Abstract
As a crucial molecular mechanism, post-translational modifications (PTMs) play critical roles in a wide range of biological processes in plants. Recent advances in mass spectrometry-based proteomic technologies have greatly accelerated the profiling and quantification of plant PTM events. Although several databases have been constructed to store plant PTM data, a resource including more plant species and more PTM types with quantitative dynamics still remains to be developed. In this paper, we present an integrative database of quantitative PTMs in plants named qPTMplants (http://qptmplants.omicsbio.info), which hosts 1 242 365 experimentally identified PTM events for 429 821 nonredundant sites on 123 551 proteins under 583 conditions for 23 PTM types in 43 plant species from 293 published studies, with 620 509 quantification events for 136 700 PTM sites on 55 361 proteins under 354 conditions. Moreover, the experimental details, such as conditions, samples, instruments and methods, were manually curated, while a variety of annotations, including the sequence and structural characteristics, were integrated into qPTMplants. Then, various search and browse functions were implemented to access the qPTMplants data in a user-friendly manner. Overall, we anticipate that the qPTMplants database will be a valuable resource for further research on PTMs in plants.
INTRODUCTION
Post-translational modification (PTM) is a biochemical process that changes the properties and extends the chemical composition of a protein by participating in the addition of chemical groups (such as phosphate, succinyl and sulfate) to specific amino acid residues or by proteolytic cleavage of the protein sequence backbone (1,2). More than 200 PTMs have been registered in UniProt, and they can regulate the structure, function and localization of proteins (3). To date, protein phosphorylation is the best studied PTM in plants, and secondly often studied PTMs are lysine modifications such as acetylation, ubiquitination and succinylation (4–6). As important modulators of protein function, cysteine modifications such as S-nitrosylation, S-sulfenylation and S-glutathionylation have received increasing attention in plants (7–9). Plant PTMs play critical roles in nearly all biological processes, such as signal transduction (4,10), metabolism (11,12), stress resistance (13,14), plant immunity (15,16) and other cellular events.
Recently, the rapid progress of high-throughput proteomic techniques has made great strides in the profiling and quantification of plant PTMs in different tissues under various conditions (17-20). For example, based on phosphoproteome and acetylome quantification, Uhrig et al. unveiled the diurnal changes of concerted phosphorylation and acetylation in seedlings, roots, leaf rosettes, flowers and siliques of Arabidopsis thaliana (21). During seed germination, Nietzel et al. characterized the oxidation state of cysteine in Arabidopsis through thiol redox proteomics (17), while He et al. evaluated the dynamic change and extensive regulation of ubiquitylation in Oryza sativa subsp. japonica (rice) by a quantitative ubiquitylomics approach (22). Through quantitative phosphoproteome and metabolome analyses, Pi et al. revealed the regulatory mechanism of GmMYB173 in soybean under salt stress (23). Additionally, Walley et al. profiled and quantified the acetylome of the Zea mays (maize) fungal-induced immune response (24). Therefore, the identification of PTMs and their dynamic changes could be used to understand the regulatory roles of PTMs in the processes of plant growth and stress responses.
Previously, numerous informative databases have been constructed to store PTM-related data in plants. For example, some general resources, such as dbPTM (25) and SysPTM (26), contain various types of PTMs identified by experiments in eukaryotes, while several PTM-specific databases, including EPSD (27), PLMD (28) and iCysMod (29), host modified sites for phosphorylation, lysine modifications and cysteine modifications in eukaryotic proteins, respectively. However, these databases only cover a limited part of the experimentally identified PTM events in plants. Furthermore, PhosPhAt (30), P3DB (31) and dbPPT (32) are dedicated to depositing phosphorylation information in plants. Moreover, several databases, including Plant PTM Viewer (33) and FAT-PTM (34), were developed to maintain and organize a flood of plant-modified substrates with accurate sites for multiple PTM types. The Plant PTM Viewer contains ∼370 000 modified sites for 19 PTM types in five plant species from 105 publications, and experimental details and available quantitative information related to PTMs are also provided (33). FAT-PTM is a database of functional analysis tools for PTMs that supports over 49 000 PTM sites for eight types of PTMs identified in Arabidopsis as well as phosphorylation dynamics data from >10 published quantitative phosphoproteomic studies (34). Although the databases mentioned above are devoted to the collection of plant PTM data, a comprehensive resource covering more plant species, more types of PTMs and their quantitative dynamics is still needed for the research community.
In this study, we developed an integrative database of quantitative PTMs in plants called qPTMplants, which hosts 123 551 published proteins with 429 821 experimentally identified nonredundant PTM sites under 583 conditions in 43 plant species for 23 types of PTMs from 293 research articles. Among them, there are 620 509 quantitative events for 136 700 PTM sites on 55 361 substrates under 354 conditions in 34 plant species from 139 published studies. In the qPTMplants database, detailed information about the PTM events was curated and provided, while annotations including the sequence and structural characteristics were also integrated. For convenient usage, qPTMplants can be searched and browsed in a user-friendly manner. Based on the curated Arabidopsis dataset, the sequence characteristics around PTM sites for different PTM types were further analyzed, while the crosstalk among diverse PTMs at the same modified residues was also studied. Taken together, qPTMplants could serve as a useful resource to further investigate the function of plant PTMs.
CONSTRUCTION AND CONTENT
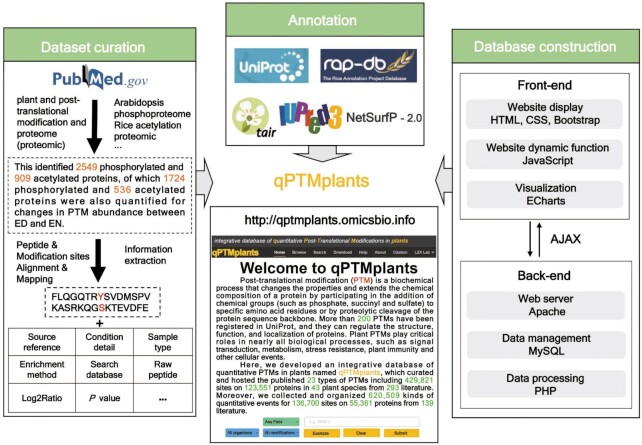
To establish a comprehensive database for PTMs in plants, published PTM datasets from the literature were integrated (Figure 1). First, we retrieved the plant PTM-related literature from PubMed published before July 2021 through a number of keywords, including plant or plant species names combined with PTM types and proteome/proteomic, such as ‘plant phosphoproteome’, ‘plant acetylation proteome’, ‘plant glycosylation proteome’, ‘plant ubiquitination proteome’ and ‘plant nitrosylation proteome’. To avoid missing plant PTM data, additional keywords such as ‘plant large-scale acetylation’, ‘plant MS acetylation’, ‘plant comprehensive analysis acetylation’ and other correlative nomenclatures were further used for the literature search. The number of related studies searched for each keyword is shown in Supplementary Table S1. We initially read the abstracts to determine whether there were PTM-omic datasets; if so or if we were unsure, then we manually checked the full texts and downloaded their corresponding supplementary files to obtain the experimentally identified PTM datasets. If there were localization probabilities available in the datasets, we used a localization probability ≥0.75 as the criterion to further screen credible PTM substrates, sites and modified peptides from the datasets. In addition, we collected detailed information from the literature, including experimental conditions, sample types, enrichment methods, mass spectrometry, reference proteome and raw peptides. For the quantitative PTM events, the log2-transformed ratio (Log2Ratio) and available P value were also manually curated.
Figure 1.
The construction procedure for the qPTMplants database.
To pinpoint the precise positions of PTM sites on the protein sequences, the collected PTM peptides and modified sites were mapped to the reference proteomes acquired from the commonly used databases of TAIR (Araport11) (35), RAP-DB (Release 2021_05) (36) and UniProt (Release 2021_03) (3) for Arabidopsis, rice and other species, respectively. For some species without reference proteomes in UniProt, such as Nicotiana benthamiana, Lotus japonicus and Actinidia deliciosa, we used their original sequences provided in the literature. During the data-mapping process, owing to the differences among the diverse reference proteomes employed in these extracted articles, some raw modified peptides could not be accurately aligned with a protein sequence in reference proteomes, and these unmapped peptides were abandoned. After the raw peptides collected were successfully matched to the protein sequence, the new PTM peptides of 15 amino acids were generated with the modified sites in the center and surrounded by 7 amino acids of upstream and downstream. Furthermore, NetSurfP (37) and IUPred2A (38) were adopted to provide structural annotations for PTM substrates. Finally, the online service of qPTMplants was implemented in PHP + MySQL + JavaScript (Figure 1).
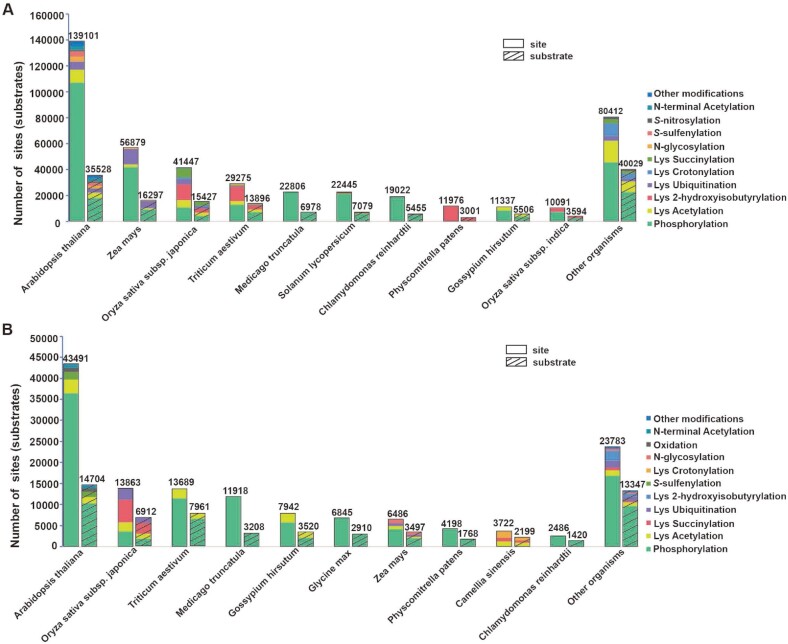
In total, 1 242 365 PTM events for 429 821 PTM sites on 123 551 proteins under 583 conditions for 23 PTM types in 43 plant species from 293 published studies were collected (Supplementary Table S2). Arabidopsis, maize and rice had the most PTM sites, including 139 101 sites in 35 528 proteins for 17 PTM types, 56 879 sites in 16 297 substrates for 4 PTM types and 41 447 sites in 15 427 proteins for 11 PTM types (Figure 2A and Supplementary Table S2). Among the types of PTMs, phosphorylation (295 559 sites, 66.45%), acetylation (40 199 sites, 9.04%), 2-hydroxyisobutyrylation (37 578 sites, 8.45%) and ubiquitination (26 796 sites, 6.02%) accounted for a large proportion of these collected PTM sites (Figure 2A and Supplementary Table S2). In addition, 620 509 quantitative events from 36 700 nonredundant PTM sites on 55 361 proteins for 13 PTM types in 34 plant species were collected, involving 43 samples and 354 different experimental conditions (Figure 2B and Supplementary Table S3). For the plant model organism Arabidopsis, there were 351 822 quantitative events for 43 289 PTM sites on 12 102 substrates for 8 PTM types, and the number of quantitative phosphorylation events was predominant in accord with that in all detected plant PTM sites (Supplementary Table S3).
Figure 2.
Data statistics of qPTMplants. (A) The distribution of site and substrate numbers of different PTM types among different organisms. (B) The number distribution of quantitative sites and substrates of different PTM types among different organisms. Lys represents the lysine residue.
USAGE
For convenient usage, qPTMplants provides multiple options, including search, browse and download, to quickly access the PTM data of plants. The help page of the website provides a detailed tutorial. Here, we describe some of these functionalities.
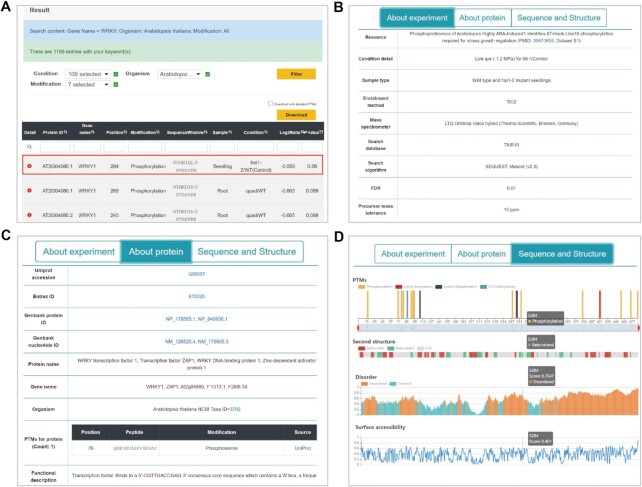
On the home page, a quick search box was provided for fast searching by protein accession, gene or protein name, function or condition (Figure 3A), while the search can be restricted by PTM type and/or organism. In addition, two search options were implemented on the search page: ‘Advanced search’ and ‘BLAST search’. In ‘Advanced search’, various keywords specified in multiple areas could be combined through the operators of ‘AND’, ‘OR’ and ‘NOT’ to perform an accurate query with up to three search keywords under specified species or PTM type of interest (Figure 3B). The option of ‘BLAST search’ was designed to find related information in qPTMplants quickly. Users can input a protein sequence in FASTA format and select the required E value to search identical or homologous proteins through the blastall program in NCBI BLAST packages (39) (Figure 3C).
Figure 3.
The search and browse options in qPTMplants. (A) Simple search function. (B) Advanced search function. (C) BLAST search function. (D) Browse by species. (E) Browse by the PTM types of the selected species. (F) Browse for proteins with the chosen PTM in selected organism. (G) Browse for conditions with the chosen PTM in selected organism.
On the browse page, all plant species in qPTMplants are listed in alphabetical order, and common species are highlighted (Figure 3D). After users click on the species of interest, the numbers of modified proteins and experimental conditions for the collected PTM types in the chosen organism are displayed in the diagram below, respectively. Meanwhile, the PTM types in the chosen species are also organized alphabetically under the statistical chart (Figure 3E). By clicking on a certain modification, the associated protein IDs (Figure 3F) and detailed conditions (Figure 3G) are shown alphabetically in the table at the bottom. Furthermore, the online server jumped to the corresponding result page based on the selection of the item from the lists for proteins or conditions (Figure 4A).
Figure 4.
The result pages for the search of a gene name. (A) The returned search results. (B) The information about the experiment. (C) The annotations about the protein. (D) The sequence and structural properties of the PTM substrate.
On the results page, the matched query results can be further filtered by condition, organism and modification. The candidate entries are displayed in the form of a table, which includes the protein ID, gene name, position, modification type, sequence window, sample type, experimental condition, Log2Ratio and P value (Figure 4A). Moreover, by clicking the download button, users can obtain a text file of all searched results, which includes the contents in the table (Supplementary Figure S1A). If users want to add labels to clearly demark PTM sites in the output file, they can check the ‘Download with labelled PTMs’ option and then click download button (Supplementary Figure S1B).
In addition, users can learn more information about the PTM sites by clicking on the ‘plus’ button. The detailed information includes the following three sections: (i) ‘About experiment’, which includes the description of source reference, detail on condition, sample type, experimental instrument and method, PRIDE accession and raw peptide (Figure 4B); (ii) ‘About protein’ shows the protein information comprising database accessions, protein and gene names, protein sequence, organism, functional description and PTMs retrieved from UniProt (Figure 4C); and (iii) ‘Sequence and Structure’ visualizes the sequence and structural features of the protein, including the PTM sites, secondary structure, disordered region and surface accessibility (Figure 4D). In this section, the diagrams can be scaled by the control element, while the details about the sequence and structure are shown through hovering over the diagram (Figure 4D).
Moreover, users can go to the download page to acquire all the PTM data for their own analysis. We guarantee that we will not collect, edit and disclose any private information sent by users to this website.
DISCUSSION
As a crucial molecular mechanism, PTMs greatly expand the complexity of the proteome and are highly involved in the regulation of numerous biological processes in plants (4,10–16). Thus, deciphering the biological functions of PTM dynamics is very important for plant growth and development. Recent advancements in proteomic technologies have greatly accelerated the high-throughput profiling and quantification of PTM events in plants, and massive amounts of plant PTM data have been generated and accumulated (17–19,40–42). Although a number of public databases have been constructed to host PTM information in the past decade (25–29,32), a more comprehensive resource for plant PTMs with a larger number of modified sites especially quantitative information, more types of PTMs and a higher coverage of species is still needed for the academic community.
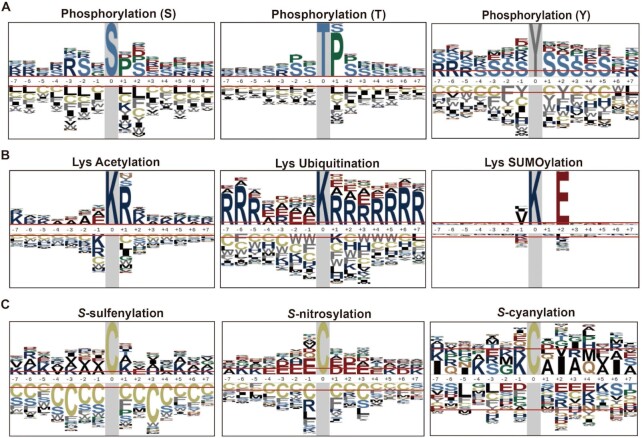
In this work, an integrative database of quantitative PTMs in plants named qPTMplants was developed, and it contains 429 821 PTM sites on 123 551 substrates for 23 PTM types from 43 plant organisms, with 620 509 quantitative events for 136 700 PTM sites on 55 361 proteins under 354 conditions. Because different catalytic enzymes can recognize specific peptides or different amino acid compositions will form diverse microenvironments (43–45), the sequence profile around the modified sites may affect the occurrence of corresponding modification. Therefore, we adopted pLogo (46) to analyze the sequence preferences of different PTMs based on the Arabidopsis dataset in qPTMplants. For phosphorylation, proline (P) and serine (S) were significantly enriched at the +1 and ±2 positions of phosphoserine and phosphothreonine, whereas arginine (R) was highly enriched at the −3 position for phosphoserine. Additionally, S residues were highly abundant surrounding phosphotyrosine (Figure 5A). These results were consistent with previous studies showing that protein phosphorylation could recognize -RXXS-, -SP- and -TP-type motifs (47–49). For three types of lysine modifications with the most modified sites, R was overrepresented at the +1 position for acetylation and highly abundant around the ubiquitination sites, suggesting that these two PTMs may mutually interplay at the same site, whereas glutamic acid (E) was significantly enriched at the +2 position for lysine SUMOylation (Figure 5B). For cysteine modifications, lysine (K) frequently appeared at positions ±5 and +1 surrounding S-sulfenylation sites, while E was overrepresented at positions −1, ±2 and ±3 for S-nitrosylation. Additionally, K was enriched at the −4 and −1 positions, and alanine (A) residues were distributed at the +1 and +3 positions for S-cyanylation (Figure 5C). In summary, different PTM types could recognize diverse sequence features in plants, whereas some PTMs occurring on the same residue appear to interact with each other (20,47–49).
Figure 5.
Sequence characteristics around the PTM sites in Arabidopsis. (A) The sequence preferences for phosphorylation. (B) Sequence preferences for three lysine modifications. (C) Sequence preferences for three cysteine modifications.
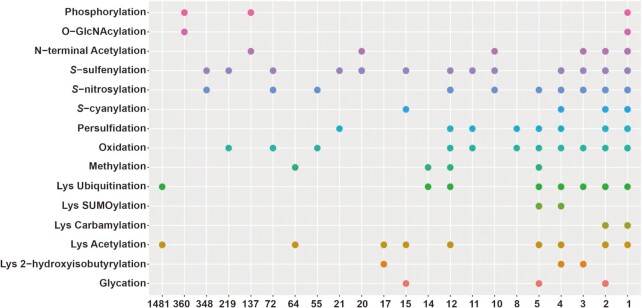
Different PTM types can occur on the same residue or multiple residues of the same protein, and these modifications may collectively modulate various biological processes in a manner named PTM crosstalk (6,50–53). From the collected PTM data of Arabidopsis in qPTMplants, 49 types of PTM crosstalk at the same residue from 6134 PTM events in 2972 sites were identified, including 25 types of crosstalk between two different PTMs and 24 types of crosstalk among multiple PTMs (Figure 6 and Supplementary Table S4). As the representative crosstalk, there were 1481 acetylation–ubiquitination sites, 360 phosphorylation–O-GlcNAcylation sites and 348 S-sulfenylation–S-nitrosylation sites in our results (Figure 6). This suggested that the competitive regulation between acetylation and ubiquitinylation, phosphorylation and O-GlcNAcylation, and S-sulfenylation and S-nitrosylation is widespread in plants. For instance, Xu et al. detected 31 vernalization-associated proteins with both O-GlcNAcylation and phosphorylation in wheat (54). The phosphorylation level of S350 on Fru-bisphosphatealdolase was reduced during vernalization, whereas the O-GlcNAcylation of S350 appeared after vernalization, indicating that the correlation of O-GlcNAcylation and phosphorylation may participate in vernalization regulation (54). Intensive plant PTM crosstalk suggested that a large proportion of serine, threonine, lysine and cysteine residues could be dynamically regulated by various types of PTMs in a complicated manner.
Figure 6.
The distribution of potential crosstalk among different PTM types in Arabidopsis and the numbers of concurrent sites are presented at the bottom.
Taken together, we constructed the qPTMplants database, which not only collected a large amount of PTM data in plants but also provided detailed annotations such as protein information, quantitative dynamics, and sequence and structural characteristics. In the future, qPTMplants will be regularly maintained and annually updated by recruiting more researchers to survey the newly published literature and manually collect the experimentally detected PTM data. In addition, more annotations, such as substrate three-dimensional structures, potential regulatory enzymes of PTMs, information on biotic and abiotic stresses, PTM networks and single-nucleotide polymorphisms, will be integrated into qPTMplants to provide more detailed and comprehensive information. We anticipate that the qPTMplants database will be a useful resource for further studies of PTMs in plants.
Supplementary Material
Contributor Information
Han Xue, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Qingfeng Zhang, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, China.
Panqin Wang, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Bijin Cao, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Chongchong Jia, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Ben Cheng, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Yuhua Shi, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Wei-Feng Guo, School of Electrical Engineering, Zhengzhou University, Zhengzhou 450001, China.
Zhenlong Wang, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
Ze-Xian Liu, State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, China.
Han Cheng, School of Life Sciences, Zhengzhou University, Zhengzhou 450001, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31601067, U2004152, 91953123, 62002329]; Guangdong Innovative and Entrepreneurial Research Team Program [2017ZT07S096]; Tip-Top Scientific and Technical Innovative Youth Talents of Guangdong Special Support Program [2019TQ05Y351]; Key Scientific and Technological Projects of Henan Province [212102310083]; China Postdoctoral Research Foundation [2021M692915]; Henan Postdoctoral Foundation [202002021].
Conflict of interest statement. None declared.
REFERENCES
- 1. Mann M., Jensen O.N.. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 2003; 21:255–261. [DOI] [PubMed] [Google Scholar]
- 2. Seo J., Lee K.J.. Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J. Biochem. Mol. Biol. 2004; 37:35–44. [DOI] [PubMed] [Google Scholar]
- 3. UniProt Consortium UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021; 49:D480–D489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yang W., Zhang W., Wang X.. Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2017; 15:4–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. He Z., Huang T., Ao K., Yan X., Huang Y.. Sumoylation, phosphorylation, and acetylation fine-tune the turnover of plant immunity components mediated by ubiquitination. Front. Plant Sci. 2017; 8:1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vu L.D., Gevaert K., De Smet I.. Protein language: post-translational modifications talking to each other. Trends Plant Sci. 2018; 23:1068–1080. [DOI] [PubMed] [Google Scholar]
- 7. Smirnoff N., Arnaud D. Hydrogen peroxide metabolism and functions in plants. New Phytol. 2019; 221:1197–1214. [DOI] [PubMed] [Google Scholar]
- 8. Martí M.C., Jiménez A., Sevilla F.. Thioredoxin network in plant mitochondria: cysteine S-posttranslational modifications and stress conditions. Front. Plant Sci. 2020; 11:571288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang D., Huo J., Zhang J., Wang C., Wang B., Fang H., Liao W.. Protein S-nitrosylation in programmed cell death in plants. Cell Mol. Life Sci. 2019; 76:1877–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Waszczak C., Akter S., Jacques S., Huang J., Messens J., Van Breusegem F.. Oxidative post-translational modifications of cysteine residues in plant signal transduction. J. Exp. Bot. 2015; 66:2923–2934. [DOI] [PubMed] [Google Scholar]
- 11. You Y., Zhang M., Yang W., Li C., Liu Y., Li C., He J., Wu W.. Starch phosphorylation and the in vivo regulation of starch metabolism and characteristics. Int. J. Biol. Macromol. 2020; 159:823–831. [DOI] [PubMed] [Google Scholar]
- 12. Gupta K.J. Protein S-nitrosylation in plants: photorespiratory metabolism and NO signaling. Sci. Signal. 2011; 4:jc1. [DOI] [PubMed] [Google Scholar]
- 13. Fang Q., Zhang J., Zhang Y., Fan N., van den Burg H.A., Huang C.F.. Regulation of aluminum resistance in Arabidopsis involves the SUMOylation of the zinc finger transcription factor STOP1. Plant Cell. 2020; 32:3921–3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mazur M.J., van den Burg H.A.. Global SUMO proteome responses guide gene regulation, mRNA biogenesis, and plant stress responses. Front. Plant Sci. 2012; 3:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ma X., Claus L.A.N., Leslie M.E., Tao K., Wu Z., Liu J., Yu X., Li B., Zhou J., Savatin D.V.et al.. Ligand-induced monoubiquitination of BIK1 regulates plant immunity. Nature. 2020; 581:199–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zavaliev R., Mohan R., Chen T., Dong X.. Formation of NPR1 condensates promotes cell survival during the plant immune response. Cell. 2020; 182:1093–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nietzel T., Mostertz J., Ruberti C., Née G., Fuchs P., Wagner S., Moseler A., Müller-Schüssele S.J., Benamar A., Poschet G.et al.. Redox-mediated kick-start of mitochondrial energy metabolism drives resource-efficient seed germination. Proc. Natl Acad. Sci. U.S.A. 2020; 117:741–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang Z., Guo G., Yang N., Pun S.S., Ho T.K.L., Ji L., Hu I., Zhang J., Burlingame A.L., Li N.. The change of gravity vector induces short-term phosphoproteomic alterations in Arabidopsis. J. Proteomics. 2020; 218:103720. [DOI] [PubMed] [Google Scholar]
- 19. Yuan B., Liu T., Cheng Y., Gao S., Li L., Cai L., Yang J., Chen J., Zhong K.. Comprehensive proteomic analysis of lysine acetylation in Nicotiana benthamiana after sensing CWMV infection. Front. Microbiol. 2021; 12:672559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhu L., Cheng H., Peng G., Wang S., Zhang Z., Ni E., Fu X., Zhuang C., Liu Z., Zhou H.. Ubiquitinome profiling reveals the landscape of ubiquitination regulation in rice young panicles. Genomics Proteomics Bioinformatics. 2020; 18:305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Uhrig R.G., Schläpfer P., Roschitzki B., Hirsch-Hoffmann M., Gruissem W.. Diurnal changes in concerted plant protein phosphorylation and acetylation in Arabidopsis organs and seedlings. Plant J. 2019; 99:176–194. [DOI] [PubMed] [Google Scholar]
- 22. He D., Li M., Damaris R.N., Bu C., Xue J., Yang P.. Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 2020; 101:1430–1447. [DOI] [PubMed] [Google Scholar]
- 23. Pi E., Zhu C., Fan W., Huang Y., Qu L., Li Y., Zhao Q., Ding F., Qiu L., Wang H.et al.. Quantitative phosphoproteomic and metabolomic analyses reveal GmMYB173 optimizes flavonoid metabolism in soybean under salt stress. Mol. Cell Proteomics. 2018; 17:1209–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Walley J.W., Shen Z., McReynolds M.R., Schmelz E.A., Briggs S.P.. Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl Acad. Sci. U.S.A. 2018; 115:210–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huang K.Y., Lee T.Y., Kao H.J., Ma C.T., Lee C.C., Lin T.H., Chang W.C., Huang H.D.. dbPTM in 2019: exploring disease association and cross-talk of post-translational modifications. Nucleic Acids Res. 2019; 47:D298–D308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li J., Jia J., Li H., Yu J., Sun H., He Y., Lv D., Yang X., Glocker M.O., Ma L.et al.. SysPTM 2.0: an updated systematic resource for post-translational modification. Database. 2014; 2014:bau025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin S., Wang C., Zhou J., Shi Y., Ruan C., Tu Y., Yao L., Peng D., Xue Y.. EPSD: a well-annotated data resource of protein phosphorylation sites in eukaryotes. Brief. Bioinform. 2021; 22:298–307. [DOI] [PubMed] [Google Scholar]
- 28. Xu H., Zhou J., Lin S., Deng W., Zhang Y., Xue Y.. PLMD: an updated data resource of protein lysine modifications. J. Genet. Genomics. 2017; 44:243–250. [DOI] [PubMed] [Google Scholar]
- 29. Wang P., Zhang Q., Li S., Cheng B., Xue H., Wei Z., Shao T., Liu Z.X., Cheng H., Wang Z.. iCysMod: an integrative database for protein cysteine modifications in eukaryotes. Brief. Bioinform. 2021; 22:bbaa400. [DOI] [PubMed] [Google Scholar]
- 30. Zulawski M., Braginets R., Schulze W.X.. PhosPhAt goes kinases—searchable protein kinase target information in the plant phosphorylation site database PhosPhAt. Nucleic Acids Res. 2013; 41:D1176–D1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yao Q., Ge H., Wu S., Zhang N., Chen W., Xu C., Gao J., Thelen J.J., Xu D. P3DB 3.0: from plant phosphorylation sites to protein networks. Nucleic Acids Res. 2014; 42:D1206–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng H., Deng W., Wang Y., Ren J., Liu Z., Xue Y.. dbPPT: a comprehensive database of protein phosphorylation in plants. Database. 2014; 2014:bau121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Willems P., Horne A., Van Parys T., Goormachtig S., De Smet I., Botzki A., Van Breusegem F., Gevaert K.. The Plant PTM Viewer, a central resource for exploring plant protein modifications. Plant J. 2019; 99:752–762. [DOI] [PubMed] [Google Scholar]
- 34. Cruz E.R., Nguyen H., Nguyen T., Wallace I.S.. Functional analysis tools for post-translational modification: a post-translational modification database for analysis of proteins and metabolic pathways. Plant J. 2019; 99:1003–1013. [DOI] [PubMed] [Google Scholar]
- 35. Lamesch P., Berardini T.Z., Li D., Swarbreck D., Wilks C., Sasidharan R., Muller R., Dreher K., Alexander D.L., Garcia-Hernandez M.et al.. The Arabidopsis Information Resource (TAIR): improved gene annotation and new tools. Nucleic Acids Res. 2012; 40:D1202–D1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sakai H., Lee S.S., Tanaka T., Numa H., Kim J., Kawahara Y., Wakimoto H., Yang C.C., Iwamoto M., Abe T.et al.. Rice Annotation Project Database (RAP-DB): an integrative and interactive database for rice genomics. Plant Cell Physiol. 2013; 54:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Klausen M.S., Jespersen M.C., Nielsen H., Jensen K.K., Jurtz V.I., Sønderby C.K., Sommer M.O.A., Winther O., Nielsen M., Petersen B.et al.. NetSurfP-2.0: improved prediction of protein structural features by integrated deep learning. Proteins. 2019; 87:520–527. [DOI] [PubMed] [Google Scholar]
- 38. Mészáros B., Erdos G., Dosztányi Z.. IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res. 2018; 46:W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., Madden T.L.. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008; 36:W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mao M., Xue Y., He Y., Zhou X., Rafique F., Hu H., Liu J., Feng L., Yang W., Li X.et al.. Systematic identification and comparative analysis of lysine succinylation between the green and white parts of chimeric leaves of Ananas comosus var. bracteatus. BMC Genomics. 2020; 21:383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cho H.Y., Wen T.N., Wang Y.T., Shih M.C.. Quantitative phosphoproteomics of protein kinase SnRK1 regulated protein phosphorylation in Arabidopsis under submergence. J. Exp. Bot. 2016; 67:2745–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rodriguez M.C., Mehta D., Tan M., Uhrig R.G.. Quantitative proteome and PTMome analysis of Arabidopsis thaliana root responses to persistent osmotic and salinity stress. Plant Cell Physiol. 2021; 62:1012–1029. [DOI] [PubMed] [Google Scholar]
- 43. Chamberlain L.H., Shipston M.J.. The physiology of protein S-acylation. Physiol. Rev. 2015; 95:341–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wible R.S., Sutter T.R.. Soft Cysteine Signaling Network: the functional significance of cysteine in protein function and the soft acids/bases thiol chemistry that facilitates cysteine modification. Chem. Res. Toxicol. 2017; 30:729–762. [DOI] [PubMed] [Google Scholar]
- 45. Deng S., Marmorstein R.. Protein N-terminal acetylation: structural basis, mechanism, versatility, and regulation. Trends Biochem. Sci. 2021; 46:15–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. O'Shea J.P., Chou M.F., Quader S.A., Ryan J.K., Church G.M., Schwartz D. pLogo: a probabilistic approach to visualizing sequence motifs. Nat. Methods. 2013; 10:1211–1212. [DOI] [PubMed] [Google Scholar]
- 47. Vlad F., Turk B.E., Peynot P., Leung J., Merlot S.. A versatile strategy to define the phosphorylation preferences of plant protein kinases and screen for putative substrates. Plant J. 2008; 55:104–117. [DOI] [PubMed] [Google Scholar]
- 48. Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., Gruissem W., Baginsky S.. Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 2009; 150:889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang X., Bian Y., Cheng K., Gu L.F., Ye M., Zou H., Sun S.S., He J.X.. A large-scale protein phosphorylation analysis reveals novel phosphorylation motifs and phosphoregulatory networks in Arabidopsis. J. Proteomics. 2013; 78:486–498. [DOI] [PubMed] [Google Scholar]
- 50. Huang R., Huang Y., Guo Y., Ji S., Lu M., Li T.. Systematic characterization and prediction of post-translational modification cross-talk between proteins. Bioinformatics. 2019; 35:2626–2633. [DOI] [PubMed] [Google Scholar]
- 51. Zhang Y., Zeng L.. Crosstalk between ubiquitination and other post-translational protein modifications in plant immunity. Plant Commun. 2020; 1:100041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Demidov D., Hesse S., Tewes A., Rutten T., Fuchs J., Ashtiyani R.K., Lein S., Fischer A., Reuter G., Houben A.. Aurora1 phosphorylation activity on histone H3 and its cross-talk with other post-translational histone modifications in Arabidopsis. Plant J. 2009; 59:221–230. [DOI] [PubMed] [Google Scholar]
- 53. Csizmok V., Forman-Kay J.D.. Complex regulatory mechanisms mediated by the interplay of multiple post-translational modifications. Curr. Opin. Struct. Biol. 2018; 48:58–67. [DOI] [PubMed] [Google Scholar]
- 54. Xu S., Xiao J., Yin F., Guo X., Xing L., Xu Y., Chong K.. The protein modifications of O-GlcNAcylation and phosphorylation mediate vernalization response for flowering in winter wheat. Plant Physiol. 2019; 180:1436–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.