Abstract
Drug discovery relies on the knowledge of not only drugs and targets, but also the comparative agents and targets. These include poor binders and non-binders for developing discovery tools, prodrugs for improved therapeutics, co-targets of therapeutic targets for multi-target strategies and off-target investigations, and the collective structure-activity and drug-likeness landscapes of enhanced drug feature. However, such valuable data are inadequately covered by the available databases. In this study, a major update of the Therapeutic Target Database, previously featured in NAR, was therefore introduced. This update includes (a) 34 861 poor binders and 12 683 non-binders of 1308 targets; (b) 534 prodrug-drug pairs for 121 targets; (c) 1127 co-targets of 672 targets regulated by 642 approved and 624 clinical trial drugs; (d) the collective structure-activity landscapes of 427 262 active agents of 1565 targets; (e) the profiles of drug-like properties of 33 598 agents of 1102 targets. Moreover, a variety of additional data and function are provided, which include the cross-links to the target structure in PDB and AlphaFold, 159 and 1658 newly emerged targets and drugs, and the advanced search function for multi-entry target sequences or drug structures. The database is accessible without login requirement at: https://idrblab.org/ttd/.
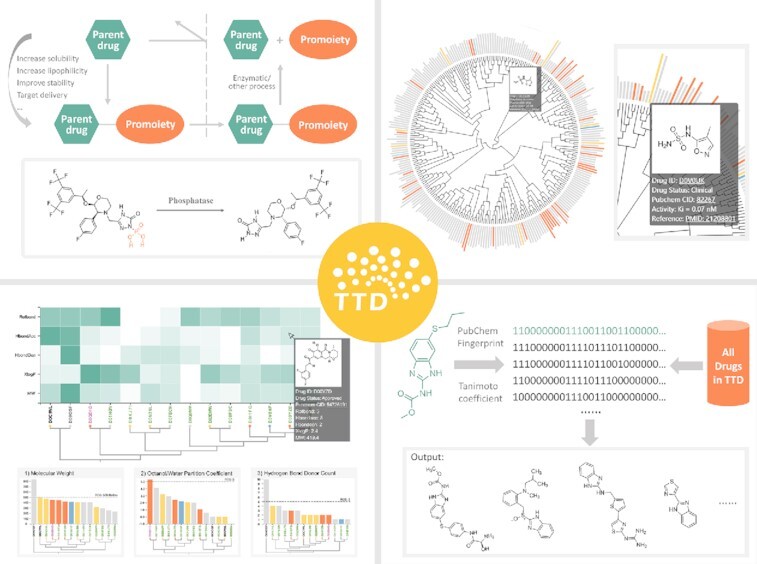
Graphical Abstract
Graphical Abstract.
Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents.
INTRODUCTION
Drug discovery is promoted by not only the knowledge of drugs (1) and their therapeutic targets (2–4), but also the comparative data with respect to other bioactive agents and other targets. Such comparative data include the knowledge of poor binders or non-binders of individual target that are useful for developing drug discovery tool of enhanced performance (5–7); the information of prodrugs that facilitates drug design by improving pharmacokinetic/pharmacodynamic features (8); the co-targets of therapeutic targets that facilitate the investigations of multi-target strategies (9), off-target (10,11) & undesired effect (9); the collective structure-activity landscapes of drugs against individual target that reveal important pharmaceutical features such as activity cliffs (12); and the drugs’ profiles of their drug-like properties that provide drug-likeness landscapes of the explored bioactive chemical space for therapeutic targets (13). Particularly, there is a rapid trend of the discovery of Artificial Intelligence (AI) tools for the drug discovery (14,15), including the AI tools for identifying bioactive compounds, and the construction of such tools requires data of poor binders and non-binders of a specific target (16). In the meantime, the existing prodrug data may inspire new ideas to avoid the drug development challenges that limit formulation option or result in undesired biopharmaceutical/pharmacokinetic performance (8). Thus, such comparative data above are urgently needed by researchers in drug discovery community. Moreover, the data of target's 3D structure are the key information for drug discovery (5). Apart from the increasing number of experimentally-resolved target crystal structures (17), advanced AI technologies (e.g. AlphaFold) have enabled the prediction of target's crystal structures of high-confidence (18,19), which requires the target-related databases, especially TTD, to include such valuable data.
While the established databases provide the comprehensive information of both drugs and targets (20–24), there is an inadequate coverage of the comparative data for the targeted agents and high-confidence 3D structures of human targets. To provide such valuable data, several major updates of Therapeutic Target Database (https://idrblab.org/ttd/) were thus introduced in this study. The first was the inclusion of > 34,800 poor binders (the target activity within the range of 50–200 μM) and >12 600 non-binders (target activity > 200 μM) for 383 and 309 successful targets (STs), 392 and 275 clinical trial targets (CTs), 137 and 91 preclinical or patented targets (PTs), and 331 and 195 research targets (RTs) respectively. Second, we added >500 prodrug-drug pairs for 91 STs, 30 CTs. Third, we provided >1100 co-targets of 423 STs and 249 CTs. These STs and CTs are targeted by 642 approved, and 624 clinical trial drugs, respectively. Fourth, we provided the 2D collective structure-activity landscapes (containing > 427 200 bioactive agents) for 444 STs, 469 CTs, 163 PTs and 489 RTs. Fifth, the drugs’ profiles of drug-like property of >33 500 agents of 435 STs, 356 CTs, 125 PTs and 186 RTs were also shown. Meanwhile, additional structural data were updated, which included the cross-links to 930 experimentally-resolved PDB structures and 1824 AlphaFold-generated structures; and 159 and 1658 newly emerged targets and drugs were also collected. Table 1 gave the statistics of targets and drugs among different database versions, and Table 2 summarized the new features and their corresponding statistics updated to the latest database. Moreover, the schema, search engine, and adopted ontology of this database were also provided in the TTD website.
Table 1.
Accumulation of drugs and their corresponding targets in the latest and previous versions of TTD database
| TTD statistics for targets and drugs | |||||||
|---|---|---|---|---|---|---|---|
| 2022 | 2020 | 2018 | 2016 | 2014 | 2012 | 2010 | |
| All targets | 3578 | 3419 | 3101 | 2589 | 2360 | 2025 | 1894 |
| Successful targets | 498 | 461 | 445 | 397 | 388 | 364 | 348 |
| Clinical trial targets | 1342 | 1191 | 1121 | 723 | 461 | 286 | 292 |
| Preclinical/patented targets | 185 | 155 | 0 | 0 | 0 | 0 | 0 |
| Research targets | 1553 | 1612 | 1535 | 1469 | 1467 | 1331 | 1254 |
| All drugs | 38 760 | 37 102 | 34 019 | 31 614 | 20 667 | 17 816 | 5028 |
| Approved drugs | 2797 | 2649 | 2544 | 2071 | 2003 | 1540 | 1514 |
| Clinical trial drugs | 10 831 | 9465 | 8103 | 7291 | 3147 | 1423 | 1212 |
| Preclinical/patented drugs | 5009 | 4845 | 0 | 0 | 0 | 0 | 0 |
| Experimental drugs | 20 123 | 20 143 | 18 923 | 17 803 | 14 856 | 14 853 | 2302 |
Table 2.
New features and their corresponding statistics added to the 2022 TTD. These new features included structure-based activity landscape of targets, profile of drug-like properties of studied targets, prodrugs together with their parent drug and target, co-targets modulated by approved or clinical trial drugs, and the poor binders and non-binders of targets
| ▪ Structure-based activity landscape of studied targets | ||||
| No. of targets with chemical structure based activity landscape | No. of drug structures | |||
| Successful | Clinical trial | Preclinical/patented | Research | |
| 444 | 469 | 163 | 489 | 427 262 |
| ▪ Drug-like properties of studied targets | ||||
| No. of targets with drug property profile | No. of drugs | |||
| Successful | Clinical trial | Preclinical/patented | Research | |
| 435 | 356 | 125 | 186 | 33 598 |
| ▪ Prodrugs together with their parent drug and target | ||||
| No. of prodrugs | Approved | Clinical trial | Preclinical/patented | Experimental |
| 146 | 79 | 9 | 300 | |
| No. of targets for prodrugs | Successful | Clinical trial | Preclinical/patented | Research |
| 91 | 30 | 1 | 1 | |
| ▪ Co-targets modulated by approved/clinical trial drugs | ||||
| No. of targets with co-targets | No. of drugs modulating co-targets | No. of co-targets | ||
| Successful | Clinical trial | Approved | Clinical trial | |
| 423 | 249 | 642 | 624 | 1127 |
| ▪ Poor binders and non-binders of studied targets | ||||
| No. of targets with poor binder(s) | No. of poor binders interacting with TTD targets | |||
| Successful | Clinical trial | Preclinical/patented | Research | |
| 383 | 392 | 137 | 331 | 34 861 |
| No. of targets with non-binder(s) | No. of non-binders interacting with TTD targets | |||
| Successful | Clinical trial | Preclinical/patented | Research | |
| 309 | 275 | 91 | 195 | 12 683 |
POOR BINDERS AND NON-BINDERS OF THERAPEUTIC TARGETS
Molecular docking is a widely-used structure-based drug discovery method (17), which employs scoring functions for scoring the binding of molecules to a target site (25). Poor binders and non-binders are useful decoy molecules for the development of the scoring functions (6). AI methods have also been extensively explored to develop bioactive molecule and pharmaceutical property screening tools, which have been primarily trained by actives (e.g. binders) and non-actives (e.g. poor binders, non-binders) (26–28). Particularly, the molecules of <10 μM activity were typically considered as inhibitors or actives (29), while those of 50–200 μM activity were reported as poor inhibitors (30,31). Meanwhile, the molecules of >200 μM activity were regarded to be inactive/of little effect (32,33). In other words, it is essential to have a conveniently-accessible resource for poor binders and non-binders of the therapeutic targets. Thus, the molecules with experimentally measured activities against each TTD target were first collected by reviewing PubMed literatures (34) using keyword combinations between target names/synonyms and ‘inhibitor’, ‘antagonist’, ‘agonist’, ‘activity’, ‘binding’, ‘affinity’, ‘IC50’, ‘Ki’, etc. Second, these PubMed literatures were manually checked to discover those containing the molecule with experimentally measured quantitative activity against any target of interest. Third, based on these collected activity values, the poor binders and non-binders were tentatively defined as of 50–200 μM (30,31) and >200 μM (32,33) activity, respectively. Using the above criteria, a total of 34 861 poor binders and 12 683 non-binders were collected for 393 and 309 STs, 392 and 275 CTs, 137 and 91 PTs, 331 and 195 RTs, respectively.
PRODRUGS
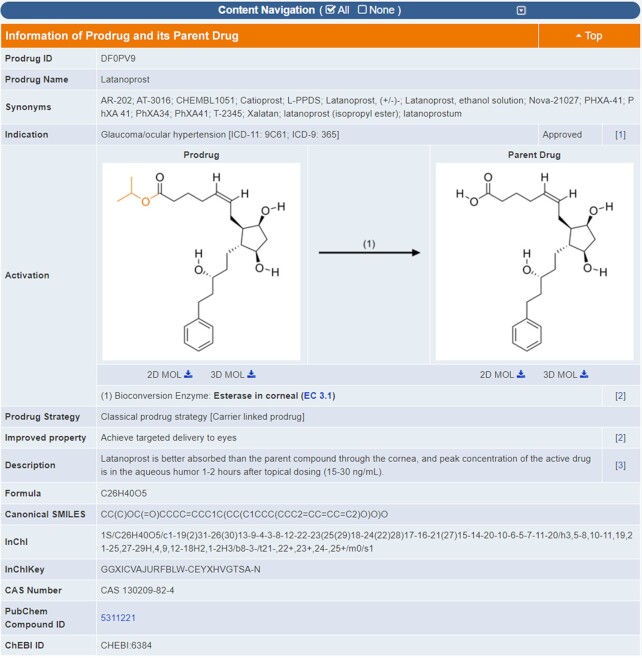
Good therapeutic drugs possess not only potent activities but also desirable pharmacokinetic and toxicological properties (35). In some cases, the drug leads may possess potent activity but poor pharmacokinetic property, which could be overcome using the prodrug strategy (8). Prodrugs are molecules modified from the parent drugs, with little or no activity but the good pharmacokinetic property, which are converted into active parent drugs inside human body via enzymatic or other process (8). Such strategy helps overcome drug discovery challenges that limit pharmacokinetic performances and drug formulation option. For instance, the prodrugs Ivemend and Gilenya were reported to improve solubility and enhance permeation, respectively (5). Therefore, a number of prodrugs were first collected by reviewing PubMed literatures (34) using various keywords such as ‘prodrug’, ‘pro-drug’, etc. Second, these literatures were manually checked to discover those containing the information of prodrug and its parent drug. Third, detailed data of a prodrug were retrieved from the literatures, which included disease indication, clinical status, prodrug strategy, improved property, bioconversion mechanism, etc. Fourth, the structures of the prodrug and its parent drug were drawn using ChemDraw based on the structures reported in each corresponding literature. As shown in Figure 1, both the detailed data and structures of prodrugs were explicitly described in the TTD prodrug page. All in all, a total of 534 prodrug-drug pairs of 91 STs and 30 CTs were collected to this update of TTD.
Figure 1.
A typical page in TTD providing prodrug information. The structures of both prodrug and its parent drug are provided along with the bioconversion enzyme or condition. Structural variation between prodrug and parent drug is highlighted in orange. The strategy for prodrug design, and the enhancement in the pharmaceutical property from parent drug to its prodrug are also described.
CO-TARGETS OF THERAPEUTIC TARGETS
Many drugs are known to interact with more macromolecular targets than their intended primary therapeutic target. In particular, a multi-target drug produces its therapeutic effect by modulating multiple targets (9). Some clinical trial drugs have been found to produce their therapeutic effects via interacting with off-targets, i.e., a macromolecular target other than their originally intended primary target (10). On the one hand, such beneficial effects of off-target have been explored for drug repurposing against complex diseases (36–39); on the other hand, off-target activity may in some instances lead to undesirable effect (40). Based on multiple targets of drugs, one can define the co-targets of a therapeutic target as the additional targets of all drugs targeting the therapeutic target. In other words, these co-targets represent both the targets co-modulated by a multi-target drug (5) and the off-target of a drug (11). Thus, those co-targets of a therapeutic target were first collected by reviewing PubMed literatures (34) by combining the target name with the keywords ‘multi-target’, ‘off-target’, ‘multiple targets’, ‘poly-pharmacology’, ‘co-targets’, ‘co-targeting’, etc. Second, all these literatures were manually checked to discover those having the information of co-targets, and the drugs of clinical importance (approved or clinical trials) that co-regulating a therapeutic target and its co-targets were also identified from literatures, company reports, and other official resources providing drug-target information. Third, detailed data of each co-target were collected to TTD and cross-linked to other reputable databases (e.g. UniProt (41) and NCBI Gene (34)). As a result, 1127 co-targets of 423 STs and 249 CTs co-modulated by 642 approved and 624 clinical trial drugs were identified and collected for this update.
COLLECTIVE STRUCTURE-ACTIVITY LANDSCAPES OF INDIVIDUAL TARGET
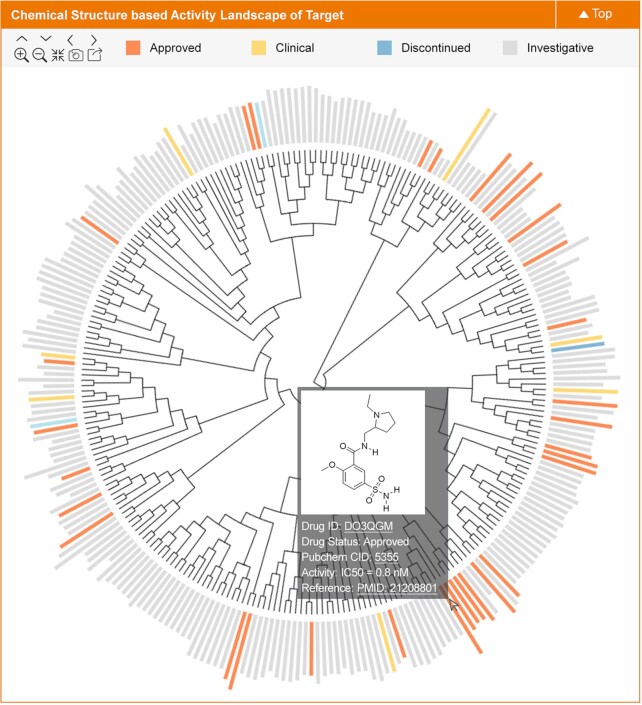
In the design of drugs against individual target, the molecular structure of the hit against a target (first molecule found to bind to the target) should be modified to optimize target binding activity (42,43). Those modified molecules, particularly the structural derivatives of a hit, largely follow certain structure-activity relationship (44), and can also lead to the dramatical activity variations, namely activity cliff (12,45,46). Such structure-activity relationships can be further evaluated by the collective structure-activity landscape of all known binders of studied target. As described in Figure 2, all known binders of a target were clustered based on their structural similarities, each binder was represented by a colored bar with its height proportional to the level of target binding activity (–log IC50, –log Ki, etc.) and color indicating each binder's clinical status (orange, yellow, blue and grey denote approved, clinical, discontinued and investigative drugs, respectively). The clustering of all binders of target was constructed using the sequential steps as follows. First, the molecular fingerprints of all binders were computed using R package ChemmineR (47). Second, the Tanimoto coefficient-based similarities among binders were computed by ChemmineR (47). Third, the complete linkage hierarchical clustering based on Euclidean distance (48) was adopted to cluster all target binders. Finally, a 2D graph was generated using the Data-Driven Documents (49), which was displayed on TTD webpage. In this update, the chemical structure-based activity landscapes of 444 STs, 469 CTs, 163 PTs and 489 RTs were provided. Figure 2 presents the 2D graph of such landscape for carbonic anhydrase VI (TTD Target ID: T06569).
Figure 2.
A typical plot in TTD showing the chemical structure-based activity landscape for a target. All known drugs of a target are clustered based on their structural similarity. Moreover, the binding activity (e.g. –log IC50, -logKi) for each drug against the target is represented by bar chart. The color of the bar indicates the highest clinical status of the corresponding drug (approved, clinical trial, etc.). Users can move the mouse over the bar to get the basic information (status, PubChem CID, activity, etc.) of each drug. The detailed drug data can be found by clicking that particular drug.
Such collective structure-activity landscape of individual target is, to the best of our knowledge, unique in the following aspects. First, each landscape in TTD is dedicated to all drugs and other binders of individual therapeutic target. Such target-specific landscape provides the overview of the structural similarity among all target-specific binders, which could help the readers to gain a quick understanding of all available binding scaffolds of a studied target. Second, such landscape gives the activities of all drugs and binders for a target along with their structural characteristics, which is useful for describing QSARs and activity cliffs. Third, this provided landscape includes the valuable information of each drug's clinical status, which demonstrated a unique perspective illustrating the relationships between drug structures and clinical development stages. Therefore, such collective structure-activity landscape of individual therapeutic target provided in TTD was of great merit for modern drug discovery.
COLLECTIVE PROFILES OF DRUG-LIKE PROPERTIES OF INDIVIDUAL TARGET
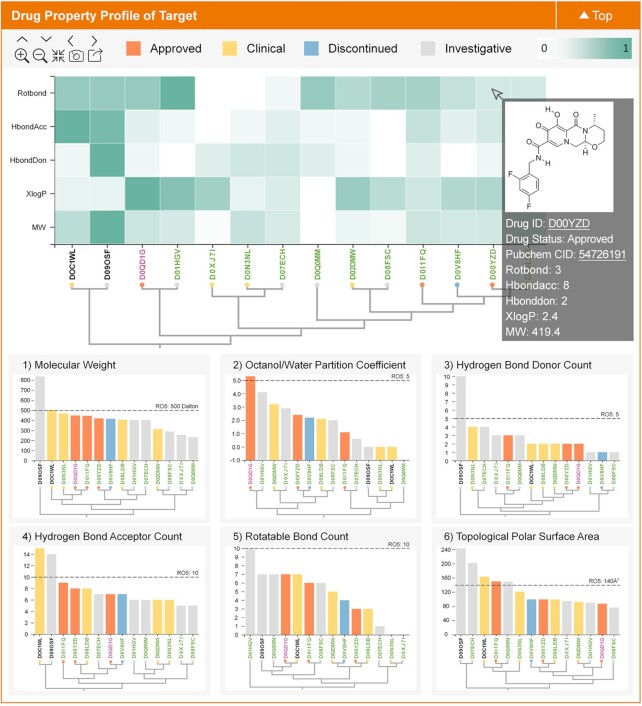
The potential of a bioactive molecule to become a drug is partly judged by the evaluation of its drug-like properties (13,50). The drug-likeness rules such as the Lipinski’s rule of five have been developed and widely used for evaluating the drug development potential of bioactive molecules (50–53). Such rules exploit drug's distinguished physicochemical property, including molecular weight and the number of hydrogen bond donors, as the basis for drug-likeness evaluations (54). The value of these drug-like properties may vary from the drugs of one target to those of another. Therefore, target-specific profiles of drug-like property may be useful for facilitating the analysis of the landscape of drug-like property for targeted therapeutics (55). As illustrated in Figure 3, the 2D profiles of the target-specific drug-like properties for those targets in TTD were provided. Particularly, all known drugs of a target were clustered based on multiple (the top plot in Figure 3) or single (six plots at the bottom of Figure 3) drug-like properties, which was displayed using the hierarchical clustering map, heatmap and bar plot. The bar color indicates the highest clinical status of the corresponding drugs (approved, clinical trial, etc.). Users can move the mouse over the bar to find the basic information (status, PubChem CID, property, etc.) of specific drugs, and the detailed information of each drug can be also found by clicking that drug. Within each graph, the known drugs of a target were clustered according to their similarities in drug-like properties, which was constructed by a process similar to that described in previous section. Each drug was represented by a vertical line with the amplitude proportional to the values of drug-like property. All in all, the profiles of 6 drug-like properties (such as molecular weight, octanol/water partition coefficient, hydrogen bond donor count, hydrogen bond acceptor count, rotatable bond count & topological polar surface area) for 435 STs, 356 CTs, 125 PTs and 186 RTs were shown. Figure 3 presents the 2D profile of drug-like property for HIV integrase (TTD Target ID: T39087).
Figure 3.
A typical plot in TTD providing the information of the drug-like property-based profile for a target. All known drugs of a target are clustered based on multiple (the top plot) or single (six plots at the bottom) drug-like properties, which is displayed using the hierarchical clustering map, heatmap and bar plot. The bar color indicates the highest clinical status of the corresponding drugs (approved, clinical trial, etc.). The user can move the mouse over the bar to find the basic data (status, PubChem CID, property, etc.) of that drug. The detailed drug data can be found by clicking that drug.
ENRICHED STRUCTURAL DATA AND ADVANCED SEARCH FUNCTION
The structures of macromolecules are important for drug discovery (56) and protein engineering or design (57). With the availability of target's 3D structures, one can employ the structure-based drug discovery methods (such as molecular docking (56,58), 3D QSAR (59,60), structure-based pharmacophore (61) and molecular dynamics simulation (62)) to identify the binders of specific target (63). The number of experimental 3D structural entries of macromolecules have increased to >180 000 (17). These nonetheless only represent a minority of known protein sequences, with 35% proteins in human proteome having structure(s) in Protein Data Bank (18). Recent progress of AI technique like AlphaFold have enabled high-confidence prediction of protein 3D structures for most human proteins (18). AlphaFold employs a deep learning architecture to predict the 3D structure of a protein from its sequence (18). Thus, the AlphaFold-generated 3D structures could greatly expand the range of targets covered by structure-based drug discovery methods (64). To have a convenient access of the structures for each TTD target, the crosslinks to PDB (providing experimentally-resolved crystal structure) and AlphaFold (describing the predicted 3D structure) were reviewed and provided in TTD, which helped to link 2754 targets to their structure data.
Sequence similarity searching is the search of proteins with similar sequences to a known target, which is useful for identifying potential targets (65) and tracing protein evolution (66). It is based on the hypothesis that proteins of similar sequences have similar functions (67). Drug similarity searching is the search of small molecules with similar structures as that of a known drug, which is useful for finding molecules with similar activities or drug-like properties (68). TTD and other databases (41,69) have already provided target similarity and drug similarity searching facilities. Nonetheless, during practical applications, multiple proteins or chemical libraries are frequently searched and analyzed for the potential target and bioactive molecule. In other words, there is a need for the facilities that can support multi-entry target and drug similarity searching. Therefore, a multi-entry target similarity searching and a multi-entry drug similarity searching facility was introduced, where the users can upload a file of multiple protein sequences or multiple molecular structures for finding TTD targets or drugs that are similar in sequence or structure. Particularly, the target similarity searching is based on the BLAST algorithm. Input with one protein sequence or a batch upload of multiple sequences for similarity search is now available in the latest version of TTD. The identified targets are ranked according to the BLAST outcomes. Moreover, the drug similarity searching is based on Tanimoto coefficients. The compound structure is first converted to PubChem Fingerprint by PaDEL-descriptors (70), and the similarity between input compound and TTD drugs was then calculated.
CONCLUDING REMARKS
With the rapid advances in modern drug discovery (71–75), there is an explosion of publications on revealing the mechanism underlying both disease and therapeutics (76–78), which in turn lead to the accumulation of huge amount of data for drug discovery. The expanded coverage of these data in TTD and other established databases collectively provide the enriched resources for drug discovery and the development of drug identification tool. The enriched data further enhance the ability to analyze and explore these derived data. Drug discovery efforts have benefited from this cycle of technology advancements, expanded knowledge and data, enhanced capabilities for the exploration of these derived data, and the advancements to the next round of the cycle. TTD and other established databases (79–81) will continue to update the new pharmaceutical data and play enhanced facilitating roles in current drug discovery efforts.
Contributor Information
Ying Zhou, State Key Laboratory for Diagnosis and Treatment of Infectious Disease, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang Provincial Key Laboratory for Drug Clinical Research and Evaluation, The First Affiliated Hospital, Zhejiang University, 79 QingChun Road, Hangzhou, Zhejiang 310000, China.
Yintao Zhang, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, China.
Xichen Lian, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, China.
Fengcheng Li, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, China.
Chaoxin Wang, Department of Computer Science, Kansas State University, Manhattan 66506, USA.
Feng Zhu, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou 310058, China; Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University, Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare, Hangzhou 330110, China.
Yunqing Qiu, State Key Laboratory for Diagnosis and Treatment of Infectious Disease, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, Zhejiang Provincial Key Laboratory for Drug Clinical Research and Evaluation, The First Affiliated Hospital, Zhejiang University, 79 QingChun Road, Hangzhou, Zhejiang 310000, China.
Yuzong Chen, State Key Laboratory of Chemical Oncogenomics, Key Laboratory of Chemical Biology, The Graduate School at Shenzhen, Tsinghua University, Shenzhen 518055, China; Qian Xuesen Collaborative Research Center of Astrochemistry and Space Life Sciences, Institute of Drug Discovery Technology, Ningbo University, Ningbo 315211, China.
FUNDING
Scientific Research Grant of Ningbo University [215-432000282]; Ningbo Top Talent Project [215-432094250]; Zhejiang Provincial Science and Technology Department [2020C03046]; National Natural Science Foundation of China [81971982, 81872798, U1909208]; Natural Science Foundation of Zhejiang Province [LR21H300001]; Leading Talent of the ‘Ten Thousand Plan’ – National High-Level Talents Special Support Plan of China; Fundamental Research Fund for Central Universities [2018QNA7023]; ‘Double Top-Class’ University Project [181201*194232101]; Key R&D Program of Zhejiang Province [2020C03010]; Alibaba-Zhejiang University Joint Research Center of Future Digital Healthcare; Alibaba Cloud; Information Technology Center of Zhejiang University. Funding for open access charge: National Natural Science Foundation of China [81872798].
Conflict of interest statement. None declared.
REFERENCES
- 1. Shih H.P., Zhang X., Aronov A.M.. Drug discovery effectiveness from the standpoint of therapeutic mechanisms and indications. Nat. Rev. Drug Discov. 2018; 17:19–33. [DOI] [PubMed] [Google Scholar]
- 2. Santos R., Ursu O., Gaulton A., Bento A.P., Donadi R.S., Bologa C.G., Karlsson A., Al-Lazikani B., Hersey A., Oprea T.I.et al.. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017; 16:19–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Licursi V., Conte F., Fiscon G., Paci P.. MIENTURNET: an interactive web tool for microRNA-target enrichment and network-based analysis. BMC Bioinformatics. 2019; 20:545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yin J., Li X., Li F., Lu Y., Zeng S., Zhu F.. Identification of the key target profiles underlying the drugs of narrow therapeutic index for treating cancer and cardiovascular disease. Comput. Struct. Biotechnol. J. 2021; 19:2318–2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bajusz D., Wade W.S., Satala G., Bojarski A.J., Ilas J., Ebner J., Grebien F., Papp H., Jakab F., Douangamath A.et al.. Exploring protein hotspots by optimized fragment pharmacophores. Nat. Commun. 2021; 12:3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seeliger D., de Groot B.L.. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J. Comput. Aided Mol. Des. 2010; 24:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xue W., Yang F., Wang P., Zheng G., Chen Y., Yao X., Zhu F.. What contributes to serotonin-norepinephrine reuptake inhibitors' dual-targeting mechanism? The key role of transmembrane domain 6 in human serotonin and norepinephrine transporters revealed by molecular dynamics simulation. ACS Chem. Neurosci. 2018; 9:1128–1140. [DOI] [PubMed] [Google Scholar]
- 8. Rautio J., Meanwell N.A., Di L., Hageman M.J.. The expanding role of prodrugs in contemporary drug design and development. Nat. Rev. Drug Discov. 2018; 17:559–587. [DOI] [PubMed] [Google Scholar]
- 9. Tao L., Zhu F., Xu F., Chen Z., Jiang Y.Y., Chen Y.Z.. Co-targeting cancer drug escape pathways confers clinical advantage for multi-target anticancer drugs. Pharmacol. Res. 2015; 102:123–131. [DOI] [PubMed] [Google Scholar]
- 10. Lin A., Giuliano C.J., Palladino A., John K.M., Abramowicz C., Yuan M.L., Sausville E.L., Lukow D.A., Liu L., Chait A.R.et al.. Off-target toxicity is a common mechanism of action of cancer drugs undergoing clinical trials. Sci. Transl. Med. 2019; 11:eaaw8412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kieber-Emmons T., Monzavi-Karbassi B., Hutchins L.F., Pennisi A., Makhoul I.. Harnessing benefit from targeting tumor associated carbohydrate antigens. Hum. Vaccin. Immunother. 2017; 13:323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Stumpfe D., Bajorath J.. Exploring activity cliffs in medicinal chemistry. J. Med. Chem. 2012; 55:2932–2942. [DOI] [PubMed] [Google Scholar]
- 13. Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L.. Quantifying the chemical beauty of drugs. Nat. Chem. 2012; 4:90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hong J., Luo Y., Mou M., Fu J., Zhang Y., Xue W., Xie T., Tao L., Lou Y., Zhu F.. Convolutional neural network-based annotation of bacterial type IV secretion system effectors with enhanced accuracy and reduced false discovery. Brief. Bioinform. 2020; 21:1825–1836. [DOI] [PubMed] [Google Scholar]
- 15. Hong J., Luo Y., Zhang Y., Ying J., Xue W., Xie T., Tao L., Zhu F.. Protein functional annotation of simultaneously improved stability, accuracy and false discovery rate achieved by a sequence-based deep learning. Brief. Bioinform. 2020; 21:1437–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shen W., Zeng X., Zhu F., Wang Y., Qin C., Tan Y., Jiang Y., Chen Y.. Out-of-the-box deep learning prediction of pharmaceutical properties by broadly learned knowledge-based molecular representations. Nat. Mach. Intell. 2021; 3:334–343. [Google Scholar]
- 17. Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M.et al.. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021; 49:D437–D451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with AlphaFold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tunyasuvunakool K., Adler J., Wu Z., Green T., Zielinski M., Zidek A., Bridgland A., Cowie A., Meyer C., Laydon A.et al.. Highly accurate protein structure prediction for the human proteome. Nature. 2021; 596:590–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z.et al.. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018; 46:D1074–D1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang Y., Zhang S., Li F., Zhou Y., Zhang Y., Wang Z., Zhang R., Zhu J., Ren Y., Tan Y.et al.. Therapeutic target database 2020: enriched resource for facilitating research and early development of targeted therapeutics. Nucleic Acids Res. 2020; 48:D1031–D1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Avram S., Bologa C.G., Holmes J., Bocci G., Wilson T.B., Nguyen D.T., Curpan R., Halip L., Bora A., Yang J.J.et al.. DrugCentral 2021 supports drug discovery and repositioning. Nucleic Acids Res. 2021; 49:D1160–D1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Armstrong J.F., Faccenda E., Harding S.D., Pawson A.J., Southan C., Sharman J.L., Campo B., Cavanagh D.R., Alexander S.P.H., Davenport A.P.et al.. The IUPHAR/BPS Guide to Pharmacology in 2020: extending immunopharmacology content and introducing the IUPHAR/MMV guide to malaria pharmacology. Nucleic. Acids. Res. 2020; 48:D1006–D1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yang Q., Wang Y., Zhang Y., Li F., Xia W., Zhou Y., Qiu Y., Li H., Zhu F.. NOREVA: enhanced normalization and evaluation of time-course and multi-class metabolomic data. Nucleic Acids Res. 2020; 48:W436–W448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Warren G.L., Andrews C.W., Capelli A.M., Clarke B., LaLonde J., Lambert M.H., Lindvall M., Nevins N., Semus S.F., Senger S.et al.. A critical assessment of docking programs and scoring functions. J. Med. Chem. 2006; 49:5912–5931. [DOI] [PubMed] [Google Scholar]
- 26. Xue Y., Li Z.R., Yap C.W., Sun L.Z., Chen X., Chen Y.Z.. Effect of molecular descriptor feature selection in support vector machine classification of pharmacokinetic and toxicological properties of chemical agents. J. Chem. Inf. Comput. Sci. 2004; 44:1630–1638. [DOI] [PubMed] [Google Scholar]
- 27. Han L.Y., Ma X.H., Lin H.H., Jia J., Zhu F., Xue Y., Li Z.R., Cao Z.W., Ji Z.L., Chen Y.Z.. A support vector machines approach for virtual screening of active compounds of single and multiple mechanisms from large libraries at an improved hit-rate and enrichment factor. J. Mol. Graph. Model. 2008; 26:1276–1286. [DOI] [PubMed] [Google Scholar]
- 28. Zhavoronkov A., Ivanenkov Y.A., Aliper A., Veselov M.S., Aladinskiy V.A., Aladinskaya A.V., Terentiev V.A., Polykovskiy D.A., Kuznetsov M.D., Asadulaev A.et al.. Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nat. Biotechnol. 2019; 37:1038–1040. [DOI] [PubMed] [Google Scholar]
- 29. Bender A., Cortes-Ciriano I.. Artificial intelligence in drug discovery: what is realistic, what are illusions? Part 2: a discussion of chemical and biological data. Drug Discov. Today. 2021; 26:1040–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thorndike J., Gaumont Y., Kisliuk R.L., Sirotnak F.M., Murthy B.R., Nair M.G., Piper J.R.. Inhibition of glycinamide ribonucleotide formyltransferase and other folate enzymes by homofolate polyglutamates in human lymphoma and murine leukemia cell extracts. Cancer Res. 1989; 49:158–163. [PubMed] [Google Scholar]
- 31. Wang B.H., Ternai B., Polya G.. Specific inhibition of cyclic AMP-dependent protein kinase by warangalone and robustic acid. Phytochemistry. 1997; 44:787–796. [DOI] [PubMed] [Google Scholar]
- 32. Beckmann-Knopp S., Rietbrock S., Weyhenmeyer R., Bocker R.H., Beckurts K.T., Lang W., Hunz M., Fuhr U.. Inhibitory effects of silibinin on cytochrome P-450 enzymes in human liver microsomes. Pharmacol. Toxicol. 2000; 86:250–256. [DOI] [PubMed] [Google Scholar]
- 33. Kwon J.Y., Jeong H.W., Kim H.K., Kang K.H., Chang Y.H., Bae K.S., Choi J.D., Lee U.C., Son K.H., Kwon B.M.. Cis-fumagillin, a new methionine aminopeptidase (type 2) inhibitor produced by Penicillium sp. F2757. J. Antibiot. 2000; 53:799–806. [DOI] [PubMed] [Google Scholar]
- 34. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Roberts S.A. Drug metabolism and pharmacokinetics in drug discovery. Curr. Opin. Drug. Discov. Dev. 2003; 6:66–80. [PubMed] [Google Scholar]
- 36. Fiscon G., Paci P.. SAveRUNNER: an R-based tool for drug repurposing. BMC Bioinformatics. 2021; 22:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fiscon G., Conte F., Farina L., Paci P.. SAveRUNNER: a network-based algorithm for drug repurposing and its application to COVID-19. PLoS Comput. Biol. 2021; 17:e1008686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kumar S., Jang C., Subedi L., Kim S.Y., Kim M.H.. Repurposing of FDA approved ring systems through bi-directional target-ring system dual screening. Sci. Rep. 2020; 10:21133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tang J., Fu J., Wang Y., Luo Y., Yang Q., Li B., Tu G., Hong J., Cui X., Chen Y.et al.. Simultaneous improvement in the precision, accuracy, and robustness of label-free proteome quantification by optimizing data manipulation chains. Mol. Cell. Proteomics. 2019; 18:1683–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lounkine E., Keiser M.J., Whitebread S., Mikhailov D., Hamon J., Jenkins J.L., Lavan P., Weber E., Doak A.K., Cote S.et al.. Large-scale prediction and testing of drug activity on side-effect targets. Nature. 2012; 486:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. UniProt, C. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu T., Nagle A., Kuhen K., Gagaring K., Borboa R., Francek C., Chen Z., Plouffe D., Goh A., Lakshminarayana S.B.et al.. Imidazolopiperazines: hit to lead optimization of new antimalarial agents. J. Med. Chem. 2011; 54:5116–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Teli M.K., Kumar S., Yadav D.K., Kim M.H.. In silico identification of prolyl hydroxylase inhibitor by per-residue energy decomposition-based pharmacophore approach. J. Cell. Biochem. 2021; 122:1098–1112. [DOI] [PubMed] [Google Scholar]
- 44. Martinez A., Alonso M., Castro A., Dorronsoro I., Gelpi J.L., Luque F.J., Perez C., Moreno F.J.. SAR and 3D-QSAR studies on thiadiazolidinone derivatives: exploration of structural requirements for glycogen synthase kinase 3 inhibitors. J. Med. Chem. 2005; 48:7103–7112. [DOI] [PubMed] [Google Scholar]
- 45. Hu H., Bajorath J.. Systematic exploration of activity cliffs containing privileged substructures. Mol. Pharm. 2020; 17:979–989. [DOI] [PubMed] [Google Scholar]
- 46. Hu H., Bajorath J.. Activity cliffs produced by single-atom modification of active compounds: Systematic identification and rationalization based on X-ray structures. Eur. J. Med. Chem. 2020; 207:112846. [DOI] [PubMed] [Google Scholar]
- 47. Cao Y., Charisi A., Cheng L.C., Jiang T., Girke T.. ChemmineR: a compound mining framework for R. Bioinformatics. 2008; 24:1733–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim M.J., Ahn E.Y., Hwang W., Lee Y., Lee E.Y., Lee E.B., Song Y.W., Park J.K.. Association between fever pattern and clinical manifestations of adult-onset Still's disease: unbiased analysis using hierarchical clustering. Clin. Exp. Rheumatol. 2018; 36:74–79. [PubMed] [Google Scholar]
- 49. Bostock M., Ogievetsky V., Heer J.. D(3): data-driven documents. IEEE Trans. Vis. Comput. Graph. 2011; 17:2301–2309. [DOI] [PubMed] [Google Scholar]
- 50. Leeson P.D., Springthorpe B.. The influence of drug-like concepts on decision-making in medicinal chemistry. Nat. Rev. Drug Discov. 2007; 6:881–890. [DOI] [PubMed] [Google Scholar]
- 51. Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J.. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug. Deliv. Rev. 2001; 46:3–26. [DOI] [PubMed] [Google Scholar]
- 52. Lipinski C.A. Lead- and drug-like compounds: the rule-of-five revolution. Drug Discov. Today Technol. 2004; 1:337–341. [DOI] [PubMed] [Google Scholar]
- 53. Li F., Zhou Y., Zhang X., Tang J., Yang Q., Zhang Y., Luo Y., Hu J., Xue W., Qiu Y.et al.. SSizer: determining the sample sufficiency for comparative biological study. J. Mol. Biol. 2020; 432:3411–3421. [DOI] [PubMed] [Google Scholar]
- 54. Lipinski C.A. Drug-like properties and the causes of poor solubility and poor permeability. J. Pharmacol. Toxicol. Methods. 2000; 44:235–249. [DOI] [PubMed] [Google Scholar]
- 55. Leeson P.D., Bento A.P., Gaulton A., Hersey A., Manners E.J., Radoux C.J., Leach A.R.. Target-based evaluation of ‘drug-like’ properties and ligand efficiencies. J. Med. Chem. 2021; 64:7210–7230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gorgulla C., Boeszoermenyi A., Wang Z.F., Fischer P.D., Coote P.W., Padmanabha Das K.M., Malets Y.S., Radchenko D.S., Moroz Y.S., Scott D.A.et al.. An open-source drug discovery platform enables ultra-large virtual screens. Nature. 2020; 580:663–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Taujale R., Venkat A., Huang L.C., Zhou Z., Yeung W., Rasheed K.M., Li S., Edison A.S., Moremen K.W., Kannan N.. Deep evolutionary analysis reveals the design principles of fold A glycosyltransferases. Elife. 2020; 9:e54532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Friesner R.A., Murphy R.B., Repasky M.P., Frye L.L., Greenwood J.R., Halgren T.A., Sanschagrin P.C., Mainz D.T.. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006; 49:6177–6196. [DOI] [PubMed] [Google Scholar]
- 59. Verma J., Khedkar V.M., Coutinho E.C.. 3D-QSAR in drug design—a review. Curr. Top. Med. Chem. 2010; 10:95–115. [DOI] [PubMed] [Google Scholar]
- 60. Huang L.C., Yeung W., Wang Y., Cheng H., Venkat A., Li S., Ma P., Rasheed K., Kannan N.. Quantitative structure-mutation-activity relationship tests (QSMART) model for protein kinase inhibitor response prediction. BMC Bioinformatics. 2020; 21:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rella M., Rushworth C.A., Guy J.L., Turner A.J., Langer T., Jackson R.M.. Structure-based pharmacophore design and virtual screening for novel angiotensin converting enzyme 2 inhibitors. J. Chem. Inf. Model. 2006; 46:708–716. [DOI] [PubMed] [Google Scholar]
- 62. Herrera-Nieto P., Perez A., De Fabritiis G.. Small molecule modulation of intrinsically disordered proteins using molecular dynamics simulations. J. Chem. Inf. Model. 2020; 60:5003–5010. [DOI] [PubMed] [Google Scholar]
- 63. Lee S.H., Ahn S., Kim M.H.. Comparing a query compound with drug target classes using 3D-chemical similarity. Int. J. Mol. Sci. 2020; 21:4208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Skalic M., Sabbadin D., Sattarov B., Sciabola S., De Fabritiis G.. From target to drug: generative modeling for the multimodal structure-based ligand design. Mol. Pharm. 2019; 16:4282–4291. [DOI] [PubMed] [Google Scholar]
- 65. Li Y.H., Li X.X., Hong J.J., Wang Y.X., Fu J.B., Yang H., Yu C.Y., Li F.C., Hu J., Xue W.W.et al.. Clinical trials, progression-speed differentiating features and swiftness rule of the innovative targets of first-in-class drugs. Brief. Bioinform. 2020; 21:649–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kwon A., Scott S., Taujale R., Yeung W., Eyers P.A., Kannan N.. Tracing the origin and evolution of pseudokinases across the tree of life. Sci. Signal. 2019; 12:eaav3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Whisstock J.C., Lesk A.M.. Prediction of protein function from protein sequence and structure. Q. Rev. Biophys. 2003; 36:307–340. [DOI] [PubMed] [Google Scholar]
- 68. Azad A.K.M., Dinarvand M., Nematollahi A., Swift J., Lutze-Mann L., Vafaee F.. A comprehensive integrated drug similarity resource for in-silico drug repositioning and beyond. Brief. Bioinform. 2021; 22:bbaa126. [DOI] [PubMed] [Google Scholar]
- 69. Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B.et al.. PubChem in 2021: new data content and improved web interfaces. Nucleic Acids Res. 2021; 49:D1388–D1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yap C.W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011; 32:1466–1474. [DOI] [PubMed] [Google Scholar]
- 71. Failli M., Paananen J., Fortino V.. ThETA: transcriptome-driven efficacy estimates for gene-based TArget discovery. Bioinformatics. 2020; 36:4214–4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Paananen J., Fortino V.. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2020; 21:1937–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Fortino V., Wisgrill L., Werner P., Suomela S., Linder N., Jalonen E., Suomalainen A., Marwah V., Kero M., Pesonen M.et al.. Machine-learning-driven biomarker discovery for the discrimination between allergic and irritant contact dermatitis. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:33474–33485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tang J., Fu J., Wang Y., Li B., Li Y., Yang Q., Cui X., Hong J., Li X., Chen Y.et al.. ANPELA: analysis and performance assessment of the label-free quantification workflow for metaproteomic studies. Brief. Bioinform. 2020; 21:621–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Li B., Tang J., Yang Q., Li S., Cui X., Li Y., Chen Y., Xue W., Li X., Zhu F.. NOREVA: normalization and evaluation of MS-based metabolomics data. Nucleic Acids Res. 2017; 45:W162–W170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Naveja J.J., Stumpfe D., Medina-Franco J.L., Bajorath J.. Exploration of target synergy in cancer treatment by cell-based screening assay and network propagation analysis. J. Chem. Inf. Model. 2019; 59:3072–3079. [DOI] [PubMed] [Google Scholar]
- 77. Jimenez J., Sabbadin D., Cuzzolin A., Martinez-Rosell G., Gora J., Manchester J., Duca J., De Fabritiis G.. PathwayMap: molecular pathway association with self-normalizing neural networks. J. Chem. Inf. Model. 2019; 59:1172–1181. [DOI] [PubMed] [Google Scholar]
- 78. Yang Q., Li B., Tang J., Cui X., Wang Y., Li X., Hu J., Chen Y., Xue W., Lou Y.et al.. Consistent gene signature of schizophrenia identified by a novel feature selection strategy from comprehensive sets of transcriptomic data. Brief. Bioinform. 2020; 21:1058–1068. [DOI] [PubMed] [Google Scholar]
- 79. Yin J., Li F., Zhou Y., Mou M., Lu Y., Chen K., Xue J., Luo Y., Fu J., He X.et al.. INTEDE: interactome of drug-metabolizing enzymes. Nucleic Acids Res. 2021; 49:D1233–D1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J., Yang Q., Fu T., Zhang X., Cui X., Tu G.et al.. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018; 46:D1121–D1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yin J., Sun W., Li F., Hong J., Li X., Zhou Y., Lu Y., Liu M., Zhang X., Chen N.et al.. VARIDT 1.0: variability of drug transporter database. Nucleic Acids Res. 2020; 48:D1042–D1050. [DOI] [PMC free article] [PubMed] [Google Scholar]