Abstract
RNA-seq has been widely used in experimental studies and produced a massive amount of data deposited in public databases. New biological insights can be obtained by retrospective analyses of previously published data. However, the barrier to efficiently utilize these data remains high, especially for those who lack bioinformatics skills and computational resources. We present MetazExp (https://bioinfo.njau.edu.cn/metazExp), a database for gene expression and alternative splicing profiles based on 53 615 uniformly processed publicly available RNA-seq samples from 72 metazoan species. The gene expression and alternative splicing profiles can be conveniently queried by gene IDs, symbols, functional terms and sequence similarity. Users can flexibly customize experimental groups to perform differential and specific expression and alternative splicing analyses. A suite of data visualization tools and comprehensive links with external databases allow users to efficiently explore the results and gain insights. In conclusion, MetazExp is a valuable resource for the research community to efficiently utilize the vast public RNA-seq datasets.
INTRODUCTION
Over the last decade, RNA-sequencing (RNA-seq) has become a routine technique in biological studies. It is widely used to capture digital signals of abundances of RNA sequence features from which one can estimate overall gene expression, transcript expression, as well as relative abundance of alternatively spliced transcripts (1). Hypotheses regarding specific genes can be generated by sequencing RNA samples from different conditions, such as disease status, experimental treatments, and genotypes. Numerous studies have produced a massive amount of RNA-seq data deposited in the public space, such as the Sequence Read Archive (SRA) database at NCBI, the European Nucleotide Archive (ENA) at EBI and the Sequence Read Archive at DDBJ. Retrospective analyses of large collections of RNA-seq data can lead to new biological insights (2,3). However, these nucleotide archives are designed as data archival repositories to store raw sequence reads. Although gene expression information may be available in user supplied summary formats at the Gene Expression Omnibus (GEO) (4), the heterogeneity in data processing methods prohibits meaningful comparisons, significantly limiting utilization of these sequence archives.
The large volume of data and the requirement for specialized computational resources and skills create a barrier for experimental biologists who wish to explore public repositories. Efforts have been made to simplify the access to public RNA-seq data by creating unified resources and databases. For example, the latest iteration of the recount database (recount3) uniformly processed >750 000 RNA-seq samples in humans and mice, enabling secondary analyses of RNA-seq datasets across different studies (5). RNA-seq data in the GTEx (Genotype-Tissue Expression) and TCGA (The Cancer Genome Atlas) have also been uniformly processed to provide normalized gene expression data (6). VastDB provided detailed profiling of human, mouse and chicken genes across multiple cell and tissue types (7). In livestock animals, the ASlive database developed by our group processed 4166 RNA-seq experiments in five major agricultural animal species to estimate alternative splicing and allowed users to explore differences in alternative splicing across tissues and species (8). In the MeDAS database, 2232 RNA-seq datasets across multiple metazoan species were re-analyzed to call alternative splicing events to enable studies of alternative splicing in development (9). All of these databases have become increasingly useful as exploratory and hypothesis generating tools. However, these databases represent either a small number of model species or are otherwise more specialized in scopes.
Here we present MetazExp (Figure 1), an online resource and analysis platform that builds upon 53 615 publicly available RNA-seq samples of 72 species across 17 orders. There are four important features of MetazExp that distinguishes itself from other databases. First, it processed by far the largest number of samples and species. Second, all samples were manually curated to label their tissues and experimental conditions. Third, it covered both gene expression and alternative splicing. Fourth, a wide range of analysis functions and visualization tools were implemented, making MetazExp a one-stop resource to utilize the diverse RNA-seq data present in public space.
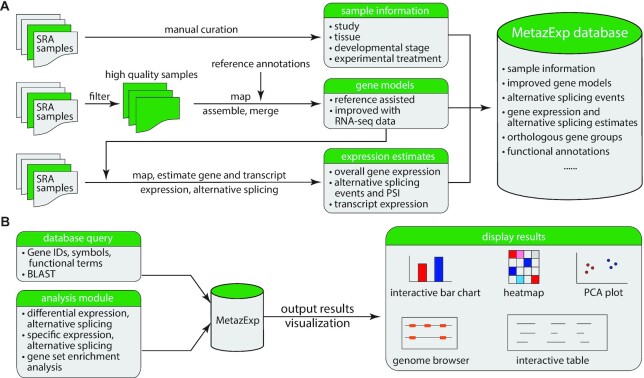
Figure 1.
Overview of MetazExp. (A) Data processing outline and structure of the database. (B) Searching MetazExp and comparative analysis using MetazExp generate a variety of output and visualization to allow better utilization of the database.
MATERIALS AND METHODS
RNA-seq data collection
A total of 72 metazoan species covering 17 orders were contained in MetazExp (Table 1). We queried the SRA, downloading RNA-seq data generated from Illumina platforms due to their dominance and high base-calling accuracy. A total of 53 615 RNA-seq experiments containing ∼175.6 tera bases were collected for construction of the database. These data were derived from 3080 studies covering different strains, genotypes, tissues, developmental stages and experimental conditions (Supplementary Table S1). As expected, the two most represented species were the model organisms Drosophila melanogaster (fruit flies) and Caenorhabditis elegans (nematodes). Other popular species included honeybees, mosquitos, and water fleas. The quality of metadata varied substantially. We therefore further manually curated sample information focusing on strain, genotype, tissue, development stage and experimental conditions based on information embedded in the abstract, description, and publications. The manual curation process consisted of three steps. First, we parsed existing meta data labels programmatically. Second, one reviewer reviewed all existing information and filled in missing information inferred from abstract, study description, and publications. Third, a second reviewer reviewed all previous information by the submitter and by the first reviewer.
Table 1.
Summary of metazoan genomes and RNA-seq experiments collected in MetazExp.
| Class | Species | Annotation database | Volume (GB) | Study | Experiments | Run |
|---|---|---|---|---|---|---|
| Brachiopoda | Lingula anatina | Ensembl | 58.93 | 1 | 16 | 16 |
| Chelicerata | Ixodes scapularis | Ensembl | 484.58 | 25 | 129 | 189 |
| Tetranychus urticae | Ensembl | 529.37 | 17 | 119 | 132 | |
| Cnidaria | Nematostella vectensis | Ensembl | 1373.85 | 32 | 978 | 1463 |
| Coleoptera | Anoplophora glabripennis | Ensembl | 316.34 | 10 | 54 | 55 |
| Dendroctonus ponderosae | Ensembl | 363.05 | 6 | 94 | 94 | |
| Tribolium castaneum | Ensembl | 2217.59 | 46 | 902 | 935 | |
| Crustacea | Daphnia magna | Ensembl | 2941.55 | 27 | 1024 | 1025 |
| Daphnia pulex | Ensembl | 1064.81 | 18 | 237 | 239 | |
| Ctenophora | Mnemiopsis leidyi | Ensembl | 796.45 | 10 | 172 | 185 |
| Diptera | Aedes albopictus | Ensembl | 2911.93 | 44 | 474 | 509 |
| Aedes aegypti | Ensembl | 7121.16 | 109 | 1907 | 1975 | |
| Anopheles arabiensis | Ensembl | 489.54 | 5 | 121 | 121 | |
| Anopheles dirus | Ensembl | 156.24 | 7 | 23 | 23 | |
| Anopheles funestus | Ensembl | 179.12 | 5 | 24 | 25 | |
| Anopheles gambiae | Ensembl | 3616.64 | 99 | 691 | 855 | |
| Anopheles merus | Ensembl | 169.08 | 4 | 44 | 45 | |
| Anopheles minimus | Ensembl | 36.76 | 2 | 11 | 11 | |
| Anopheles sinensis | Ensembl | 51.70 | 3 | 10 | 10 | |
| Anopheles stephensi | Ensembl | 594.51 | 23 | 139 | 152 | |
| Culex quinquefasciatus | Ensembl | 287.13 | 13 | 50 | 50 | |
| Culicoides sonorensis | Ensembl | 174.36 | 3 | 38 | 38 | |
| Drosophila ananassae | Ensembl | 319.16 | 12 | 215 | 330 | |
| Drosophila grimshawi | Ensembl | 262.72 | 1 | 199 | 303 | |
| Drosophila melanogaster | Ensembl | 70067.54 | 1158 | 25 672 | 27 751 | |
| Drosophila mojavensis | Ensembl | 509.99 | 12 | 169 | 234 | |
| Drosophila pseudoobscura | Ensembl | 2043.96 | 26 | 496 | 554 | |
| Drosophila sechellia | Ensembl | 282.70 | 17 | 107 | 109 | |
| Drosophila simulans | Ensembl | 1292.55 | 47 | 443 | 455 | |
| Drosophila virilis | Ensembl | 359.79 | 22 | 180 | 238 | |
| Drosophila yakuba | Ensembl | 600.69 | 26 | 168 | 227 | |
| Glossina austeni | Ensembl | 63.64 | 1 | 4 | 4 | |
| Glossina brevipalpis | Ensembl | 75.25 | 1 | 8 | 8 | |
| Glossina fuscipes | Ensembl | 53.89 | 1 | 6 | 6 | |
| Glossina morsitans | Ensembl | 530.46 | 20 | 136 | 136 | |
| Glossina pallidipes | Ensembl | 143.84 | 2 | 22 | 22 | |
| Glossina palpalis | Ensembl | 176.74 | 2 | 22 | 22 | |
| Lucilia cuprina | Ensembl | 253.58 | 2 | 22 | 22 | |
| Lutzomyia longipalpis | Ensembl | 161.39 | 5 | 61 | 70 | |
| Mayetiola destructor | Ensembl | 233.13 | 4 | 31 | 31 | |
| Musca domestica | Ensembl | 719.12 | 17 | 142 | 142 | |
| Phlebotomus papatasi | Ensembl | 234.55 | 3 | 154 | 154 | |
| Stomoxys calcitrans | Ensembl | 135.60 | 1 | 7 | 7 | |
| Teleopsis dalmanni | Ensembl | 67.67 | 2 | 18 | 18 | |
| Echinodermata | Strongylocentrotus purpuratus | Ensembl | 1044.15 | 16 | 294 | 295 |
| Hemiptera | Acyrthosiphon pisum | Ensembl | 2068.17 | 38 | 442 | 442 |
| Bemisia tabaci | Refseq | 828.28 | 22 | 196 | 202 | |
| Cimex lectularius | Ensembl | 269.11 | 8 | 35 | 35 | |
| Nilaparvata lugens | Ensembl | 140.50 | 12 | 31 | 31 | |
| Rhodnius prolixus | Ensembl | 289.87 | 8 | 40 | 40 | |
| Hymenoptera | Apis mellifera | Ensembl | 11022.15 | 142 | 2345 | 2457 |
| Nasonia vitripennis | Ensembl | 263.80 | 11 | 86 | 128 | |
| Solenopsis invicta | Ensembl | 873.55 | 17 | 210 | 230 | |
| Bombus terrestris | Ensembl | 991.26 | 19 | 321 | 321 | |
| Isoptera | Zootermopsis nevadensis | Ensembl | 392.02 | 6 | 71 | 73 |
| Lepidoptera | Bombyx mori | Ensembl | 6725.34 | 121 | 959 | 981 |
| Heliconius melpomene | Ensembl | 936.32 | 12 | 155 | 156 | |
| Helicoverpa armigera | Refseq | 712.46 | 16 | 114 | 119 | |
| Melitaea cinxia | Ensembl | 654.89 | 4 | 432 | 643 | |
| Plutella xylostella | Refseq | 716.55 | 18 | 88 | 89 | |
| Mollusca | Crassostrea gigas | Ensembl | 2696.20 | 45 | 828 | 901 |
| Biomphalaria glabrata | Ensembl | 476.87 | 9 | 127 | 127 | |
| Octopus bimaculoides | Ensembl | 323.12 | 3 | 117 | 208 | |
| Myriapoda | Strigamia maritima | Ensembl | 220.68 | 4 | 13 | 13 |
| Brugia malayi | Ensembl | 1601.12 | 13 | 249 | 250 | |
| Caenorhabditis elegans | Ensembl | 34256.93 | 588 | 9225 | 10 084 | |
| Onchocerca volvulus | Ensembl | 104.13 | 2 | 21 | 21 | |
| Pristionchus pacificus | Ensembl | 475.57 | 12 | 204 | 205 | |
| Strongyloides ratti | Ensembl | 112.70 | 4 | 22 | 22 | |
| Platyhelminthes | Schistosoma mansoni | Ensembl | 2653.00 | 31 | 1143 | 1144 |
| Porifera | Amphimedon queenslandica | Ensembl | 178.09 | 6 | 298 | 341 |
| Rotifera | Adineta vaga | Ensembl | 74.16 | 2 | 10 | 10 |
| Total | 17 5623 | 3080 | 53 615 | 58 558 |
Gene model improvement
The genome sequences and reference annotations of 69 and 3 species were obtained from the Ensembl (10) and RefSeq (11) databases, respectively (Table 1). Except for a few well annotated genomes, the annotations of most species were largely incomplete. For example, in 33 species, no alternatively spliced transcripts were annotated in multi-exon genes (Supplementary Table S2). We adopted a previously described procedure (8) to improve genome annotation and obtain a uniform annotation for each species against which mapping will be performed. Briefly, high coverage RNA-seq data curated manually were mapped to the reference genome using HISAT2 (12). We defined high coverage, high quality data as those that were paired end, at least 100 bp in read length, at least 50% unique mapping rate and at least a certain sequencing depth. The requirement for sequencing depth varied depending on data availability in each species. In Drosophila melanogaster, we required that at least 4G bases were sequenced. The alignments produced by HISAT2 were assembled into reference guided gene models in GTF format using StringTie2 (13). The resulting GTFs were compared iteratively with the merged GTF using cuffcompare (1). In each iteration, novel multi-exonic transcripts that were at least 200 bp long with at least 2x coverage per transcript and 1× per exon for all exons were merged to the GTF. Finally, all unannotated transcripts must occur in at least three experiments and account for at least 50% experiments of any tissue or at least one-third of all experiments.
The improvement of gene models was substantial. Splice junctions and exons on average increased by 11.62% and 17.49% respectively relative to reference annotations (Supplementary Table S2). The average proportion of multi-exonic genes with alternatively spliced transcript isoforms increased from 8.97% to 33.39% (Supplementary Table S2). The average number of isoforms per multi-exonic gene increased from 1.2 to 1.63.
Estimation of gene expression
RNA-seq reads from all experiments were then aligned to the improved reference annotation for each species using HISAT2, after which StringTie2 was used to estimate gene expression levels. Both transcripts per million (TPM) and fragments per kilo base per million mapped reads (FPKM) were obtained and can be chosen by users for custom analyses.
Calling alternative splicing events and estimating PSI
rMATS (14) was used to call five classic alternative splicing types including alternative 5′ splice sites (A5SS), skipped exon (SE), mutually exclusive exons (MXE), retained intron (RI), and alternative 3′ splice sites (A3SS). SE generally was the most abundant type, accounting for 29.79% on average. Among the identified alternative splicing events, 88.62% were derived from gene annotations while 11.38% were novel and discovered from read alignments by rMATS. PSI (percentage spliced in) was used to quantify alternative splicing. We considered PSI based on counts of junction reads only (JC) and counts of both junction and exonic reads (JCEC) as reported by rMATS.
Orthologous gene group identification and functional annotations
To explore conservation of gene expression and alternative splicing, we identified orthologous gene groups based on the longest protein sequences of genes using orthofinder (15). Functional annotations of genes were obtained by two approaches depending on data sources. For species from Ensembl, the gene ontology terms were obtained directly from the reference annotations. For RefSeq genomes, Blast2GO was used to obtain gene ontology annotations (16,17). Interproscan was used to obtain protein families and conservative domains (18,19). Protein sequences were submitted to KAAS (KEGG automatic annotation server) to compare against the manually curated KEGG GENES database. KAAS returned KO (KEGG Orthology) assignments and generated KEGG pathways (20).
Differential expression and alternative splicing analyses
An important feature MetazExp offers is the capability to select samples from different experimental conditions (tissues, developmental stages, stress treatments) and compare their gene expression and alternative splicing profiles. Two commonly used differential expression methods, DESeq2 (21) and edgeR (22), were used to compare overall gene expression while statistical models implemented in rMATS were used to compare alternative splicing. When possible (study not completely confounded with treatment), batch effects due to studies were adjusted using DESeq2 or edgeR. In addition to comparing expression and alternative splicing across two groups of samples, condition specific (e.g. tissue specific) expression or alternative splicing was identified if a gene's expression or PSI was higher/lower than all other conditions.
Enrichment analyses
MetazExp implements a flexible enrichment analysis procedure based on the R package ClusterProfiler (23), including the hypergeometric test and the Gene Set Enrichment Analysis (GSEA) (24). Both gene ontology (GO) and KEGG pathways were tested for enrichment.
In summary, we manually curated the metadata of the diverse RNA-seq samples, implemented a wide variety of popular analytical and visualization tools, making MetazExp a versatile platform to efficiently utilize public RNA-seq data in metazoan.
RESULTS
Querying the database
MetazExp is hosted at https://bioinfo.njau.edu.cn/metazExp. There are nine popular metazoan species on the front page to allow quick access. Additional species can be easily accessed by navigating through an interactive searchable table listing all species. For each species, MetazExp provides five access points to utilize the resource, including the summary, search, blast, comparison and specificity pages.
In the summary page for each species, users can obtain an overview of the data and download expression data for each experiment. An interactive table is provided to display study and experimental information with links to download expression data from MetazExp and view metadata at SRA.
There are two ways to initiate a query to the database. In the search page, users can search for genes by gene identifiers, symbols, Pfam and GO annotations (Figure 2A) or by listing genes in pathways (Figure 2B). Alternatively, genes can be searched by sequence similarity in the blast page (Figure 2C), which is useful when looking for orthologous genes. The search result is displayed in a concise interactive table containing basic information for the genes with links to external databases to further expand gene expression information (Figure 2D).
Figure 2.
Accessing MetazExp. MetazExp can be accessed through two primary querying methods. (A) On the search page, the search box contains multiple text search options to look for specific genes, genes in a protein family, genes within a gene ontology term or a KEGG pathway. (B) An example is shown for the pathway search result, where the circadian rhythm KEGG pathway in Drosophila is displayed. Red boxes indicate genes that are present in the database. (C) Alternatively, MetazExp can be accessed by blast searching a nucleotide or protein sequence. (D) Search result is displayed in an interactive table with links to external databases and to MetazExp to retrieve expression information.
Visualizing gene expression information
MetazExp contains rich information on the diversity of overall gene expression and alternative splicing for each gene across many experimental conditions in SRA that we manually curated. Database searches based on keyword text and sequence similarity, analysis of differential or specific expression can all result in an interactive table, which contains several identifying details and links to gene expression information.
The gene expression page contains several sections. First, basic information of the gene is listed at the top of the page, including genome position, gene symbol, orthology and various functional annotations such as Pfam, GO and KEGG pathways, all with links to external databases. Notably, users can open a popup window to explore orthologous gene expression in other species, an important feature that is uncommon in other databases. Second, a genome browser was implemented to allow users to explore the gene, transcripts and alternative splicing in its genomic context (Figure 3A). Third, a functional structure graph is displayed to illustrate positions of Pfam domains. Fourth, gene expression or alternative splicing across samples were displayed by a hierarchical and interactive bar chart (Figure 3B). Each bar represents an experimental group in the bar chart and can be further expanded to show the diversity of expression among the same treatment group. As TPM and FPKM are roughly independent of sequence coverage, the bar chart offers a quick and approximate visualization of relative expression between and within treatment groups. Finally, each gene expression page contains two tables respectively to list associated transcripts and alternative splicing events with links to show further details. Importantly, the impact of alternative splicing events can be visualized with their relative positions to protein domains.
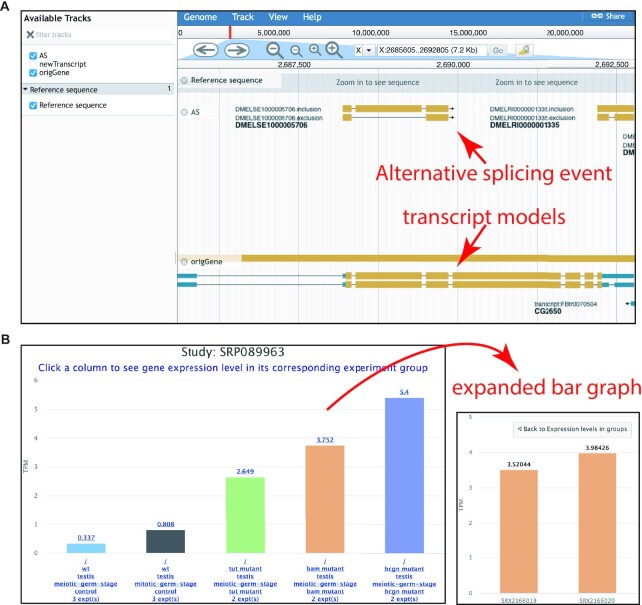
Figure 3.
Visualizing data in the MetazExp. (A) Genome browser showing alternative splicing events and transcript models in the gene per in Drosophila. (B) Interactive bar chart showing gene expression differences across different genotypes and conditions. The bar chart can be clicked to expand to visualize expression in replicates (experiments).
Differential and specific expression analysis
A key feature of MetazExp is its ability to run analyses comparing experimental groups. In the comparison page, users can select RNA-seq experiments of interest to perform differential expression or alternative splicing analysis. We implemented and allowed users to choose several popular methods including the DESeq2 and edgeR for differential gene expression and MATS_LRT, rMATS_unpaired, rMATS_paried for differential splicing analyses. In addition, the hypergeometric test and the GSEA were implemented on MetazExp to analyze functional enrichments for clusters of genes. As these analyses take time (generally within 20 min), users will be asked to provide an email address to confirm submission and receive notification of completion and retrieve results. To help the users understand what the analyses do, we provide an example result page to show actual result from an example dataset. Additional instructions and details about the database and analyses can be found on the Help and FAQ pages.
In the result page, there are three sections to display the differential expression analysis. The first section is a principal component analysis (PCA) and a heat map illustrating clustering of samples based on global gene expression. The second section contains two bar charts for GO and pathway enrichment analysis on the differentially expressed genes. Finally, the differentially expressed genes are listed in an interactive table including key analytical results such as mean expression levels in treatment groups, fold-change and Q-value. Users can click on links to open new pages to explore the details of gene expression. Similar visualizations are also produced for differential alternative splicing analysis.
In addition to pair-wise comparison of gene expression, MetazExp also allows users to identify genes with condition specific high and low expression. Users must select at least 4 experimental groups from which all pair-wise comparisons are made to discover condition specific expression, defined as genes whose expression was higher or lower than all other conditions. Similar to differential expression or alternative splicing analyses, the results are presented with various forms of visualization and summary including a PCA plot and a heatmap, a bar chart summarizing genes in each group, tables listing enriched GO terms and KEGG pathways and all specifically expressed or alternatively spliced genes in an interactive table. Importantly, all results including tables and graphs can be downloaded in a single tarball.
A case study to explore tissue-specific gene expression and alternative splicing in silkworm
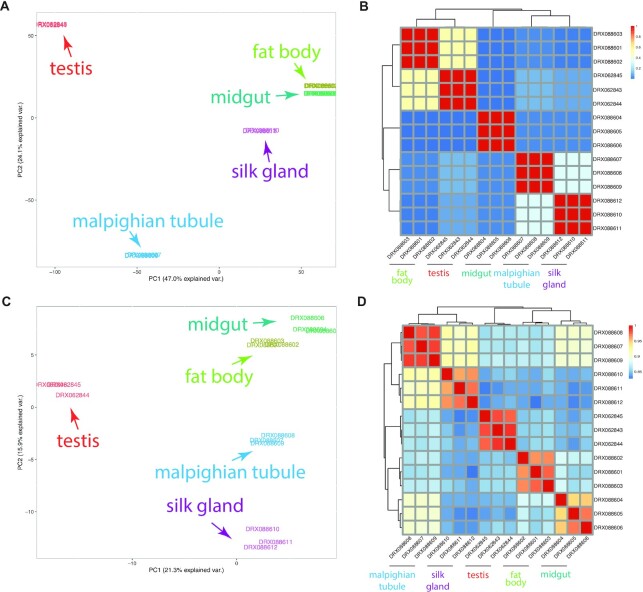
To illustrate the power of MetazExp, we present here a case study to explore tissue-specific gene expression and alternative splicing in Bombyx mori (silkworm) based on a published RNA-seq study (SRA bioproject accession DRP003401) (25). The entire analysis with 15 samples, three replicates for each of five tissues including testis, midgut, fat body, Malpighian tubule and silk gland, completed in 8 min on the server. We retrieved the result page following the link sent by email from the server. The samples were well clustered into five groups respectively corresponding to the five tissues based on both gene expression (Figure 4A and B) and alternative splicing profiles (Figure 4C and D), suggesting that both overall gene expression and alternative splicing profiles contained signatures of tissue specific patterns.
Figure 4.
Case study using the MetazExp. A tissue specific expression and alternative splicing analysis was performed with RNA-seq data in the SRA using MetazExp. (A) PCA plot of overall gene expression for 15 samples covering five tissues in silkworms. (B) Hierarchical clustering based on overall gene expression. (C Same data but plotted for PCA of alternative splicing. (D) Hierarchical clustering based on alternative splicing (PSI).
DESeq2 and MATS_LRT with default parameters were used to detect specifically expressed genes and alternative splicing events. A total of 7409 tissue-specifically highly or lowly expressed genes and 72 tissue-specifically alternative splicing events were identified. In these genes, testis-specific highly or lowly expressed genes and alternative splicing events were the most frequent. MetazExp reported an important glycolytic enzyme gene, BmEno2 (the corresponding ID is BGIBMGA002337 in Ensembl), which was specifically expressed in testis with an average FPKM value of 271.356. This result was reported and confirmed by RT-PCR in the study that initially produced these RNA-seq data (25).
Hypergeometry test with default parameters were used to perform enrichment analysis of specifically expressed and alternatively spliced genes. We found the specifically expressed genes in five tissues were enriched on 61 GO terms and 43 KEGG pathways. The enrichment analysis results revealed functional differences in tissue-specifically expressed genes (Supplementary Tables S3 and S4). The tissue-specifically spliced genes were only enriched on two GO terms and one KEGG pathway (Supplementary Tables S5 and S6). These tissue-specifically expressed and spliced genes as well as related analyses, not reported by the original study (25) that generated the RNA-seq data, may generate new testable hypotheses involved in silkworm growth and development.
CONCLUSIONS
In summary, we have shown that MetazExp is by far the most comprehensive database and analysis platform for gene expression analysis to date. It allows users to search gene expression and alternative splicing profiles and perform analyses comparing treatment groups, and provides various visualizations to facilitate exploration of complex datasets. Thus, MetazExp may serve as an important hypothesis generating and data exploratory engine for further functional studies.
DATA AVAILABILITY
All data are available at: https://bioinfo.njau.edu.cn/metazExp, and all codes available at: https://github.com/qgg-lab/metazExp-pipeline.
Supplementary Material
ACKNOWLEDGEMENTS
We acknowledge the works of all the genome and RNA-seq data producers.
Contributor Information
Jinding Liu, College of Information Management, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China; Research Center for Correlation of Domain Knowledge, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China; Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA.
Fei Yin, Research Center for Correlation of Domain Knowledge, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China.
Kun Lang, College of Information Management, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China; Research Center for Correlation of Domain Knowledge, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China.
Wencai Jie, State Key Laboratory of Pharmaceutical Biotechnology, School of Life Sciences, Nanjing University, Nanjing, Jiangsu 210023, China.
Suxu Tan, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA.
Rongjing Duan, Academy for Advanced Interdisciplinary Studies, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China.
Shuiqing Huang, College of Information Management, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China; Research Center for Correlation of Domain Knowledge, Nanjing Agricultural University, Nanjing, Jiangsu 210095, China.
Wen Huang, Department of Animal Science, Michigan State University, East Lansing, MI 48824, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundamental Research Funds for the Central Universities [KYXK2021006 to S.H.]; USDA Hatch Project [MICL02560 to W.H.]; Michigan State University (to W.H.). Funding for open access charge: Michigan State University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L.. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010; 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lachmann A., Torre D., Keenan A.B., Jagodnik K.M., Lee H.J., Wang L., Silverstein M.C., Ma’ayan A.. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018; 9:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Collado-Torres L., Nellore A., Kammers K., Ellis S.E., Taub M.A., Hansen K.D., Jaffe A.E., Langmead B., Leek J.T.. Reproducible RNA-seq analysis using recount2. Nat. Biotechnol. 2017; 35:319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M.et al.. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013; 41:D991–D995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wilks C., Zheng S.C., Chen F.Y., Charles R., Solomon B., Ling J.P., Imada E.L., Zhang D., Joseph L., Leek J.T.et al.. recount3: summaries and queries for large-scale RNA-seq expression and splicing. 2021; bioRxiv doi:23 May 2021, preprint: not peer reviewed 10.1101/2021.05.21.445138. [DOI] [PMC free article] [PubMed]
- 6. Wang Q., Armenia J., Zhang C., Penson A.V., Reznik E., Zhang L., Minet T., Ochoa A., Gross B.E., Iacobuzio-Donahue C.A.et al.. Unifying cancer and normal RNA sequencing data from different sources. Sci Data. 2018; 5:180061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tapial J., Ha K.C.H., Sterne-Weiler T., Gohr A., Braunschweig U., Hermoso-Pulido A., Quesnel-Vallieres M., Permanyer J., Sodaei R., Marquez Y.et al.. An atlas of alternative splicing profiles and functional associations reveals new regulatory programs and genes that simultaneously express multiple major isoforms. Genome Res. 2017; 27:1759–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu J., Tan S., Huang S., Huang W.. ASlive: a database for alternative splicing atlas in livestock animals. BMC Genomics. 2020; 21:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Z., Zhang Y., Bush S.J., Tang C., Chen L., Zhang D., Urrutia A.O., Lin J.W., Chen L.. MeDAS: a metazoan developmental alternative splicing database. Nucleic Acids Res. 2021; 49:D144–D150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Howe K.L., Contreras-Moreira B., De Silva N., Maslen G., Akanni W., Allen J., Alvarez-Jarreta J., Barba M., Bolser D.M., Cambell L.et al.. Ensembl Genomes 2020-enabling non-vertebrate genomic research. Nucleic Acids Res. 2020; 48:D689–D695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. O’Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D.et al.. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016; 44:D733–D745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L.. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019; 37:907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kovaka S., Zimin A.V., Pertea G.M., Razaghi R., Salzberg S.L., Pertea M.. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019; 20:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shen S., Park J.W., Lu Z.X., Lin L., Henry M.D., Wu Y.N., Zhou Q., Xing Y.. rMATS: robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:E5593–E5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Emms D.M., Kelly S.. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019; 20:238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Conesa A., Gotz S.. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics. 2008; 2008:619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gene Ontology C. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021; 49:D325–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G.et al.. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014; 30:1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kanehisa M., Furumichi M., Sato Y., Ishiguro-Watanabe M., Tanabe M.. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021; 49:D545–D551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Robinson M.D., McCarthy D.J., Smyth G.K.. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010; 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yu G., Wang L.G., Yan G.R., He Q.Y.. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015; 31:608–609. [DOI] [PubMed] [Google Scholar]
- 24. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kikuchi A., Nakazato T., Ito K., Nojima Y., Yokoyama T., Iwabuchi K., Bono H., Toyoda A., Fujiyama A., Sato R.et al.. Identification of functional enolase genes of the silkworm Bombyx mori from public databases with a combination of dry and wet bench processes. BMC Genomics. 2017; 18:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available at: https://bioinfo.njau.edu.cn/metazExp, and all codes available at: https://github.com/qgg-lab/metazExp-pipeline.