Abstract
Literature-described targets of herbal ingredients have been explored to facilitate the mechanistic study of herbs, as well as the new drug discovery. Though several databases provided similar information, the majority of them are limited to literatures before 2010 and need to be updated urgently. HIT 2.0 was here constructed as the latest curated dataset focusing on Herbal Ingredients’ Targets covering PubMed literatures 2000–2020. Currently, HIT 2.0 hosts 10 031 compound-target activity pairs with quality indicators between 2208 targets and 1237 ingredients from more than 1250 reputable herbs. The molecular targets cover those genes/proteins being directly/indirectly activated/inhibited, protein binders, and enzymes substrates or products. Also included are those genes regulated under the treatment of individual ingredient. Crosslinks were made to databases of TTD, DrugBank, KEGG, PDB, UniProt, Pfam, NCBI, TCM-ID and others. More importantly, HIT enables automatic Target-mining and My-target curation from daily released PubMed literatures. Thus, users can retrieve and download the latest abstracts containing potential targets for interested compounds, even for those not yet covered in HIT. Further, users can log into ‘My-target’ system, to curate personal target-profiling on line based on retrieved abstracts. HIT can be accessible at http://hit2.badd-cao.net.
INTRODUCTION
Being a rich source of drug candidates, herbal active ingredients play a critical role in the development of new drugs. From 1981 to 2019, 33.6% of the drugs approved by the FDA were reported to be derived from natural products or their derivatives (1). To better understand the interaction between herbal compounds and molecular targets, the first herbal-ingredient-target database, HIT, was established in 2010 via manual curation of 1301 literature-described targets for herbal compound from 3250 literatures, with convenient links to therapeutic targets database (TTD) and Drugbank etc (2,3). The target information of HIT has been extensively exploited to study the mechanism of natural compounds, as well as to make discoveries from herbal medicine. For instance, based on HIT, Luo Y et al. revealed the therapeutic mechanism of cryptotanshinone in the treatment of liver cancer (4). And Wang et al. identified potential targets for asthma according to the clinical efficacy of TCM formulations (5).
During the past ten years (2011–2021), there was an explosive increase in the studies on the natural ingredients and their targets. A number of databases were then constructed covering herbal compound and target interactions. Notably, a nice exemplary database for natural products, NPASS (6), provides experimentally-determined quantitative activity records for natural products, including nearly 2,000 herbal ingredient-target pairs for about 700 herbal ingredients. Other herbal databases also included important information of herbal ingredient-target pairs (7–11), but most of them downloaded and incorporated early HIT data, appended with predicted targets, such as HERB, TCMID, TCMSP and SymMap (7–10). These databases have greatly enriched the target diversity for herbal ingredients. However, the literature-described targets of herbal ingredients being carefully curated, though valuable, remain limited and need to be updated urgently and regularly.
Yet those fresh evidences of ingredient-target interactions may be published in everyday-released literatures, while the traditional databases have difficulties in keeping track of the latest results. Users often need to take great efforts manually searching the up-to-date literatures not yet covered by current databases and dig targets out to tie up with database items, which is highly time-consuming. Thus, it is necessary to propose a new platform to provide not only the regularly updated targets, but also the real-time checking for potential target from daily-released papers.
Here, we introduced such a platform of HIT 2.0 for the above purpose. In this version, advanced text mining algorithms and rigorous curation were comprehensively employed. In addition to calibrating HIT 1.0 data from literatures between 2000 and 2010, our curation team made a complete refreshment by adding literatures between 2010 and 2020, resulting in almost twice the data abundancy as before, plus new features of target confidence indicators. More importantly, the text-mining system of compound-target suggestion is now open to users, where researchers can retrieve the most-related literatures via ‘Target-mining’ for any natural compounds. At last, HIT 2.0 provides an on-line function of ‘My-target’ for personal curation and downloading.
MATERIALS AND METHODS
Data source
Similar to HIT 1.0, the ingredient information was sourced from the widely used TCM-ID database of the 2020 updated version (12), which covers 2751 herbs and 7375 herb ingredients. Compound aliases were derived from the Chemical Abstracts Service (CAS). The same set of 59 keywords was used as that in HIT 1.0 to describe interactions between compounds and molecular targets (Supplementary Table S1). Some keywords are nouns describing the interaction (Type A), while the others (Type B) are phrases describing the specific effect, such as ‘inhibit the activity of’ proteins.
Text mining
Text mining of NLP was used to identify the targets of herbal ingredients in the literature, with workflow being illustrated as follows (Figure 1):
Figure 1.
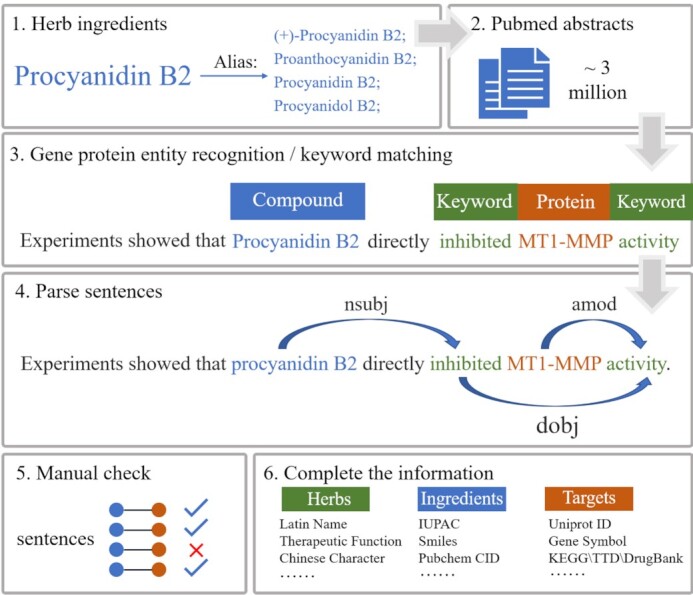
Workflow of HIT 2.0. (1, 2) Retrieve PubMed using different names of herbal ingredients. (3) Mine PubMed abstracts to identify gene/protein entities. (4) Detect whether ‘compound’, ‘gene’ and ‘keyword’ are in a directional dependency tree path. 5&6. Manual check and complete the information.
Step1: Retrieve abstracts in PubMed containing keywords of the name/alias of herb ingredients.
Step2: Annotate gene/protein entities in the abstract using PubTator Central (13), an automated annotation platform for biomedical entities. These genes/proteins may be targets of ingredients, and only abstracts containing gene/protein entities will be retained.
Step3: Parse the full abstract into sentences. Screen out those sentences based on either of below two rules. Rule1: ‘Compound name’ AND ‘any word in type A’ AND ‘Gene’. For instance, the sentence ‘EGCG is a novel Hsp90 inhibitor’. Rule2: ‘Compound name’ AND ‘any word in type B interaction’ AND ‘any word in type B effect’ AND ‘Gene’. For instance, the sentence ‘Procyanidin B2 directly inhibited MT1-MMP activity.’
Step4: Use the Stanford Parser engine (14) to extract sentence syntactic structure and grammatical relations (15). Only those sentences are retained where ‘compound’, ‘gene’ and ‘keyword’ are detected in a directional dependency tree path.
Manual curation
Finally, a curation team of 11 Ph.D. candidates reviewed 17 000 sentences, and each item was double checked by at least two candidates. The entries with consistence from two curators were remained. While those of disagreement were reviewed by a third curator, with the final result being decided by a majority vote.
RESULTS
The construction of HIT 2.0 was based on literature mining and manual curation. In the part of literature mining, PubMed abstracts were firstly retrieved containing keywords of the name/alias of herbal ingredients. Then the abstract was parsed into sentences and only those sentences containing herbal ingredients, genes, and keywords were kept for further curation. For convenience, an online platform was set up where each curator has their own account. Any time after log-in, curators can review the PubMed tasks completed, and to-be-completed. Now the whole platform has been opened as ‘Target-mining’ and ‘My-target’ system, enabling users to identify those most related literatures and keep close tracking of the latest targets for their interested compounds.
Data updating
In this version, more advanced text-mining algorithms of Natural language processing (NLP) and rigorous manual curation were applied using the same set of keywords as that of HIT 1.0 (16). NLP has been widely applied to biomedical text mining. Dependent syntactic analysis enables sentence structure parsing to highlight the relationships between medical entities (15). The NLP algorithm is used in HIT 2.0 to determine whether compounds may interact with genes/proteins. Initially, 7100 abstracts were obtained from PubMed after text-mining system. After the on-line curation, a total of 10 031 compound-target pairs were produced, involving 2208 molecular targets and 1,237 compounds from 1250 herbs. Interestingly, 56 miRNA genes have been included into our targeting list, mainly with modes of up/down regulated genes.
The types of compound-target interactions cover 10 categories: indirectly inhibit/activate, up/down-regulated gene, directly inhibit/activate, binders, enzyme substrates, enzyme products and other. Quality indicators of compound-target interactions have been developed covering the nature of the interaction, the number of literatures supporting the same pair, and the citing reports of each literature. Based on above, users can choose those preferred for further analysis. Each compound-target pair can be viewed with a key description parsed from the sourcing literature. Compounds are linked to PubChem (17) and ChEMBL (18) databases. As for targets, Crosslinks have been made to databases of TTD (2), DrugBank (3), KEGG (19), PDB (20), UniProt (21), Pfam (22), NCBI, TCM-ID (12) and others for more detailed information.
Compared to similar herb databases containing literature-described targets, HIT 2.0 has doubled the data abundancy of HIT 1.0, forming a nice complement to the previous databases. Currently, it is also the largest database in terms of curated ingredient-target pairs for herbs, as Table 1 shows.
Table 1.
Overview of the literature-described targets from peering databases
| Published year | Literature-described targets | Herbal ingredients | Herbal ingredient-target activity pairs | Sourcing literatures | |
|---|---|---|---|---|---|
| HIT 2.0 | 2208 | 1237 | 10 031 | 7100 PubMed abstracts | |
| HIT | 2011 | 1301 | 586 | 5208 | 3250 PubMed abstracts |
| HERB | 2020 | 1241 | 370 | 4815 | 1966 PubMed abstracts |
| TCMID | 2013 | 680 | / | / | 4500 Chinese Literatures |
| NPASS | 2017 | 464 | 719 | 1936 | 1288 PubMed abstracts |
| TCMSP | 2014 | 3311 predicted by SysDT | 2 9 384 | 8 4 260 predicted by SysDT | / |
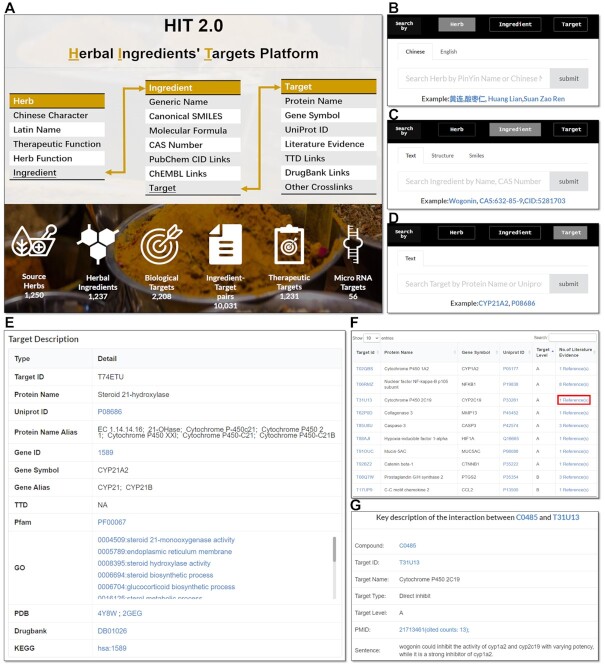
HIT 2.0 allows keyword search and compound similarity search. The searching interface and results pages are illustrated in Figure 2. Keyword search is available for herb information [Chinese pinyin, Chinese characters, Latin name and English name], ingredients information [different names, CAS number and CID number] and target information [gene/protein name, gene symbol, UniProt ID]. Auto-completion and fuzzy search are supported for keyword search. Besides that, similarity search can also be made via compound structures, with a Tanimoto coefficient above 70% as a cut-off. Both the SMILES formula and the artificially drawn structure by build-in software Ketcher (https://lifescience.opensource.epam.com/ketcher/) can be used as input.
Figure 2.
Searching and resulting pages in HIT 2.0. (A) Database structure and data statistics. (B) Herbs can be searched via keywords such as Chinese Pinyin, Chinese characters and Latin names. (C) Herbal ingredients can be searched via structure similarity or keywords of name, CID and CAS number. (D) Targets can be searched via keywords of gene/protein name, gene symbol and Uniprot ID. (E) Detailed information of the targets. (F) Additional targets of the compound. (G) ‘ Literature evidence ’ provides the key descriptions parsed from sourcing literatures.
Target-mining and My-target curation
As PubMed literatures are released every day, there is a constant need for researchers to check the latest evidences after 2020 for their interested compound. So far, no software becomes available to mine the literatures containing potential target-ingredient interactions. For the convenience of curation, two options have been provided for users.
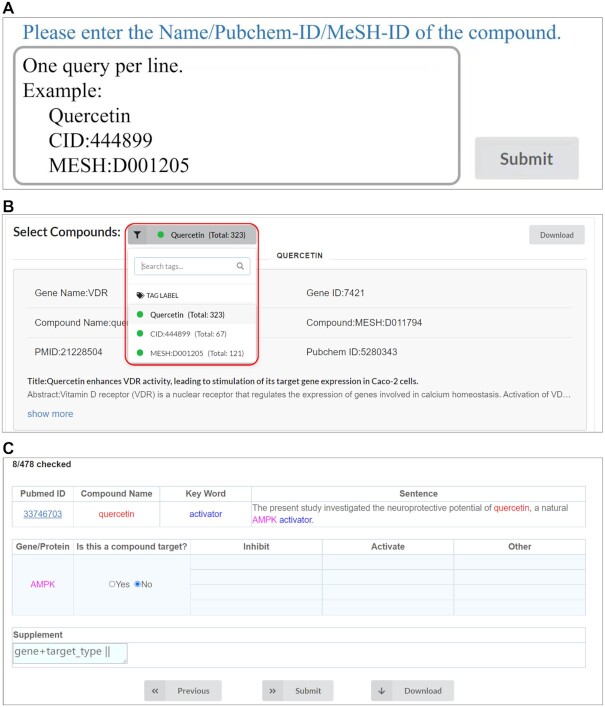
Target-mining function was built-in enabling users to retrieve related PubMed abstracts published from 2010 till the daily updates for any compounds (Figure 3A and B). It was realized that the abstracts identified by text-mining may contain false positives. Thus HIT 2.0 provides on-line My-target curation function to further check the detailed sentences in sourcing abstracts (Figure 3C). The sentences containing interesting entities have been highlighted in different colors, so that key items can be easily spotted together with the interaction types. A link to PubMed with full abstract is also provided.
Figure 3.
Target-mining and My-target curation system. (A) The interface of Target-mining function. Compound name, MeSH ID and Pubchem ID can be submitted to retrieve potential targets. (B) PubMed abstract retrieved by Target-mining. (C) The interface of My-target curation system.
In brief, ‘Target-mining’ was designed to efficiently and precisely retrieve those most-related abstracts based on our empirically derived rules combined with advanced text-mining algorithm. Via this, users can retrieve and download the latest literatures for local checking. Alternatively, users can choose ‘My-target curation’ to make curation on-line. Each user has their own account. Anytime after log-in, users can review the tasks completed and continue to finish the full task.
DISCUSSION
Complement to other drug and drug-target databases, herbal databases containing literature-described targets have served as the primary source for mechanistic study of natural products by providing rich sets of information between medicinal herbs, active compounds and molecular targets under different experimental conditions. With a steady literature accumulation describing new evidences in the past decade, however, the previous versions need to make timely updating to meet up with the pressing needs. In this paper, HIT 2.0 was thus constructed to maintain a regular updating by adding another ten years, marking as the latest and the largest one regarding curated ingredient-target pairs for herbs. Meanwhile, we set up Target-mining and My-target curation system based on technology of natural processing language, allowing researchers to keep tracking the latest evidences and curate personal targets of interested compounds at convenient time. The launch of HIT 2.0 will be an important addition to bridge herbs ingredients and FDA approved drugs via molecular targets and may facilitate the discovery of new druggable molecules, as well as to identify potential therapeutic targets.
One key technology applied in HIT 2.0 was the auto text-mining tools. Currently, there are several annotating tools to recognize the biomedical entities, such as PubTator Central (13), HunFlair (23), and ScispaCy (24). Among them, PubTator Central was developed by NCBI to annotate PubMed abstracts automatically and allows users to download annotations in bulk via PubMed ID lists. In addition, the annotated genes have direct links to the Gene database, which allows HIT 2.0 to access more details of the targets without being installed. Considering the overall convenience, simplicity and complexity, PubTator central was chosen for HIT 2.0.
Meanwhile, the last decade has witnessed an extensive application of molecular targets into functional explanation for herbal compounds. Though the reliability of literature-evidenced targets may be more accurate than computationally-predicted targets, it is aware that the results collected into HIT are mostly from in vitro rather than in vivo experiments. In fact, these natural compounds are likely metabolized into different forms which may change the targeting profiles (25). Furthermore, the biological activity of an ingredient is often related to not only the group of molecular targets, but also the network interactions among targets. In this sense, HIT 2.0 may serve as a valuable start in deriving collective functions of herbal components, particularly to herbs and herbal formulas. Towards this direction, HIT will continue to enrich the data abundancy and subsequent analysis to maintain high-quality resources for domain research.
Supplementary Material
Contributor Information
Deyu Yan, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Genhui Zheng, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Caicui Wang, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Zikun Chen, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Tiantian Mao, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Jian Gao, International Human Phenome Institutes (Shanghai), Shanghai, China; Department of Thoracic Surgery, Fudan University Shanghai Cancer Center, Shanghai, China.
Yu Yan, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Xiangyi Chen, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Xuejie Ji, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Jinyu Yu, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Saifeng Mo, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Haonan Wen, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Wenhao Han, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Mengdi Zhou, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Yuan Wang, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Jun Wang, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Kailin Tang, Dept. of Gastroenterology, Shanghai Tenth People's Hospital, School of Life Sciences and Technology, Tongji University, Shanghai 200092, China.
Zhiwei Cao, School of Life Sciences, Fudan University, Shanghai 200092, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Key R&D Program of China [2017YFC1700200, 2019YFA0905900]; National Natural Science Foundation of China [32070657, 81830080]. Funding for open access charge: National Key R&D Program of China [2017YFC1700200].
Conflict of interest statement. None declared.
REFERENCES
- 1. Newman D.J., Cragg G.M.. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020; 83:770–803. [DOI] [PubMed] [Google Scholar]
- 2. Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y.et al.. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020; 48:D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo Y., Song L., Wang X., Huang Y., Liu Y., Wang Q., Hong M., Yuan Z.. Uncovering the mechanisms of cryptotanshinone as a therapeutic agent against hepatocellular carcinoma. Front. Pharmacol. 2020; 11:1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang Y., Chen Y.J., Xiang C., Jiang G.W., Xu Y.D., Yin L.M., Zhou D.D., Liu Y.Y., Yang Y.Q.. Discovery of potential asthma targets based on the clinical efficacy of Traditional Chinese Medicine formulas. J. Ethnopharmacol. 2020; 252:112635. [DOI] [PubMed] [Google Scholar]
- 6. Zeng X., Zhang P., He W., Qin C., Chen S., Tao L., Wang Y., Tan Y., Gao D., Wang B.et al.. NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. 2018; 46:D1217–D1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang S., Dong L., Liu L., Guo J., Zhao L., Zhang J., Bu D., Liu X., Huo P., Cao W.et al.. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021; 49:D1197–D1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., Wen C.. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018; 46:D1117–D1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y.et al.. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminform. 2014; 6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu Y., Zhang F., Yang K., Fang S., Bu D., Li H., Sun L., Hu H., Gao K., Wang W.et al.. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019; 47:D1110–D1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang R.Z., Yu S.J., Bai H., Ning K.. TCM-Mesh: The database and analytical system for network pharmacology analysis for TCM preparations. Sci. Rep. 2017; 7:2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen X., Zhou H., Liu Y.B., Wang J.F., Li H., Ung C.Y., Han L.Y., Cao Z.W., Chen Y.Z.. Database of traditional Chinese medicine and its application to studies of mechanism and to prescription validation. Br. J. Pharmacol. 2006; 149:1092–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wei C.H., Allot A., Leaman R., Lu Z.. PubTator central: automated concept annotation for biomedical full text articles. Nucleic Acids Res. 2019; 47:W587–W593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marneffe M.-C., MacCartney B., Manning C.. Generating Typed Dependency Parses from Phrase Structure Parses. 2006; http://www.lrec-conf.org/proceedings/lrec2006/pdf/440_pdf.pdf. [Google Scholar]
- 15. Kveler K., Starosvetsky E., Ziv-Kenet A., Kalugny Y., Gorelik Y., Shalev-Malul G., Aizenbud-Reshef N., Dubovik T., Briller M., Campbell J.et al.. Immune-centric network of cytokines and cells in disease context identified by computational mining of PubMed. Nat. Biotechnol. 2018; 36:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ye H., Ye L., Kang H., Zhang D., Tao L., Tang K., Liu X., Zhu R., Liu Q., Chen Y.Z.et al.. HIT: linking herbal active ingredients to targets. Nucleic Acids Res. 2011; 39:D1055–D1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mendez D., Gaulton A., Bento A.P., Chambers J., De Veij M., Felix E., Magarinos M.P., Mosquera J.F., Mutowo P., Nowotka M.et al.. ChEMBL: towards direct deposition of bioassay data. Nucleic Acids Res. 2019; 47:D930–D940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M.. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021; 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M.et al.. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021; 49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. UniProt C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Weber L., Sanger M., Munchmeyer J., Habibi M., Leser U., Akbik A.. HunFlair: an easy-to-use tool for state-of-the-art biomedical named entity recognition. Bioinformatics. 2021; 37:2792–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neumann M., King D., Beltagy I., Ammar W.. Proceedings of the 18th BioNLP Workshop and Shared Task. 2019; https://aclanthology.org/W19-50. [Google Scholar]
- 25. Kang H., Tang K., Liu Q., Sun Y., Huang Q., Zhu R., Gao J., Zhang D., Huang C., Cao Z.. HIM-herbal ingredients in-vivo metabolism database. J. Cheminform. 2013; 5:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.