Abstract
Mapping gene interactions within tissues/cell types plays a crucial role in understanding the genetic basis of human physiology and disease. Tissue functional gene networks (FGNs) are essential models for mapping complex gene interactions. We present TissueNexus, a database of 49 human tissue/cell line FGNs constructed by integrating heterogeneous genomic data. We adopted an advanced machine learning approach for data integration because Bayesian classifiers, which is the main approach used for constructing existing tissue gene networks, cannot capture the interaction and nonlinearity of genomic features well. A total of 1,341 RNA-seq datasets containing 52,087 samples were integrated for all of these networks. Because the tissue label for RNA-seq data may be annotated with different names or be missing, we performed intensive hand-curation to improve quality. We further developed a user-friendly database for network search, visualization, and functional analysis. We illustrate the application of TissueNexus in prioritizing disease genes. The database is publicly available at https://www.diseaselinks.com/TissueNexus/.
INTRODUCTION
Functional gene networks (FGNs) (1–7) are an essential model for mapping the functional interaction landscape among genes. In an FGN, the node represents a gene and the edge weight indicates the co-functional probability that two genes participate in the same biological pathway (9). The evidence that supports the co-functional relationship of two genes includes diverse interaction data, such as regulatory relationship, coexpression, protein-protein interaction, etc. (1,6,8). Constructed by integrating heterogeneous genomic data, FGNs are a type of composite network, which differs from coexpression or regulatory networks where there is only one type of edges. For example, the edge in a regulatory network represents transcriptional factor-target gene binding. FGNs have been continuously developed and applied successfully to solving fundamental biological and biomedical questions, including novel interaction discovery and disease gene prioritization (1,10–13). Early efforts in this field are devoted to constructing global (i.e. not tissue-wise) gene networks (2). Because gene interactions may be remodeled in different tissues, tissue networks are later constructed, examples of which are included in the GIANT (1), diseaseQUEST (5) and BaiHui databases (8).
In the broader context of human gene networks including coexpression and regulatory networks, RNA-seq data have been successfully applied to network modeling. For example, using the expression data generated by the Genotype-Tissue Expression (GTEx) consortium, coexpression networks for 35 human tissues are constructed, providing a rich resource for understanding gene regulation and function (14). The gene regulatory networks built with the GTEx expression data suggest the tissue specificity of transcriptional control (15). Further, sample-specific regulatory networks are constructed for yeast and lymphoblastoid cell lines (16). A database of sample-specific regulatory networks for human tissues is later developed (17,18). In the area of FGNs, RNA-seq data have also been used but in a limited number of studies. One example is the HumanNet database (13), for which RNA-seq expression data are used to build the network.
For FGNs, existing networks have mainly been built with Bayesian Classifiers (BCs) (1,4,5,8), and a few are built using Bayesian-based likelihood (13). Both methods have the limitation of not considering the dependence among features, and they cannot capture the nonlinearity in real data well (19,20). Because the functional interaction between genes is complex and have nonlinear nature, leveraging more advanced machine learning models may improve the accuracy of networks.
In this work, we improve the construction of human tissue FGNs as follows. First, rather than using BCs, we adopt XGBoost, which is a state-of-the-art machine learning method and can capture nonlinearity in data (21–23), to predict functional interactions between genes. Another reason for choosing XGBoost is that it can be scaled to large datasets; this is important in practice because the number of training samples (gene pairs) is on the order of millions. In addition, because tissue labels of public genomic data may be annotated with different names or be missing, we performed intensive hand-curation of the tissue labels of the input expression data. Thus, we obtained a set of RNA-seq expression datasets for each tissue.
With the proposed approach, we constructed TissueNexus, a compendium of 49 tissue/cell line FGNs. What distinguishes our networks from existing FGNs is summarized in Table 1. The major features of TissueNexus include the following. (i) The tissue labels of the input gene expression data are intensively hand-curated. The curation of a large number of RNA-seq datasets is extremely labor-intensive and has not previously been available. (ii) The networks are constructed by integrating by far the largest number of RNA-seq datasets (n = 1,341) and samples (n = 52,087). (iii) The networks are constructed using an advanced machine learning approach, namely, XGBoost, which can result in more accurate networks as will be shown in the Construction and analysis of TissueNexus networks section.
Table 1.
Comparison of TissueNexus with existing functional gene network databases
| Databases | Tissues | Gene expression technology | Tissue gene expression | #Datasetsa (all/expression) | #Samples | Methods | References |
|---|---|---|---|---|---|---|---|
| mouseNet | Not tissue-wise | Microarray | No | 340/333 | 13,634 | Bayesian | (2) |
| classifier | |||||||
| diseaseQUEST | Multiple tissues | Microarray | No | 174/169 | 2,736 | Bayesian | (5) |
| classifier | |||||||
| GIANT | Multiple tissues | Microarray | No | 987/980 | 38,000 | Bayesian | (1) |
| classifier | |||||||
| HumanNet | Not tissue-wise | Microarray (125 datasets) | No | NAb/158 | 16,220 | Bayesian | (6) |
| RNA-seq (33 datasets) | likelihood | ||||||
| BaiHui | Brain only | Microarray | Yes | 216/213 | 4,688 | Bayesian | (8) |
| classifier | |||||||
| TissueNexus | Multiple tissues | RNA-seq | Yes | 1,345/1,341 | 52,087 | XGBoost | This work |
aIn the Datasets column, all and expression indicate the numbers of all genomic datasets and only the gene expression datasets used to build functional networks in each database, respectively. Expression is the dominant data type used to construct networks.
bThe number of all integrated datasets is not provided in the original paper.
We deployed a web server for users to query genes and visualize functional interactions between the queried gene and its neighbors for each individual tissue. To gain functional insight into the network, we implemented functional analysis tools for the network including Gene Ontology enrichment and disease enrichment. We also provide functional annotations of the queried gene such as annotations of associated diseases/traits and drugs. We illustrate the application of TissueNexus to prioritizing risk genes of complex diseases and demonstrate its better performance over existing networks.
MATERIALS AND METHODS
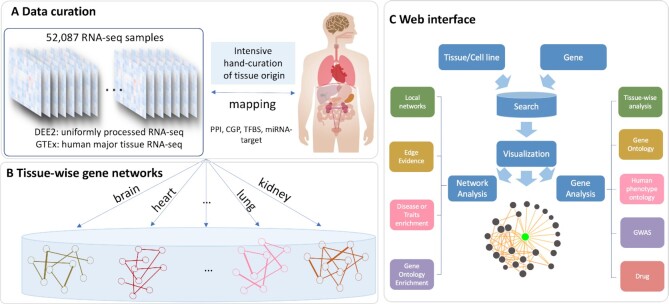
An overview of TissueNexus is presented in Figure 1. The methods employed to construct the networks and the instructions for using the web server are described below.
Figure 1.
Overview of the TissueNexus database. (A) Hand-curation of tissue labels for human RNA-seq samples of the DEE2 database. The gene expression data in the GTEx portal are also included. A total of 1,341 RNA-seq datasets containing 52,087 samples were obtained for 49 tissues or cell lines. (B) Construction of functional gene networks for each tissue or cell line. The networks are stored in the MySql database. (C) Network search, visualization and analysis. After querying a gene, its local network containing top-connected neighbors is obtained and visualized. The web server also provides a series of analyses for the network and the queried gene, including analyses of Gene Ontology enrichment, disease enrichment, evidence supporting the functional relationship of gene pairs, expression level across tissues, annotation of associated diseases/traits, and annotation of associated drugs.
Curation and processing of RNA-seq data for each tissue/cell line
The Digital Expression Explorer 2 (DEE2) (24) is a database of RNA-seq gene expression data generated by applying a unified pipeline to the raw RNA-seq reads in the short read archive (SRA) database (25). In DEE2, the RNA-seq data are organized into datasets, with each dataset containing gene expression measurements of multiple different samples. The human RNA-seq data along with the description of sample information including tissue origins were downloaded. We also downloaded gene expression data of different tissues from the GTEx portal (version:v8, https://gtexportal.org/home/datasets). Gene expression is measured in fragments per kilobase of exon model per million mapped reads (FPKM) values. Because the tissue labels of the expression data may be annotated with different names or be missing, we manually read the descriptions of individual samples and performed intensive hand-curation of the tissue label of the input expression data. Thus, we assigned a tissue or cell line label to each sample. Because the Pearson correlation coefficients (PCCs) of pairs of genes in each dataset need to be calculated as features and PCCs could be spurious if the number of samples is small, we retained only the datasets containing at least 10 samples following the practice applied in the previous work (26). In each dataset, lowly expressed genes (i.e. those with FPKM values < 0.1 in more than 90% samples as used in (26)) were removed. This preprocessing procedure was applied to the data of each tissue. Finally, we obtained RNA-seq gene expression data for 49 tissues. The numbers of the RNA-seq datasets and samples for each tissue are shown in Table 2. The details of these datasets are available at https://diseaselinks.com/TissueNexus/data.php.
Table 2.
The numbers of human RNA-seq datasets and samples integrated for each tissue or cell line
| #Tissues | #Datasets | Proportion of datasets% | #Samples | Proportion of samples% |
|---|---|---|---|---|
| A549 | 13 | 1.0 | 264 | 0.5 |
| Acute lymphoblastic leukemia | 46 | 3.4 | 1250 | 2.4 |
| Acute myeloid leukemia | 20 | 1.5 | 1435 | 2.8 |
| Adipose | 12 | 0.9 | 1856 | 3.6 |
| B lymphocyte | 33 | 2.5 | 768 | 1.5 |
| Bladder | 12 | 0.9 | 254 | 0.5 |
| Blood | 112 | 8.4 | 7619 | 14.6 |
| Bone | 41 | 3.1 | 1383 | 2.7 |
| Bone marrow | 31 | 2.3 | 1210 | 2.3 |
| Brain | 48 | 3.6 | 4459 | 8.6 |
| Breast | 71 | 5.3 | 2477 | 4.8 |
| Bronchial epithelium | 14 | 1.0 | 373 | 0.7 |
| Bronchus | 16 | 1.2 | 501 | 1.0 |
| Chronic myeloid leukemia | 11 | 0.8 | 765 | 1.5 |
| Colon | 57 | 4.3 | 1988 | 3.8 |
| Embryo | 28 | 2.1 | 610 | 1.2 |
| Embryonic stem cell | 30 | 2.2 | 743 | 1.4 |
| Epidermis | 27 | 2.0 | 909 | 1.7 |
| Forebrain | 15 | 1.1 | 540 | 1.0 |
| Frontal cortex | 14 | 1.0 | 630 | 1.2 |
| Glia | 18 | 1.3 | 568 | 1.1 |
| Glioma | 13 | 1.0 | 406 | 0.8 |
| Hct116 | 20 | 1.5 | 375 | 0.7 |
| Heart | 13 | 1.0 | 1621 | 3.1 |
| Hek293 | 28 | 2.1 | 580 | 1.1 |
| Hela | 23 | 1.7 | 430 | 0.8 |
| Intestinal epithelium | 16 | 1.2 | 288 | 0.6 |
| Intestine | 15 | 1.1 | 326 | 0.6 |
| Kidney | 30 | 2.2 | 605 | 1.2 |
| Liver | 35 | 2.6 | 1304 | 2.5 |
| Lung | 49 | 3.7 | 1902 | 3.7 |
| Macrophage | 17 | 1.3 | 446 | 0.9 |
| Mammary gland | 12 | 0.9 | 315 | 0.6 |
| Mcf7 | 35 | 2.6 | 722 | 1.4 |
| Melanocyte | 29 | 2.2 | 962 | 1.8 |
| Neural progenitor | 13 | 1.0 | 302 | 0.6 |
| Neural stem cell | 17 | 1.3 | 560 | 1.1 |
| Neuron | 29 | 2.2 | 683 | 1.3 |
| Non-small cell lung cancer | 31 | 2.3 | 1097 | 2.1 |
| Ovary | 15 | 1.1 | 477 | 0.9 |
| Pancreas | 22 | 1.6 | 873 | 1.7 |
| Prostate | 36 | 2.7 | 915 | 1.8 |
| Serum | 10 | 0.7 | 179 | 0.3 |
| Skin | 55 | 4.1 | 3349 | 6.4 |
| Skin fibroblast | 19 | 1.4 | 371 | 0.7 |
| T lymphocyte | 36 | 2.7 | 1241 | 2.4 |
| Umbilical cord | 15 | 1.1 | 366 | 0.7 |
| Uterine cervix | 26 | 1.9 | 496 | 1.0 |
| Urinary bladder | 13 | 1.0 | 294 | 0.6 |
Construction and analysis of TissueNexus networks
Feature calculation. Genomic features are calculated as input for machine learning approaches to build gene networks. Because we predict pairwise gene-gene relationships, pairwise features must be used. For each RNA-seq dataset, the PCC of gene pairs was calculated and used as a feature. Therefore, the number of expression-based features is equal to that of RNA-seq datasets for each tissue. Taking the liver tissue with 35 RNA-seq datasets as an example, the number of RNA-seq expression features is 35 (Table 2). In addition to expression-based features, we also integrate six pairwise genomic features, which are obtained from the GIANT website (1), including shared 3′ UTR microRNA binding motifs, the cooccurrence of transcription factor binding sites, chemical and genetic perturbations, and three protein-protein interaction (PPI) features from MINT (27), IntAct (28) and BioGRID (29), respectively. For each tissue, gene expression is the dominant feature type used to construct the network (Table 1).
Network construction and performance
For each tissue, the tissue-wise RNA-seq features and the six general interaction features are integrated to build the FGN using XGBoost (Figure 1A; see details in Supplementary Note 1). The evidence supporting the functional relationship of gene pairs comes from both RNA-seq and the general features. Only the expressed genes in each tissue are used to build the network. In the network, the node represents a gene and the edge weight represents the probability (in the range of [0, 1]) that two genes take participate in the same biological process or pathway.
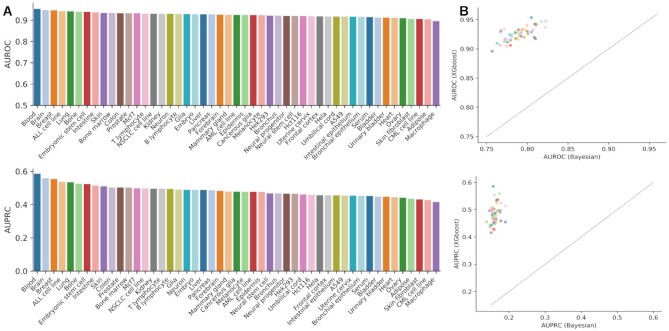
We evaluate the performance of each functional gene network based on 5-fold cross-validation. To avoid overestimation of performance, the cross-validation is conducted based on gene holdout rather than edge holdout; that is, genes are split into 5-fold to make sure that any gene used for training models during cross-validation will not occur in the holdout set. The FGNs based on XGBoost are accurate, with AUROC = 0.9249 ± 0.0119 and AUPRC = 0.4817 ± 0.0355 across all tissues; the AUROC and AUPRC for each network is shown in Figure 2 A. A comparison based on exactly the same data shows that XGBoost outperforms Bayesian classifiers which have AUROC = 0.7893 ± 0.0159 and AUPRC = 0.1612 ± 0.0103 (Figure 2B). Further, we investigated whether the networks achieved high precision at low recall values. We considered three low recall values = 0.05, 0.1 and 0.2. The precisions for each tissue network at these three low values are shown in Supplementary Figure S1. At recall = 0.05, these networks show appreciably high precision values ranging from 0.8625 to 0.9253 across all tissues, with a mean = 0.890 and standard deviation = 0.014. When the recall increases to 0.1 and 0.2, respectively, the precision decreases correspondingly (Supplementary Figure S1). We find that the number of interacting genes of the same gene vary across tissues and the edge weight of the same pair of genes also change from tissue to tissue; most interactions are shared between tissues and some belong to one of the tissues compared (Supplementary Figure S2); this result suggests the importance of tissue context when analyzing functional interactions between genes.
Figure 2.
Performance of tissue functional gene networks. (A) Network performance of each network based on 5-fold cross-validation. The cross-validation is conducted based on gene holdout rather than edge holdout; that is, genes are split into 5-folds to make sure that any gene used for training models during cross-validation will not occur in the holdout set. (B) Comparison of XGBoost with Bayesian classifiers in constructing tissue functional gene networks.
We investigate the influence of feature datasets on tissue functional gene networks. We observe that restricting RNA-seq data to the tissue of interest improves network performance compared to integrating RNA-seq data also from other tissues (Supplementary Figure S3). We test the performance of the network built with only RNA-seq features. The AUROC and AUPRC values across all tissues are 0.7901 ± 0.0237 and 0.1857 ± 0.0295, respectively, which are lower than that based on all features (Supplementary Figure S4). We also observe that, for each tissue, the network performance is correlated with the number of RNA-seq datasets of the tissue of interest (Supplementary Figure S5).
We compare the coverage of the networks with existing human RNA-seq based gene networks, including coexpression networks in GTEx-TSN (14), regulatory networks in GTEx-PANDA (15), and sample-specific regulatory networks in GRAND (17,18). We observe that GTEx-PANDA has higher coverage than GTEx-TSN. For some tissues, the coverages of TissueNexus networks are higher than that in GTEx-PANDA; for the other tissues, GTEx-PANDA networks have higher coverages (Supplementary Table S1). The coverages of GRAND networks vary across individual samples and the mean coverage of each tissue network is roughly on the same order of magnitude of GTEx-PANDA or TissueNexus networks.
Implementation
TissueNexus has been implemented with a mixture of programming languages on the Apache HTTP server (v2.4.41). The HTTPS protocol is deployed on TissueNexus to support secure communication over computer networks. The main web interface is implemented using PhP (v7.4.3) and HTML5. As the full database contains tens of millions of records (gene pairs), the network data of each tissue or cell line are stored in the open-source MySql (v8.0.25) database to make the database robust and scalable. The network of the queried gene is stored in json format, which enables efficient communication between the server and client when coupled with Ajax. The d3 JavaScript package is used to visualize the network of each query gene. A sliding bar is designed to dynamically visualize subnetworks containing only the edge with weights higher than the threshold set by the sliding bar. The queried network in json format is stored as HTTP cookies, thus enabling highly efficient thresholding of the network. In addition, considering the widespread application of mobile devices, the web server is also designed to be mobile-friendly.
DATABASE OVERVIEW
Database content
TissueNexus is a database of FGNs for 49 human tissues or cell lines, which are constructed by integrating hand-curated tissue functional genomic data. The web server provides functions to query functional interactions between genes for each individual tissue, visualize the network, viewing the evidence of each interaction, investigate biological functions, and download the network. The web interface mainly consists of three modules: (i) tissue and gene input, (ii) network visualization and (iii) network and gene analysis, which are summarized in Figure 1 and detailed below.
Tissue and gene input
On the Home page, users can select one of the 49 tissues/cell lines from the pull-down menu. To input gene symbols, the autocomplete function is implemented so that candidate gene symbols will pop up when one or more characters are typed in by users. In addition, we also implemented the function to query multiple genes at a time. Users can input multiple genes separated by a comma. Then, when the Search button is clicked, users will be guided to the result page for the queried gene and network.
Network visualization
On the result page, the local network containing the queried gene and its top-connected neighbors will be shown. For the sake of visualizability, only the top 25 connected genes as well as the between-neighbor connections are shown. Users can use the sliding bar to determine the threshold of the edge weight and only connections with weights higher than the threshold will be displayed.
Network and gene analysis
Functional analysis and annotation are implemented for the network and the queried gene (Figure 1C), as detailed below.
Network analysis
First, the weight of the top neighbors connected to the queried gene is presented. Second, to help users to gain functional insight into the network, Gene Ontology enrichment analysis (GOEA) is performed using GOTermFinder (30). The enrichment analysis is based on the most recent versions of the Gene Ontology database (2021-09-01 version, http://current.geneontology.org/ontology/go-basic.obo) and functional gene annotations (2021-09-01 version, http://geneontology.org/gene-associations/goa_human.gaf.gz). Third, a disease enrichment approach established in our previous work (8) is applied to test whether a disease or trait is overrepresented in the network. To help users understand whether a given edge is supported by RNA-seq based features or other general features or both, we estimated the contribution of each dataset to the functional relationship of gene pairs using a Bayesian approach, of which the details are described on https://humanbase.readthedocs.io/en/latest/functional-networks.html#evidence. The evidence of the edge on each dataset is provided on the web server.
Gene analysis
This panel provides five types of annotations for the queried gene, including expression levels across tissues, annotation in the GO database (http://ftp.ncbi.nlm.nih.gov/gene/DATA/gene2go.gz, 2021-5-21), annotation of human phenotype ontology (Jun 2021 release), annotation of disease/traits based on the GWAS Catalog database (2020-11-20), and annotation of drugs based on DGIdb (2021-May).
An example search
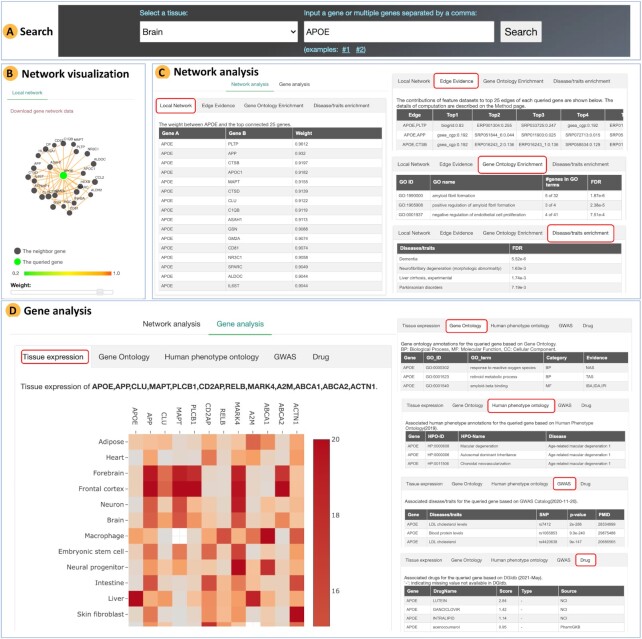
An example of the usage of our database is provided by searching the brain network for APOE, a gene related to multiple diseases/traits, such as blood lipid levels and Alzheimer’s disease (AD) (31–33). An overview of this example search is presented in Figure 3.
Figure 3.
Overview of an example search. The brain network is searched for APOE. (A) Selecting brain tissue and inputting the APOE gene. (B) Visualization of the local network of APOE containing the top-connected genes. (C) Functional analysis of the local network, including Gene Ontology and disease enrichment. (D) Annotation to APOE, including expression level across tissues and annotation of GO terms, phenotypes, diseases/traits, and drugs.
First, we select the brain tissue, and we type in APOE on the Home page (Figure 3A).
Second, on the result page, the local network of APOE containing the top-connected genes is visualized. The edge color indicates the weight between two genes. Users can use the sliding bar below the network to adjust the threshold of edge weight (Figure 3B).
Third, we can investigate the network via the functional analysis implemented on the server. These analyses consist of two parts: network analysis and gene analysis (Figure 3C). The network analysis panel includes submenus. The weight between APOE and each neighbor is shown under the local network menu. For example, MAPT is functionally related to APOE with weight = 0.9155; these two genes are known to interact with each other, supporting the high weight between them. The network is enriched in biological processes such as the amyloid fibril formation (GO:1990000, false discovery rate (FDR) = 1.8 × 10-6), negative regulation of endothelial cell proliferation (GO:0001937, FDR = 7.5 × 10-4). Some of these processes, e.g. amyloid fibril formation, are associated with AD, suggesting that the network is biologically meaningful because APOE is a known genetic risk factor of AD. Based on disease enrichment, it is observed that the network is associated with dementia (FDR = 5.52×10-6) and neurofibrillary degeneration (FDR = 1.63 × 10-3).
The gene analysis panel provides expression profiles across tissues and functional annotation for the queried gene based on publicly available databases. As shown in Figure 3D, APOE is most highly expressed in the liver and is associated with multiple disease/traits such as LDL cholesterol levels. The associated drugs, such as lutein and ganciclovir, are also provided.
Application to prioritizing disease risk genes
Gene networks can be applied to many subsequent analysis, such as gene function prediction, novel interaction discovery, and disease gene prediction, etc. The focus of this work is not the application of networks but presenting a resource of tissue FGNs for the community. Here we illustrate the application of FGNs in prioritizing disease risk genes. We test the performance of our networks in predicting disease risk genes using the approach established in previous work (1,4). Briefly, this method first extracts network weights as features to characterize each gene, and then adopts machine learning methods to build a model to distinguish disease-associated genes (positives) from non-disease genes (negatives) (see the method details in Supplementary Note 2). We obtain disease genes from GWAS and Online Mendelian Inheritance in Man (OMIM). For GWAS, following the previous study (1), the genes achieving genome-wide significance (P < 5.0 ×10-8) in the GWAS Catalog database (downloaded on 2020-11-20) are treated as positives. Negatives are randomly sampled after excluding positives. The tree-based method ExtraTree is used to build the model.
We consider a number of diseases representing a wide spectrum of pathological mechanisms, including cancers (breast cancer (BC), prostate cancer (PC), colorectal cancer (CC), lung cancer (LC), melanoma skin cancer (MSC)), neurological degeneration disease (Alzheimer’s disease (AD), Parkinson disease (AD)), psychiatric disorders (autism spectrum disorder (ASD), major depression disorder (MDD), and schizophrenia (SZ)), heart disease (atrial fibrillation (AF)), and metabolic disorders (type 2 diabetes (T2D) and obesity (OB)). The genes for these diseases are deposited to the Zenodo repository (https://zenodo.org/record/5553579). Based on the above-described method, we evaluate the performance of these networks in predicting disease risk genes using 5-fold cross-validation. During each fold, the features corresponding to the left out genes are also removed to avoid overestimation of the performance. The AUROC and AUPRC are used as the performance metrics. For each disease, a relevant tissue network is selected to construct the feature matrix. For example, because Parkinson’s disease is pathologically rooted in the brain, the brain network is selected. As shown in Figure 4, the networks accurately predict disease risk genes according to AUROC and AUPRC.
Figure 4.
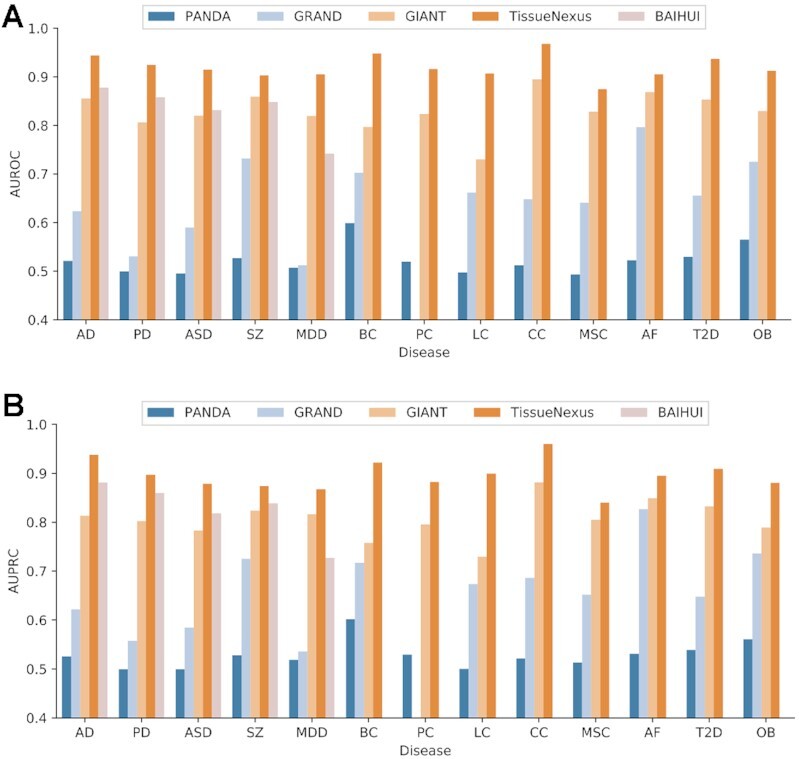
Comparison of TissueNexus to existing tissue gene networks based on their performance in predicting risk genes for different types of diseases. (A) AUROC. (B) AUPRC. (Notes: The BaiHui database contains only a brain gene network and is therefore compared on only brain disorders; For the GRAND database, Because it contains multiple sample-specific networks for each tissue, we calculate the mean and standard deviation of AUROC and AUPRC. The mean of AUROC and AUPRC is presented, with the standard deviation provided in Supplementary Table S2; GRAND does not contain networks for prostate so that prostate cancer gene prediction is not performed on GRAND). Disease abbreviations: AD (Alzheimer’s disease), PD (Parkinson’s disease), ASD (autism spectrum disorder), SZ (schizophrenia), MDD (major depression disorder), BC (breast cancer), PC (prostate cancer), LC (lung cancer), CC: (colon cancer), MSC (melanoma skin cancer), AF (atrial fibrillation), T2D (type 2 diabetes), OB (obesity).
We compare our networks with the tissue FGNs in the GIANT and BaiHui databases, the regulatory networks in the GTEx-PANDA and GRAND databases (diseaseQUEST and mouseNet are not compared because they are not human networks. GTEx-TSN networks are not compared, because their low coverage results in a highly sparse feature matrix; the high sparsity matrix further makes the network not able to predict the disease gene well and makes it unfair to compare with other networks with high coverage. The BaiHui database contains only a brain gene network and is therefore compared only on brain disorders). We find that TissueNexus perform better than GIANT, BaiHui, GTEx-PANDA and GRAND (Figure 4). In addition, we compare these networks using independent test genes from the DisGeNet database (34). Briefly, for these diseases, we obtain additional risk genes from DisGeNet. We find that TissueNexus achieves better performance for most of the diseases (Supplementary Figure S6). Furthermore, we also compare our networks with general integrated networks including PCNet (7), STRING (35) and HumanNet (6). We find that TissueNexus networks achieve overall better performance (Supplementary Figure S7).
We investigate whether disease gene prediction could be improved by using relevant tissue networks. We analyze T2D and AD as case studies. The pancreas and brain networks are used as the disease-relevant tissue networks for T2D and AD, respectively. For T2D, we compare the pancreas network to all other tissue networks. We find that the pancreas network ranks in the first place among all 49 networks (Supplementary Figure S8A), suggesting that the prediction model based on relevant tissues is more accurate than using other tissue networks. For AD, we compared the brain network to all other tissue networks. The brain network ranks in the fourth place (the top 8%) among all 49 networks (Supplementary Figure S8B). Although the brain network does not rank in the first place, the percentile of the rank (8%) is appreciable, especially considering the complexity of AD, the cross talk among tissues, and the noise in genomic data. This analysis suggests that the network of relevant tissues is more informative in predicting risk genes than other tissue networks.
In summary, these results suggest that our networks could be valuable for prioritizing disease risk genes.
DISCUSSION AND FUTURE DIRECTIONS
FGNs represent essential models for mapping the functional interaction landscape among genes. To help understand the functional interaction in different tissues, we built a compendium of FGNs for 49 human tissue-/cell lines. Our comparison shows that the interaction partners of the same gene across different tissues are different and that the weight of the same pair of genes also varies across tissues. This finding indicates the remodeling of functional gene network in human tissues. The differences among these networks could be further explored to understand the molecular basis of tissues or cell types. In a broader context, tissues gene networks have been constructed in other studies, such as the tissue coexpression networks (14), tissue-specific regulatory networks (15), and sample-specific regulatory networks (16). FGNs are different from coexpression or regulatory networks, because the edge in an FGN represents the co-functional probability that two genes participate in the same biological pathway (9). In contrast, the edge in coexpression networks represents the correlation between gene expression profiles and the edge in a regulatory network means transcriptional factor-target gene binding.
Motivated by the study of sample-specific regulatory networks (16–18), a natural and meaningful extension of our work is to build FGNs for individual samples. To this end, the merits of the method used in building sample-specific regulatory networks could be leveraged and novel methods that take into account the specific nature of functional gene networks need to be established. Further, a challenge we would face is how to evaluate the accuracy of sample-specific networks, due partly to the difference for networks from individuals to individuals and the dynamic nature of the networks.
Regarding the application of tissue FGNs to disease gene prediction, we have shown that FGNs are promising for predicting risk genes. However, because the human body is a system and interactions exist between tissues, it is possible that including multiple tissues that are related to the disease could further improve risk gene prediction. We plan to study this question in the future. In addition, it needs to be noted that tissue gene networks-based prediction of disease genes could be affected by multiple factors, including the above-mentioned between-tissue interactions, the noise in genomic data, and the false positives of disease genes. Designing novel approaches to address these issues could potentially improve disease gene prediction and benefit subsequent applications such as drug development.
Our database could be improved in several ways in the future. First, except for some cell line networks, most networks in this database are built for major organs or tissues. As human tissues are composed of heterogeneous cell types that carry out different functions, we plan to extend this work to build cell type networks by integrating single cell RNA-seq data (36–38). Second, while this work focuses on presenting the networks as a rich resource for the community, mining these networks using advanced machine learning approaches may deepen our understanding of gene functions. Such network mining approaches will be implemented in our subsequent work to better exploit the networks.
In summary, we present TissueNexus as a rich resource of 49 tissue-/cell line functional gene networks. We illustrate its application in prioritizing disease risk genes. It is expected that these networks will contribute to the understanding of gene functions and complex diseases, and will become a valuable resource in the field.
DATA AVAILABILITY
The web interface of the database is available at https://www.diseaselinks.com/TissueNexus/. This website is freely accessible to all users, without the need for logging on or a password.
Supplementary Material
Contributor Information
Cui-Xiang Lin, School of Computer Science and Engineering, Central South University, Changsha, Hunan 410083, P.R. China; Hunan Provincial Key Lab on Bioinformatics, Central South University, Changsha, Hunan 410083, P.R. China.
Hong-Dong Li, School of Computer Science and Engineering, Central South University, Changsha, Hunan 410083, P.R. China; Hunan Provincial Key Lab on Bioinformatics, Central South University, Changsha, Hunan 410083, P.R. China.
Chao Deng, School of Computer Science and Engineering, Central South University, Changsha, Hunan 410083, P.R. China; Hunan Provincial Key Lab on Bioinformatics, Central South University, Changsha, Hunan 410083, P.R. China.
Yuanfang Guan, Department of Computational Medicine and Bioinformatics, University of Michigan, Ann Arbor, MI 48109, USA.
Jianxin Wang, School of Computer Science and Engineering, Central South University, Changsha, Hunan 410083, P.R. China; Hunan Provincial Key Lab on Bioinformatics, Central South University, Changsha, Hunan 410083, P.R. China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [U1909208, 61972423, 61732009]; 111 Project [B18059]; Hunan Provincial Science and Technology Program [2018WK4001]. Funding for open access charge: National Natural Science Foundation of China [U1909208].
Conflict of interest statement. None declared.
REFERENCES
- 1. Greene C.S., Krishnan A., Wong A.K., Ricciotti E., Zelaya R.A., Himmelstein D.S., Zhang R., Hartmann B.M., Zaslavsky E., Sealfon S.C.et al.. Understanding multicellular function and disease with human tissue-specific networks. Nat. Genet. 2015; 47:569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Guan Y., Myers C.L., Lu R., Lemischka I.R., Bult C.J., Troyanskaya O.G.. A genomewide functional network for the laboratory mouse. PLoS Comput. Biol. 2008; 4:e1000165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Guan Y., Ackert-Bicknell C.L., Kell B., Troyanskaya O.G., Hibbs M.A.. Functional genomics complements quantitative genetics in identifying disease-gene associations. PLoS Comput. Biol. 2010; 6:e1000991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guan Y., Gorenshteyn D., Burmeister M., Wong A.K., Schimenti J.C., Handel M.A., Bult C.J., Hibbs M.A., Troyanskaya O.G.. Tissue-specific functional networks for prioritizing phenotype and disease genes. PLoS Comput. Biol. 2012; 8:e1002694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yao V., Kaletsky R., Keyes W., Mor D.E., Wong A.K., Sohrabi S., Murphy C.T., Troyanskaya O.G.. An integrative tissue-network approach to identify and test human disease genes. Nat. Biotechnol. 2018; 36:1091–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hwang S., Kim C.Y., Yang S., Kim E., Hart T., Marcotte E.M., Lee I.. HumanNet v2: human gene networks for disease research. Nucleic Acids Res. 2019; 47:D573–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Huang J.K., Carlin D.E., Yu M.K., Zhang W., Kreisberg J.F., Tamayo P., Ideker T.. Systematic evaluation of molecular networks for discovery of disease genes. Cell Systems. 2018; 6:484–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li H.-D., Bai T., Sandford E., Burmeister M., Guan Y.. BaiHui: cross-species brain-specific network built with hundreds of hand-curated datasets. Bioinformatics. 2019; 35:2486–2488. [DOI] [PubMed] [Google Scholar]
- 9. Troyanskaya O.G., Dolinski K., Owen A.B., Altman R.B., Botstein D.. A Bayesian framework for combining heterogeneous data sources for gene function prediction (in Saccharomyces cerevisiae). Proc. Natl. Acad. Sci. U.S.A. 2003; 100:8348–8353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Recla J.M., Robledo R.F., Gatti D.M., Bult C.J., Churchill G.A., Chesler E.J.. Precise genetic mapping and integrative bioinformatics in diversity outbred mice reveals hydin as a novel pain gene. Mamm. Genome. 2014; 25:211–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hu J., Wan J., Hackler L. Jr, Zack D.J., Qian J.. Computational analysis of tissue-specific gene networks: application to murine retinal functional studies. Bioinformatics. 2010; 26:2289–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ata S.K., Wu M., Fang Y., Ou-Yang L., Kwoh C.K., Li X.L.. Recent advances in network-based methods for disease gene prediction. Brief. Bioinform. 2020; 22:bbaa303. [DOI] [PubMed] [Google Scholar]
- 13. Huang X., Liu H., Li X., Guan L., Li J., Tellier L., Yang H., Wang J., Zhang J.. Revealing Alzheimer’s disease genes spectrum in the whole-genome by machine learning. BMC Neurol. 2018; 18:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pierson E.GTEx Consortium GTEx Consortium Koller D., Battle A., Mostafavi S., Ardlie K.G., Getz G., Wright F.A., Kellis M., Volpi S.et al.. Sharing and specificity of co-expression networks across 35 human tissues. PLoS Comput. Biol. 2015; 11:e1004220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sonawane A.R., Platig J., Fagny M., Chen C.Y., Paulson J.N., Lopes-Ramos C.M., DeMeo D.L., Quackenbush J., Glass K., Kuijjer M.L.. Understanding tissue-specific gene regulation. Cell Rep. 2017; 21:1077–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuijjer M.L., Tung M.G., Yuan G., Quackenbush J., Glass K.. Estimating sample-specific regulatory networks. iScience. 2019; 14:226–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lopes-Ramos C.M., Chen C.Y., Kuijjer M.L., Paulson J.N., Sonawane A.R., Fagny M., Platig J., Glass K., Quackenbush J., DeMeo D.L.. Sex differences in gene expression and regulatory networks across 29 human tissues. Cell Rep. 2020; 31:107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ben Guebila M., Lopes-Ramos C.M., Weighill D., Sonawane A.R., Burkholz R., Shamsaei B., Platig J., Glass K., Kuijjer M.L., Quackenbush J.. GRAND: a database of gene regulatory network models across human conditions. Nucleic Acids Res. 2021; gkab778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dojer N., Bednarz P., Podsiadlo A., Wilczynski B.. BNFinder2:faster Bayesian network learning and Bayesian classification. Bioinformatics. 2013; 29:2068–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huttenhower C., Hibbs M., Myers C., Troyanskaya O.G.. A scalable method for integration and functional analysis of multiple microarray datasets. Bioinformatics. 2006; 22:2890–2897. [DOI] [PubMed] [Google Scholar]
- 21. Chen T., Guestrin C.. XGBoost: a scalable tree boosting system. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. 2016; 785–794. [Google Scholar]
- 22. Zheng R., Li M., Chen X., Wu F.X., Pan Y., Wang J.. BiXGBoost: a scalable, flexible boosting-based method for reconstructing gene regulatory networks. Bioinformatics. 2019; 35:1893–1900. [DOI] [PubMed] [Google Scholar]
- 23. Ogunleye A., Wang Q.G.. XGBoost model for chronic kidney disease diagnosis. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020; 17:2131–2140. [DOI] [PubMed] [Google Scholar]
- 24. Ziemann M., Kaspi A., El-Osta A.. Digital expression explorer 2: a repository of uniformly processed RNA sequencing data. Gigascience. 2019; 8:giz022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leinonen R., Sugawara H., Shumway M.International Nucleotide Sequence Database Collaboration . The sequence read archive. Nucleic Acids Res. 2011; 39:D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li H.D., Menon R., Govindarajoo B., Panwar B., Zhang Y., Omenn G.S., Guan Y.. Functional networks of highest-connected splice isoforms: from the chromosome 17 human proteome project. J. Proteome. Res. 2015; 14:3484–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ceol A., Chatr Aryamontri A., Licata L., Peluso D., Briganti L., Perfetto L., Castagnoli L., Cesareni G.. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010; 38:D532–D539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Orchard S., Ammari M., Aranda B., Breuza L., Briganti L., Broackes-Carter F., Campbell N.H., Chavali G., Chen C., del-Toro N.et al.. The MIntAct project–IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014; 42:D358–D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oughtred R., Rust J., Chang C., Breitkreutz B.J., Stark C., Willems A., Boucher L., Leung G., Kolas N., Tyers M.et al.. The BioGRID database: a comprehensive biomedical resource of curated protein, genetic, and chemical interactions. Protein Sci. 2021; 30:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyle E.I., Weng S., Gollub J., Jin H., Botstein D., Cherry J.M., Sherlock G.. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004; 20:3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Serrano-Pozo A., Das S., Hyman B.T.. APOE and Alzheimer’s disease: advances in genetics, pathophysiology, and therapeutic approaches. Lancet Neurol. 2021; 20:68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zalocusky K.A., Nelson M.R., Huang Y.. An Alzheimer’s-disease-protective APOE mutation. Nat. Med. 2019; 25:1648–1649. [DOI] [PubMed] [Google Scholar]
- 33. Haddy N., De Bacquer D., Chemaly M.M., Maurice M., Ehnholm C., Evans A., Sans S., Do Carmo Martins M., De Backer G.et al.. The importance of plasma apolipoprotein E concentration in addition to its common polymorphism on inter-individual variation in lipid levels: results from Apo Europe. Eur. J. Hum. Genet. 2002; 10:841–850. [DOI] [PubMed] [Google Scholar]
- 34. Pinero J., Bravo A., Queralt-Rosinach N., Gutierrez-Sacristan A., Deu-Pons J., Centeno E., Garcia-Garcia J., Sanz F., Furlong L.I.. DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res. 2017; 45:D833–D839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P.et al.. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heaton H., Talman A.M., Knights A., Imaz M., Gaffney D.J., Durbin R., Hemberg M., Lawniczak M.. Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods. 2020; 17:615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X., Wang K., Lyu Y., Pan H., Zhang J., Stambolian D., Susztak K., Reilly M.P., Hu G., Li M.. Deep learning enables accurate clustering with batch effect removal in single-cell RNA-seq analysis. Nat. Commun. 2020; 11:2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao T., Lyu S., Lu G., Juan L., Zeng X., Wei Z., Hao J., Peng J.. SC2disease: a manually curated database of single-cell transcriptome for human diseases. Nucleic Acids Res. 2021; 49:D1413–D1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The web interface of the database is available at https://www.diseaselinks.com/TissueNexus/. This website is freely accessible to all users, without the need for logging on or a password.