Abstract
LncRNAWiki, a knowledgebase of human long non-coding RNAs (lncRNAs), has been rapidly expanded by incorporating more experimentally validated lncRNAs. Since it was built based on MediaWiki as its database system, it fails to manage data in a structured way and is ineffective to support systematic exploration of lncRNAs. Here we present LncRNAWiki 2.0 (https://ngdc.cncb.ac.cn/lncrnawiki), which is significantly improved with enhanced database system and curation model. In LncRNAWiki 2.0, all contents are organized in a structured manner powered by MySQL/Java and curators are able to submit/edit annotations based on the curation model that includes a wider range of annotation items. Moreover, it is equipped with popular online tools to help users identify lncRNAs with potentially important functions, and provides more user-friendly web interfaces to facilitate data curation, retrieval and visualization. Consequently, LncRNAWiki 2.0 incorporates a total of 2512 lncRNAs and 106 242 associations for disease, function, drug, interacting partner, molecular signature, experimental sample, CRISPR design, etc., thus providing a comprehensive and up-to-date resource of functionally annotated lncRNAs in human.
INTRODUCTION
LncRNAWiki, officially released in 2015, is a knowledgebase that harnesses community collective knowledge in collecting, editing and annotating human lncRNAs (1). Considering the increasing volume of human lncRNAs accumulated over the past several years (1–7), LncRNAWiki has been rapidly developed by incorporating a growing collection of functional lncRNAs and their annotations (8–11). Currently, >2000 experimentally validated human lncRNAs have been curated (compared to <100 lncRNAs in 2015) in LncRNAWiki, providing up-to-date knowledge of human lncRNAs for the global scientific community.
Despite these efforts, LncRNAWiki was originally built based on MediaWiki and accordingly has severe limitations on managing structured data and achieving customized functionalities, together making it unfeasible to support in-depth exploration of molecular functions and biological roles of lncRNAs. Here, we present an updated version, LncRNAWiki 2.0 (https://ngdc.cncb.ac.cn/lncrnawiki) that is greatly improved with enhanced database system and curation model (Table 1). Specifically, it is re-built based on MySQL/Java and thus is capable to organize all contents in a structured manner. Moreover, it is significantly updated by adopting a new curation model that incorporates a wider range of annotation items. Curation quality is well controlled by recruiting experts to review the curation results and allowing any user to report errors. Based on the new curation model and structured database system, LncRNAWiki 2.0 incorporates a larger number of human lncRNAs and their associations with experimental evidence and also provides several online tools for lncRNA ID conversion, sequence search and function prediction. Different from existing knowledgebases with specific focuses on disease association (e.g. LncRNADisease (12), LncR2metasta (13) and Lnc2Cancer (14)), interaction (e.g. NPInter (15), LncCeRBase (16), LncRNA2Target (17), LncACTdb (18) and LncTarD (19)), biological function (e.g. LncTarD (19) and LncR2metasta (13)), clinical information (e.g. D-lnc (20) and LncR2metasta (13)), subcellular localization (e.g. RNALocate (21) and LncSLdb (22)) and development (e.g. Dynamic-BM (23)), LncRNAWiki 2.0, coupled with community curation, includes more comprehensive annotation items in terms of conservation, experimental sample, clinical information, biological function, molecular signature, regulator, target and CRISPR design (Table S1).
Table 1.
Comparison between LncRNAWiki 2.0 and 1.0
| Item | Version 2.0 | Version 1.0 | |
|---|---|---|---|
| Curation | Curation model | 10 sections/41 subjects | NA |
| Submission/Edit | Yes | Yes | |
| Error report | Yes | NA | |
| Review | Yes | NA | |
| Data | Functional lncRNAs | 2512 | 86 |
| Disease associations | 13 395 | NA | |
| Functional associations | 12 650 | NA | |
| Molecular signatures | 18 840 | NA | |
| Interacting partners | 4093 | NA | |
| Biological contexts | 10 | NA | |
| Drugs | 1065 | NA | |
| CRISPR design | 587 sgRNAs | NA | |
| Tool | Function prediction | In silico prediction | NA |
| ID conversion | Yes | NA | |
| BLAST | Yes | Yes | |
| System | Database | MySQL/Java | MediaWiki/PHP |
| Search/Browse | Enhanced by different data items | By lncRNA | |
| Statistics | Yes | NA | |
| Download | Customized download | Yes | |
IMPLEMENTATION
LncRNAWiki 2.0 was implemented based on Spring Boot (https://spring.io/projects/spring-boot/), MySQL (https://www.mysql.com) and Apache Tomcat Server (https://tomcat.apache.org). Web interfaces were developed by HTML5, CSS3, AJAX (Asynchronous JavaScript and XML), JQuery and Bootstrap (https://getbootstrap.com). Meanwhile, data visualization was rendered by HighCharts (https://www.highcharts.com.cn), ECharts (https://echarts.apache.org), Plotly.js (https://plotly.com) and DataTables (https://datatables.net). Web tools were set up by HTML widgets, NCBI BLAST+ and R packages, including plumber (https://www.rplumber.io/), ggplot2 (http://had.co.nz/ggplot2/) and clusterProfiler (24).
CURATION MODEL
LncRNAWiki 2.0 provides community curation functionality with standardized curation model and friendly web interfaces for information submission, edit, review and error report (Figure 1). Basically, registered users are regarded as curators (including community curators as well as expert curators), and are allowed to submit and edit/update annotations of newly reported or existing lncRNAs. Most importantly, based on controlled vocabularies and descriptive terms, we built a standardized curation model involving 41 subjects, which can be classified into 10 sections, namely, basic information, publication, conservation, experimental sample, clinical information, biological function, molecular signature, regulator, target, and CRISPR design. To ensure high-quality curation, expert curators are recruited to review and check these submissions/edits, and only reviewed annotations with literature support can be incorporated into LncRNAWiki 2.0. Notably, any user could report errors in the lncRNA page, which can be achieved conveniently just by clicking on ‘Report’ in each section without registration. When any error report is sent out, LncRNAWiki 2.0 is able to automatically notify expert curators to review and check the reported error, with the aim to ensure curation quality in LncRNAWiki 2.0 that is significantly improved in contrast to the previous version.
Figure 1.

Community curation workflow of LncRNAWiki 2.0. Based on the standardized curation model, registered users submit/edit/update lncRNA annotations according to published literature, which will be checked and reviewed by expert curators. On the other hand, to ensure curation quality, any user is allowed to report errors in the lncRNA page without registration, which will also be checked and revised by expert curators.
DATA INTEGRATION AND CURATION
Based on the informative curation model that consists a series of essential annotation items for lncRNAs and their associations as mentioned above, LncRNAWiki 2.0 is able to provide a comprehensive picture of experimentally validated and functionally annotated lncRNAs from diverse aspects. Toward this end, LncRNAWiki 2.0 provides a large collection of lncRNA knowledge curated from published literatures and integrated from existing databases, including RNALocate (21), NPInter (15), LncRNADisease (12), Lnc2Cancer (14), LncSLdb (22), Dynamic-BM (23), LncReg (25), D-lnc (20), lncRNA2Target (17), LncCeRBase (16), LncR2metasta (13), EWAS Atlas (26), LncACTdb (18), LncTarD (19) and CRISPRlnc (27) as well as LncRNAWiki 1.0 (1). Furthermore, we adopt the following procedures to ensure high-quality annotations are incorporated in LncRNAWiki 2.0: (i) unify the lncRNA names with the gene symbol-alias conversion table from HGNC (2021.4.23 version) (28); (ii) standardize the associated vocabularies (e.g. disease names, tissue/cell line names, experimental methods) to merge annotations derived from different sources; (iii) exclude the retracted publications (due to paper mill) and their annotations; (iv) remove the redundant and questionable/controversial annotations and (v) correct any other errors. As a result, LncRNAWiki 2.0 incorporates a larger collection of 2512 experimentally studied lncRNAs (in contrast to 86 lncRNAs in version 1), and integrates a wider range of 106 242 associations for disease (13 395), function (12 650), drug (1065), interacting partner (4093), molecular signature (18 840), experimental sample (49 691), CRISPR design (587), etc.
Based on these comprehensive annotations, LncRNAWiki 2.0 enables users to obtain an up-to-date landscape of literature-reported lncRNAs as well as their reported frequency (Figure 2). Notably, among the 2512 lncRNAs, several lncRNAs have been documented by a large number of publications, and the top 20 extensively studied lncRNAs with more publications are MALAT1, HOTAIR, H19, MEG3, UCA1, PVT1, NEAT1, GAS5, CDKN2B-AS1, XIST, TUG1, KCNQ1OT1, HULC, PCAT1, CCAT1, CYTOR, DANCR, SNHG1, HOTTIP and AFAP1-AS1 (Figure 2A). Among them, H19, MEG3, NEAT1, GAS5, HOTAIR, PCAT1 and KCNQ1OT1 are ubiquitously expressed and have been studied in multiple biological contexts; MALAT1, H19, MEG3, HOTAIR and PVT1 are involved in a variety of diseases; HOTAIR, H19, MALAT1, MEG3 and GAS5 are revealed to be sensitive to different drugs; MALAT1, H19, HOTAIR, MEG3, UCA1 and XIST function in diverse biological processes or pathways and tend to play quite essential roles. At the same time, HOTAIR, MALAT1, NEAT1, MEG3, UCA1 and H19 are found to interact with a large number of targets and are regulated by many different regulators. Even though, these lncRNAs still deserve systematic and in-depth studies based on known information to deepen our knowledge of lncRNAs.
Figure 2.
Extensively studied lncRNAs and frequently surveyed items. (A) Top 20 extensively studied lncRNAs by publication count. These lncRNAs reach out top 20 with larger number of total publications. For each subject, counts of lncRNA association items are normalized and presented as blue or red, indicating low or high counts. (B) Top 3 frequently surveyed items by lncRNA count.
In addition, frequently surveyed biological contexts, diseases, drugs, primary regulators, targets and extensively investigated biological functions could be easily retrieved and accessed (Figure 2B). According to the current knowledge in LncRNAWiki 2.0, frequently surveyed biological contexts are subcellular localization, disease and trait; frequently surveyed diseases are liver cancer, leukemia and colorectal cancer; and frequently surveyed drugs are panobinostat, cisplatin and docetaxel, ELAVL1, STAT3 and EWSR1 are found to be frequently surveyed primary regulators and miR-106a-3p, EZH2 and CDKN1A are primary targets of lncRNAs in human. Additionally, the top 3 closely associated pathways are PI3K/AKT signaling pathway, Wnt/beta-catenin signaling pathway and MAPK signaling pathway; the top 3 closely associated biological processes are cell growth, apoptosis and epithelial-mesenchymal transition; and the top 3 closely associated functional mechanisms are ceRNA, transcriptional regulation and epigenetic regulation. More detailed statistics are publicly available at https://ngdc.cncb.ac.cn/lncrnawiki/statistics.
DATA MANAGEMENT
To enable community curation and provide associated functionalities, LncRNAWiki 2.0 is implemented based on MySQL/Java and significantly improved in database system to facilitate data management and analysis. Friendly web interfaces are accordingly developed to ease data management, including data submission, edit, review, error report, as well as browse, search, download and statistics. Specifically, the submission functionality is provided for community curation (as mentioned above), in which registered users could submit data and annotations for any lncRNA(s) of interest. As a result, all lncRNAs and their associations are summarized and presented as a tabular format, which could be browsed by preset groups or with customized filters and easily exported in csv format in the browse page. Additionally, detailed annotations for each lncRNA are presented in a structured manner and publicly accessible and downloadable. Meanwhile, lncRNAs and their associations could be retrieved through global search in the homepage of LncRNAWiki as well as the browse page by specifying any keyword (e.g. tissue/cell line, drug, target/regulator, PMID). Based on the well-structured curation model and powered by the enhanced database system, the update version of LncRNAWiki presents a series of data management web interfaces for all collected lncRNAs and annotations.
ONLINE ANALYSIS TOOLS
In addition, LncRNAWiki 2.0 is equipped with online analysis tools for data analysis, including function prediction, ID conversion and BLAST (https://ngdc.cncb.ac.cn/lncrnawiki/tool). It can ease users to convert the IDs from one database to other databases and investigate the lncRNA sequence similarity by BLAST. Particularly, to facilitate users to identify lncRNAs with potentially important functions, function prediction is achieved by associating with interacting partners, which are derived from manual curation and co-expressed genes sourced from LncExpDB (7).
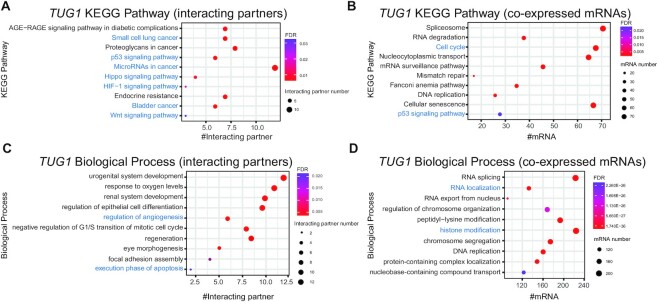
Take TUG1 for example, function prediction based on curated interacting partners and co-expressed mRNAs indicates that TUG1 may be involved in many other pathways and biological processes (Figure 3). Annotations derived from LncTarD (19), Lnc2Cancer (14), LncR2metasta (13), LncCeRBase (16) and literature curation show that TUG1 is associated with 19 pathways and 24 biological processes (https://ngdc.cncb.ac.cn/lncrnawiki/lncrna?symbol=TUG1). Among these annotations, eight pathways (small cell lung cancer, p53 signaling pathway, microRNAs in cancer, hippo signaling pathway, HIF-1 signaling pathway, bladder cancer, Wnt signaling pathway and cell cycle) are captured by function prediction based on curated interacting partners (Figure 3A) and co-expressed mRNAs (Figure 3B). Moreover, TUG1 is predicted to be associated with other pathways such as proteoglycans in cancer pathway, spliceosome and RNA degradation (Figure 3A and B). On the other hand, four manually curated biological processes (apoptosis, angiogenesis, RNA localization, and histone modification) are yielded by function prediction (Figure 3C and D). Also, TUG1 is predicted to participate in many other biological processes such as response to oxygen levels, regulation of epithelial cell differentiation, RNA export, and chromosome organization (Figure 3C and D). Therefore, LncRNAWiki 2.0 can greatly facilitate users to explore potential function for any given lncRNA.
Figure 3.
In silico prediction of TUG1 biological functions. KEGG pathways are predicted based on curated interacting partners (targets and regulators) (A) and co-expressed mRNAs (B). Also, biological processes are predicted based on curated interacting partners (targets and regulators) (C) and co-expressed mRNAs (D). It is noted that pathways/biological processes in blue have been experimentally validated.
DISCUSSION AND FUTURE DEVELOPMENTS
In this study, we present an updated release of LncRNAWiki 2.0, which is significantly updated by enhanced curation model and database system. It features standardized curation model, multifaceted quality control, comprehensive and up-to-date lncRNA knowledge, user-friendly web interfaces and powerful online analysis functionalities. Compared to the previous version, it is more convenient to provide community-curated annotations and organize all contents in terms of different topics. At the same time, curation quality is well controlled with expert curator review and community error report. Most importantly, LncRNAWiki 2.0 houses a large number of 2512 lncRNAs with 106 242 associations supported by 7703 publications, and thus provides a comprehensive and up-to-date landscape of experimentally validated human lncRNAs and their annotations. Moreover, function prediction results provide more insights into the investigation of lncRNAs’ molecular functions and biological roles.
As an important resource of the National Genomics Data Center (11), LncRNAWiki 2.0, in close partnership with LncBook (https://ngdc.cncb.ac.cn/lncbook) (6) and LncExpDB (https://ngdc.cncb.ac.cn/lncexpdb) (7), is devoted to serving as an open-access, community-contributed resource of human lncRNAs. Therefore, future directions include frequent curation of newly published articles, improvement of the curation model by adding more annotation items, and optimization of web interfaces to be friendlier and more interactive. Considering multi-omics analysis as a powerful strategy to characterize functional lncRNAs and elucidate the potential molecular mechanisms (29), we plan to develop tools for multi-omics annotation and visualization of lncRNAs. Given that there are many expert lncRNA databases covered by RNAcentral (30), we also expect to collaborate with RNAcentral members to standardize and integrate diverse lncRNA annotations. Meanwhile, we sincerely invite worldwide scientists, particularly authors with recent publications, to participate in community curation by providing annotations for any lncRNA of interest, with the aim to build LncRNAWiki 2.0 into a valuable resource covering more comprehensive lncRNAs and their annotations and thus to provide high-quality curated knowledge for lncRNA research.
DATA AVAILABILITY
LncRNAWiki 2.0 is freely available online at https://ngdc.cncb.ac.cn/lncrnawiki.
Supplementary Material
ACKNOWLEDGEMENTS
We thank a number of users for reporting bugs and providing suggestions as well as Zhuojing Fan for her valuable help on web interface design.
Contributor Information
Lin Liu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Zhao Li, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Chang Liu, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Dong Zou, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China.
Qianpeng Li, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Changrui Feng, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Wei Jing, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Sicheng Luo, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China; Sino-Danish College, University of Chinese Academy of Sciences, Beijing 100049, China.
Zhang Zhang, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
Lina Ma, National Genomics Data Center & CAS Key Laboratory of Genome Sciences and Information, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China; China National Center for Bioinformation, Beijing 100101, China; University of Chinese Academy of Sciences, Beijing 100049, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDINGS
Strategic Priority Research Program of the Chinese Academy of Sciences [XDB38030400, XDA19050302]; Youth Innovation Promotion Association of Chinese Academy of Sciences [2019104]; National Natural Science Foundation of China [32030021, 31871328]. Funding for open access charge: Strategic Priority Research Program of the Chinese Academy of Sciences.
Conflict of interest statement. None declared.
REFERENCES
- 1. Ma L., Li A., Zou D., Xu X., Xia L., Yu J., Bajic V.B., Zhang Z.. LncRNAWiki: harnessing community knowledge in collaborative curation of human long non-coding RNAs. Nucleic Acids Res. 2015; 43:D187–D192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Derrien T., Johnson R., Bussotti G., Tanzer A., Djebali S., Tilgner H., Guernec G., Martin D., Merkel A., Knowles D.G.et al.. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. 2012; 22:1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Volders P.J., Verheggen K., Menschaert G., Vandepoele K., Martens L., Vandesompele J., Mestdagh P.. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015; 43:D174–D180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Iyer M.K., Niknafs Y.S., Malik R., Singhal U., Sahu A., Hosono Y., Barrette T.R., Prensner J.R., Evans J.R., Zhao S.et al.. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015; 47:199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhao Y., Li H., Fang S., Kang Y., Wu W., Hao Y., Li Z., Bu D., Sun N., Zhang M.Q.et al.. NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res. 2016; 44:D203–D208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma L., Cao J., Liu L., Du Q., Li Z., Zou D., Bajic V.B., Zhang Z.. LncBook: a curated knowledgebase of human long non-coding RNAs. Nucleic Acids Res. 2019; 47:D128–D134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z., Liu L., Jiang S., Li Q., Feng C., Du Q., Zou D., Xiao J., Zhang Z., Ma L.. LncExpDB: an expression database of human long non-coding RNAs. Nucleic Acids Res. 2021; 49:D962–D968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. BIG Data Center Members The BIG Data Center: from deposition to integration to translation. Nucleic Acids Res. 2017; 45:D18–D24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. BIG Data Center Members Database Resources of the BIG Data Center in 2018. Nucleic Acids Res. 2018; 46:D14–D20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. BIG Data Center Members Database Resources of the BIG Data Center in 2019. Nucleic Acids Res. 2019; 47:D8–D14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. National Genomics Data Center Members and Partners Database Resources of the National Genomics Data Center in 2020. Nucleic Acids Res. 2020; 48:D24–D33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bao Z., Yang Z., Huang Z., Zhou Y., Cui Q., Dong D. LncRNADisease 2.0: an updated database of long non-coding RNA-associated diseases. Nucleic Acids Res. 2019; 47:D1034–D1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S., He X., Zhang R., Deng W.. LncR2metasta: a manually curated database for experimentally supported lncRNAs during various cancer metastatic events. Brief Bioinform. 2021; 22:bbaa178. [DOI] [PubMed] [Google Scholar]
- 14. Gao Y., Shang S., Guo S., Li X., Zhou H., Liu H., Sun Y., Wang J., Wang P., Zhi H.et al.. Lnc2Cancer 3.0: an updated resource for experimentally supported lncRNA/circRNA cancer associations and web tools based on RNA-seq and scRNA-seq data. Nucleic Acids Res. 2021; 49:D1251–D1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Teng X., Chen X., Xue H., Tang Y., Zhang P., Kang Q., Hao Y., Chen R., Zhao Y., He S.. NPInter v4.0: an integrated database of ncRNA interactions. Nucleic Acids Res. 2020; 48:D160–D165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pian C., Zhang G., Tu T., Ma X., Li F.. LncCeRBase: a database of experimentally validated human competing endogenous long non-coding RNAs. Database (Oxford). 2019; 2019:baz090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng L., Wang P., Tian R., Wang S., Guo Q., Luo M., Zhou W., Liu G., Jiang H., Jiang Q.. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res. 2019; 47:D140–D144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang P., Li X., Gao Y., Guo Q., Wang Y., Fang Y., Ma X., Zhi H., Zhou D., Shen W.et al.. LncACTdb 2.0: an updated database of experimentally supported ceRNA interactions curated from low- and high-throughput experiments. Nucleic Acids Res. 2019; 47:D121–D127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhao H., Shi J., Zhang Y., Xie A., Yu L., Zhang C., Lei J., Xu H., Leng Z., Li T.et al.. LncTarD: a manually-curated database of experimentally-supported functional lncRNA-target regulations in human diseases. Nucleic Acids Res. 2020; 48:D118–D126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jiang W., Qu Y., Yang Q., Ma X., Meng Q., Xu J., Liu X., Wang S.. D-lnc: a comprehensive database and analytical platform to dissect the modification of drugs on lncRNA expression. RNA Biol. 2019; 16:1586–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang T., Tan P., Wang L., Jin N., Li Y., Zhang L., Yang H., Hu Z., Zhang L., Hu C.et al.. RNALocate: a resource for RNA subcellular localizations. Nucleic Acids Res. 2017; 45:D135–D138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wen X., Gao L., Guo X., Li X., Huang X., Wang Y., Xu H., He R., Jia C., Liang F.. lncSLdb: a resource for long non-coding RNA subcellular localization. Database (Oxford). 2018; 2018:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui Y., Chen X., Niu Y., Wang D., Luo H., Fan Z., Wang D., Wu W., Teng X., He S.et al.. Dynamic-BM: multispecies Dynamic BodyMap database from temporal RNA-seq data. Brief. Bioinform. 2018; 19:1302–1309. [DOI] [PubMed] [Google Scholar]
- 24. Yu G., Wang L.G., Han Y., He Q.Y.. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012; 16:284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou Z., Shen Y., Khan M.R., Li A.. LncReg: a reference resource for lncRNA-associated regulatory networks. Database (Oxford). 2015; 2015:bav083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M., Zou D., Li Z., Gao R., Sang J., Zhang Y., Li R., Xia L., Zhang T., Niu G.et al.. EWAS Atlas: a curated knowledgebase of epigenome-wide association studies. Nucleic Acids Res. 2019; 47:D983–D988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen W., Zhang G., Li J., Zhang X., Huang S., Xiang S., Hu X., Liu C.. CRISPRlnc: a manually curated database of validated sgRNAs for lncRNAs. Nucleic Acids Res. 2019; 47:D63–D68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Braschi B., Denny P., Gray K., Jones T., Seal R., Tweedie S., Yates B., Bruford E.. Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res. 2019; 47:D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Q., Li Z., Feng C., Jiang S., Zhang Z., Ma L.. Multi-omics annotation of human long non-coding RNAs. Biochem. Soc. Trans. 2020; 48:1545–1556. [DOI] [PubMed] [Google Scholar]
- 30. RNAcentral Consortium RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res. 2021; 49:D212–D220. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LncRNAWiki 2.0 is freely available online at https://ngdc.cncb.ac.cn/lncrnawiki.