Abstract
DNA methylation is known to be the most stable epigenetic modification and has been extensively studied in relation to cell differentiation, development, X chromosome inactivation and disease. Allele-specific DNA methylation (ASM) is a well-established mechanism for genomic imprinting and regulates imprinted gene expression. Previous studies have confirmed that certain special regions with ASM are susceptible and closely related to human carcinogenesis and plant development. In addition, recent studies have proven ASM to be an effective tumour marker. However, research on the functions of ASM in diseases and development is still extremely scarce. Here, we collected 4400 BS-Seq datasets and 1598 corresponding RNA-Seq datasets from 47 species, including human and mouse, to establish a comprehensive ASM database. We obtained the data on DNA methylation level, ASM and allele-specific expressed genes (ASEGs) and further analysed the ASM/ASEG distribution patterns of these species. In-depth ASM distribution analysis and differential methylation analysis conducted in nine cancer types showed results consistent with the reported changes in ASM in key tumour genes and revealed several potential ASM tumour-related genes. Finally, integrating these results, we constructed the first well-resourced and comprehensive ASM database for 47 species (ASMdb, www.dna-asmdb.com).
INTRODUCTION
DNA methylation is an important epigenetic modification that plays a key role in cell differentiation (1,2), development (3,4), ageing (5), genomic imprinting (6,7), X chromosome inactivation (8,9) and disease (10,11). Bisulfite sequencing (BS-Seq) is a method for detecting DNA methylation at single-base resolution on the genome scale by converting nonmethylated cytosines into thymines and has substantially improved the study of DNA methylation (12).
Diploidy normally affords protection against the deleterious effects of recessive mutations. Nevertheless, the functional haploid state eliminates this protection, making single genomic or epigenetic changes dysfunctional. Owing to this feature of haplotypes, imprinted genes are susceptible targets for many animal and plant diseases, and the destruction of imprinting can lead to cell dysfunction (13). Imprinting is mainly related to allele-specific DNA methylation (ASM), and different methylation patterns in alleles can lead to different phenotypes, such as diseases and even different therapeutic and drug responses to diseases (7,14–16).
Recent reports have shown that ASM is increased in some cancers, such as lymphoma and myeloma (17), and ASM can serve as an effective tumour marker and plays important roles in the development of seeds and seedlings (15,18–20). Studies have revealed that the loss of maternal allele methylation of insulin-like growth factor II (IGF2) is associated with increased expression of growth-promoting genes in Wilms tumour (21). In breast cancer, the specific up regulation of imprinted genes such as HM13 is due to the loss of DNA methylation (22). Furthermore, the risk of ductal carcinoma in situ (DCIS) increased with higher KvDMR-ICR2 (KvDMR imprinting control region 2) methylation and lower PLAGL1/ZAC1 methylation (23). Therefore, research on ASM in diseases, especially in cancer, is extremely urgent and necessary.
However, due to the past lack of ASM detection tools before, research on ASM at the genome scale is greatly limited. In recent years, several ASM detection tools, such as MethHaplo (24), MethPipe (25), MONOD2 (20), DAMEfinder (26) and CpelAsm (27), have been developed for ASM study. This progress has made it possible to carry out ASM research at the genome scale. Compared with other ASM detection tools, MethHaplo, developed by our laboratory, can perform ASM detection through methylation sequence assembly without relying on heterozygous SNP information. By this means, we can obtain genome-wide ASM results, especially in regions where heterozygous SNPs are not enriched. This superiority of MethHaplo enables us to carry out ASM-related research more comprehensively in the whole genome, such as seeking potential ASMs in promoter or intergenic regions and exploring the mechanism of ASM in cancer.
Therefore, we collected 5,998 Gene Expression Omnibus (GEO) (28) samples (including 4400 BS-Seq data and 1598 RNA-Seq data) from 47 species, including Homo sapiens and Mus musculus, and performed DNA methylation, ASM and allele-specific expressed gene (ASEG) analyses of the corresponding samples. Using the results from these analyses, we constructed a well-resourced, comprehensive database (ASMdb) that not only contains ASM results from multiple species but also provides ASEG results from the corresponding RNA-Seq datasets.
In addition, to provide more information about DNA methylation and ASM in cancer, we compiled the data on DNA methylation in humans and performed further analysis of differential DNA methylation and high-frequency ASM for cancer and normal data in nine tissues, including liver and lung, with sufficient data samples. We hope that these specific analysis results could facilitate research on ASM in cancer. We are firmly convinced that this comprehensive multispecies ASM database could provide a good vision for the analysis of ASM and promote research on various aspects of ASM.
MATERIALS AND METHODS
Database implementation
The database was organized using MySQL (version 5.7.26), and the web interface was developed using HTML with JavaScript (Figure 1). The ‘Meth Browser’ module was constructed with JBrowse (release 1.16.6) (29), which could show single-base DNA methylation level and ASM and allow exploration of methylation patterns. The database has a convenient web interface to facilitate searching, browsing and downloading the DNA methylation data.
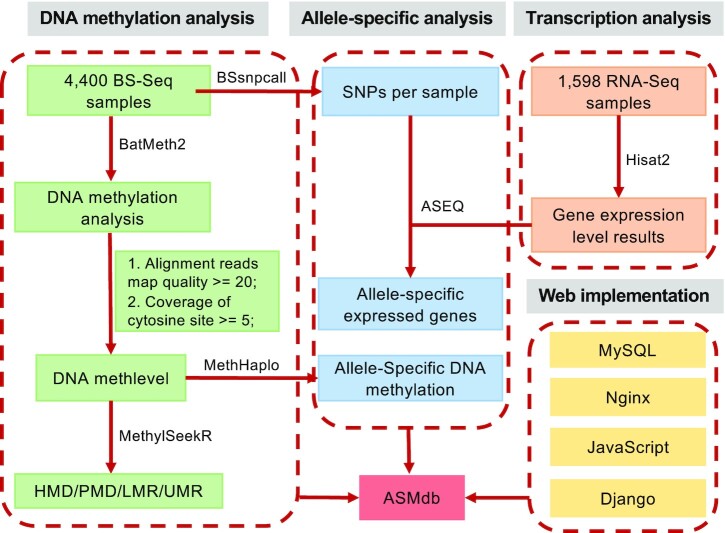
Figure 1.
Procedure used for ASMdb construction. The ASMdb database was constructed with MySQL and Django tools. BatMeth2 was used to map BS-Seq data, calculate the DNA methylation level and visualize the methylation patterns. MethHaplo was used to detect allele-specific DNA methylation. Hisat2 was used for RNA-Seq data mapping. ASEQ was used to detect allele-specific expressed genes. For annotation purposes, we used MethylSeekR to divide the genome into four categories of regions: unmethylated regions (UMRs), low-methylation regions (LMRs), partially methylated domains (PMDs) and highly methylated domains (HMDs) according to the methylation level.
Data collection
BS-Seq is currently the most common technique for detecting single-base DNA methylation at the genome-wide scale. To construct a comprehensive allele-specific DNA methylation database, we searched the NCBI GEO database, downloaded all available whole-genome bisulfite sequencing data by October 2019, and filtered the low-quality data. Finally, 4400 (out of 5014) BS-Seq DNA methylation datasets and 1598 (out of 1819) corresponding RNA-Seq datasets were used (Table 1, Supplementary Table S1). These datasets originated from 47 species, including Homo sapiens, Mus musculus, Arabidopsis thaliana and Oryza sativa (Figure 2A and B). The database also shows the distribution of human methylation data in various tissues (Figure 2C).
Table 1.
Statistics of BS-Seq and RNA-Seq datasets in ASMdb
| Species | BS-Seq | RNA-Seq | ||||
|---|---|---|---|---|---|---|
| Projects | Samples | Categories | Projects | Samples | Categories | |
| Homo sapiens | 174 | 1484 | 417 | 41 | 758 | 105 |
| Mus musculus | 227 | 2026 | 681 | 55 | 575 | 162 |
| Arabidopsis thaliana | 40 | 416 | 198 | 15 | 140 | 40 |
| Danio rerio | 2 | 48 | 11 | 1 | 3 | 3 |
| Macaca mulatta | 4 | 39 | 12 | 1 | 16 | 8 |
| Pan troglodytes | 1 | 38 | 8 | 1 | 16 | 8 |
| Marchantia polymorpha | 2 | 29 | 2 | 0 | 0 | 0 |
| Solanum lycopersicum | 4 | 23 | 14 | 2 | 10 | 5 |
| Harpegnathos saltator | 1 | 20 | 8 | 0 | 0 | 0 |
| Oryza sativa | 2 | 20 | 7 | 2 | 11 | 5 |
| Others | 67 | 257 | 142 | 22 | 69 | 43 |
| Total | 524 | 4400 | 1500 | 140 | 1598 | 379 |
Note. Categories represent different tissues, stages, or conditions.
Figure 2.
Overview of ASMdb. (A) Main species included in ASMdb. (B) Proportion of BS-Seq data from various species in ASMdb. (C) Proportion of BS-Seq data from each tissue in humans. (D) Main functional modules in ASMdb. (E) An example of a genome browser screenshot around the FOXD3 gene region in human liver tissue (chr1:63321858–63325268, 3.41 kb).
Processing of DNA methylation data
The trimming of low-quality reads and artificial sequences was performed with Fastp (30). The parameters of Fastp are as follows: the window size option shared by sliding (-W) is set to 4, the mean quality requirement option shared by sliding (-M) is set to 20, the quality threshold for a qualified base (-q) is set to 15, the percentage of bases allowed to be unqualified (-u) is set to 40%, one read's N base number (-n) is set to 5, and the threshold for the low complexity filter (-Y) is set to 0. The clean reads were mapped to the corresponding reference genomes (Supplementary Table S2) using BatMeth2 (31), and the SAM files were converted to the BAM format with SAMtools (32). DNA methylation calling was performed with the Calmeth function in the BatMeth2 package (31). Sequences with a map quality score lower than 20 were filtered out, and cytosine sites with coverage of 5 or more were considered effective methylation sites for further analysis.
Filtering data with low bisulfite conversion rate
Considering that most of the WGBS data did not include spike-in sequences for bisulfite conversion rate estimation, we tried several commonly used methods to evaluate the bisulfite conversion rate: (i) calculating the methylation level of mitochondria in mammalian humans and mice; (ii) calculating the methylation level of CHG in animals; (iii) calculating the methylation level of mammalian telomere repeat CCCATT (33) and (iv) calculating the methylation level of chloroplasts in Arabidopsis thaliana. Because there are few reports about DNA methylation in mitochondria (34–36), we did not use this method to calculate bisulfite conversion. We kept the data for which the bisulfite conversion rate of the CHG method was >95% or that of the chloroplast method was >98%. After data filtering, we obtained 1484 (out of 1656) sets of high-quality human methylation data, 2026 (out of 2329) sets of mouse data, 287 (out of 384) sets of other animal data and 416 (out of 458) sets of Arabidopsis thaliana data, with a total of 4400 (out of 5014) sets of high-quality DNA methylation data.
Identification of ASM
The DNA methylation level was calculated with BatMeth2 software (31). ASM was identified by MethHaplo (24) with the default parameters. In the ASM detection process, all the totally methylated (methylation level > 0.9) and totally unmethylated (methylation level < 0.1) sites were removed first, and only partially methylated cytosine sites, denoted as effective sites, were retained for haplotype region identification.
Identification of allele-specific expressed genes (ASEGs)
The raw reads from the RNA-Seq data were trimmed as clean reads using Fastp with the default parameters. The clean reads were mapped to the corresponding reference genome using Hisat2 (37), and SAMtools was used to sort the BAM file. The SNP information used for ASEG detection was derived from the BS-Seq corresponding to the RNA-Seq data. The ASEGs were detected by ASEQ (38).
Identification of HMD/PMD/LMR/UMR
DNA methylation files containing the coverage and methylation level were obtained using BatMeth2. According to the methylation level, each BS-Seq sample was divided into partially methylated domains (PMDs), low-methylation regions (LMRs) and unmethylated regions (UMRs) using MethylSeekR (39). We removed the gap regions with continuous ‘N’ in the genome, and the remaining regions were called highly methylated domains (HMDs) (40). The details of the scripts are shown in the ‘Help’ module of ASMdb.
High-frequency ASM related gene (ASMG) and ASEG analysis
In each sample, we counted the number of ASM loci in each gene and the upstream 3 kb range of the gene and then added up the frequency of the gene covering ASM in all samples. Finally, we identified the 100 ASM genes with the highest frequencies. Similarly, we calculated the ASEGs with the highest frequency in each sample, which were called high-frequency ASEGs.
Differential DNA methylation genes between cancer and normal tissues
We screened the DNA methylation data on human cancer and corresponding normal tissues and obtained the methylation data of 8 tissues (no differential methylation was detected in ovary data), including the brain, liver and lung. Then, we used the Wilcoxon rank-sum test to perform differential DNA methylation analysis (41). Finally, we performed a screen to identify genes whose P-value less than 0.01 and whose absolute value of the difference in the DNA methylation level was greater than 0.1 (P-value < 0.01 and |meth.diff| > 0.1). In addition, our database allows the user to set different P-values and differential DNA methylation thresholds to filter the differentially methylated genes.
RESULTS
Web interface
A user-friendly web interface (Figures 2 and 3) is provided to allow users to query the database through multiple modules: (i) ‘Meth Browser’, a genome browser for browsing and searching single-base DNA methylation level, ASM, SNP, HMD/PMD/LMR/UMR, and other chromosome methylation states (Figure 2E); (ii) ‘Analysis/Function’ (Figure 2D), a retrieval module for the online illustration of the ASM and ASEGs in specific samples, the DNA methylation profile in various species, the DNA methylation profile of gene promoters and gene bodies in different samples, differential DNA methylation in cancer, and high-frequency ASMG and ASEG in cancer; (iii) ‘DataSets’, a module that shows all the datasets in the ASMdb and the statistical results from the corresponding data; (iv) ‘Tools’, a module that contains the related tools for DNA methylation analysis and (v) ‘Help’ and ‘About Us’, modules with detailed documentation and tutorials. For more detailed instructions, we provide a PDF document (https://www.dna-asmdb.com/download/ASMdb-tutorial.pdf).
Figure 3.
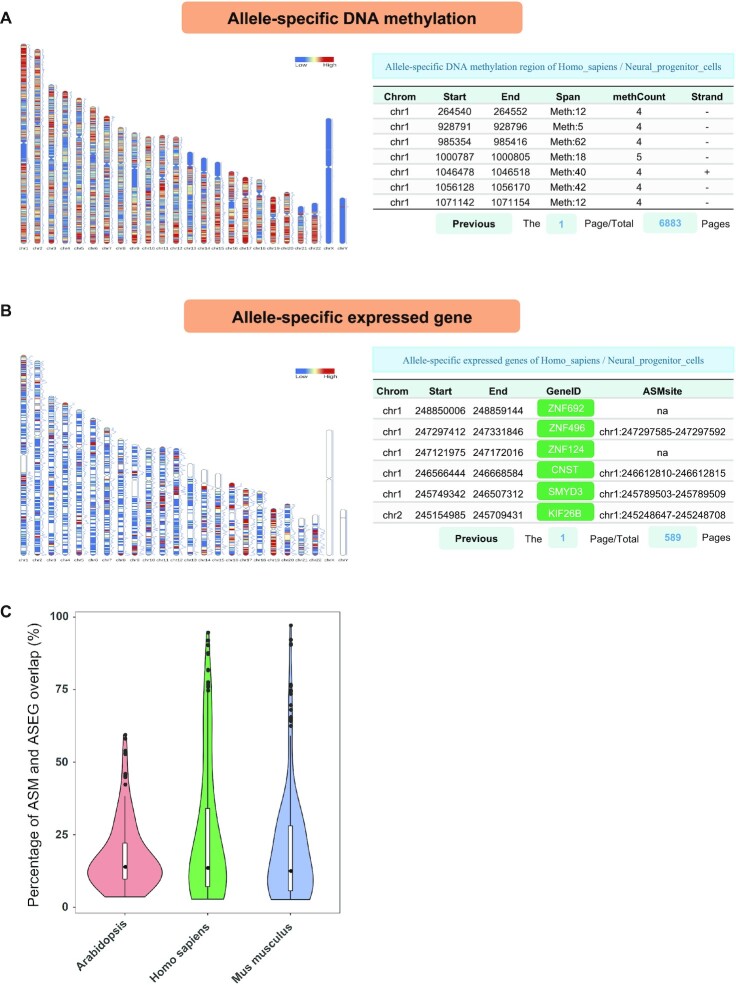
Allele-specific analysis. (A) The distribution of ASM on chromosomes and the list of ASM obtained from human neural progenitor cells. (B) The distribution of ASEGs on chromosomes and the list of ASEGs obtained from human neural progenitor cells. (C) The overlap between ASM and ASEG. We calculated the percentage of ASEGs overlapping with ASM obtained from each methylation dataset and with ASEGs obtained from the corresponding RNA-Seq dataset. For statistical credibility, we removed the data with fewer than 500 ASM or ASEGs.
ASMdb genome browser
The genome browser was developed using JBrowse (version 1.16.6), which provides a user-friendly and convenient interface for browsing single-base DNA methylation levels and ASM. Users are able to select specific genomic regions to view the associated DNA methylation patterns in diverse samples. Moreover, JBrowse plugins, such as ‘Methylation Plugin’ and ‘ScreenShot Plugin’ (42), provide additional models for displaying DNA methylation information and give users the abilities to save and download the results in PDF/PNG format. Additionally, the ASM results per sample detected from BS-Seq data are shown as an independent track in the genome browser. Users can search in the ASMdb Meth Browser based on its genomic location or the gene symbol. Associated tracks, such as HMD/PMD/LMR/UMR, SNPs, gene expression levels, CpG islands and RefSeq genes, are shown in the genome browser. To better illustrate the genome browser, an example showing the DNA methylation distribution around the FOXD3 gene in the genome browser is presented in Figure 2E. Studies have shown that FOXD3 has a momentous impact in a variety of cancers, including liver cancer, and its expression in cancer is regulated by DNA methylation (43–45). The results of our genome browser revealed an apparent difference in DNA methylation levels between liver cancer and normal liver tissues. Moreover, users can upload local data for viewing, and the browser allows the uploading of files in the format supported by JBrowse, such as bigWig and BED.
Function of ASMdb
Allele-specific related analysis
The ‘Allele-specific DNA methylation’ page displays the heat map of ASM density on chromosomes and ASM information for each sample as well as a description of the sample (link to the genome browser) and sample ID (link to the NCBI). This page also presents the ASM list, which includes information on the chromosome location, the length of the ASM regions, the number of ASM cytosine sites in each ASM region and the length of this ASM region (Figure 3A).
The ‘Allele-specific expressed genes’ page provides the heat map of ASEG distribution on chromosomes in each sample, as well as the list of ASEG information. The ASEG table includes details of the gene list and ASM region near each gene (within 3 kb) (Figure 3B). Users can query the ASEG genes in the specified sample and check whether ASM occurs near those genes. The ASM and ASEG analysis results provided by this database can be helpful for allele-specific related research. Moreover, we found that in some regions where ASM is detected, ASEG is usually also detected (average percentage in humans: 25.02%; average percentage in mice: 19.78%; average percentage of Arabidopsis: 17.41%) (Figure 3C).
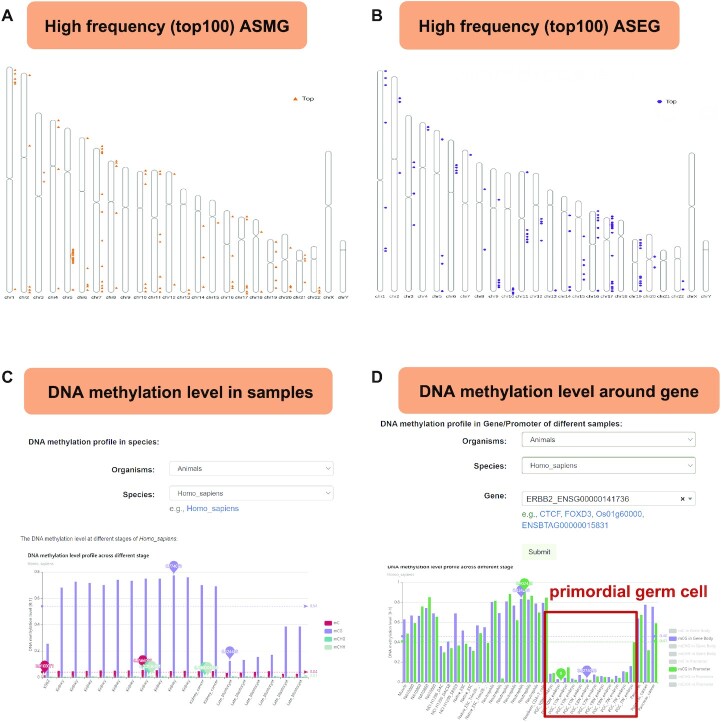
The ‘High-frequency allele genes in species’ page provides high-frequency ASMG and high-frequency ASEG in each species. In addition, we exhibit examples that show the high-frequency ASMG and ASEG information from all human BS-Seq data (Figure 4A and B).
Figure 4.
Screenshots of representative functional modules in ASMdb. (A) The distribution of high-frequency ASMG on chromosomes in humans. (B) The distribution of high-frequency ASEG on chromosomes in humans. (C) An example of the average DNA methylation level profile across samples from humans. (D) DNA methylation profile around the ERBB2 gene across samples from humans. The red box highlights the DNA methylation level of primordial germ cells.
DNA methylation profile
A query on the ‘Sample DNA methylation profile’ page displays a bar plot or histogram of the DNA methylation levels across all samples. This page provides a DNA methylation table with a description of each sample (link to the genome browser), including the sample ID (link to NCBI) and the mC, mCG, mCHG and mCHH methylation levels (Figure 4C).
A query on the ‘Gene meth profile across samples’ page displays a bar plot or histogram of the DNA methylation profiles of the gene body or promoter across different samples. This page provides information regarding the location of the gene and the corresponding DNA methylation level (Figure 4D). We can view the DNA methylation level of the ERBB2 gene in different tissues. Figure 4D shows that the average DNA methylation levels of ERBB2 in primordial germ cells (PGCs) were significantly lower than those in other tissues. Such results provide useful information to users for the study of DNA methylation.
Allele-specific DNA methylation in cancer
ASMdb provides the DNA methylation level distribution of each gene promoter and gene body in cancer and normal tissues in human BS-Seq data (Figure 5A). This database allows the selection of different cancer types as well as corresponding DNA methylation data (Table 2, Supplementary Table S3). For instance, previous studies have shown that ERBB2 is closely related to overall survival in lung cancer (46). Consistently, there is an obvious difference in the DNA methylation level between normal lung and lung cancer tissue around the ERBB2 gene (Figure 5A and B). To obtain the gene expression level in cancer, ASMdb provides an association analysis with GEPIA2 (47). The expression of ERBB2 in lung cancer tissue was significantly higher than that in normal tissue (Figure 5A). Moreover, the methylation level of the promoter of the ERBB2 gene was significantly decreased in cancer tissue (Figure 5B). This is consistent with our understanding that genes with high methylation levels in the promoter region have low expression levels. Overall, using this function, we were able to view the DNA methylation levels in different tissues and under different conditions.
Figure 5.
The ERBB2 gene was used as an example to show the representative functional modules in ASMdb. (A) The location information of ERBB2 and its expression level from the GEPIA2 database. (B) The DNA methylation levels of the ERBB2 gene in normal and cancer samples. The red box indicates the differential DNA methylation level around the promoter. (C) Differential DNA methylation genes detected between lung cancer and normal lung data. The red box indicates that ERBB2 was detected as a significantly differentially methylated gene in lung cancer.
Table 2.
Analysis of the corresponding disease types in ASMdb tissues
| Tissues | Disease Type(s) | Tissues | Disease type(s) |
|---|---|---|---|
| Blood | • ALL | Brain | • Alzheimer |
| • AML3 | • Cancer | ||
| • CLL | • Schizophrenia | ||
| • Colon-cancer | |||
| • Lung-cancer | |||
| Breast | • Cancer | Colon | • Cancer |
| Liver | • Cancer | Lung | • Cancer |
| Prostate | • Cancer | Pancreas | • Cancer |
Differentially methylated genes in cancer
To further explore the differentially methylated genes in cancer, we screened the DNA methylation data related to cancer and then performed differential DNA methylation analysis with the corresponding normal DNA methylation data. We found that the promoter region of the ERBB2 gene in lung cancer showed obvious differences in DNA methylation levels (Figure 5C).
High-frequency ASMG and ASEG in representative cancers
We counted the high-frequency ASMG and ASEG in representative cancers. The results of the high-frequency ASMG distribution on chromosomes in liver cancer and normal liver tissues are shown in Figure 6A. The results of high-frequency ASEG distribution on chromosomes in lung cancer and lung normal tissues are shown in Figure 6B. Moreover, ASMdb provides a list of high-frequency ASMG and ASEG in cancer and corresponding normal tissues.
Figure 6.
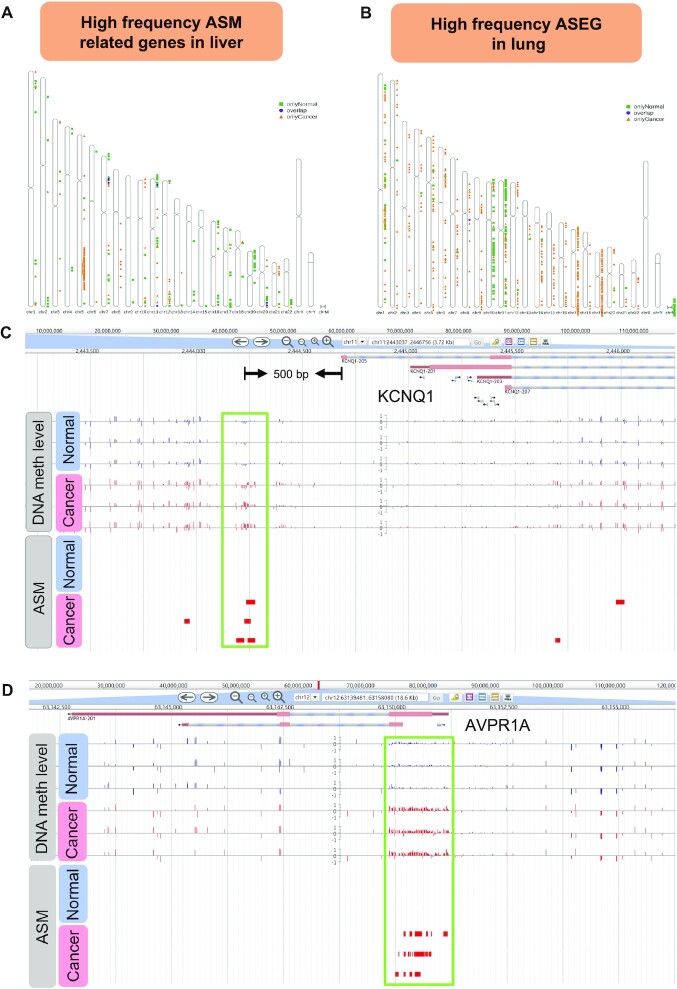
Application examples of ASMdb. (A) The distribution of high-frequency ASM-related genes in liver cancer and normal data. (B) The distribution of high-frequency ASEG in lung cancer and normal data. (C) Genome browser screenshot of the KCNQ1 gene in human liver cancer and normal data. The green box highlights the differential DNA methylation levels and ASM between cancer and normal data. (D) Genome browser screenshot of the AVPR1A gene in human liver cancer and normal data. The green box highlights the differential DNA methylation levels and ASM between cancer and normal data.
Functional examples
Combined with previous studies and the related results in our database, we demonstrated two functional examples. Previous studies indicated that the KCNQ1 gene is a known imprinted gene that plays a key role in liver cancer, breast cancer and other cancers (48–50). In ASMdb, KCNQ1 was detected as a high frequency ASMG and ASEG in liver cancer. According to the DNA methylation level and ASM distribution of the gene, we observed a significant DNA methylation difference in the gene promoter. Interestingly, ASM was found among cancer samples only in the promoter of KCNQ1 (Figure 6C). The ASM distribution in the promoter may play an essential role in the allele-specific expression of KCNQ1.
Additionally, we detected that the AVPR1A gene has a high frequency of ASM distribution in liver cancer (Figure 6D), implying an association between AVPR1A and liver cancer. Although studies have found that AVPR1A is related to the occurrence of prostate cancer and thyroid cancer (51,52), the gene has not been reported in liver cancer. The results of the differential enrichment of ASM and the DNA methylation level of the AVPR1A gene in liver cancer indicate the potential significance of the AVPR1A gene in liver cancer.
DISCUSSION AND FUTURE DIRECTIONS
The first allele-specific DNA methylation databases
In this study, we developed the ASMdb database, which can serve as a comprehensive resource on allele-specific DNA methylation in diverse organisms. Currently, there are some existing databases for DNA methylation mainly based on BeadChip data, which do not provide comprehensive information on genome-wide DNA methylation and allele-specific DNA methylation based on high-throughput sequencing data. For example, MethHC 2.0 (53) provides only human BeadChip data, MethBank 3.0 (54) contains DNA methylation BeadChip data from humans and mice as well as 354 WGBS-Seq data from seven species, and Pancan-meQTL (55) is a database of DNA methylation BeadChip data for the analysis of DNA methylation and SNP associations in cancer. ASMdb is a comprehensive and valuable allele-specific DNA methylation database containing 5998 high-throughput datasets, including BS-Seq and RNA-Seq data. ASMdb provides DNA methylation and ASM results for each data point and analysis results on cancer data, including genes with differential DNA methylation and high-frequency ASMG/ASEG.
Future directions
In the future, we will continue to update ASMdb as follows: (i) During the ASMdb database development stage, we have collected BS-Seq data from before October 2019. In the future, we will further collect and analyse DNA methylation data from different sources and species. (ii) We will provide additional online functions on the website based on user feedback. We promise that ASMdb will be kept up to date to ensure that its value as a user-friendly allele-specific DNA methylation database. We expect that ASMdb will contribute to research on DNA methylation and ASM in cellular function.
ABBREVIATION
| ASEG | Allele-specific expressed gene |
| ASM | Allele-specific DNA methylation |
| ASMG | Allele-specific DNA methylation related gene |
| BS-Seq | Bisulfite sequencing |
| DCIS | Ductal carcinoma in situ |
| GEO | Gene Expression Omnibus |
| HMD | Highly methylated domain |
| ICR2 | Imprinting control region 2 |
| IGF2 | Insulin-like growth factor II |
| LMR | Lowly methylated region |
| PGC | Primordial germ cell |
| PMD | Partially methylated domain |
| UMR | Unmethylated region |
DATA AVAILABILITY
ASMdb is a database with online and open access, available at https://www.dna-asmdb.com. Any constructive comments and suggestions are welcome to send to Prof. Guoliang Li at email address guoliang.li@mail.hzau.edu.cn.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Mr. Hao Liu from the National Key Laboratory of Crop Genetic Improvement for essential help in running the high-throughput computing clusters. We thank the group members for providing feedback on the database. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributor Information
Qiangwei Zhou, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Pengpeng Guan, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Zhixian Zhu, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Sheng Cheng, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Cong Zhou, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Huanhuan Wang, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Qian Xu, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
Wing-kin Sung, Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China; Department of Computer Science, National University of Singapore, Singapore 117417, Singapore; Department of Computational and Systems Biology, Genome Institute of Singapore, Singapore 138672, Singapore.
Guoliang Li, National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China; Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31771402, 31970590]; National Key Research and Development Program of China [2018YFC1604000]; Fundamental Research Funds for the Central Universities [2662017PY116]. Funding for open access charge: National Natural Science Foundation of China [31771402, 31970590]; National Key Research and Development Program of China [2018YFC1604000]; Fundamental Research Funds for the Central Universities [2662017PY116].
Conflict of interest statement. None declared.
REFERENCES
- 1. Laurent L., Wong E., Li G., Huynh T., Tsirigos A., Ong C.T., Low H.M., Kin Sung K.W., Rigoutsos I., Loring J.et al.. Dynamic changes in the human methylome during differentiation. Genome Res. 2010; 20:320–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bock C., Beerman I., Lien W.H., Smith Z.D., Gu H., Boyle P., Gnirke A., Fuchs E., Rossi D.J., Meissner A.. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol. Cell. 2012; 47:633–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Doi A., Park I.H., Wen B., Murakami P., Aryee M.J., Irizarry R., Herb B., Ladd-Acosta C., Rho J., Loewer S.et al.. Differential methylation of tissue- and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat. Genet. 2009; 41:1350–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo Y., Lu X., Xie H.. Dynamic Alu methylation during normal development, aging, and tumorigenesis. Biomed. Res. Int. 2014; 2014:784706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jones M.J., Goodman S.J., Kobor M.S.. DNA methylation and healthy human aging. Aging Cell. 2015; 14:924–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barlow D.P., Bartolomei M.S.. Genomic imprinting in mammals. Cold Spring Harb. Perspect. Biol. 2014; 6:a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tucci V., Isles A.R., Kelsey G., Ferguson-Smith A.C., Erice Imprinting G.. Genomic imprinting and physiological processes in mammals. Cell. 2019; 176:952–965. [DOI] [PubMed] [Google Scholar]
- 8. Hall E., Volkov P., Dayeh T., Esguerra J.L., Salo S., Eliasson L., Ronn T., Bacos K., Ling C.. Sex differences in the genome-wide DNA methylation pattern and impact on gene expression, microRNA levels and insulin secretion in human pancreatic islets. Genome Biol. 2014; 15:522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luijk R., Wu H., Ward-Caviness C.K., Hannon E., Carnero-Montoro E., Min J.L., Mandaviya P., Muller-Nurasyid M., Mei H., van der Maarel S.M.et al.. Autosomal genetic variation is associated with DNA methylation in regions variably escaping X-chromosome inactivation. Nat. Commun. 2018; 9:3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Robertson K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005; 6:597–610. [DOI] [PubMed] [Google Scholar]
- 11. Yang X., Han H., De Carvalho D.D., Lay F.D., Jones P.A., Liang G.. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014; 26:577–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Frommer M., McDonald L.E., Millar D.S., Collis C.M., Watt F., Grigg G.W., Molloy P.L., Paul C.L.. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dolinoy D.C., Das R., Weidman J.R., Jirtle R.L.. Metastable epialleles, imprinting, and the fetal origins of adult diseases. Pediatr. Res. 2007; 61:30R–37R. [DOI] [PubMed] [Google Scholar]
- 14. Farhadova S., Gomez-Velazquez M., Feil R.. Stability and lability of parental methylation imprints in development and disease. Genes (Basel). 2019; 10:999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsieh T.F., Shin J., Uzawa R., Silva P., Cohen S., Bauer M.J., Hashimoto M., Kirkbride R.C., Harada J.J., Zilberman D.et al.. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:1755–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim D.H., Maher E.R.. Genomic imprinting syndromes and cancer. Adv. Genet. 2010; 70:145–175. [DOI] [PubMed] [Google Scholar]
- 17. Do C., Dumont E.L.P., Salas M., Castano A., Mujahed H., Maldonado L., Singh A., DaSilva-Arnold S.C., Bhagat G., Lehman S.et al.. Allele-specific DNA methylation is increased in cancers and its dense mapping in normal plus neoplastic cells increases the yield of disease-associated regulatory SNPs. Genome Biol. 2020; 21:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du M., Luo M., Zhang R., Finnegan E.J., Koltunow A.M.. Imprinting in rice: the role of DNA and histone methylation in modulating parent-of-origin specific expression and determining transcript start sites. Plant J. 2014; 79:232–242. [DOI] [PubMed] [Google Scholar]
- 19. Zhang H., Zhang Z., Liu X., Duan H., Xiang T., He Q., Su Z., Wu H., Liang Z.. DNA methylation haplotype block markers efficiently discriminate follicular thyroid carcinoma from follicular adenoma. J. Clin. Endocrinol. Metab. 2021; 106:1011–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guo S., Diep D., Plongthongkum N., Fung H.L., Zhang K., Zhang K.. Identification of methylation haplotype blocks aids in deconvolution of heterogeneous tissue samples and tumor tissue-of-origin mapping from plasma DNA. Nat. Genet. 2017; 49:635–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ravenel J.D., Broman K.W., Perlman E.J., Niemitz E.L., Jayawardena T.M., Bell D.W., Haber D.A., Uejima H., Feinberg A.P.. Loss of imprinting of insulin-like growth factor-II (IGF2) gene in distinguishing specific biologic subtypes of Wilms tumor. J. Natl. Cancer Inst. 2001; 93:1698–1703. [DOI] [PubMed] [Google Scholar]
- 22. Goovaerts T., Steyaert S., Vandenbussche C.A., Galle J., Thas O., Van Criekinge W., De Meyer T.. A comprehensive overview of genomic imprinting in breast and its deregulation in cancer. Nat. Commun. 2018; 9:4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harrison K., Hoad G., Scott P., Simpson L., Horgan G.W., Smyth E., Heys S.D., Haggarty P.. Breast cancer risk and imprinting methylation in blood. Clin Epigenetics. 2015; 7:92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou Q., Wang Z., Li J., Sung W.K., Li G.. MethHaplo: combining allele-specific DNA methylation and SNPs for haplotype region identification. BMC Bioinformatics. 2020; 21:451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song Q., Decato B., Hong E.E., Zhou M., Fang F., Qu J., Garvin T., Kessler M., Zhou J., Smith A.D.. A reference methylome database and analysis pipeline to facilitate integrative and comparative epigenomics. PLoS One. 2013; 8:e81148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Orjuela S., Machlab D., Menigatti M., Marra G., Robinson M.D.. DAMEfinder: a method to detect differential allele-specific methylation. Epigenet. Chromatin. 2020; 13:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abante J., Fang Y., Feinberg A.P., Goutsias J.. Detection of haplotype-dependent allele-specific DNA methylation in WGBS data. Nat. Commun. 2020; 11:5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Buels R., Yao E., Diesh C.M., Hayes R.D., Munoz-Torres M., Helt G., Goodstein D.M., Elsik C.G., Lewis S.E., Stein L.et al.. JBrowse: a dynamic web platform for genome visualization and analysis. Genome Biol. 2016; 17:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen S., Zhou Y., Chen Y., Gu J.. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018; 34:i884–i890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Q., Lim J.Q., Sung W.K., Li G.. An integrated package for bisulfite DNA methylation data analysis with Indel-sensitive mapping. BMC Bioinformatics. 2019; 20:47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Processing Genome Project Data . The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009; 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhou J., Zhao M., Sun Z., Wu F., Liu Y., Liu X., He Z., He Q., He Q.. BCREval: a computational method to estimate the bisulfite conversion ratio in WGBS. BMC Bioinformatics. 2020; 21:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. van der Wijst M.G., van Tilburg A.Y., Ruiters M.H., Rots M.G.. Experimental mitochondria-targeted DNA methylation identifies GpC methylation, not CpG methylation, as potential regulator of mitochondrial gene expression. Sci. Rep. 2017; 7:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Breton C.V., Song A.Y., Xiao J., Kim S.J., Mehta H.H., Wan J., Yen K., Sioutas C., Lurmann F., Xue S.et al.. Effects of air pollution on mitochondrial function, mitochondrial DNA methylation, and mitochondrial peptide expression. Mitochondrion. 2019; 46:22–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sirard M.A. Distribution and dynamics of mitochondrial DNA methylation in oocytes, embryos and granulosa cells. Sci. Rep. 2019; 9:11937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Romanel A., Lago S., Prandi D., Sboner A., Demichelis F.. ASEQ: fast allele-specific studies from next-generation sequencing data. BMC Med Genomics. 2015; 8:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Burger L., Gaidatzis D., Schubeler D., Stadler M.B.. Identification of active regulatory regions from DNA methylation data. Nucleic. Acids. Res. 2013; 41:e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Salhab A., Nordstrom K., Gasparoni G., Kattler K., Ebert P., Ramirez F., Arrigoni L., Muller F., Polansky J.K., Cadenas C.et al.. A comprehensive analysis of 195 DNA methylomes reveals shared and cell-specific features of partially methylated domains. Genome Biol. 2018; 19:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mann H.B., Whitney D.R.. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947; 18:50–60. [Google Scholar]
- 42. Hofmeister B.T., Schmitz R.J.. Enhanced JBrowse plugins for epigenomics data visualization. BMC Bioinformatics. 2018; 19:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He G.Y., Hu J.L., Zhou L., Zhu X.H., Xin S.N., Zhang D., Lu G.F., Liao W.T., Ding Y.Q., Liang L.. The FOXD3/miR-214/MED19 axis suppresses tumour growth and metastasis in human colorectal cancer. Br. J. Cancer. 2016; 115:1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sarkar S., O’Connell M.R., Okugawa Y., Lee B.S., Toiyama Y., Kusunoki M., Daboval R.D., Goel A., Singh P.. FOXD3 regulates CSC marker, DCLK1-S, and invasive potential: prognostic implications in colon cancer. Mol. Cancer Res. 2017; 15:1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. He G., Hu S., Zhang D., Wu P., Zhu X., Xin S., Lu G., Ding Y., Liang L.. Hypermethylation of FOXD3 suppresses cell proliferation, invasion and metastasis in hepatocellular carcinoma. Exp. Mol. Pathol. 2015; 99:374–382. [DOI] [PubMed] [Google Scholar]
- 46. Zhang X., Gao C., Liu L., Zhou C., Liu C., Li J., Zhuang J., Sun C.. DNA methylation-based diagnostic and prognostic biomarkers of nonsmoking lung adenocarcinoma patients. J. Cell. Biochem. 2019; 120:13520–13530. [DOI] [PubMed] [Google Scholar]
- 47. Tang Z., Kang B., Li C., Chen T., Zhang Z.. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic. Acids. Res. 2019; 47:W556–W560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bjornsson H.T., Brown L.J., Fallin M.D., Rongione M.A., Bibikova M., Wickham E., Fan J.B., Feinberg A.P.. Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. J. Natl. Cancer Inst. 2007; 99:1270–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rapetti-Mauss R., Bustos V., Thomas W., McBryan J., Harvey H., Lajczak N., Madden S.F., Pellissier B., Borgese F., Soriani O.et al.. Bidirectional KCNQ1:beta-catenin interaction drives colorectal cancer cell differentiation. PNAS. 2017; 114:4159–4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fan H., Zhang M., Liu W.. Hypermethylated KCNQ1 acts as a tumor suppressor in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 2018; 503:3100–3107. [DOI] [PubMed] [Google Scholar]
- 51. Zhao N., Peacock S.O., Lo C.H., Heidman L.M., Rice M.A., Fahrenholtz C.D., Greene A.M., Magani F., Copello V.A., Martinez M.J.et al.. Arginine vasopressin receptor 1a is a therapeutic target for castration-resistant prostate cancer. Sci. Transl. Med. 2019; 11:eaaw4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Shen Y., Dong S., Liu J., Zhang L., Zhang J., Zhou H., Dong W.. Identification of potential biomarkers for thyroid cancer using bioinformatics strategy: a study based on GEO datasets. Biomed. Res. Int. 2020; 2020:9710421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huang H.Y., Li J., Tang Y., Huang Y.X., Chen Y.G., Xie Y.Y., Zhou Z.Y., Chen X.Y., Ding S.Y., Luo M.F.et al.. MethHC 2.0: information repository of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 2021; 49:D1268–D1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li R., Liang F., Li M., Zou D., Sun S., Zhao Y., Zhao W., Bao Y., Xiao J., Zhang Z.. MethBank 3.0: a database of DNA methylomes across a variety of species. Nucleic Acids Res. 2018; 46:D288–D295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gong J., Wan H., Mei S., Ruan H., Zhang Z., Liu C., Guo A.Y., Diao L., Miao X., Han L.. Pancan-meQTL: a database to systematically evaluate the effects of genetic variants on methylation in human cancer. Nucleic Acids Res. 2019; 47:D1066–D1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ASMdb is a database with online and open access, available at https://www.dna-asmdb.com. Any constructive comments and suggestions are welcome to send to Prof. Guoliang Li at email address guoliang.li@mail.hzau.edu.cn.