of open-access NGS submissions in early years, the last decade became a tsunami compared to the initial flood of data (Figure 1). Our goal is to ensure NIH-funded research data is findable, accessible, interoperable and reusable (FAIR) (2) to facilitate the unique opportunities that arise from scientific inquiry at the massive (petabyte) scale of SRA data. The National Institutes of Health (NIH) Science and Technology Research Infrastructure for Discovery, Experimentation, and Sustainability (STRIDES) Initiative (https://datascience.nih.gov/strides) supports access to nearly 14 million augmented SRA records. A two-year effort made all the Sequence Read Archive (SRA) records publicly available on two cloud platforms, retrievable in multiple formats, along with associated metadata in both Google Cloud Platform BigQuery and Amazon Web Services Athena (see https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud/).
Figure 1.
Growth of SRA over the last decade. Through September 2021 Public Access contains approximately 25.6 Petabase pairs originating from over 14.8 million publicly available runs averaging 1.7 Gbp per run, 0.83 GB per run, 9.6 million spots per run and 187 bp per spot.
User access is our priority, and it is widely appreciated that because NGS ‘[base] quality scores require considerable space to store and transmit, they are a major bottleneck in any sequence analysis pipeline’ (3). Surprisingly, the same authors show that eliminating most base quality scores can yield increased genotyping accuracy compared to that obtained employing quality scores even when coverage is not high. Yet, we recognize as have others that ‘some samples will always be worth storing in their raw DNA sequence form’ (4). Our goal is to serve all user data applications with options for efficient retrieval, storage, and computational needs. To this end, we introduce additional normalized working files without submitted base quality scores, but with read quality flags that allow user retrieval of files with simplified scores supplied in a manner compatible with applications expecting the scores (SRA Lite). The existing (larger) normalized files with submitted base quality scores (SRA original) are available with free access through Amazon Web Services Registry of Open Data (https://registry.opendata.aws/ncbi-sra/). For details on cloud platform SRA retrieval see (5).
Recently available from cloud platforms are taxonomic analyses of all public SRA records as determined by our SRA Tax Analysis Tool (STAT) (6, https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud-based-taxonomy-analysis-table/).
STAT Results
The data are available for download or interactive querying via BigQuery and Athena. (https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud-based-examples/). STAT assigns taxonomic identity at the read level, integrating these signals conservatively to the spot, and this data is reported in the Taxonomy Analysis Table (‘tax_analysis’) (https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud-based-taxonomy-analysis-table/). We draw attention to the reported columns ‘self_count’ and ‘total_count’. The first (‘self count’) is the total number of spots assigned the NCBI Taxonomy ID (TaxId) column entry (‘tax id’) for the row of the identified accession. Column ‘total count’ by comparison, is a sum of all spots assigned to the identified taxonomic node equivalent to the total number of both self and all child nodes; in essence the total number of spots assigned to a least common ancestor ‘tax id’. It is important to note that results are proportional to the size of each sequenced genome, and this must be considered against the genomic complexity of the sequenced sample in coming to conclusions based on hit abundance.
STAT challenges
Over the years we have recognized some general limitations of STAT analysis that are consequential for users to understand.
Reference input contamination
NCBI reference genomes (“RefSeq”, 7) are used as input to build our 32 base k-mer (i.e. k = 32) taxonomic database, and contamination (8), or even biological complexity, represents a challenge. We chose a simple heuristic to preferentially assign identical k-mers found in multiple superkingdoms where one of those is eukaryote, favoring placement either in the non-eukaryotic superkingdom at the warranted level of specificity, or the cellular organism root. For example, imagine a k-mer chosen for the database using human reference sequence as input that derived from a bacterial contaminant. When merging the eukaryotic and bacterial databases the same k-mer may be derived from the reference bacterial genome. In such a case we choose to preferentially remove the k-mer from the eukaryotic database and leave it in the bacterial database. Human endogenous retroviral sequences (9) may likewise give rise to an identical k-mer in eukaryotic and viral databases that would be relegated to viruses.
Under-represented and highly variant virus input
RefSeq suffers from a significant lack of representative viral genomes. Moreover, many viral sequence records represent myriad variants of biomedical interest, such as evolving strains whether during a pandemic, or through endemic history. For these reasons, we chose to augment our input genomic reference sequences with records extracted from NCBI BLAST® ‘nt/nr’ assigned a TaxId whose lineage root is the superkingdom ‘Viruses’ (https://ftp.ncbi.nlm.nih.gov/blast/documents/blastdb.html). This likely results in a small or modest amount of ‘noise’ as the cost incurred for significantly increasing breadth and utility of viral taxonomic identification.
Heterologous updates
Both the k-mer database and processing that generates the taxonomic analysis report are periodically updated but resource costs limit our ability to reanalyse all or even significant portions of SRA while simultaneously analysing the steadily increasing wave of new submissions. While we prioritize keeping apace of submissions, the Taxonomy Analysis Information table (‘tax_analysis_info’) contains summary information about the technical details of each analysis, particularly the columns ‘parser_version’ and ‘aligns_to_version’ (https://www.ncbi.nlm.nih.gov/sra/docs/sra-cloud-based-taxonomy-analysis-information-table/). Future migration of the STAT pipeline entirely to cloud-based infrastructure may soon allow us to bring conformity to results.
Taxonomic model
The NCBI Taxonomy database ‘includes organism names and classifications for every sequence in the nucleotide and protein sequence databases of the International Nucleotide Sequence Database Collaboration (INSDC)’ (10). Most species are missing sequence representatives (11), and when a taxon lacks a sequence representative in Genbank (or RefSeq) it will also be absent from our STAT k-mer database since the input sequences are drawn only from these sources (6). Further, while phylogenetic, the Taxonomy tree is not solely based on sequence similarity (10,11). In both building the k-mer database and reporting results, STAT assumes strictly molecular based phylogenetic relationships between the taxonomically assigned k-mers. Any divergence from this model will impact results.
STAT opportunities
Several opportunities led to extensions of STAT utility.
SARS-CoV-2 Detection Tool
At the outset of the current SARS-CoV-2 pandemic we realized that by using just the virus superkingdom STAT k-mer database a simple tool could be made able to quickly determine if an unassembled NGS sample is likely to contain SARS-CoV-2. While the database contains diagnostic k-mers spanning the entire superkingdom, the tool only reports content from within the taxonomic Family node Coronaviridae (https://www.ncbi.nlm.nih.gov/sra/docs/sra-detection-tool, 6).
STAT Human Sequence Removal Tool
Submitters want to eliminate the potential for unintended inclusion of human sequence in clinical NGS patient samples targeting pathogens which could personally identify an anonymized research subject. We employ our existing STAT tools with a human k-mer database, adding a step to remove reads identified as human from the submission (6). After completing a submission, submitters can contact the SRA group (sra@ncbi.nlm.nih.gov) to request all project submissions be subject to human ‘contaminant’ removal. Alternatively, users can ‘filter’ their data before submitting to SRA by using the Docker or GitHub snapshots of the STAT Human Sequence Removal Tool. The docker image has over 100,000 pulls since its introduction.
Species-specific resource
After submitting our STAT manuscript (6) to the BioRxiv preprint server we were contacted by a graduate student seeking help to develop quantitative PCR (qPCR) for investigating the role of a human microbiota bacterial species in health. Together we realized that the effort building a taxonomic database to identify k-mers diagnostic for a given taxon resulted in an unexpected resource. Extracting from the database >2000 bacterium-specific 32 base k-mers, followed by ad hoc testing verified that these were unique sequences only identifying the target bacterium when 100% identical. Mapping some, or all, of these k-mers is likely to yield several species-specific amplifiable regions suitable for qPCR.
CONCLUSION
For the Sequence Read Archive, NCBI is pursuing the widest availability and reliability possible for users seeking archival data, metadata, and the results of STAT taxonomic analyses through cloud platform storage. The ability to easily query taxonomic content intrinsic to SRA records for subset selection should facilitate scientific inquiry at the petabyte scale. We hope that access to all public SRA records free of charge via the AWS Registry of Open Data (https://registry.opendata.aws/ncbi-sra/) will encourage greater use by the scientific community to meet the continued growth of submissions.
DATA AVAILABILITY
The STAT core source code is available as an open-source GitHub repository (https://github.com/ncbi/ngs-tools/tree/tax).
The STAT Human Sequence Removal Tool is available as:
A docker image found at DockerHub (https://hub.docker.com/r/ncbi/sra-human-scrubber).
An open-source code GitHub repository (https://github.com/ncbi/sra-human-scrubber).
The SARS-CoV-2-detection-tool is a docker image available on DockerHub (https://hub.docker.com/r/ncbi/sars-cov-2-detection-tool).
ACKNOWLEDGEMENTS
Vadim Zalunin, Alex Efremov, and Andrey Kochergin for building, maintaining, and improving the STAT pipeline. Dr. Isabel F. Escapa for suggesting the possibility of a species-specific resource. Susan J. Roberts for indispensable editing assistance.
Contributor Information
Kenneth Katz, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Oleg Shutov, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Richard Lapoint, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Michael Kimelman, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
J Rodney Brister, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
Christopher O’Sullivan, National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894, USA.
FUNDING
Intramural Research Program of the National Library of Medicine, National Institutes of Health. Funding for open access charge: Intramural Research Program of the National Library of Medicine, National Institutes of Health.
Conflict of Interest. None declared.
This paper is linked to: doi:10.1093/nar/gkab1112.
REFERENCES
- 1. Kodama Y., Shumway M., Leinonen R.International Nucleotide Sequence Database Collaboration . The Sequence Read Archive: explosive growth of sequencing data. Nucleic Acids Res. 2012; 40:D54–D56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilkinson M.D., Dumontier M., Aalbersberg I.J., Appleton G., Axton M., Baak A., Blomberg N., Boiten J.W., da Silva Santos L.B., Bourne P.E.et al.. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data. 2016; 15:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yu Y.W., Yorukoglu D., Peng J., Berger B.. Quality score compression improves genotyping accuracy. Nat. Biotechnol. 2015; 33:240–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonfield J.K., Mahoney M.V.. Compression of FASTQ and SAM format sequencing data. PLoS One. 2013; 8:e59190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sayers E.W., Bolton E.E., Brister J.R., Canese K., Chan J., Comeau D.C., Connor R., Funk K., Chris C., Kim S.et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021; 10.1093/nar/gkab1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Katz K.S., Shutov O., Lapoint R., Kimelman M., Brister J.R., O’Sullivan C. STAT: A fast, scalable, MinHash-based k-mer tool to assess Sequence Read Archive next generation sequence submissions. Genome Biol. 2021; 22:270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Steinegger M., Salzberg S.L.. Terminating contamination: large-scale search identifies more than 2,000,000 contaminated entries in GenBank. Genome Biol. 2020; 21:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nelson P.N., Carnegie P.R., Martin J., Davari Ejtehadi H., Hooley P., Roden D., Rowland-Jones S., Warren P., Astley J., Murray P.G.. Demystified. Human endogenous retroviruses. Mol. Pathol. 2003; 56:11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Federhen S. The NCBI Taxonomy database. Nucleic. Acids. Res. 2012; 40:D136–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., Mcveigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The STAT core source code is available as an open-source GitHub repository (https://github.com/ncbi/ngs-tools/tree/tax).
The STAT Human Sequence Removal Tool is available as:
A docker image found at DockerHub (https://hub.docker.com/r/ncbi/sra-human-scrubber).
An open-source code GitHub repository (https://github.com/ncbi/sra-human-scrubber).
The SARS-CoV-2-detection-tool is a docker image available on DockerHub (https://hub.docker.com/r/ncbi/sars-cov-2-detection-tool).