Abstract
5-Methylcytosine (m5C) is one of the most prevalent covalent modifications on RNA. It is known to regulate a broad variety of RNA functions, including nuclear export, RNA stability and translation. Here, we present m5C-Atlas, a database for comprehensive collection and annotation of RNA 5-methylcytosine. The database contains 166 540 m5C sites in 13 species identified from 5 base-resolution epitranscriptome profiling technologies. Moreover, condition-specific methylation levels are quantified from 351 RNA bisulfite sequencing samples gathered from 22 different studies via an integrative pipeline. The database also presents several novel features, such as the evolutionary conservation of a m5C locus, its association with SNPs, and any relevance to RNA secondary structure. All m5C-atlas data are accessible through a user-friendly interface, in which the m5C epitranscriptomes can be freely explored, shared, and annotated with putative post-transcriptional mechanisms (e.g. RBP intermolecular interaction with RNA, microRNA interaction and splicing sites). Together, these resources offer unprecedented opportunities for exploring m5C epitranscriptomes. The m5C-Atlas database is freely accessible at https://www.xjtlu.edu.cn/biologicalsciences/m5c-atlas.
INTRODUCTION
To date, >170 ribonucleotide modifications have been identified in various RNA types. These modifications affect transcript functions and regulate gene expression, in part by influencing the intramolecular interaction of RNA with RNA binding proteins through changing the local RNA 3D structures (1). Although a large number of RNA modifications have been identified, the underlying molecular mechanisms remain largely unclear. As one of the most common post-transcriptionally modified bases, 5-methylcytosine (m5C) is a dynamic RNA marker found in most eukaryotes, prokaryotes and archaea (2). High throughput and biochemical studies have shown that m5C is widely distributed over all RNA species and plays essential roles in RNA biology. For example, m5C regulates ribosome synthesis and processing by altering the conformation of rRNA (3), thereby affecting translation fidelity (4). m5C sites on tRNA are evolutionarily conserved and contribute to maintenance of tertiary structure (5,6). m5C has also been reported to function in mRNA, influencing its turnover (7,8), export from nucleus (9) and translation (5,10). Additionally, aberrant levels of RNA cytosine methylation are implicated in various disease states (8,11–13). While extensive research has been carried out, the full regulatory functions of RNA m5C remain unclear.
High-throughput technologies for RNA m5C detection
With recent advances in high-throughput sequencing (HTS) technology, the amount of data on RNA methylation is huge and continues to grow. Several HTS technologies have been developed to detect RNA m5C methylation, including m5C immunoprecipitation sequencing (m5C-RIP-seq) (7,14), m5C RNA bisulfite sequencing (m5C BS-seq) (15), m5C methylation individual nucleotide resolution crosslinking immunoprecipitation (m5C-miCLIP) (16,17), 5-azacytidine-mediated RNA immunoprecipitation (5-Aza-IP) (18) and TET-Assisted peroxotungstate oxidation sequencing (TAWO-Seq) (19). Among them, m5C-RIP-seq applies anti-m5C antibodies to enrich the methylation-containing fragments, yielding low resolution methylation peaks (from 50 bp to 150 bp). Similar to the design of RIP-Seq, 5-Aza-IP incorporates cytidine nucleoside 5-azacytidine into RNA, and uses antibodies to capture the methyltransferase that is covalently linked with 5-azacytidine residues. m5C-miCLIP and photo-crosslinking-assisted m5C sequencing (PA-m5C) are technologies that permit the detection of m5C at single nucleotide resolution (20). A key limitation of 5-Aza-IP, miCLIP and PA-m5C lies in the requirement for overexpression of methyltransferases in the cell to interact with the modified bases, restricting detection to only a subset of sites targeted by the specific writer. RNA bisulfite sequencing (RNA BS-seq) has been a gold standard technique in the detection of cytosine modification, because it is a reverse transcription-based method which permits methylation quantification at m5C sites. TAWO-Seq is another technique using chemical conversion: it applies peroxotungstate oxidation to distinguish hm5C from m5C. However, the reverse transcription-based methods still face some technical limitations, such as the incomplete conversion of unmethylated cytosine (21), false positive sites confounded by RNA secondary structures (22), and artifacts introduced by different filtering methods during data processing. Overall, these HTS technologies have their own unique sets of advantages and shortcomings in RNA m5C detection.
Construction of m5C-Atlas
To date, several publicly available databases, such as MODOMICS and RMBase, have been curated for epitranscriptomics. Among them, MODOMICS is a database of RNA modification with a focus on the chemical structures of modified ribonucleotides, corresponding biosynthetic pathways and RNA modification enzymes (1). RMBase is an epitranscriptome database that contains 1 397 000 modification sites detected by HTS techniques, covering multiple types of modifications such as m6A, pseudouridine and m5C (23). Both databases contain information on m5C with different biological and technical perspectives. However, these databases are not specifically developed for m5C and only contain a fraction of the available information on RNA cytosine methylation. We have therefore constructed m5C-Atlas (Figure 1), the first comprehensive database exclusively for RNA 5-methylcytosine, to help decipher the m5C epitranscriptomes.
Figure 1.
The design of m5C-Atlas. m5C-Atlas features a high-confidence collection of m5C sites and quantitative profiles in multiple species. Through a rigorous filtering process, most potential false positive sites analyzed by pipeline were removed. It also provides conservation information among three vertebrate species and the disease-association of individual m5C sites
Compared with the existing epitranscriptome databases, m5C-Atlas features a high-confidence collection of reliable m5C sites from single base resolution technologies (see Table 1). Relative to other low-resolution technologies (eg. MeRIP-Seq), the base-resolution methods offer superior accuracy and reliability. The methylation levels of the putative m5C sites were further quantified from various BS-Seq samples derived from different cell lines and tissues. The collected methylation sites span diverse species, including animal, plant, microorganisms and viruses (see Table 2). Besides basic gene annotations, m5C-Atlas also provides a rich set of functional annotations, such as the evolutionary conservation of the modification sites between vertebrates (human, mouse and zebrafish), overlap with RNA binding protein (RBP), miRNA and splicing junctions, and any single nucleotide polymorphism (SNP) associated with the loss of the m5C methylation locus. In addition, the stem region of RNA secondary structure was predicted by RNAfold using sequences of mature RNA transcripts and annotated for user (24).
Table 1.
Comparison of m5C-Atlas with other databases
| m5C-Atlas | MODOMICS | RMBase | |
|---|---|---|---|
| High-accuracy m5C sites | More Complete | Limited Volume | Incomplete |
| Detection methods & species | 6 &13 | LC–MS | 1 & 3 |
| Quantitative methylation profiles | Yes | - | - |
| Matched gene expression profiles | Yes | ||
| Conservation in vertebrates | Yes | - | - |
| Viral m5C sites | Yes | - | - |
| RNA modification pathways | - | Yes | - |
| Condition-specific m5C profiles | Yes | - | - |
| Putative secondary structure | Yes | - | - |
| Post-transcriptional annotations | Yes | - | Yes |
Table 2.
Contents of m5C-Atlas database
| Species | Site number (mRNA) | Cell line/tissue | condition/treatment | Quantitative profiles | Basic annotation | Disease association | Conservation in vertebrates |
|---|---|---|---|---|---|---|---|
| Homo sapiens | 124 105 | 22 | 91 | Yes | Yes | Yes | Yes |
| Mus musculus | 16 279 | 17 | 13 | Yes | Yes | Yes | Yes |
| Danio rerio | 7846 | 1 | 7 | Yes | Yes | Yes | Yes |
| Drosophila melanogaster | 5421 | 2 | 3 | Yes | Yes | Yes | - |
| Arabidopsis thaliana | 684 | 6 | 22 | Yes | Yes | Yes | - |
| Saccharomyces cerevisiae | 1539 | 1 | 2 | Yes | Yes | - | - |
| Brassica rapa | 21 | 1 | 2 | Yes | Yes | - | - |
| Caenorhabditis elegans | 4 | 1 | 1 | Yes | Yes | - | - |
| Ginkgo biloba | 5 | 1 | 2 | Yes | Yes | - | - |
| Murine leukemia virus | 5 | 1 | 1 | Yes | Yes | - | - |
| Triticum turgidum subsp | 17 | 1 | 2 | Yes | Yes | - | - |
| Nannochloropsis oculata | 39 | 1 | 2 | - | - | - | - |
| Human immunodeficiency virus | 31 | 2 | 1 | - | - | - | - |
MATERIALS AND METHODS
High-confidence collection of reliable m5C sites
High-confidence m5C sites were collected from 22 datasets in the NCBI GEO database (25), covering 13 species, such as human, mouse, zebrafish, fly, Arabidopsis, yeast and viruses. The other positional information was also downloaded from original articles or relevant GEO datasets. Moreover, in order to report the m5C sites on tRNA and rRNA, the processed data were download from the NCBI GEO database (18), supplementary material (26) and previous studies (23) directly. A total of 351 bisulfite sequencing samples were obtained and analyzed for filtering high-confidence m5C sites (Supplementary Table S1). The raw m5C bisulfite sequencing data were directly downloaded from the NCBI GEO database. Adaptor contaminations and low-quality nucleotides were trimmed by Trim Galore with parameter setting -stringency 1 -length 35 (27). The clean reads were mapped to reference genomes by an RNA BS-seq alignment tool with default parameters, meRanGh available with meRanTK version 1.2.1b (28). The unique mapped reads were then selected by Samtools (samtools view -F 12 -q 30) and the reads with mapping quality larger than 30 were used to call candidate m5C sites by meRanCall (-md 5 -sc 10 -ei 0.1 -cr 0.99 -fdr 0.01 -bed63 -np -gref). The false discovery rate (FDR) of methylated cytosines were controlled at 0.01. Besides, all m5C sites were filtered by the minimal coverage of 30 (the detail scripts and pipelines for processing the raw data were available in Supplementary Material). In addition, the IVT m5C sites were used as the negative data to filter false positive sites (29).
Although the meRanTK-based pipeline is classic, recent studies provided more stringent pipelines to process BS-seq on mRNA (21) and tRNA (26), respectively. To improve the reliability of m5C sites, the stringent pipeline was used to process the BS-seq data also. By this advanced filter pipeline from Huang et al.(21) the site calling for different species were performed with parameters: -c 20 -C 3 -r 0.1 -p 0.05 –cutoff 3 –CR gene –method binomial. The m5C sites identified by this optimized pipeline are available for download on the download page.
Secondary structure annotation
RNA secondary structures are essential for RNA stability and function. The high temperature during bisulfite treatment during DNA BS-Seq can cause severe RNA degradation. Thus, the temperature of the treatment process for RNA is lower than DNA, so that incomplete conversion is a major issue of the technique. We used the RNA secondary structure prediction software RNAfold to infer the secondary structure of the region where the m5C sites were located (MEA 0.1, -T 70). This information was annotated to suggest potential thermal stable stem structure for user, since the hybridized regions of nucleic acids are often resistant to bisulfite conversion (21,22).
Quantitative profiles of putative m5C sites
The coverage number and cytosine count were recorded from candidate m5C site files as calculated by meRanCall. The methylation levels of m5C sites in different conditions, treatments, cell lines or tissues were also calculated and recorded. In addition, the gene expression levels in different samples were quantified from BAM files of the aligned BS-Seq by StringTie (gene expression levels in TPM).
Conservation of m5C sites in vertebrates
In the cross-species comparative analysis, we used the LiftOver tool from the UCSC genome browser to map the m5C locus of a single species to the homologous coordinates of the target species (30,31). If the mapped sites also have m5C modifications in the target species, then these methylation sites will be considered to have methylation conservation between the two species. In addition, for the human m5C sites, the conservation level of putative sites was calculated and shown in m5C-Atlas. Two types of conservation scores, phastCons (32) and fitCons (33), were used to represent the degree of evolutionary conservation under the genomic region of the corresponding RNA methylation site.
Basic annotation for m5C sites
In addition to basic gene and transcript annotation information (34), splice sites, miRNA target sites and RNA binding protein (RBP) binding sites were also integrated into m5C-Atlas to help understand the potential functions and regulation of m5C in various aspects (see Supplementary Table S2). The miRNA target sites and RBP intermolecular interaction with RNA information were obtained from starBase (35) and POSTAR (36) respectively. The splicing site information were obtained from the UCSC database (37).
Potential involvement of individual m5C sites in pathogenesis
By comparison with previous databases (RMVar (38) and RMDisease (39)), it is assumed that the disease caused by single-base mutation may be induced by the loss of m5C methylation, the instability of RNA structure, function changes or changes in downstream interactions due to the changes in cytosine position caused by mutation. Therefore, the analysis of diseases caused by modified cytosine can implicate the potential involvement of a single m5C site in pathogenesis. The site mutation and disease data used in this analysis were obtained from dbSNP database (40).
Database and web interface implementation
MySQL was used to store and manage metadata in m5C-Atlas. Hypertext Markup Language (HTML), Cascading Style Sheets (CSS) and Hypertext Preprocessor (PHP) were used to build the displayed Web interface. The Jbrowse genome browser was used for interactive exploration and visualization of related records (41).
RESULT
m5C-Atlas aggregates a total of 166,540 high-reliability sites (see Table 2 and Supplementary Table S2). The coverage of each site is greater than 30 and high-stringency filtration was performed through stringent pipeline and other methods. These sites cover 13 species, including human (134 649 sites), mouse (16 279 sites), zebrafish (7846 sites), fly (5421 sites), Arabidopsis (684 sites), Saccharomyces cerevisiae (1539 sites), Brassica rapa (21 sites), Caenorhabditis elegans (4 sites), Ginkgo biloba (5 sites), Nannochloropsis oculata (39 sites), Triticum turgidum subsp (17 sites) and two viruses, Human immunodeficiency virus (31 sites), Murine leukemia virus (5 sites). For human m5C data, three distinct techniques were used to support it, including MeRIP-seq, Aza-IP-seq, and BS-seq.
Quantitative profiles were estimated over 206 experimental conditions (different cell lines, tissues or experimental treatments), which were gathered from 351 high-throughput sequencing samples including the methylation level of sites, the number of cytosines, and the coverage number. Among them, there are 177 human m5C modification high-throughput sequencing samples, including 22 cell lines/tissues, and 91 different experimental treatments or conditions. For 69 mouse high-throughput sequencing samples, 17 cell lines/tissues and 13 different experimental treatments or conditions were provided in m5C-Atlas (for the other 11 species, please refer to Table 2). The BAM files of high-throughput samples are used to obtain gene expression through StringTie, and are matched to each site according to the condition, cell line and treatment, providing a reference for the user's analysis and processing.
Basic annotations for 11 species, including gene annotation information for 11 species, RBP information for five species, splicing site information for four species, and miRNA information for three species were all generated (Supplementary Table S2). Disease association information (SNP) caused by single m5C site mutation was also analyzed and displayed in 5 species. Moreover, conservation analysis was performed among human, mouse and zebrafish to identify the conserved m5C sites between two vertebrate species (see Table 2).
The main function of Atlas is to collect, reprocess, and display high-quality location information for users. Users can search the gene/gene interval of interest on the homepage to obtain the m5C methylation data of this gene/gene interval. Users can also click on different modules (mRNA, tRNA, rRNA), select different species, organelles, and annotation information under different modules to perform range screening to view m5C methylation site information. In addition, all information can be downloaded from the download module.
Case study on lncRNA: XIST
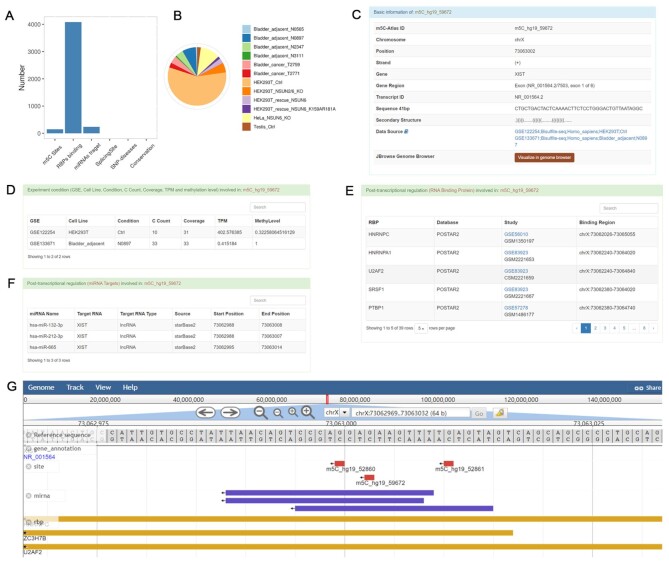
XIST is a long non-coding RNA (lncRNA) expressed in the X inactivation center (XIC). After transcription, XIST is not translated into protein, but instead silences gene transcription on one of the two X chromosomes of female mammals (42,43). Previous studies have identified m5C on functionally important region of XIST, which can regulate the interaction between XIST and the chromatin-associated protein complex, PRC2 (44). Searching by gene name, XIST, at the front page of m5C-Atlas database, returns 147 m5C sites and statistical graphs (Figure 2A and B). The field of species and tissue/cell line allows users to select organisms and the supporting cellular conditions. Meanwhile, users can query the specific m5C sites and then filter the associated RBP, miRNA, splicing site or SNP to investigate on specific functional annotation. Detailed information of the sites will be displayed after clicking on one of the site ID (m5C_hg19_59672 for example). The basic information section includes the site ID number, chromosome, position, strand, gene, gene region, transcript ID, 41 bp reference sequence, secondary structure and data source information (Figure 2C). The experimental condition section reports one or more biological samples under which the site is m5C modifiable. For our example (m5C_hg19_59672), the table shows that the site was observed to be m5C modifiable under two experimental conditions (Figure 2D). The GSE number, cell line, condition, cytosine count, coverage, TPM and methylation level of each experimental condition are all listed in the table. Other annotation information is listed after the experimental condition part, including RBP, miRNA-RNA interaction, splicing site, SNP and multi-technical support (the number of technologies that can detect this site). For site m5C_hg19_59672, the database shows that it lies within the interaction range of 39 RBPs and 3 miRNAs (Figure 2F and E, respectively). These information, in turn, can be used to evaluate the interaction between m5C sites and other post-transcriptional regulators. User can also click ‘Visualize in genome browser’ (Figure 2C) to display the Jbrowse (Figure 2G).
Figure 2.
The m5C site of lncRNA XIST and related information. (A) The bar chart showing the number of m5C sites on the XIST gene, RBP, miRNAs target, splicing site, SNP diseases, and conservation site. (B) 147 m5C sites records of XIST were detected and statistic in different conditions and cell lines/tissues. (C) Basic information of m5C_hg19_59672 site on XIST. (D) Different experimental conditions that can detect the m5C site. (E) RNA binding protein part provides the information that the RBP interaction region covers this m5C site. (F) miRNA target part provides the information that the miRNA interaction region covers this m5C site. (G) Jbrowse of m5C_hg19_59672 site.
Case study on protein-coding gene: NECTIN2
NECTIN2 is an immunoglobulin-like molecular protein. It plays a fundamental role in the formation of adhesion and tight junctions between epithelial cells and fibroblasts (45). As an immune molecular ligand, NECTIN2 is upregulated on the surface of virus-infected cells and cancer cells, thereby activating CD226 to mediate the recognition of toxic lymphocytes and the killing of abnormal cells (46). A total of five records related to ‘NECTIN2’ are returned after querying its gene name at the front page of the m5C-Atlas. These records can be further selected by filtering the associated RBP, miRNA, splice site, and SNP information. Detailed information will be returned after clicking the specific m5C sites. The website will be redirected to the source page of the GEO database after selecting the samples under specific cellular conditions. In addition, it is convenient to find that the selected site was identified in 93 different-condition experiments under the experimental condition section. The query page provides users a summary of m5C status on NECTIN2, suggesting that cytosine methylation may have a potential role in regulating the function of the mRNA transcripts of NECTIN2, since >90 experimental conditions were found this m5C site. In the section of technique annotation, the site has also been reported as m5C-modifiable under certain conditions of the RBS-seq technology which increases confidence that the locus is truly m5C modifiable. After examine its associated post-transcriptional regulation for RBP and splicing site, three RBPs and one splicing site were found to overlap or be close to this m5C site. Interestingly, the m5C formation at this site is silenced by a known disease-related SNP, which might indicate that the site is involved in the pathogenesis of uterine corpus endometrial carcinoma.
Case study on protein-coding gene: HDGF
Hepatoma-derived growth factor (HDGF) is a hairpin-binding growth factor associate with several cancer types including breast, lung and pancreatic cancers (47–49). As an oncogene, HDGF was identified that relates to metastatic tumour progression by stabilizing the mRNA through the binding of m5C modified sites at 3’ UTR to YBX1 and NSUN2 (8). Searching for the gene name ‘HDGF’ at the front page of the m5C-Atlas database, a total of 84 records related to ‘HDGF’ are returned. These records can be further filtered by RBP, miRNA, splice site, and SNP information. After clicking m5C sites (here we choose site m5C_hg19_67285), more information of this site will be provided. Under experiment condition part, the site is identified to appear in 45 different-condition experiments with high coverage. In the conservation annotation part, this site has been identified to conserve with the site of mouse (chr3:87914991+). Moreover, the phastCons and fitCons of this site has also been calculated and displayed. In post-transcriptional regulation for RBP and splicing site parts, 16 RBPs and one splicing site were found to be associate with this m5C site.
CONCLUSION
With recent advances in HTS technologies, the transcriptomic profiles of RNA modifications under different biological conditions have been revealed. As one of the most prevalent post transcriptional modifications on RNA, 5-methylcytosine has received much attention during the last few years. Many biological processes, including cell development and carcinogenesis (8), have been linked to both the topology and dynamics of RNA m5C. Despite its important roles in RNA biology, a database for RNA m5C is still lacking. Here, we utilized advanced pipelines, secondary structure annotation, IVT, multiple technologies overlapping and stringent method which put forward previously to analyze the data to present m5C-Atlas, a new and comprehensive knowledgebase for deciphering the m5C epitranscriptome.
We collected quantitative data of m5C methylation sites from 13 species and re-processed the RNA bisulfite sequencing datasets using two protocols: a classic pipeline and a stringent pipeline. The reference sites are merged from Aza-IP, miCLIP and RBS-seq, enabling exploration of the consistency between different techniques. In addition, m5C-Atlas provides insights into the functions of individual m5C sites via functional annotations such as conservation in vertebrate species and the association with the diseases-related SNPs. Other transcriptomic data, such as RBP, miRNA, and splicing sites were all incorporated into the m5C-Atlas, and are available both as results tables and as tracks on a genome browser. These resources will provide researchers with new opportunities to study the function of m5C epitranscriptomes.
Although the m5C-Atlas is a comprehensive database for 5-methylcytosine, there are still some limitations should be improved in the current version, 1), the data quality for each samples or species is unequal, for example, the raw data for Brassica rapa, Caenorhabditis elegans, Ginkgo biloba, Nannochloropsis oculata and Triticum turgidum subsp contained rRNA information, which reduced the mRNA information in sequencing, resulting the less m5C sites were reported on mRNA; 2), the current pipeline for tRNA m5C calling is still a challenge, the sequences of tRNA isodecoders are highly similar, and the current BS-seq is difficult to map m5C sites to exact tRNA; 3), The annotation information of some species, such as Nannochloropsis oculata, are incomplete.
Supplementary Material
ACKNOWLEDGEMENTS
Author's contributions: K. Chen and Z. Wei conceived the idea; J. Ma processed the epitranscriptome datasets and implemented the functional annotations; B. Song. constructed the web interface; J. Ma drafted the manuscript. All authors read, critically revised and approved the final manuscript. We thank for the Jianheng Liu from Sun Yat-sen University for his help in the data processing and the discussion for tRNA m5C BS-seq.
Contributor Information
Jiongming Ma, Key Laboratory of Gastrointestinal Cancer (Fujian Medical University), Ministry of Education, School of Basic Medical Sciences, Fujian Medical University, Fuzhou 350004, China; Department of Biological Sciences, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China.
Bowen Song, Department of Mathematical Sciences, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China; Institute of Systems, Molecular and Integrative Biology, University of Liverpool, L69 7ZB, Liverpool, UK.
Zhen Wei, Department of Biological Sciences, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China; Institute of Ageing & Chronic Disease, University of Liverpool, L69 7ZB, Liverpool, UK.
Daiyun Huang, Department of Biological Sciences, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China; Department of Computer Science, University of Liverpool, L69 7ZB, Liverpool, UK.
Yuxin Zhang, Department of Biological Sciences, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China.
Jionglong Su, School of AI and Advanced Computing, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China.
João Pedro de Magalhães, Institute of Ageing & Chronic Disease, University of Liverpool, L69 7ZB, Liverpool, UK.
Daniel J Rigden, Institute of Systems, Molecular and Integrative Biology, University of Liverpool, L69 7ZB, Liverpool, UK.
Jia Meng, Department of Biological Sciences, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China; AI University Research Centre, Xi’an Jiaotong-Liverpool University, Suzhou, Jiangsu 215123, China; Institute of Systems, Molecular and Integrative Biology, University of Liverpool, L69 7ZB, Liverpool, UK.
Kunqi Chen, Key Laboratory of Gastrointestinal Cancer (Fujian Medical University), Ministry of Education, School of Basic Medical Sciences, Fujian Medical University, Fuzhou 350004, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31671373]; XJTLU Key Program Special Fund [KSF-T-01, KSF-E-51 and KSF-P-02]. Funding for open access charge: School of Basic Medical Sciences, Fujian Medical University.
Conflict of interest statement. None declared.
REFERENCES
- 1. Boccaletto P., Machnicka M.A., Purta E., Piatkowski P., Baginski B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A.et al.. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018; 46:D303–D307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen Y.S., Yang W.L., Zhao Y.L., Yang Y.G.. Dynamic transcriptomic m(5) C and its regulatory role in RNA processing. Wiley Interdiscip. Rev. RNA. 2021; 12:e1639. [DOI] [PubMed] [Google Scholar]
- 3. Sharma S., Yang J., Watzinger P., Kotter P., Entian K.D.. Yeast Nop2 and Rcm1 methylate C2870 and C2278 of the 25S rRNA, respectively. Nucleic Acids Res. 2013; 41:9062–9076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heissenberger C., Liendl L., Nagelreiter F., Gonskikh Y., Yang G., Stelzer E.M., Krammer T.L., Micutkova L., Vogt S., Kreil D.P.et al.. Loss of the ribosomal RNA methyltransferase NSUN5 impairs global protein synthesis and normal growth. Nucleic Acids Res. 2019; 47:11807–11825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tuorto F., Liebers R., Musch T., Schaefer M., Hofmann S., Kellner S., Frye M., Helm M., Stoecklin G., Lyko F.. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat. Struct. Mol. Biol. 2012; 19:900–905. [DOI] [PubMed] [Google Scholar]
- 6. Motorin Y., Helm M.. tRNA stabilization by modified nucleotides. Biochemistry. 2010; 49:4934–4944. [DOI] [PubMed] [Google Scholar]
- 7. Cui X., Liang Z., Shen L., Zhang Q., Bao S., Geng Y., Zhang B., Leo V., Vardy L.A., Lu T.et al.. 5-Methylcytosine RNA methylation in arabidopsis thaliana. Mol. Plant. 2017; 10:1387–1399. [DOI] [PubMed] [Google Scholar]
- 8. Chen X., Li A., Sun B.F., Yang Y., Han Y.N., Yuan X., Chen R.X., Wei W.S., Liu Y., Gao C.C.et al.. 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 2019; 21:978–990. [DOI] [PubMed] [Google Scholar]
- 9. Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W.et al.. 5-methylcytosine promotes mRNA export - NSUN2 as the methyltransferase and ALYREF as an m(5)C reader. Cell Res. 2017; 27:606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chan C.T., Dyavaiah M., DeMott M.S., Taghizadeh K., Dedon P.C., Begley T.J.. A quantitative systems approach reveals dynamic control of tRNA modifications during cellular stress. PLoS Genet. 2010; 6:e1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Blanco S., Bandiera R., Popis M., Hussain S., Lombard P., Aleksic J., Sajini A., Tanna H., Cortes-Garrido R., Gkatza N.et al.. Stem cell function and stress response are controlled by protein synthesis. Nature. 2016; 534:335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blanco S., Dietmann S., Flores J.V., Hussain S., Kutter C., Humphreys P., Lukk M., Lombard P., Treps L., Popis M.et al.. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J. 2014; 33:2020–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Flores J.V., Cordero-Espinoza L., Oeztuerk-Winder F., Andersson-Rolf A., Selmi T., Blanco S., Tailor J., Dietmann S., Frye M.. Cytosine-5 RNA methylation regulates neural stem cell differentiation and motility. Stem Cell Rep. 2017; 8:112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang L., Perrera V., Saplaoura E., Apelt F., Bahin M., Kramdi A., Olas J., Mueller-Roeber B., Sokolowska E., Zhang W.et al.. m(5)C methylation guides systemic transport of messenger RNA over graft junctions in plants. Curr. Biol. 2019; 29:2465–2476. [DOI] [PubMed] [Google Scholar]
- 15. Schaefer M., Pollex T., Hanna K., Lyko F.. RNA cytosine methylation analysis by bisulfite sequencing. Nucleic Acids Res. 2009; 37:e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Van Haute L., Dietmann S., Kremer L., Hussain S., Pearce S.F., Powell C.A., Rorbach J., Lantaff R., Blanco S., Sauer S.et al.. Deficient methylation and formylation of mt-tRNA(Met) wobble cytosine in a patient carrying mutations in NSUN3. Nat. Commun. 2016; 7:12039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hussain S., Sajini A.A., Blanco S., Dietmann S., Lombard P., Sugimoto Y., Paramor M., Gleeson J.G., Odom D.T., Ule J.et al.. NSun2-mediated cytosine-5 methylation of vault noncoding RNA determines its processing into regulatory small RNAs. Cell Rep. 2013; 4:255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Khoddami V., Cairns B.R.. Identification of direct targets and modified bases of RNA cytosine methyltransferases. Nat. Biotechnol. 2013; 31:458–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan F., Bi Y., Siejka-Zielinska P., Zhou Y.L., Zhang X.X., Song C.X.. Bisulfite-free and base-resolution analysis of 5-methylcytidine and 5-hydroxymethylcytidine in RNA with peroxotungstate. Chem. Commun. (Camb.). 2019; 55:2328–2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Courtney D.G., Tsai K., Bogerd H.P., Kennedy E.M., Law B.A., Emery A., Swanstrom R., Holley C.L., Cullen B.R.. Epitranscriptomic addition of m(5)C to HIV-1 transcripts regulates viral gene expression. Cell Host Microbe. 2019; 26:217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang T., Chen W., Liu J., Gu N., Zhang R.. Genome-wide identification of mRNA 5-methylcytosine in mammals. Nat. Struct. Mol. Biol. 2019; 26:380–388. [DOI] [PubMed] [Google Scholar]
- 22. Amort T., Rieder D., Wille A., Khokhlova-Cubberley D., Riml C., Trixl L., Jia X.Y., Micura R., Lusser A.. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017; 18:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xuan J.J., Sun W.J., Lin P.H., Zhou K.R., Liu S., Zheng L.L., Qu L.H., Yang J.H.. RMBase v2.0: deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018; 46:D327–D334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sayers E.W., Beck J., Bolton E.E., Bourexis D., Brister J.R., Canese K., Comeau D.C., Funk K., Kim S., Klimke W.et al.. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2021; 49:D10–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liu J., Huang T., Zhang Y., Zhao T., Zhao X., Chen W., Zhang R.. Sequence-and structure-selective mRNA m5C methylation by NSUN6 in animals. Natl. Sci. Rev. 2021; 8:nwaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Storvall H., Ramskold D., Sandberg R.. Efficient and comprehensive representation of uniqueness for next-generation sequencing by minimum unique length analyses. PLoS One. 2013; 8:e53822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rieder D., Amort T., Kugler E., Lusser A., Trajanoski Z.. meRanTK: methylated RNA analysis ToolKit. Bioinformatics. 2016; 32:782–785. [DOI] [PubMed] [Google Scholar]
- 29. Zhang Z., Chen T., Chen H.X., Xie Y.Y., Chen L.Q., Zhao Y.L., Liu B.D., Jin L., Zhang W., Liu C.et al.. Systematic calibration of epitranscriptomic maps using a synthetic modification-free RNA library. Nat. Methods. 2021; 18:1213–1222. [DOI] [PubMed] [Google Scholar]
- 30. Haeussler M., Zweig A.S., Tyner C., Speir M.L., Rosenbloom K.R., Raney B.J., Lee C.M., Lee B.T., Hinrichs A.S., Gonzalez J.N.et al.. The UCSC Genome Browser database: 2019 update. Nucleic Acids Res. 2019; 47:D853–D858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Song B., Chen K., Tang Y., Wei Z., Su J., de Magalhães J.P., Rigden D.J., Meng J.. ConsRM: collection and large-scale prediction of the evolutionarily conserved RNA methylation sites, with implications for the functional epitranscriptome. Brief. Bioinform. 2021; 22:bbab088. [DOI] [PubMed] [Google Scholar]
- 32. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S.et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gulko B., Hubisz M.J., Gronau I., Siepel A.. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015; 47:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yu G., Wang L.G., He Q.Y.. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015; 31:2382–2383. [DOI] [PubMed] [Google Scholar]
- 35. Li J.H., Liu S., Zhou H., Qu L.H., Yang J.H.. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014; 42:D92–D97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu Y., Xu G., Yang Y.T., Xu Z., Chen X., Shi B., Xie D., Lu Z.J., Wang P.. POSTAR2: deciphering the post-transcriptional regulatory logics. Nucleic Acids Res. 2019; 47:D203–D211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lawrence M., Huber W., Pages H., Aboyoun P., Carlson M., Gentleman R., Morgan M.T., Carey V.J.. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013; 9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Luo X., Li H., Liang J., Zhao Q., Xie Y., Ren J., Zuo Z.. RMVar: an updated database of functional variants involved in RNA modifications. Nucleic Acids Res. 2021; 49:D1405–D1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen K., Song B., Tang Y., Wei Z., Xu Q., Su J., de Magalhães J.P., Rigden D.J., Meng J.. RMDisease: a database of genetic variants that affect RNA modifications, with implications for epitranscriptome pathogenesis. Nucleic Acids Res. 2021; 49:D1396–D1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Skinner M.E., Uzilov A.V., Stein L.D., Mungall C.J., Holmes I.H.. JBrowse: a next-generation genome browser. Genome Res. 2009; 19:1630–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dinescu S., Ignat S., Lazar A.D., Constantin C., Neagu M., Costache M.. Epitranscriptomic signatures in lncRNAs and their possible roles in cancer. Genes. 2019; 10:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Brown C.J., Ballabio A., Rupert J.L., Lafreniere R.G., Grompe M., Tonlorenzi R., Willard H.F.. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature. 1991; 349:38–44. [DOI] [PubMed] [Google Scholar]
- 44. Amort T., Souliere M.F., Wille A., Jia X.Y., Fiegl H., Worle H., Micura R., Lusser A.. Long non-coding RNAs as targets for cytosine methylation. RNA Biol. 2013; 10:1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Takai Y., Miyoshi J., Ikeda W., Ogita H.. Nectins and nectin-like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell biology. 2008; 9:603–615. [DOI] [PubMed] [Google Scholar]
- 46. Molfetta R., Milito N.D., Zitti B., Lecce M., Fionda C., Cippitelli M., Santoni A., Paolini R.. The Ubiquitin-proteasome pathway regulates Nectin2/CD112 expression and impairs NK cell recognition and killing. Eur. J. Immunol. 2019; 49:873–883. [DOI] [PubMed] [Google Scholar]
- 47. Chen S.C., Kung M.L., Hu T.H., Chen H.Y., Wu J.C., Kuo H.M., Tsai H.E., Lin Y.W., Wen Z.H., Liu J.K.et al.. Hepatoma-derived growth factor regulates breast cancer cell invasion by modulating epithelial–mesenchymal transition. J. Pathol. 2012; 228:158–169. [DOI] [PubMed] [Google Scholar]
- 48. Ren H., Tang X., Lee J.J., Feng L., Everett A.D., Hong W.K., Khuri F.R., Mao L.. Expression of hepatoma-derived growth factor is a strong prognostic predictor for patients with early-stage non-small-cell lung cancer. J. Clin. Oncol. 2004; 22:3230–3237. [DOI] [PubMed] [Google Scholar]
- 49. Uyama H., Tomita Y., Nakamura H., Nakamori S., Zhang B., Hoshida Y., Enomoto H., Okuda Y., Sakon M., Aozasa K.et al.. Hepatoma-derived growth factor is a novel prognostic factor for patients with pancreatic cancer. Clin. Cancer Res. 2006; 12:6043–6048. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.