Abstract
Ruminant Genome Database (RGD; http://animal.nwsuaf.edu.cn/RGD) provides visualization and analysis tools for ruminant comparative genomics and functional annotations. As more high-quality ruminant genome assemblies have become available, we have redesigned the user interface, integrated and expanded multi-omics data, and developed novel features to improve the database. The new version, RGD v2.0, houses 78 ruminant genomes; 110-species synteny alignments for major livestock (including cattle, sheep, goat) and wild ungulates; 21 012 orthologous gene clusters with Gene Ontology and pathway annotation; ∼8 600 000 conserved elements; and ∼1 000 000 cis-regulatory elements by utilizing 1053 epigenomic data sets. The transcriptome data in RGD v2.0 has nearly doubled, currently with 1936 RNA-seq data sets, and 155 174 phenotypic data sets have been newly added. New and updated features include: (i) The UCSC Genome Browser, BLAT, BLAST and Table Browser tools were updated for six available ruminant livestock species. (ii) The LiftOver tool was newly introduced into our browser to allow coordinate conversion between different ruminant assemblies. And (iii) tissue specificity index, tau, was calculated to facilitate batch screening of specifically expressed genes. The enhanced genome annotations and improved functionality in RGD v2.0 will be useful for study of genome evolution, environmental adaption, livestock breeding and biomedicine.
INTRODUCTION
Ruminants are one of the most successful and ecologically important herbivorous animal groups on Earth, exhibiting diverse morphologies (e.g. different headgear, body size, and tooth) (1) and great adaptations to various ecological environments (e.g. polar region, Tibet plateau, desert steppe and tropical rainforest) (2). In addition, the ruminants include several important livestock species: cattle, zebu, yak, buffalo, sheep, goats and reindeer, which have prominent contributions to the prosperity of agriculture and civilization (3). Therefore, ruminants are excellent models for the study of environmental adaptation and agronomic traits.
Ruminantia has nearly 200 extant species in six different families (Tragulidae, Antilocapridae, Giraffidae, Cervidae, Moschidae, and Bovidae) and each family has distinct characteristics (4). In 2019, the Ruminant Genome Project (RGP) that we launched released 47 ruminant genomes (5). This batch of data was also used to build the RGD v1.0 database in the same time (5). Since then, improvements in sequencing and computational technologies have enabled researchers to generate a growing wealth of genomics, epigenetics, transcriptomics and breeding data for ruminant species (6–9). Nowadays, nearly 80 ruminant genomes have become available, and 12 of them assembled based on third-generation sequencing platform (10). In addition, the Functional Annotation of Animal Genomes (FAANG) consortium (6) and more public works have generated genome-wide data on RNA-seq (7,11–13), chromatin modification and chromatin accessibility (14–20). Application of comparative genomics methods to the new datasets, and full utilization of thousands of human regulatory datasets will provide a powerful basis to form rich annotations to all aligned ruminant genomes, and identify clade or species-specific mutations.
In the update, we performed genome alignments of 110 high-quality ruminant and outgroup genomes, including 29 genomes based on third-generation sequencing platform. Some key outgroup node species, such as platypus, tuatara, chicken, tropical clawed frog, coelacanth, zebrafish, and sea lamprey, were newly added to perform the alignments. RGD v2.0 also has greatly improved expression data and functional element annotation. We expanded RNA-seq data from 1151 to 1936 samples, which is currently the largest ruminant expression atlas spanning eight ruminant species. Ruminant epigenomic data were also expanded from 32 to 220 data sets. In addition, 833 human epigenomic data sets were mapped to the ruminant genomes. These datasets providing various information for functional element annotation. We also added phenotypic data including QTL and GWAS regions from cattle, sheep and goats. The RGD will be continuously updated, and provides useful resources for ruminant research communities. Here, we describe the data and tools currently available in RGD v2.0.
NEW DATA AND VISUALIZATIONS
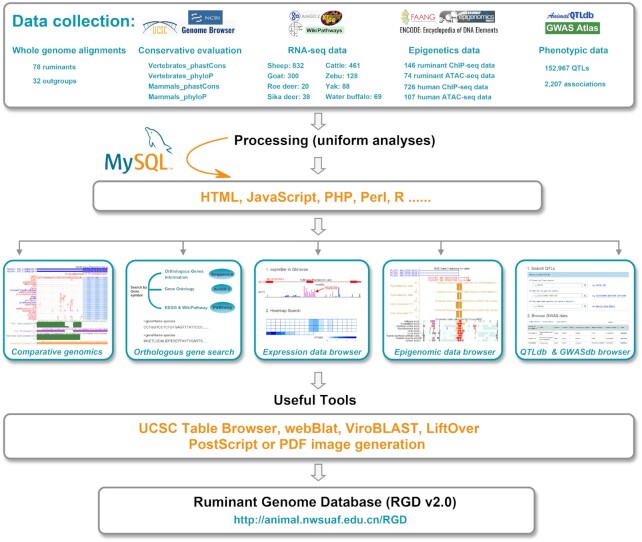
We redesigned the RGD interface and divided the home page into 11 modules, including (i) ‘the UCSC Genome Browser (hereafter referred to as ‘Genome Browser’)’ (21), (ii) ‘Expression’, (iii) ‘Epigenome’, (iv) ‘Orthologous Gene’, (v) ‘Comparative Genomics’, (vi) ‘QTLdb’, (vii) ‘GWASdb’, (viii) ‘BLAT Tool’ (22), (ix) ‘BLAST Tool’ (23), (x) ‘Table Download’ and (xi) ‘LiftOver’ (21). Each module provides detailed pages to access available data and tools. A wealth of new data is available in RGD v2.0. The main updates include three new 110-way alignments, identification and annotation of orthologous genes, expansion of regulatory and transcriptome data, and newly added phenotypic data, etc. We upgraded the BLAST tool and newly introduced the ‘Table Browser’ and ‘LiftOver’ tools (21). RGD uses CodeIgniter framework and MySQL as the database managements system to achieve interactive data mining and visualization. The database construction pipeline is shown in Figure 1. Below, we describe the currently available data and tools, with a focus on new data and features.
Figure 1.
The processing pipeline used to construct RGD v2.0. RGD v2.0 integrated comparative genomics, transcriptomics, epigenomics and phenotypic data. Users can search a gene symbol or a genomic region to get results in an interactive table and graph.
New assemblies for genome browser
The current Genome Browser hosts seven genome assemblies, including four newly added ones, Oar_rambouillet_v1.0 (sheep), BosGru3.0 (yak), UOA_Brahman_1 (zebu), and UOA_WB_1 (water buffalo), covering major ruminant livestock species. The standard basic information, including assembly chromosome and scaffold names, gap locations, NCBI gene annotations and GC percent in 5-base windows were also imported into the database.
110-species alignments and conservation scores
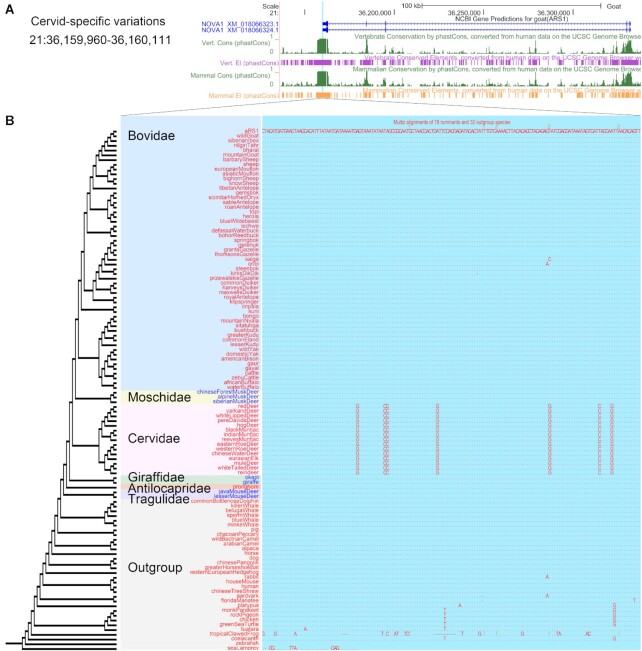
Genome-wide synteny is a fundamental guide to locate genes, identify conserved elements, reveal similarities and differences among species, and trace the course of evolution. In RGD v1.0, we released 67-species alignments (including 55 ruminants and 12 outgroup species) against goat genome assembly (ARS1), which was the most continuous assembly in Ruminantia at that time. With the rapid development of third-generation sequencing technology, genome quality has been greatly improved. In this work, we collected the best assembled genome versions of 78 ruminants and 32 outgroup species (Supplementary Table S1), which are usually used as reference genomes in NCBI, to perform multiple alignments using Last and Multiz softwares. In RGD v2.0, we have created three new multiple alignment tracks that feature these 110 species (Supplementary Table S1): one for cattle (ARS-UCD1.2_Btau5.0.1Y), one for sheep (Oar_rambouillet_v1.0_addY), and one for goat (ARS1) (Figure 2A and B). Users can find sequence differences among species by zooming in the alignments to the base level (Figure 2B and Supplementary Figure S1A). Beyond this, tracks can be configured to activate coding sequence codon display. The coding sequences of 110-way alignments can be translated into amino acids (Supplementary Figure S1A). We used the 110-species synteny data to identify the orthologous genes of ruminants and outgroup species, and provided up to 21 012 orthologous genes for each species. Each gene was annotated with the corresponding Gene Ontology (GO) ID (24) and pathway. Users can enter a gene symbol to get the results of three parts: coding sequence and protein sequence for each species, detailed classification (Molecular Function, Cellular Component and Biological Process) of GO, and KEGG (25) & WikiPathways (26). Users can click ‘GO ID’ to link to AmiGO 2 database (24), or click the ‘Pathway show’ to get a detailed pathway figure.
Figure 2.
An example of Cervidae-specific diverged sites in the 3’UTR of the NOVA1 gene. (A) NOVA1 gene structure and conserved elements in 100 vertebrates and 30 mammals. (B) Cervidae uniquely divergent sites present in NOVA1. We display the new tracks of 110-species alignments and conserved elements. And 29 genomes were assembled based on third-generation sequencing platform.
Both phastCons (27) and phyloP (28) conservation scores in mammalian and vertebrate evolutionary scales also have been newly added for the three major livestock (cattle, sheep, and goat) (Supplementary Figure S1B). We downloaded four conservation scores files (phastCons100way, phastCons30way, phyloP100way and phyloP30way) and two conserved elements files (phastConsElements100way and phastConsElements30way) based on human (hg38) genome from the UCSC Genome Browser (21), and converted the human genome coordinates to the cattle, sheep, and goat using LiftOver with -minMatch = 0.2. In total, 8 576 390 and 2 536 600 conserved elements across vertebrates and mammals were identified, respectively (Supplementary Table S2).
Epigenomic data browser
More than 90% of the common variants associated with complex traits are located in intergenic and intronic regions (29,30). Prediction of potential regulatory elements will help to identify the function of mutations in evolutionary and population genetics studies (31). With continuous effort on curating the published data, RGD v2.0 now contains 220 epigenomic data sets for ruminants (188 new ones), including 196 for cattle, 23 for sheep and one for goat (Supplementary Table S3). The downloaded reads were first aligned to their respective reference genomes using BWA (version 0.7.17-r1188) (32). Then samtools (33) was used to remove low-quality and multiple-mapping reads with the option ‘‘-q 20’’. Peaks were called by MACS v.2.1.1 (34), retaining only data with a P-value < 1e−5. Due to relatively more available epigenomic data for cattle tissues, we applied ChromHMM software (v1.22) (35) to discover chromatin states in this species. First, gene annotation file was used to build libraries of subsequent chromatin state annotations by using subroutine ‘ConvertGenetable’. Then reads counts of each epigenome were calculated in non-overlapping 200 bp bins to mark signal enrichment regions by using subroutine ‘BinarizeBed’. Finally, we trained several models with varying numbers of states and chose 15-state model since it captured all key chromatin interactions. We display the ruminant histone ChIP-seq and ATAC-seq data in Genome Browser using −log10 (P-value) as the height of the y-axis (Supplementary Figure S2A). And these data are prioritized to identify the ruminant regulatory regions (promoters, enhancers, and silencers). A total of 965 844 (genome proportion: 13.73%) cis-regulatory elements from cattle and 166 848 (genome proportion: 2.34%) cis-regulatory elements from sheep were identified (Supplementary Table S4). We did not compute statistics for goat since there was only one sample. Compared with other published species, such as mouse (genome proportion: 12.6%) (31), and pig (genome proportion: 17.38%) (36), the regulatory data of sheep and goats are far from saturated.
As a supplement, 833 human epigenomes (Supplementary Table S5) containing four core histone marks (H3K4me3: 192, H3K27ac: 141, H3K4me1: 196 and H3K4me3: 197) from the Roadmap Epigenomics Project (37) and one open chromatin marker (ATAC-seq: 107) from the Encyclopedia of DNA Elements (ENCODE) project (38) are mapped on the cattle, sheep and goat reference genomes by LiftOver tool (21) with the optimal minMatch threshold 0.2 (31). We retained only regions with exact reciprocal mapping back to the human genome according the published method (31). We found that ∼80% of human regulatory data can be mapped to the genomes of cattle, sheep, and goats. The promoter regions have the higher recall rates (82−87%) than any other signal regions (68−85%), indicating that promoters are the most conserved of the signal regions (Supplementary Table S6). The mapped genome coverages for cattle, sheep, and goat were 28%, 21% and 21%, respectively, indicating that the ruminant regulatory elements can be well recovered by human data (Supplementary Table S6). In RGD v1.0, we used the read coverage to display the mapped human regulatory data, while in RGD v2.0 we adjusted the display mode of human regulatory data to the peak files, and also used -log10 (P-value) as y-axis (Supplementary Figure S2B), making these data more standardized. The human core histone marks and one open chromatin marker combining all collected human tissues and cell lines are shown in five different colors signal tracks (Supplementary Figure S2B).
Expression data browser
Genome-wide expression analysis of RNA-seq data is a prerequisite for gene function researches. In RGD v1.0, we have developed a geneHeatmap tool to display the gene expression data. In the new version, we added two new common species, zebu and water buffalo. The RNA-seq samples were expanded from 1151 to 1936 (sheep: 832, cattle: 461, goat: 300, zebu: 128, yak: 88, water buffalo: 69, sika deer: 38 and roe deer: 20; Supplementary Table S7). We downloaded the raw reads from the NCBI Sequence Read Archive (SRA) and removed adaptor sequences and low-quality reads using Trimmomatic (version 0.36) (39). The high-quality reads were mapped to their respective reference genomes with STAR (version 2.5.1) (40), then unaligned reads were extracted for further mapping by HISAT2 (version 2.0.3-beta) to improve reads utilization (41). The Picard tool (version 2.1.1) was used to merge the two bam files. We computed Fragments Per Kilobase per Million mapped reads (FPKM) values for each gene by StringTie (version1.3.4) (42). The tissue specificity index, tau (43), was newly added using in-house Perl scripts according to the suggestions by the user community. We provide download of expression matrix files for eight ruminant species, which can facilitate screening genes of interest according to the tau value and tissue expression level.
In RGD v2.0, the transcriptome data covered more abundant tissue/cell types and more developmental stages. We classified these samples first by repetition, followed by tissue/cell type, and finally by organ system (Supplementary Table S7). Therefore, users can easily find gene expression patterns and tissues with high expression abundance (Supplementary Figure S3). We also added some groups that researchers often compare to analyze their functions. For instance, one of the most important economic traits of goat is cashmere production. We provide gene expressions of skin samples of Dazu black goats distributed in southern China, as well as Mongolian cashmere goats distributed in northern China, which can help study genes participating in cashmere production. Other groups, such as samples at high and low altitudes; mammary gland at early lactation, late lactation, and dry period; and endometrium with high fertility and low fertility, etc., can also provide valuable information for animal production traits and adaptability.
The color depth of each heatmap cell represents the average FPKM value of each tissue. Users can view the detailed numbers as the pointer hovers over the heatmap cell. The search results can be downloaded in CSV format. The expression data is also organized into the tracks of the Genome Browser, named ‘expreBar’ (Supplementary Figure S4A). Users can click a specific gene to link to a more detailed visualization page that generates a box-whisker plot showing the range of expression levels across samples (Supplementary Figure S4B).
QTLdb and GWASdb browser
Information about reported phenotypic data is newly available in the RGD v2.0. We downloaded the 165 353 QTLs from AnimalQTLdb (44), and filtered entries that were not anchored to genome chromosome. Finally, 150 617 cattle QTLs associated with 630 agronomic traits and 2350 sheep QTLs associated with 121 agronomic traits were retained. We provide three ways to retrieve QTLdb: search by gene symbol, find QTLs by genome location, and find associated genes by a trait name or a keyword. We also integrated 2207 associations from GWAS Atlas (8), 1630 for cattle, 539 for sheep and 38 for goat. We converted genomic coordinates of SNPs to the newer assemblies using LiftOver by default parameters. Users can browse the GWAS data by any keywords. These data can help users associate phenotypes and genotypes to further confirm user's interested genes.
Gene information search
In RGD v1.0, we have provided basic gene information search for four main livestock (goat, sheep, cattle, and yak), including genomic location, CDS length, transcript profile, relevant GO ID and GO terms, and KEGG pathways. In RGD v2.0, we upgraded this module to full database retrieval on the home page, and optimized the user's search method, which can identify redundant spaces in the input text. Users can enter a gene symbol, a NCBI gene ID, or an Ensembl gene ID, to view all available information. The search results include eight main contents: (i) Gene Summary, (ii) Gene Structure and Regulation, (iii) Gene Expression, (iv) Quantitative Trait Locus, (v) Orthologous Gene, (vi) Gene Ontology, (vii) KEGG and WikiPathway, and (viii) Genome Browser. The results are presented in a classical tabular format and can be linked to Genome Browser and external databases.
SOFTWARE AND TOOL IMPROVEMENTS
BLAT and BLAST
RGD v1.0 created a webBLAT server (22) and wwwBLAST based on the goat (ARS1), sheep (Oar_v4.0), and cattle (ARS-UCD1.2_Btau5.0.1Y) genome. In RGD v2.0, we added four new genomes (sheep: Oar_rambouillet_v1.0_addY, zebu: UOA_Brahman_1, yak: BosGru3.0 and water buffalo: UOA_WB_1), extending the genome assemblies to all available ruminant livestock species. We also introduced ViroBLAST (23) to replace the outmoded wwwBLAST. The current alignment tools can help users retrieve homologous DNA, RNA, and protein sequences faster. And the webBLAT tool can return a list of links to all genome locations by aligning with the input sequence, which can then be displayed in Genome Browser to facilitate the access to more sequence features.
Table Download
RGD v2.0 has a UCSC Table Browser (21) tool for retrieving raw data in various format and performing intersections and unions between data in different tracks. Users can select clade, genome, assembly, group, track, table, regions of interest, output format and define output file name to extract subsets of the Genome Browser quickly and easily. We also wrote an additional module (Table Download) to extract other data in the database, including information of orthologous genes, gene expressions, QTLs and GWAS. Users can realize batch download by entering gene symbol or NCBI gene ID list.
LiftOver tool
The LiftOver tool (21) is available in RGD v2.0, which can realize coordinate conversion between different ruminant assemblies. We generated 33 chain files to support the following genome conversion: four genome versions of cattle (ARS_UCD1.2, Btau_5.0.1, UMD_3.1.1 and UMD_3.1), three genome versions of sheep (Oar_rambouillet_v1.0, Oar_v4.0 and Oar_v3.1), three genome versions of goat (ARS1, CHIR_2.0 and CHIR_1.0), the water buffalo genome (UOA_WB_1), as well as the human genome (Hg38.p13). Users can perform online coordinate conversions between different ruminant genome assemblies, and facilitate migration of human data to ruminant species.
Access to data and pipeline
All sample information including the SRA accession numbers can be found in the ‘Sample Info’ of each corresponding section of the RGD interface. And all data can be accessed in the ‘Downloads’ section. The processing pipeline and the full details of materials and methods are shown in the ‘About & Manual’ section. Our database can be accessed online at http://animal.nwsuaf.edu.cn/code/index.php/RGD.
DISCUSSION AND FUTURE PLANS
To date, the functional element annotations are still rare in ruminants, especially in wild species. Using comparative genomic approaches to build a bridge connecting these data, can effectively illustrate the role of genes and mutations in species evolution, population genetics and animal production. The previous researches have proved that RGD v1.0 can effectively help users to locate the causal and functional mutations, such as cervid-specific variations controlling antler regeneration (45), reindeer-specific variations involved in vitamin D metabolism (46), and giraffe-specific variations exhibiting exceptional hypertension resistance (47). With the reducing cost and growing power of sequencing, a wealth of multi-omics data including genomics, transcriptomics, and epigenetics have been accumulated. Here, we have fully upgraded the database, used a unified pipeline to process and analyze the data, migrated a large number of human regulatory data, and ultimately, provided rich genomic annotations for ruminant species.
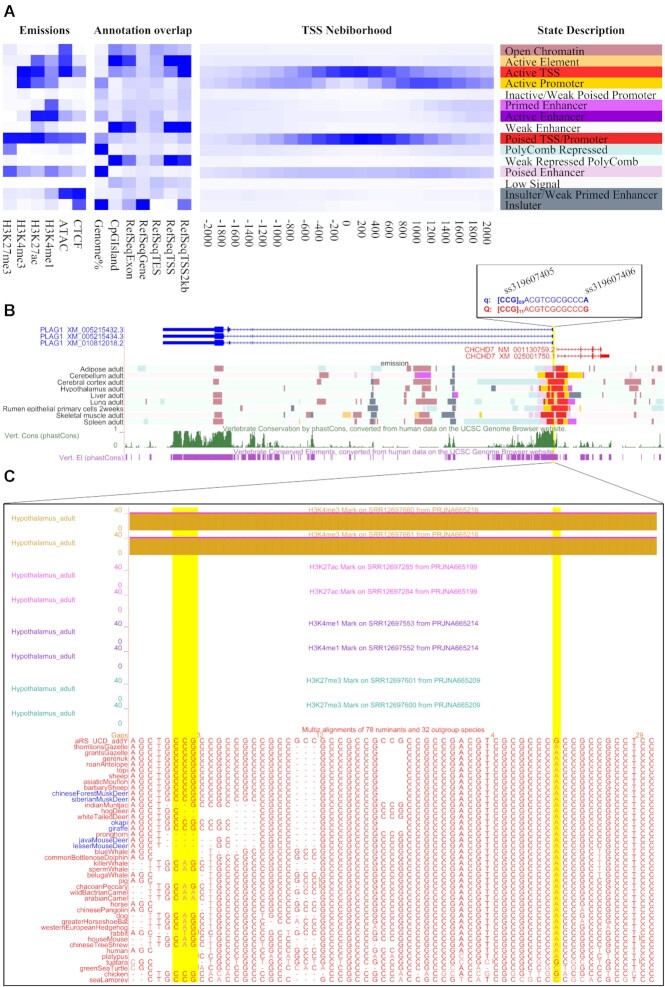
For example, we first observed bovine PLAG1 gene, which has been proved by many studies to influence the stature (weight and height) of cattle (48–50). Two candidate variations, ss319607405 (14:23375648–23375650), and ss319607406 (14:23375692), located in the PLAG1-CHCHD7 intergenic region, have been reported the most likely cis-acting causal variants by influencing bidirectional promoter strength (48). By searching the bovine genome browser hosted in RGD v2.0, we found that both these two variations were located in the active transcriptional start site (TSS) and promoter regions (Figure 3A and B), and also affected a highly conserved element with high phastCons scores (Figure 3B). We further examined the regulatory signals of various tissues and found that H3K4me3 maker exhibited strong signals in adipose, alveolar macrophages, bESC, blastocyst, cerebellum, cerebral cortex, hypothalamus, liver, lung, rumen epithelial primary cells, skeletal muscle, spleen, testis, and trophectoderm (Figure 3C, and Supplementary Figure S5), consistent with the published verification results of luciferase reporter assays (48). Multiple sequence alignments also showed a tandem repeat of CCG copies at ss319607405 locus (Figure 3C), which may affect transcriptional regulation by altering binding of nuclear factors. There are also another two examples that show evidence of regulation through RGD v2.0, (i) a 504-bp deletion at ∼14 kb downstream of the FGF5 gene, that previously has been reported to be involved in cashmere production, affects enhancer activity (51,52), and (ii) mutations in FSHR gene that are associated with litter size in Hu sheep, regulate core promoter activity (53) (Supplementary Figure S6 and S7). Therefore, RGD v2.0 is also of great value in ruminant livestock.
Figure 3.
Application of the regulatory data and multiple genome alignments to identify the key variations of PLAG1 gene. (A) The heat map on the left shows the emission probabilities of 15-chromatin state identified by ChromHMM based on the data of six epigenomic markers (H3K4me3, H3K27ac, H3K4me1, H3K27me3, CTCF and ATAC-seq) from nine bovine tissues. The heat maps in the middle and right show the overlap enrichment for various genomic annotations. (B) Two candidate variations, ss319607405 (14:23375648–23375650), and ss319607406 (14:23375692), were located in the active transcriptional start site (TSS) and promoter regions, and overlapped with a highly conserved element. (C) Detailed regulatory signals and multiple sequence alignments around these two variations.
In summary, RGD v2.0 currently hosts 3664 tracks, including 1442 new ones, 2183 updated tracks and 39 tracks maintained the original state (Supplementary Table S8). RGD v2.0 covers almost all available high-quality multi-omics data of ruminant species, and contains ∼2.98 TB of data. We will continuously update the RGD v2.0, and plan to collect all variation data of ruminant species, including SNPs, indels and Structure Variations (SVs) data in future. This comprehensive data resource will enable researchers to be easily applied to the biological, evolutionary, ecological and husbandry studies.
Supplementary Material
ACKNOWLEDGEMENTS
We thank High Performance Computing (HPC) Center of Northwest A&F University (NWAFU) for providing computing resources.
Contributor Information
Weiwei Fu, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China; Center for Ruminant Genetics and Evolution, Northwest A&F University, Yangling 712100, China.
Rui Wang, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China; Center for Ruminant Genetics and Evolution, Northwest A&F University, Yangling 712100, China.
Hojjat Asadollahpour Nanaei, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China; Center for Ruminant Genetics and Evolution, Northwest A&F University, Yangling 712100, China.
Jinxin Wang, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China; Center for Ruminant Genetics and Evolution, Northwest A&F University, Yangling 712100, China.
Dexiang Hu, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China; Center for Ruminant Genetics and Evolution, Northwest A&F University, Yangling 712100, China.
Yu Jiang, Key Laboratory of Animal Genetics, Breeding and Reproduction of Shaanxi Province, College of Animal Science and Technology, Northwest A&F University, Yangling 712100, China; Center for Ruminant Genetics and Evolution, Northwest A&F University, Yangling 712100, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31822052]; National Thousand Youth Talents Plan [Z111021502]; Shaanxi Province Provincial Agricultural special funds [K3370220015]. Funding for open access charge: National Natural Science Foundation of China [31822052].
Conflict of interest statement. None declared.
REFERENCES
- 1. Hofmann R.R. Evolutionary steps of ecophysiological adaptation and diversification of ruminants: a comparative view of their digestive system. Oecologia. 1989; 78:443–457. [DOI] [PubMed] [Google Scholar]
- 2. Hackmann T.J., Spain J.N.. Invited review: ruminant ecology and evolution: perspectives useful to ruminant livestock research and production. J. Dairy Sci. 2010; 93:1320–1334. [DOI] [PubMed] [Google Scholar]
- 3. Larson G., Burger J.. A population genetics view of animal domestication. Trends Genet. 2013; 29:197–205. [DOI] [PubMed] [Google Scholar]
- 4. Hassanin A., Douzery E.J.. Molecular and morphological phylogenies of ruminantia and the alternative position of the moschidae. Syst. Biol. 2003; 52:206–228. [DOI] [PubMed] [Google Scholar]
- 5. Chen L., Qiu Q., Jiang Y., Wang K., Lin Z., Li Z., Bibi F., Yang Y., Wang J., Nie W.et al.. Large-scale ruminant genome sequencing provides insights into their evolution and distinct traits. Science. 2019; 364:eaav6202. [DOI] [PubMed] [Google Scholar]
- 6. Kern C., Wang Y., Xu X., Pan Z., Halstead M., Chanthavixay G., Saelao P., Waters S., Xiang R., Chamberlain A.et al.. Functional annotations of three domestic animal genomes provide vital resources for comparative and agricultural research. Nat. Commun. 2021; 12:1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fang L., Cai W., Liu S., Canela-Xandri O., Gao Y., Jiang J., Rawlik K., Li B., Schroeder S.G., Rosen B.D.et al.. Comprehensive analyses of 723 transcriptomes enhance genetic and biological interpretations for complex traits in cattle. Genome Res. 2020; 30:790–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tian D., Wang P., Tang B., Teng X., Li C., Liu X., Zou D., Song S., Zhang Z.. GWAS Atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res. 2020; 48:D927–D932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hu Z.L., Park C.A., Reecy J.M.. Building a livestock genetic and genomic information knowledgebase through integrative developments of Animal QTLdb and CorrDB. Nucleic Acids Res. 2019; 47:D701–D710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schoch C.L., Ciufo S., Domrachev M., Hotton C.L., Kannan S., Khovanskaya R., Leipe D., McVeigh R., O’Neill K., Robbertse B.et al.. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database (Oxford). 2020; 2020:baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tang Q., Gu Y., Zhou X., Jin L., Guan J., Liu R., Li J., Long K., Tian S., Che T.et al.. Comparative transcriptomics of 5 high-altitude vertebrates and their low-altitude relatives. Gigascience. 2017; 6:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Malmuthuge N., Liang G., Guan L.L.. Regulation of rumen development in neonatal ruminants through microbial metagenomes and host transcriptomes. Genome Biol. 2019; 20:172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nguyen L.T., Reverter A., Cánovas A., Venus B., Anderson S.T., Islas-Trejo A., Dias M.M., Crawford N.F., Lehnert S.A., Medrano J.F.et al.. STAT6, PBX2, and PBRM1 emerge as predicted regulators of 452 differentially expressed genes associated with puberty in Brahman heifers. Front Genet. 2018; 9:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ishibashi M., Ikeda S., Minami N.. Comparative analysis of histone H3K4me3 modifications between blastocysts and somatic tissues in cattle. Sci. Rep. 2021; 11:8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ming H., Sun J., Pasquariello R., Gatenby L., Herrick J.R., Yuan Y., Pinto C.R., Bondioli K.R., Krisher R.L., Jiang Z.. The landscape of accessible chromatin in bovine oocytes and early embryos. Epigenetics. 2021; 16:300–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C.H., Gao Y., Jadhav U., Hung H.H., Holton K.M., Grodzinsky A.J., Shivdasani R.A., Lassar A.B.. Creb5 establishes the competence for Prg4 expression in articular cartilage. Commun. Biol. 2021; 4:332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fang L., Liu S., Liu M., Kang X., Lin S., Li B., Connor E.E., Baldwin R.L.t., Tenesa A., Ma L.et al.. Functional annotation of the cattle genome through systematic discovery and characterization of chromatin states and butyrate-induced variations. BMC Biol. 2019; 17:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan X., Cai Y., Li Z., Chen X., Heller R., Wang N., Wang Y., Zhao C., Wang Y., Xu H.et al.. Modes of genetic adaptations underlying functional innovations in the rumen. Sci. China Life Sci. 2021; 64:1–21. [DOI] [PubMed] [Google Scholar]
- 19. Org T., Hensen K., Kreevan R., Mark E., Sarv O., Andreson R., Jaakma Ü., Salumets A., Kurg A.. Genome-wide histone modification profiling of inner cell mass and trophectoderm of bovine blastocysts by RAT-ChIP. PLoS One. 2019; 14:e0225801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Naval-Sanchez M., Nguyen Q., McWilliam S., Porto-Neto L.R., Tellam R., Vuocolo T., Reverter A., Perez-Enciso M., Brauning R., Clarke S.et al.. Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds. Nat. Commun. 2018; 9:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Navarro Gonzalez J., Zweig A.S., Speir M.L., Schmelter D., Rosenbloom K.R., Raney B.J., Powell C.C., Nassar L.R., Maulding N.D., Lee C.M.et al.. The UCSC Genome Browser database: 2021 update. Nucleic Acids Res. 2021; 49:D1046–D1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002; 12:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Deng W., Nickle D.C., Learn G.H., Maust B., Mullins J.I.. ViroBLAST: a stand-alone BLAST web server for flexible queries of multiple databases and user's datasets. Bioinformatics. 2007; 23:2334–2336. [DOI] [PubMed] [Google Scholar]
- 24. Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S.. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009; 25:288–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K.. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017; 45:D353–D361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martens M., Ammar A., Riutta A., Waagmeester A., Slenter D.N., Hanspers K., Miller A.R., Digles D., Lopes E.N., Ehrhart F.et al.. WikiPathways: connecting communities. Nucleic Acids Res. 2021; 49:D613–D621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou Y., Liang Y., Lynch K.H., Dennis J.J., Wishart D.S.. PHAST: a fast phage search tool. Nucleic Acids Res. 2011; 39:W347–W352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ward L.D., Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016; 44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farh K.K., Marson A., Zhu J., Kleinewietfeld M., Housley W.J., Beik S., Shoresh N., Whitton H., Ryan R.J., Shishkin A.A.et al.. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015; 518:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nguyen Q.H., Tellam R.L., Naval-Sanchez M., Porto-Neto L.R., Barendse W., Reverter A., Hayes B., Kijas J., Dalrymple B.P.. Mammalian genomic regulatory regions predicted by utilizing human genomics, transcriptomics, and epigenetics data. Gigascience. 2018; 7:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ernst J., Kellis M.. ChromHMM: automating chromatin-state discovery and characterization. Nat. Methods. 2012; 9:215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhao Y., Hou Y., Xu Y., Luan Y., Zhou H., Qi X., Hu M., Wang D., Wang Z., Fu Y.et al.. A compendium and comparative epigenomics analysis of cis-regulatory elements in the pig genome. Nat. Commun. 2021; 12:2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M.J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004; 306:636–640. [DOI] [PubMed] [Google Scholar]
- 39. Bolger A.M., Lohse M., Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014; 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kim D., Langmead B., Salzberg S.L.. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 2015; 12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pertea M., Kim D., Pertea G.M., Leek J.T., Salzberg S.L.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016; 11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kryuchkova-Mostacci N., Robinson-Rechavi M.. A benchmark of gene expression tissue-specificity metrics. Brief. Bioinform. 2017; 18:205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hu Z.L., Park C.A., Wu X.L., Reecy J.M.. Animal QTLdb: an improved database tool for livestock animal QTL/association data dissemination in the post-genome era. Nucleic Acids Res. 2013; 41:D871–D879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Y., Zhang C., Wang N., Li Z., Heller R., Liu R., Zhao Y., Han J., Pan X., Zheng Z.et al.. Genetic basis of ruminant headgear and rapid antler regeneration. Science. 2019; 364:eaav6335. [DOI] [PubMed] [Google Scholar]
- 46. Lin Z., Chen L., Chen X., Zhong Y., Yang Y., Xia W., Liu C., Zhu W., Wang H., Yan B.et al.. Biological adaptations in the Arctic cervid, the reindeer (Rangifer tarandus). Science. 2019; 364:eaav6312. [DOI] [PubMed] [Google Scholar]
- 47. Liu C., Gao J., Cui X., Li Z., Chen L., Yuan Y., Zhang Y., Mei L., Zhao L., Cai D.et al.. A towering genome: experimentally validated adaptations to high blood pressure and extreme stature in the giraffe. Sci. Adv. 2021; 7:eabe9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Karim L., Takeda H., Lin L., Druet T., Arias J.A., Baurain D., Cambisano N., Davis S.R., Farnir F., Grisart B.et al.. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 2011; 43:405–413. [DOI] [PubMed] [Google Scholar]
- 49. Bouwman A.C., Daetwyler H.D., Chamberlain A.J., Ponce C.H., Sargolzaei M., Schenkel F.S., Sahana G., Govignon-Gion A., Boitard S., Dolezal M.et al.. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018; 50:362–367. [DOI] [PubMed] [Google Scholar]
- 50. Takasuga A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016; 87:159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang X., Cai B., Zhou J., Zhu H., Niu Y., Ma B., Yu H., Lei A., Yan H., Shen Q.et al.. Disruption of FGF5 in cashmere goats using CRISPR/Cas9 results in more secondary hair follicles and longer fibers. PLoS One. 2016; 11:e0164640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cai Y., Fu W., Cai D., Heller R., Zheng Z., Wen J., Li H., Wang X., Alshawi A., Sun Z.et al.. Ancient genomes reveal the evolutionary history and origin of cashmere-producing goats in China. Mol. Biol. Evol. 2020; 37:2099–2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Du X., Guo J., Cao Q.Y., Yao W., Li Q.F.. A haplotype variant of Hu sheep follicle-stimulating hormone receptor promoter region decreases transcriptional activity. Anim. Genet. 2019; 50:407–411. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.