Abstract
PhylomeDB is a unique knowledge base providing public access to minable and browsable catalogues of pre-computed genome-wide collections of annotated sequences, alignments and phylogenies (i.e. phylomes) of homologous genes, as well as to their corresponding phylogeny-based orthology and paralogy relationships. In addition, PhylomeDB trees and alignments can be downloaded for further processing to detect and date gene duplication events, infer past events of inter-species hybridization and horizontal gene transfer, as well as to uncover footprints of selection, introgression, gene conversion, or other relevant evolutionary processes in the genes and organisms of interest. Here, we describe the latest evolution of PhylomeDB (version 5). This new version includes a newly implemented web interface and several new functionalities such as optimized searching procedures, the possibility to create user-defined phylome collections, and a fully redesigned data structure. This release also represents a significant core data expansion, with the database providing access to 534 phylomes, comprising over 8 million trees, and homology relationships for genes in over 6000 species. This makes PhylomeDB the largest and most comprehensive public repository of gene phylogenies. PhylomeDB is available at http://www.phylomedb.org.
INTRODUCTION
The evolutionary history of a group of homologous genes (i.e. a gene family) is best established through phylogenetic analysis, which results in representations of homology relationships between the gene's sequences (i.e. multiple sequence alignments), and of the inferred pattern of their past divergence (i.e. a phylogenetic tree) (1). Phylogenetic trees and multiple sequence alignments can be analyzed to delineate homology, orthology and paralogy relationships between genes or domains (2). Distinguishing orthologs (i.e. homologous genes that diverged from speciation events) from paralogs (homologous genes that diverged from duplication events) has important implications for comparing genomes across species, predicting the function of newly annotated genes, or reconstructing species relationships (3). In addition, gene trees serve to uncover relevant past evolutionary events such as gene duplication, loss or horizontal transfer, or to reveal footprints of purifying and directional selection, among others. In addition, when analyzing genome-wide collections of gene trees (i.e. a phylome) in a collective manner rather than individually, then evolutionary events at the genome and organismal level can be inferred, such as past hybridizations and polyploidizations (4–6). Similarly, by searching for specific patterns among gene trees in a phylome, the set of genes undergoing duplication or positive selection in a specific lineage can be identified, which in turn can serve to derive hypotheses about the genomic changes underlying a given evolutionary innovation (7,8). These and other applications underscore the usefulness of genome-wide collections of phylogenetic trees. However, building large collections of phylogenetic trees is computationally costly and complex in design, and many researchers with modest expertise in bioinformatics or phylogenetic reconstruction methods benefit from precomputed gene phylogenies present in public repositories, such as Ensembl, PANTHER, EGGNOG or PhylomeDB (9–12).
PhylomeDB (Phylome database) is a knowledgebase of evolutionary relationships between protein-coding genes, represented in the form of annotated phylogenetic trees and multiple sequence alignments, which are reconstructed through standardized, state-of-the-art phylogenetic pipelines. One unique feature of PhylomeDB is that its phylogenetic reconstruction pipeline uses a gene-centric approach in which each gene encoded in the genome of interest (seed genome) is sequentially used as a seed in a phylogenetic reconstruction pipeline, which reproduces the procedures a phylogeneticist would perform to reconstruct the evolutionary history of a given gene (see below for the description of the pipeline). This gene-centric approach circumvents several of the problems associated with defining gene families. By nature, gene families are inherently hierarchical, and diversify in complex ways due to gene duplication and loss (3). Alternative approaches define families by clustering a network of pairwise relations to identify densely connected sub-networks that cannot represent the actual hierarchy present in the data (2). PhylomeDB’s gene-centric approach overcomes this step and therefore results in a collection of evolutionary histories, each one taken from the perspective of a single gene. This collection fully covers the genes encoded in the seed genome and is partially redundant, with a given evolutionary (i.e. a speciation, or a gene duplication) event likely captured in several trees. Such redundancy, in turn, allows the use of consistency-based approaches in downstream evolutionary analyses, such as the detection of duplications (13), and the inference of orthology and paralogy relationships (14).
Beyond reconstructing comprehensive collections of gene trees and alignments, which can be browsed and mined, PhylomeDB annotates them with functional and evolutionary information. Firstly, protein sequences are annotated with respect to their protein domain composition using HMMER searches (15) to protein domains in PFAM (16), and GO functions through Uniprot crosslinking (17). Secondly, phylogenetic trees are automatically processed (see below) to provide a root and annotate internal nodes, which are labelled as speciation or duplication events according to the species-overlap algorithm (2). In addition, sequences at terminal nodes are annotated for taxonomy, gene and protein name, as well as other available annotations provided by source databases, if available. Finally, PhylomeDB provides orthology and paralogy calls based on the most up-to-date release of consistency-based predictions from the MetaPhOrs server (14), which extends homology relationships to over 6000 species. Phylogenetic trees, alignments, sequences and other associated information can be searched, browsed and displayed interactively. In addition, entire collections (phylomes) and relevant tables (i.e. orthology and paralogy) can be downloaded through an FTP server for bulk downstream processing. Since its first release in 2006 (18), PhylomeDB has been continuously expanded and has been enriched with new features, always in accordance with recommendations and standards set by the Quest for Orthologs consortium (19), in which PhylomeDB is a founder and active member. Here we describe the major changes in the current release (version 5) of this resource.
PHYLOMEDB CORE IMPROVEMENTS
Changes to the back end
The PhylomeDB database is currently hosted at the Barcelona Supercomputing Center (BSC-CNS), which is the third institution hosting this resource. Moving such a resource through different institutions and computer systems is not ideal, but reflects the reality of our research group and underscores the commitment of the PhylomeDB team to maintain this resource. The current version of PhylomeDB runs on a Virtual Machine with Ubuntu Linux and 16 GB of memory, where most tasks related to database and web interface operations are carried out. Other computations such as the reconstruction of new phylomes or the annotation of trees and alignments are computed in the Mare Nostrum supercomputer, one of the most powerful supercomputers in Europe. The current back end has been completely updated, with significant software package updates including Apache (version 2.4.18), PHP (version 7.0.15), MariaDB (version 10.2.22), ETE (version 3.1.1) and Python (version 3.7.4). Given the significant expansion of the database, new implementations in the sequence search algorithms have been necessary to reduce response times. Firstly, the faster Diamond v2.0.9.147 (20) algorithm instead of BLAST (21) is used for sequence searches. Secondly, the search jobs are run asynchronously in the Mare Nostrum supercomputer, thereby avoiding virtual machine memory overload.
Newly designed web interface and Improved functionalities
This upgrade of PhylomeDB includes a newly designed web interface, which has been implemented using Drupal (version 7.80), SQLite (version 3.7.3) and several JavaScript libraries, including jQuery (version 1.11.0). The tracking of page visits is provided by Google Analytics. The help and frequently asked questions section have been improved based on user's feedback. To facilitate navigation and access to relevant data, the information on gene trees, orthology predictions and alignments have been combined into a single entry page (see Figure 1), and the FTP section has been completely restructured to improve data accessibility, including direct links to the relevant FTP sections from the relevant parts in the entry pages. New search functionalities include the availability to restrict searches in subsets of the database (i.e. specific phylomes or phylome collections according to user's preferences). Search results tables can be sorted and filtered. Although phylome collections (i.e. subsets of related phylomes) have been available since version 4, this new release includes the possibility of creating user-defined collections. This provides the opportunity to registered users (registration is free) to create their own collections comprising the phylomes of their interest (i.e. those covering their organisms in focus), which streamlines searches and simplifies navigation. Additional efforts have been made to improve the navigation layout for tablets and mobile devices
Figure 1.
(A) Example of the search functionality in PhylomeDB. Users can choose among four different search approaches, shown in different tabs: Search by gene (left-most tab), in which users can search a gene tree by PhylomeID, gene name or external identifiers; Search by sequence (middle-left) in which users can provide a protein sequence which is going to be used for similarity search against sequences in PhylomeDB; Search by phylome (middle-right) in which users can search for specific phylomes among those publicly available; Finally, collection search (right-most) allows users to filter gene and phylome searches to specific collections. (B) Example of the integrated tree visualization showing the gene SHP1 from the yeast Saccharomyces cerevisiae. Several items can be distinguished: The top panel (I) allows the user to switch among available trees, including the ones containing the target protein sequence as seed as well as the ones in which that sequence is present but is not seed (i.e: collateral trees). The tool panel (II) above the tree has multiple elements: it can open a drop-down list of tree features: to interact with during the tree visualization, it can generate a hard link of the tree, download the tree image in .PNG format or download the orthology relationships within the tree in OrthoXML format (48). The tree features pop-up (III) allows the user to change the number of attributes displayed by the image. The search pop-up (IV) allows highlighting specific nodes that match the query term for different categories such as the species name. In addition, clicking on the nodes and leaves will generate a pop-up menu with multiple options such as collapsing nodes, switching sister branches, rerooting, and more. There is also a domain and sequence panel in which PFAM motifs are represented by different shapes, lengths and colors (V). They can be clicked and a direct link will redirect the motif to the original PFAM entry. Finally, the tree legend (VI) indicates the rooting strategy followed in this tree and its classification for the rest of evolutionary events.
Phylogenetic reconstruction, analysis pipelines and benchmarking
At the core of PhylomeDB lies a fully-automated phylogenetic reconstruction pipeline which is regularly updated to keep up to date with recent developments in relevant software tools. In designing this pipeline, we prioritized accuracy over speed. Each phylome is reconstructed over a set of species with available proteomes (here we refer to proteomes as the complete catalogue of sequences of protein-coding genes encoded in a species genome), of which one species - the one in focus - will act as a seed. The PhylomeDB V5 pipeline includes several software updates and technical implementations and proceeds as follows. For each (seed) gene in the seed species a BLAST search against a database containing all phylome proteomes is launched to retrieve a set of proteins with a significant similarity (e-value < 1e–05, continuous overlap over 50% of the query sequence). The number of hits used in further processing is limited to the closest 150 homologs, unless specified otherwise in the phylome description page. Then, a multiple sequence alignment is subsequently constructed using a consistency-based approach. First the set of homologous protein sequences is aligned with three different programs: MUSCLE v3.8.1551 (22), MAFFT v7.407 (23) and KALIGN v2.04 (24). These programs are run with the sequences in forward and reverse orientation, resulting in six different alignments that are combined into a consensus alignment using M-Coffee v12.0 (25). Then this alignment is trimmed using trimAl v1.4.rev15 (26) using a consistency cut-off of 0.1667 and a gap score cut-off of 0.1. The final, trimmed alignment is used to reconstruct a maximum likelihood tree using IQ-Tree v1.6.9 (27) under different models (DCmut, JTTDCMut, LG, WAG, VT, models are explored by default, unless specified otherwise). The final maximum likelihood tree is reconstructed using the best model selected based on the Bayesian information criterion (28). Finally, partition support is calculated using rapid bootstrap (1000 repetitions), as implemented in IQ-Tree. This procedure is iteratively run over all genes of the seed species until a phylome is completed.
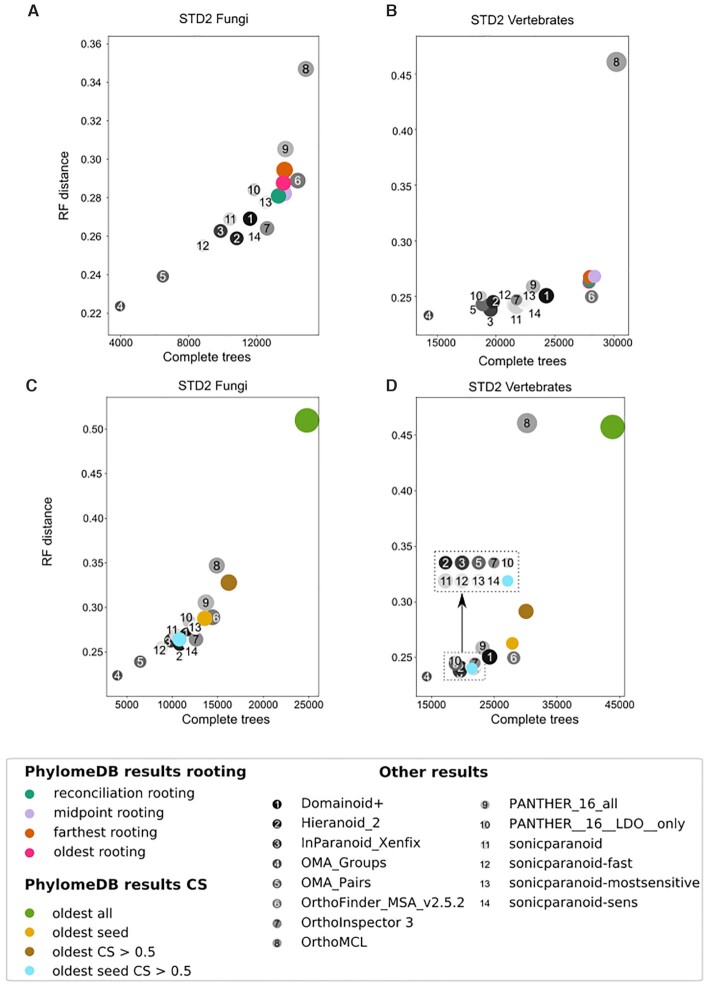
Several automated pipelines are run over entire proteomes or phylomes to precompute relevant features, these include rooting of the trees (either by out-group taxon rooting or midpoint rooting, depending on the phylome), and annotation of internal nodes as speciation or duplication events, as inferred by the species overlap algorithm. The methodology for orthology and paralogy inference has been assessed using the Quest for Orthologs (QFO) 2020 benchmarking service (29). This benchmark is based on a set of 78 selected species, covering the entire tree of life. Given that the size of the test dataset largely exceeds the standard size of phylomes in phylomeDB (12–20 species), we reconstructed 155 phylomes (totalling 1 566 697 trees), to provide a phylogenetic coverage over the species in the benchmark similar to those obtained in a standard phylome analysis. This allowed us to first assess that the changes in the tree reconstruction pipeline did not negatively impact the results as well as to explore different parameters used during orthology prediction. For instance, we studied the effect of using different tree rooting methods, such as taxon out-group rooting, midpoint rooting, reconciliation-based rooting as implemented in Treerecs (30), or rooting at the most distant sequence from the seed. Our results indicated that the four rooting methods provided similar results with respect to orthology predictions for the seed, and therefore we did not alter the rooting algorithm implemented in PhylomeDB (outgroup-based rooting, see Figure 2A and B). In addition, we studied the effects of applying a consistency-based approach as implemented in MetaPhORs (14), or of restricting orthology calls to include sequences used as a seed. Our results (Figure 2C and D) show that these two filters can serve to improve the accuracy of orthology prediction (measured as the ability of selected orthologs to reconstruct an assumed species tree, y-axis), at the cost of the number of predicted orthologs. Overall PhylomeDB produced results in line with other methods, which vary along a similar accuracy versus coverage trade-off trend.
Figure 2.
Plots depicting the results obtained in two test sets of the QFO 2020 benchmark when comparing different approaches to orthology prediction based on phylome information. Graphs represented here correspond to the generalized species tree discordance test (G_STD2) run on the set of fungal (graphs A and C) and vertebrate (graphs B and D) orthology predictions. This test compares a gene tree reconstructed based on the submitted orthology predictions to a pre-computed, binary, species tree. In the x-axis we find the number of completed gene trees obtained from the submitted orthologs and in the y-axis the robinson and foulds (RF) measure that calculates the number of shared bipartitions between the species tree and the gene tree and normalized by the total number of bipartitions in both trees. Graphs A and B compare results obtained using four different rooting methods: in pink rooting to the farthest sequence from an outgroup taxon (oldest), in orange rooting to the leaf that is farthest located from the seed, in cyan rooting based on minimizing the reconciliation cost and in purple using midpoint rooting. Graphs C and D compare different ways to filter orthology predictions. In green are all found orthologous pairs, in brown all orthologous pairs with a consistency score above 0.5, in yellow all possible orthologous pairs involving the seed protein, and in blue all orthologous pairs involving the seed and with a consistency score above 0.5. Grey coloured dots represent results obtained by other methods and were extracted from the QFO public results 2020 (https://orthology.benchmarkservice.org/proxy/). Size of the dots is relative to the number of orthologs in the dataset. Square found in graph D indicates which sets of data are found in the region as they overlap.
Dataset expansion and community project support
PhylomeDB v5 provides evolutionary computations for >23 million proteins (compared to ∼10 million in previous release), over 8 million trees (as opposed to 1.5 millions in v4) and 534 public phylomes (roughly a 4-fold increase since 2014). In addition, integration with MetaPhOrs (14) provides consistency-based orthology and paralogy relationships and expands homology relationships in PhylomeDB to over 6000 species. PhylomeDB provides support in the annotation and analysis of newly sequenced genomes, some as part of large-scale initiatives such as i5K (31), 1KFG (1000 Fungal Genomes) (http://1000.fungalgenomes.org/), or ERGA (https://www.erga-biodiversity.eu/). These are community-driven projects that partner with PhylomeDB to perform a phylome reconstruction coupled with the annotation and initial analyses of a newly sequenced genome. In these projects, phylomeDB analyses have proven useful to, among others, (i) identify potential errors in gene annotation based on comparative analyses (split genes, transposable elements, etc.), (ii) provide functional annotation based on annotated functions of orthologs and (iii) identify major genomic changes in the relevant lineages. Finally, the resulting organism-focused phylome constitutes a valuable resource for the research community working in this species (see Table 1 for a representative list of community-driven projects in PhylomeDB).
Table 1.
Representative projects where PhylomeDB has been coupled to annotation and first analysis of newly sequenced genomes
| Species (common name) | Phylomedb ID | Reference |
|---|---|---|
| Plants | ||
| Olea europaea (Olive tree) | 215–222 | (6) |
| Nicotiana benthamiana (Benth, a close relative of tobacco) | 817 | (32) |
| Beta vulgaris (Sugar beet) | 152 | (33) |
| Phaseolus vulgaris (Common bean) | 8–11 | (34) |
| Solanum commersonii (Wild potato) | 147 | (35) |
| Vertebrates | ||
| 48 bird species | 225–230 | (36) |
| Scophthalmus maximus (Turbot) | 18 | (37) |
| Panthera onca (Jaguar) | 583 and 584 | (38) |
| Lynx pardinus (Iberian lynx) | 277 and 278 | (39) |
| Other (invertebrate) animals | ||
| Cinara cedri (Cedar aphid) | 701–706 | (5) |
| Polistes canadensis (Red paper wasp) and other eusocial insects | 134–136 | (40) |
| Strigamia martitima (Centipede) | 177 | (41) |
| Daktulosphaira vitifoliae (Grape phylloxera) | 196 | (42) |
| Mytilus galloprovincialis (Mediterranean mussel) | 787 | (43) |
| Fungi | ||
| Penicillium expansum (Blue mold) | 279–283 | (44) |
| Phycomyces blakesleeanus and Mucor circinelloides | 252–255 | (45) |
| Geotrichum candidum | 233–236 | (46) |
| Candida subhashii | 777 | (47) |
First column indicates the name of the species of interest for the project, the second column lists the phylomeID for the phylomes reconstructed as part of the project and in the third column is the reference to the publication.
DATA AVAILABILITY
PhylomeDB is freely available, without registration at http://phylomedb.org/.
ACKNOWLEDGEMENTS
The authors wish to thank Miguel Angel Naranjo-Ortíz, Laia Carreté, Ernst Thuer, Ismael Collado, Andrés Garisoain, and Giacomo Mutti, as well as other members of Gabaldon's group, the Quest for Orthologs consortium and PhylomeDB users for their suggestions and feedback. The authors thankfully acknowledge the computer resources at MareNostrum and the technical support provided by Barcelona Supercomputing Center (BCV-2021-2-0004).
Contributor Information
Diego Fuentes, Barcelona Supercomputing Centre (BSC-CNS). Jordi Girona 29, 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028 Barcelona, Spain.
Manuel Molina, Barcelona Supercomputing Centre (BSC-CNS). Jordi Girona 29, 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028 Barcelona, Spain.
Uciel Chorostecki, Barcelona Supercomputing Centre (BSC-CNS). Jordi Girona 29, 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028 Barcelona, Spain.
Salvador Capella-Gutiérrez, Barcelona Supercomputing Centre (BSC-CNS). Jordi Girona 29, 08034 Barcelona, Spain.
Marina Marcet-Houben, Barcelona Supercomputing Centre (BSC-CNS). Jordi Girona 29, 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028 Barcelona, Spain.
Toni Gabaldón, Barcelona Supercomputing Centre (BSC-CNS). Jordi Girona 29, 08034 Barcelona, Spain; Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Baldiri Reixac 10, 08028 Barcelona, Spain; Catalan Institution for Research and Advanced Studies (ICREA), Barcelona, Spain.
FUNDING
Spanish Ministry of Science and Innovation (MICINN) [PGC2018-099921-B-I00], cofounded by European Regional Development Fund (ERDF); Catalan Research Agency (AGAUR) [SGR423]; European Union's Horizon 2020 research and innovation programme [ERC-2016–724173]; Gordon and Betty Moore Foundation [GBMF9742]; Instituto de Salud Carlos III [INB Grant PT17/0009/0023 – ISCIII-SGEFI/ERDF]; U.C. was funded in part through H2020 Marie Skłodowska-Curie Actions [H2020-MSCA-IF-2017-793699]; MICINN [IJC2019- 039402-I]. Funding for open access charge: ERC.
Conflict of interest statement. None declared.
REFERENCES
- 1. Gabaldón T. Evolution of proteins and proteomes: a phylogenetics approach. Evol. Bioinform. Online. 2005; 1:117693430500100004. [PMC free article] [PubMed] [Google Scholar]
- 2. Gabaldón T. Large-scale assignment of orthology: back to phylogenetics?. Genome Biol. 2008; 9:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gabaldón T., Koonin E.V.. Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 2013; 14:360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marcet-Houben M., Gabaldón T.. Beyond the whole-genome duplication: phylogenetic evidence for an ancient interspecies hybridization in the baker's yeast lineage. PLoS Biol. 2015; 13:e1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Julca I., Marcet-Houben M., Cruz F., Vargas-Chavez C., Johnston J.S., Gómez-Garrido J., Frias L., Corvelo A., Loska D., Cámara F.et al.. Phylogenomics identifies an ancestral burst of gene duplications predating the diversification of aphidomorpha. Mol. Biol. Evol. 2020; 37:730–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Julca I., Marcet-Houben M., Vargas P., Gabaldón T.. Phylogenomics of the olive tree (Olea europaea) reveals the relative contribution of ancient allo- and autopolyploidization events. BMC Biol. 2018; 16:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fernández R., Gabaldón T.. Gene gain and loss across the metazoan tree of life. Nat. Ecol. Evol. 2020; 4:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernández R., Marcet-Houben M., Legeai F., Richard G., Robin S., Wucher V., Pegueroles C., Gabaldón T., Tagu D.. Selection following gene duplication shapes recent genome evolution in the pea aphid acyrthosiphon pisum. Mol. Biol. Evol. 2020; 37:2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howe K.L., Achuthan P., Allen J., Allen J., Alvarez-Jarreta J., Amode M.R., Armean I.M., Azov A.G., Bennett R., Bhai J.et al.. Ensembl 2021. Nucleic Acids Res. 2021; 49:D884–D891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas P.D., Campbell M.J., Kejariwal A., Mi H., Karlak B., Daverman R., Diemer K., Muruganujan A., Narechania A.. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003; 13:2129–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Huerta-Cepas J., Szklarczyk D., Heller D., Hernández-Plaza A., Forslund S.K., Cook H., Mende D.R., Letunic I., Rattei T., Jensen L.J.et al.. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019; 47:D309–D314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huerta-Cepas J., Capella-Gutiérrez S., Pryszcz L.P., Marcet-Houben M., Gabaldón T.. PhylomeDB v4: zooming into the plurality of evolutionary histories of a genome. Nucleic Acids Res. 2014; 42:D897–D902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huerta-Cepas J., Dopazo H., Dopazo J., Gabaldón T.. The human phylome. Genome Biol. 2007; 8:R109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chorostecki U., Molina M., Pryszcz L.P., Gabaldón T.. MetaPhOrs 2.0: integrative, phylogeny-based inference of orthology and paralogy across the tree of life. Nucleic Acids Res. 2020; 48:W553–W557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Finn R.D., Clements J., Eddy S.R.. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011; 39:W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mistry J., Chuguransky S., Williams L., Qureshi M., Salazar G.A., Sonnhammer E.L.L., Tosatto S.C.E., Paladin L., Raj S., Richardson L.J.et al.. Pfam: the protein families database in 2021. Nucleic Acids Res. 2021; 49:D412–D419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J., O’Donovan C.. The GOA database: gene ontology annotation updates for 2015. Nucleic Acids Res. 2015; 43:D1057–D1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huerta-Cepas J., Bueno A., Dopazo J., Gabaldón T.. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008; 36:D491–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Linard B., Ebersberger I., McGlynn S.E., Glover N., Mochizuki T., Patricio M., Lecompte O., Nevers Y., Thomas P.D., Gabaldón T.et al.. Ten years of collaborative progress in the quest for orthologs. Mol. Biol. Evol. 2021; 38:3033–3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buchfink B., Xie C., Huson D.H.. Fast and sensitive protein alignment using DIAMOND. Nat. Methods. 2015; 12:59–60. [DOI] [PubMed] [Google Scholar]
- 21. Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J.. Basic local alignment search tool. J. Mol. Biol. 1990; 215:403–410. [DOI] [PubMed] [Google Scholar]
- 22. Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Katoh K. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005; 33:511–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lassmann T., Sonnhammer E.L.L.. Kalign–an accurate and fast multiple sequence alignment algorithm. BMC Bioinformatics. 2005; 6:298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wallace I.M., O’Sullivan O., Higgins D.G., Notredame C.. M-Coffee: combining multiple sequence alignment methods with T-Coffee. Nucleic Acids Res. 2006; 34:1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Capella-Gutierrez S., Silla-Martinez J.M., Gabaldon T.. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009; 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen L.-T., Schmidt H.A., Haeseler A., Minh B.Q.. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwarz G. Estimating the dimension of a model. Ann. Stat. 1978; 6:461–464. [Google Scholar]
- 29. Altenhoff A.M., Garrayo-Ventas J., Cosentino S., Emms D., Glover N.M., Hernández-Plaza A., Nevers Y., Sundesha V., Szklarczyk D., Fernández J.M.et al.. The Quest for Orthologs benchmark service and consensus calls in 2020. Nucleic Acids Res. 2020; 48:W538–W545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Comte N., Morel B., Hasić D., Guéguen L., Boussau B., Daubin V., Penel S., Scornavacca C., Gouy M., Stamatakis A.et al.. Treerecs: an integrated phylogenetic tool, from sequences to reconciliations. Bioinformatics. 2020; 36:4822–4824. [DOI] [PubMed] [Google Scholar]
- 31. i5K Consortium The i5K initiative: advancing arthropod genomics for knowledge, human health, agriculture, and the environment. J. Hered. 2013; 104:595–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schiavinato M., Marcet-Houben M., Dohm J.C., Gabaldón T., Himmelbauer H.. Parental origin of the allotetraploid tobacco Nicotiana benthamiana. Plant J. 2020; 102:541–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dohm J.C., Minoche A.E., Holtgräwe D., Capella-Gutiérrez S., Zakrzewski F., Tafer H., Rupp O., Sörensen T.R., Stracke R., Reinhardt R.et al.. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature. 2014; 505:546–549. [DOI] [PubMed] [Google Scholar]
- 34. Vlasova A., Capella-Gutiérrez S., Rendón-Anaya M., Hernández-Oñate M., Minoche A.E., Erb I., Câmara F., Prieto-Barja P., Corvelo A., Sanseverino W.et al.. Genome and transcriptome analysis of the Mesoamerican common bean and the role of gene duplications in establishing tissue and temporal specialization of genes. Genome Biol. 2016; 17:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aversano R., Contaldi F., Ercolano M.R., Grosso V., Iorizzo M., Tatino F., Xumerle L., Dal Molin A., Avanzato C., Ferrarini A.et al.. The solanum commersonii genome sequence provides insights into adaptation to stress conditions and genome evolution of wild potato relatives. Plant Cell. 2015; 27:954–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T.et al.. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014; 346:1320–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Figueras A., Robledo D., Corvelo A., Hermida M., Pereiro P., Rubiolo J.A., Gómez-Garrido J., Carreté L., Bello X., Gut M.et al.. Whole genome sequencing of turbot (Scophthalmus maximus; Pleuronectiformes): a fish adapted to demersal life. DNA Res. 2016; 23:181–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Figueiró H.V., Li G., Trindade F.J., Assis J., Pais F., Fernandes G., Santos S.H.D., Hughes G.M., Komissarov A., Antunes A.et al.. Genome-wide signatures of complex introgression and adaptive evolution in the big cats. Sci. Adv. 2017; 3:e1700299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Abascal F., Corvelo A., Cruz F., Villanueva-Cañas J.L., Vlasova A., Marcet-Houben M., Martínez-Cruz B., Cheng J.Y., Prieto P., Quesada V.et al.. Extreme genomic erosion after recurrent demographic bottlenecks in the highly endangered Iberian lynx. Genome Biol. 2016; 17:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patalano S., Vlasova A., Wyatt C., Ewels P., Camara F., Ferreira P.G., Asher C.L., Jurkowski T.P., Segonds-Pichon A., Bachman M.et al.. Molecular signatures of plastic phenotypes in two eusocial insect species with simple societies. Proc. Natl. Acad. Sci. U.S.A. 2015; 112:13970–13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chipman A.D., Ferrier D.E.K., Brena C., Qu J., Hughes D.S.T., Schröder R., Torres-Oliva M., Znassi N., Jiang H., Almeida F.C.et al.. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014; 12:e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rispe C., Legeai F., Nabity P.D., Fernández R., Arora A.K., Baa-Puyoulet P., Banfill C.R., Bao L., Barberà M., Bouallègue M.et al.. The genome sequence of the grape phylloxera provides insights into the evolution, adaptation, and invasion routes of an iconic pest. BMC Biol. 2020; 18:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gerdol M., Moreira R., Cruz F., Gómez-Garrido J., Vlasova A., Rosani U., Venier P., Naranjo-Ortiz M.A., Murgarella M., Greco S.et al.. Massive gene presence-absence variation shapes an open pan-genome in the Mediterranean mussel. Genome Biol. 2020; 21:275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ballester A.-R., Marcet-Houben M., Levin E., Sela N., Selma-Lázaro C., Carmona L., Wisniewski M., Droby S., González-Candelas L., Gabaldón T.. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant. Microbe. Interact. 2015; 28:232–248. [DOI] [PubMed] [Google Scholar]
- 45. Corrochano L.M., Kuo A., Marcet-Houben M., Polaino S., Salamov A., Villalobos-Escobedo J.M., Grimwood J., Álvarez M.I., Avalos J., Bauer D.et al.. Expansion of signal transduction pathways in fungi by extensive genome duplication. Curr. Biol. 2016; 26:1577–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Morel G., Sterck L., Swennen D., Marcet-Houben M., Onesime D., Levasseur A., Jacques N., Mallet S., Couloux A., Labadie K.et al.. Differential gene retention as an evolutionary mechanism to generate biodiversity and adaptation in yeasts. Sci. Rep. 2015; 5:11571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mixão V., Hegedűsová E., Saus E., Pryszcz L.P., Cillingová A., Nosek J., Gabaldón T.. Genome analysis of Candida subhashii reveals its hybrid nature and dual mitochondrial genome conformations. DNA Res. 2021; 28:dsab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmitt T., Messina D.N., Schreiber F., Sonnhammer E.L.L.. SeqXML and OrthoXML: standards for sequence and orthology information. Brief. Bioinform. 2011; 12:485–488. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
PhylomeDB is freely available, without registration at http://phylomedb.org/.