Abstract
Interpreting the molecular mechanism of genomic variations and their causal relationship with diseases/traits are important and challenging problems in the human genetic study. To provide comprehensive and context-specific variant annotations for biologists and clinicians, here, by systematically integrating over 4TB genomic/epigenomic profiles and frequently-used annotation databases from various biological domains, we develop a variant annotation database, called VannoPortal. In general, the database has following major features: (i) systematically integrates 40 genome-wide variant annotations and prediction scores regarding allele frequency, linkage disequilibrium, evolutionary signature, disease/trait association, tissue/cell type-specific epigenome, base-wise functional prediction, allelic imbalance and pathogenicity; (ii) equips with our recent novel index system and parallel random-sweep searching algorithms for efficient management of backend databases and information extraction; (iii) greatly expands context-dependent variant annotation to incorporate large-scale epigenomic maps and regulatory profiles (such as EpiMap) across over 33 tissue/cell types; (iv) compiles many genome-scale base-wise prediction scores for regulatory/pathogenic variant classification beyond protein-coding region; (v) enables fast retrieval and direct comparison of functional evidence among linked variants using highly interactive web panel in addition to plain table; (vi) introduces many visualization functions for more efficient identification and interpretation of functional variants in single web page. VannoPortal is freely available at http://mulinlab.org/vportal.
INTRODUCTION
Genome-wide association studies (GWASs) and large-scale genome sequencing studies have uncovered many genetic variants and somatic mutations associated with different human diseases/traits, yet interpreting the molecular mechanisms of these genomic variations and their causal relationships with disease/trait development is challenging (1,2). As the growing volume of functional genomic/epigenomic profiling across a large number of human tissue/cell types, such as the Encyclopedia of DNA Elements (ENCODE) Project (3), Roadmap Epigenomics Project (4) and the International Human Epigenome Consortium (IHEC) Project (5), context-dependent fine-mapping of causal variants and identifying fine-grained molecular phenotypes that mediate the effect between an investigated variant and a particular disease/trait become practical. In addition, a number of computational tools have been developed to predict the regulatory potential or pathogenicity of variant genome-widely (6,7), such as the pioneer algorithm GWAVA (8) and the disease-specific model DIVAN (9), which significantly facilitates the characterization of genomic variants at single base level for interpretation of disease/trait development.
Despite the great effort of international projects in generating, processing, and distributing large amounts of genome/epigenome sequencing data and functional annotations, biologists and clinicians nowadays face great difficulties to curate, collect and compare variant information from different resources, and sometimes even need to download huge annotation files or manually calculate prediction scores. Several variant annotation databases, such as UCSC Variant Annotation Integrator (10), Ensembl Variation Database (11), VarSome (12) and Bystro (13), provide convenient avenues to inspect the genomic and phenotypic features of given variants, but they barely provide genomic effects of variants in linkage disequilibrium (LD) with the single variant being queried and offer limited annotations for non-coding variants. Besides, the overwhelming growth of tissue/cell type-specific and disease/trait-specific variant annotations enables evidence-driven prioritization of candidate causal/pathogenic variants in particular conditions. Unfortunately, existing databases like RegulomeDB (14) and HaploReg (15) often fail to incorporate the latest context-dependent annotations and genome-scale functional predictions which are crucial for drawing biologically meaningful conclusions from investigated variants.
In this work, by systematically integrating large-scale tissue/cell type-specific genomic/epigenomic profiles, base-wise functional prediction scores, and frequently-used annotation databases from various biological domains, we develop a novel variant annotation database VannoPortal for biologists and clinicians to efficiently retrieve comprehensive and context-specific features, including variant basic information, evolutionary annotation, disease/trait association, variant regulatory potential, and variant pathogenicity. VannoPortal leverages multiscale orthogonal evidences to support the functionality or pathogenicity of queried variants. It significantly enlarges the annotation scope to almost all possible substitutions of a small variant in the human reference genome, and make efforts to improve the interpretability of variant annotations by using many intuitive visualizations and interactive web components. VannoPortal is free and open to all users without login and registration at http://mulinlab.org/vportal.
MATERIALS AND METHODS
Variant basic information and allele frequency
Allele information of known single nucleotide variations (SNVs) and insertions/deletions (indels) were collected from gnomAD r2.0.2 (16), 1000 Genomes Project phase 3 (17), and dbSNP b151 (18). For SNV, alleles are enumerated if only genomic coordinate is provided based on human reference genome. For customized alleles which are conflict with human reference genome or are absent in known variant databases, only region-level annotation is supported. We applied a Java library Jannovar v0.30 (19) to annotate gene and transcript information. Commonly-used allele frequency information for worldwide populations were downloaded from 1000 Genomes Project phase 3 and gnomAD r2.0.2. We also incorporated allele frequencies from other genome sequencing or genotyping projects, including GenomeAsia (20), jMorp (21), ABraOM (22), UK10K project (23), UK Biobank (24), etc. CrossMap (25) was used to convert genome coordinates between GRCh37 and GRCh38 when the annotation is not provided for a certain genome assembly version.
Evolutionary information
Most base-wise conservation scores were extracted from CADD v1.4 (26), including PhyloP (27), phastCons (28), GERP (29), fitCons (30), and bStatistic (31) except for SiPhy (32). Similar to CADD score, we calculated the ‘PHRED-scaled’ score for each of these scores by taking the rank in order of magnitude, which makes them comparable to each other. For each score, a likely conserved signal was defined once the ‘PHRED-scaled’ score was >10. Based on genotypes from 1000 Genomes Project phase 3, variant-level positive selection scores were calculated and classified according to the description of our dbPSHP (33) and 1000 Genomes Selection Browser (34).
Disease/trait association
LD information for five super populations (AFR, AMR, EAS, EUR, SAS) were calculated using genotypes from 1000 Genomes Project phase 3. Disease/trait-associated variants were collected from The NHGRI-EBI GWAS Catalog v1.0.2 (35). The likely disease/trait-causal variants were downloaded from our CAUSALdb v1.1 (36). Expression quantitative trait loci (eQTL) and splicing quantitative trait loci (sQTL) of 54 human tissue/cell types were downloaded from GTEx v8 (37), and information for other types of molecular trait quantitative trait loci (xQTL) were collected from our QTLbase v1.2 (38). VarNote random-sweep searching algorithm (39) was used to extract annotations and filter linked variants in LD.
Regulatory potential
Context-dependent regulatory variant prediction scores were integrated from cepip (40), GenoSkyline-Plus (41), FUN-LDA (42), GenoNet (43), and FitCons2 (44) for 127 tissue/cell types. The combined score of tissue/cell type-specific regulatory potential was calculated by rank product. Based on consolidated and imputed epigenomes of 127 human tissue/cell types from Roadmap Epigenomics (4) and 869 samples from EpiMap (45), we intersected each query variant with narrow peaks of histone marks, transcription factor (TF) (measured by chromatin immunoprecipitation sequencing (ChIP-Seq)) and open chromatin (measured by DNase I hypersensitive sites sequencing (DNase-Seq) or transposase accessible chromatin sequencing (ATAC-seq)) using VarNote random access function. Significant 5 kb Hi-C interactions of 60 tissue/cell types were borrowed from our GWAS4D (46), and a virtual 4C diagram anchored at query variant locus was plotted using CHiCP (47). Motif information for 136 TFs was collected from CIS-BP (48), JASPAR (49), and ENCODE-motifs (50). Binding affinity effect changes between different alleles of query variant were estimated according to our previous method (51). TF binding ChIP-seq significant peaks in different tissue/cell types were systematically integrated from CistromeDB (52), DeepBlueR (53), GTRD (54) and EpiMap (45). We also incorporated allelic imbalance evidence of chromatin accessibility and TF binding from multiple studies (55,56).
Pathogenicity
Genome-scale base-wise prediction scores of pathogenic and cancer driver regulatory variants were downloaded from RegBase-PAT and RegBase-CAN (7). According to the Youden's J statistics derived from trained model for each tool, query variants can be classified as likely pathogenic or neutral properties. Nonsynonymous SNV pathogenicity scores were downloaded from dbNSFP V4.1a (57). Prediction scores for splicing-altering potential were retrieved from dbscSNV (58), S-CAP (59), and SpliceAI (60). ClinVar was used to annotate genomic variation and its relationship to human health (61). COSMIC (62) and ICGC (63) aggregated mutation datasets were adopted to annotate somatic recurrence in cancers. Finally, CIViC was used to annotate mutation-dependent effects on cancer drug treatment (64).
Database design and annotation retrieve strategy
VannoPortal is built on a Java-based web framework. Several interactive web pages are implemented by D3.js, jQuery and related JavaScript modules. To ensure fast retrieval of relevant information from huge annotation databases, each annotation file was concerted to BED, VCF or 1-based tabular text file, then compressed and indexed by VarNote. The parallel random-sweep searching or independent random-access strategies of VarNote were used to ensure a highly efficient query.
RESULTS
Data summary of VannoPortal
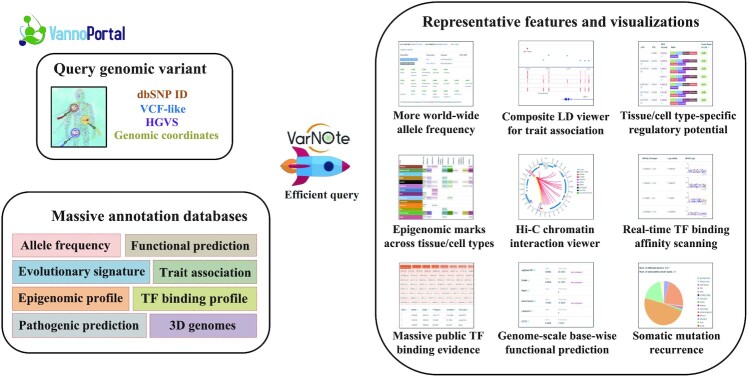
By systematically integrating genomic/epigenomic profiles and variant annotations from various biological domains, the initial version of VannoPortal contains 40 independent variant-level and region-level information archived in over 4TB indexed annotation files (Supplementary Table S1). To simplify biological interpretation, VannoPortal classified these annotations into five major categories including variant basic information, evolutionary annotation, disease/trait association, variant regulatory potential, and variant pathogenicity. Specifically, (i) in ‘variant basic information’ annotation, VannoPortal reports the genomic attributes, affected genes and transcripts and worldwide allele frequencies. In addition to the 1000 Genomes project (17) and gnomAD (16), VannoPortal also incorporates allele frequency information from other genome sequencing projects. (ii) In ‘evolutionary annotation’, VannoPortal provides comprehensive aggregation of 11 base-wise conservation scores and 13 variant-level score regarding positive/negative selection in recent human evolution, which could benefit the identification of functional variants from an evolutionary perspective. (iii) In ‘disease/trait association’, VannoPortal collects disease/trait-associated signals and credible variants identified by GWAS, and molecular trait QTLs across most of human tissue/cell types. By leveraging population-specific LD information, this disease/trait association evidence can be easily compared among correlated variants in VannoPortal. (iv) Since interpreting the non-coding regulatory variants is challenging, VannoPortal comprehensively integrates large-scale tissue/cell type-specific epigenomes and functional predictions in the ‘regulatory potential’ section. For example, context-dependent prioritization of regulatory potential among high LD variants enables the identification of potentially causal regulatory variants in phenotypically relevant tissue/cell types; Mapping variant locus to critical histone marks, chromatin states and TF binding sites across hundreds of tissue/cell type-specific samples, from Roadmap Epigenomics (4) or EpiMap (45) projects, will greatly facilitate the grasp of regulatory code underlying the investigated variant; Linking variant to its target genes or affected regulators can further pinpoint the molecular mechanism and direct functional follow-up. (v5) Finally, in ‘variant pathogenicity’ annotation, VannoPortal not only includes deleterious scores for missense and splicing-altering variants, it also summarizes multiple genome-scale predictions and evidence to interpret pathogenic variants for disease progression and targeted therapy (Figure 1).
Figure 1.
Database architecture, function structure and representative features.
Advanced features of VannoPortal over existing databases
The key design principle of VannoPortal is to avoid simple aggregation of exiting annotation databases, and to advocate evidence-driven interpretation and prioritization. To this end, VannoPortal has the following distinctive features and improvements compared with existing databases. First, VannoPortal is equipped with our recent novel index system and parallel random-sweep searching algorithms for efficient management of backend databases and information extraction (39). It only takes seconds to randomly access or screen terabyte-level annotation datasets for each independent query. Particularly, VannoPortal allows fast retrieval and direct comparison of functional annotations among variants in LD by providing several interactive panels, while existing databases, such as Ensembl Variation Database (11) and VarSome (12), only annotate single variant with suboptimal efficiency. Second, VannoPortal incorporates many base-wise and genome-scale features to annotate SNVs and indels, which enlarges the annotation scope to almost all possible substitutions of small variants in the human reference genome. Whereas limited information for variants outside protein-coding regions was provided by most of existing databases. Third, VannoPortal provides genome-wide, multiscale and orthogonal evidences regarding whether a variant is functional. For example, to evaluate whether a given variant has regulatory, pathogenic or cancer-driver potential, multiple prediction scores and phenotypic evidence were reported. Fourth, compared to commonly-used HaploReg (15) and RegulomeDB (14) for regulatory variant annotation, VannoPortal greatly expands context-dependent variant annotation to incorporate large-scale epigenomic maps and regulatory profiles across over 33 tissue/cell types and thousands of biosamples. Finally, VannoPortal focuses more on the interpretability of variant annotations rather than information enumeration. For instance, all genome-scale prediction scores were transformed to comparable values and then were classified into meaningful variant consequences.
Database usage
VannoPortal accepts many query formats, including dbSNP ID, VCF-like, HGVS and even only genomic coordinates.Both GRCh37/hg19 and GRCh38/hg38 of human genome assembly are supported. For known SNVs and indels, VannoPortal will automatically extract all allele information from the backend database and provide allele-specific annotation switching if multiple alternative alleles are reported. For rare, somatic or unobserved SNVs and indels, VannoPortal allows customized alleles in several region-level annotation sections. The query result page of VannoPortal incorporates five major annotation sections (including variant basic information, evolution, phenotype, regulatory potential, pathogenicity) as well as several sub-categories in each section. The navigation bar displays the annotation hit status for a query variant on each of sub-categories. By clicking the name of the sub-category, the page will scroll to the detailed panel of the corresponding item (Figure 2).
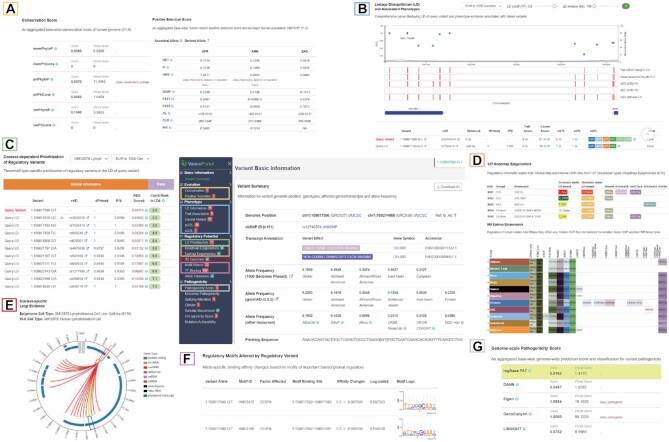
Figure 2.
Result page and distinctive web components of VannoPortal. (A) Conservation scores and positive selection scores in the ‘Evolution’ panel. (B) A composite viewer showing LD structure, disease/trait association tracks and evidence table in the ‘Phenotype’ panel. (C) Tissue/cell type-specific regulatory variant prioritization function in the ‘Regulatory potential’ panel. (D) Two rich tables displaying critical histone marks, chromatin states, and TF binding sites across hundreds of tissue/cell type-specific samples at variant locus, from Roadmap Epigenomics or EpiMap projects. (E) A circular plot showing significant 5 kb Hi-C chromatin interactions between variant locus and its target region. (F) Real-time motif scanning table for the predicted allele-specific effect of TF binding. (G) Genome-scale pathogenic scores in the ‘Pathogenicity’ panel.
In the left panel of the result page: (i) ‘Variant basic information’ panel shows genomic position, allele information, dbSNP ID, transcript annotation and allele frequencies from different populations. The page can be redirected to the original database page for details once clicking on different arrowhead links (Figure 2). (ii) ‘Evolution’ panel reports base-wise conservation scores and variant-level scores regarding positive/negative selection in recent human evolution. Note that the scores beyond empirical cutoffs were labeled as ‘likely conserved’ or ‘likely influenced by selection or population history’ or other noteworthy signatures (Figure 2A). (iii) ‘Phenotype’ panel incorporates an interactive LD viewer along with some disease/trait association tracks, including disease/trait-associated evidence and eQTL/sQTL/xQTL hits. Users can click each variant in the plot or vertical bar in the evidence tracks to check the summary information of supporting evidence. By selecting the dropdown list or dragging the slider bar, users can adjust the population, LD r2 cutoff and LD window size to filter out variants. As the LD threshold changes, the bottom table lists the LD information and the number of supporting evidences for all correlated variants (Figure 2B). More detailed information for disease/trait associations is displayed in separate table viewers. (iv) ‘Regulatory potential’ panel systematically demonstrates tissue/cell type-specific functional predictions, epigenomic signals and TF binding evidence in different aspects. By assigning a desired tissue/cell type and adjusting LD parameters, the query variant can be prioritized together with all linked variants, and a combined ranking score based on the five state-of-the-art prediction scores can be calculated for each of the variants within the LD region (Figure 2C). Importantly, in two rich table viewers, users can comprehensively grasp the chromatin states and epigenomic features at variant locus across 127 Roadmap Epigenomics tissue/cell types and 869 EpiMap samples. Clicking on each tissue name can unfold the view to cell type level in the EpiMap viewers (Figure 2D). In addition, according to user-selected tissue/cell type, an interactive circular plot displays the topmost significant 5 kb chromatin interactions anchored at the variant-contained locus (Figure 2E). When users click on each interaction arc, chromatin marks within the interacted 5 kb bins can be displayed. Last, users can easily check the predicted changes in TF binding affinity through real-time motif scanning, TF binding evidence of public ChIP-seq peaks, and the allele-specific footprint events in several rich table viewers (Figure 2F). (v) ‘Pathogenicity’ panel enumerates many genome-scale pathogenic prediction scores, deleterious scores for missense and splicing-altering variants, as well as cancer driver prediction scores for somatic mutations (Figure 2G). According to the classification of each prediction score, users can easily determine whether the query variant is likely pathogenic in a certain context. Known health-associated evidence and therapeutic implications are also listed in separate tables. Finally, users can download all of the functional predictions and annotation information for each query variant by simply clicking the download button at the top right of the result page or by RESTful API.
Case studies
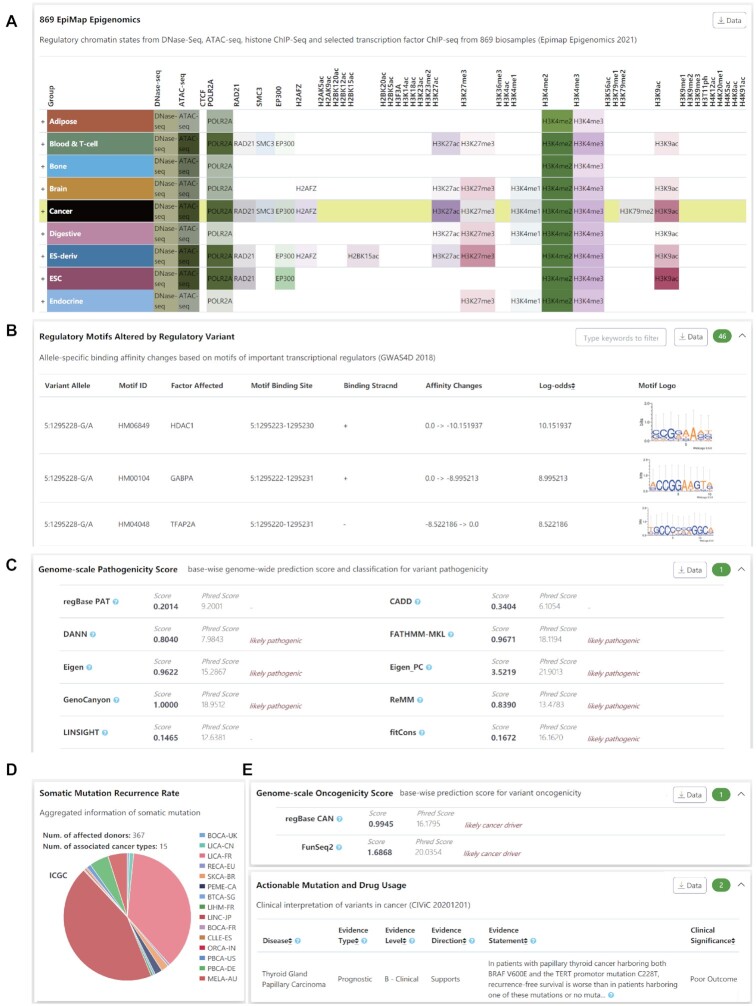
To investigate the reliability and practicality of VannoPortal for identifying potentially causal variants in different scenarios of genetic study, we exemplified several classical or novel loci according to published results. (i) For common regulatory variants revealed by GWAS, we used an experimentally validated causal variant rs12740374, which alters plasma low-density lipoprotein cholesterol (LDL-C) by modulating hepatic very low-density lipoprotein secretion (65), to test whether VannoPortal could precisely annotate the variant effect. Consistent with the reported findings, VannoPortal reveals many lines of evidence for the causality of cholesterol traits and molecular trait QTLs (Supplementary Figure S1A). In the context of cholesterol trait-relevant cell type HepG2, VannoPortal successfully prioritizes rs12740374 as a top regulatory variant with the highest combined score among LD variants (Supplementary Figure S1B). Epigenomic annotations also demonstrate that rs12740374 is located in the active chromatin and harbors EP300 and cohesion binding signals across many tissue/cell types (Supplementary Figure S1C). Notably, in agreement with published results (65), VannoPortal motif scanning result shows that rs12740374 may create a CEBPA transcription factor binding site (Supplementary Figure S1D). (ii) We also examined a low-frequency variant rs74956615 associated with coronavirus disease 19 (COVID-19) (66–68). This variant has been documented to confer risk for critical illness of COVID-19 near the gene that encodes tyrosine kinase 2 (TYK2). Based on the LD of the EUR population, VannoPortal can link this variant to a TYK2 missense variant rs34536443 (r2 = 0.8332) which significantly associate with the susceptibility of many autoimmune diseases (Supplementary Figure S2A). Searching on rs34536443 reveals that it can affect different isoforms of TYK2, and its minor C allele is totally absent in the East Asian population (Supplementary Figure S2B). Both conservation scores and pathogenicity scores from VannoPortal also support the likely damaging role of this variant (Supplementary Figure S2C–E). (iii) For rare pathogenic variants, we queried rs12565 which was previously found to cause cardiovascular diseases by altering the recruitment of REST to target gene NPPA (69). Interestingly, this non-coding variant exhibits very high conservation scores (Supplementary Figure S3A) and obtains active chromatin states in only heart tissues, including open chromatin marked by DNase-seq peak and histone modifications of H3K4me3, H3K4me1, and H3K27ac (Supplementary Figure S3B). Both public TF ChIP-seq data and motif scanning results indicate that rs12565 may modulate the binding affinity of REST (Supplementary Figure S3C, D). In addition, genome-scale pathogenicity scores from VannoPortal consistently show that this non-coding variant is likely pathogenic (Supplementary Figure S3E). (iv) For somatic cancer-driver mutation, we inspected a well-known pan-cancer mutation chr5:g.1295228:G > A (GRCh37, rs1242535815) in –124bp upstream of TERT promoter which reactivates TERT expression by recruitment of the TF GABP (70). The oncogenicity and regulatory mechanism underlying this mutation are well supported by VannoPortal, such as overactive chromatin states in cancers (Figure 3A), increased HDAC1 and GABPA bindings (Figure 3B), as well as many lines of cancer-driven evidence and therapeutic implications (Figure 3C–E).
Figure 3.
Supporting evidence from VannoPortal for the regulatory potential and cancer-driven roles of chr5:g.1295228:G > A (GRCh37, rs1242535815). (A) rs1242535815 overlaps active chromatin states (e.g. DNase-seq and ATAC-seq), histone marks (e.g. H3K27ac, H3K4me2, H3K4me3 and H3K9ac) and TF binding sites (e.g. POLR2A, RAD21 and SMC3) across many human tissues, particularly in cancers. (B) rs1242535815 A allele potentially increases the binding affinity of HDAC1 and GABPA. (C) rs1242535815 is a likely pathogenic mutation supported by many genome-scale base-wise pathogenicity prediction methods. (D) rs1242535815 is a highly recurrent mutation in cancer patients. (E) rs1242535815 is a likely cancer driver mutation supported by regBase-CAN and other tools, and it could be a prognostic marker in cancer therapy.
CONCLUSIONS
VannoPortal systematically incorporates lots of new genome-scale and context-dependent variant annotation resources from various biological domains, particularly for variants outside of protein-coding regions. It focuses more on the interpretability of variant annotations instead of simple aggregation of known information using many intuitive visualizations and interactive web components, and enables direct comparison of some functional evidence (e.g. disease/trait association, tissue/cell type-specific regulatory potential) between query variant and its linked ones without multi-round queries. Along with the rapid evolution of advanced biotechnologies and new genetic findings (71,72), VannoPortal will continue to update the existing annotation databases and introduce more advanced features, such as prioritization of target genes for non-coding regulatory variants, integration of more prediction scores for variant affecting post-transcriptional and translational processes, support of large variant annotation, and incorporation of genetic-based translational medicine data (73,74). Given the suboptimal assumption of independence between the base positions of the sequence motif, we will combine large-scale tissue/cell type-specific open chromatin profiles (e.g. DNase-Seq and ATAC-seq) and powerful statistical methods (e.g. gkm-SVM (75) and KMAC (76)) to annotate the most plausible TFs associated with regulatory variants. We believe that this novel platform will benefit researchers to interrogate the biological functions of genome variations and create significant impacts in the era of human genetics and genomics.
Supplementary Material
Contributor Information
Dandan Huang, Department of Bioinformatics, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Department of Biochemistry and Molecular Biology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Yao Zhou, Department of Bioinformatics, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Department of Pharmacology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Xianfu Yi, School of Biomedical Engineering, Tianjin Medical University, Tianjin 300070, China.
Xutong Fan, Department of Bioinformatics, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Department of Pharmacology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Jianhua Wang, Department of Bioinformatics, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Department of Pharmacology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China.
Hongcheng Yao, Centre for PanorOmic Sciences-Genomics and Bioinformatics Cores, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong SAR 999077, China.
Pak Chung Sham, Centre for PanorOmic Sciences-Genomics and Bioinformatics Cores, LKS Faculty of Medicine, The University of Hong Kong, Hong Kong SAR 999077, China.
Jihui Hao, Department of Pancreatic Cancer, Tianjin Medical University Cancer Institute and Hospital, National Clinical Research Center for Cancer, Key Laboratory of Cancer Prevention and Therapy, Tianjin's Clinical Research Center for Cancer, Tianjin 300060, China.
Kexin Chen, Department of Epidemiology and Biostatistics, Tianjin Key Laboratory of Molecular Cancer Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300060, China.
Mulin Jun Li, Department of Bioinformatics, The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, School of Basic Medical Sciences, National Clinical Research Center for Cancer, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300070, China; Department of Pharmacology, School of Basic Medical Sciences, Tianjin Medical University, Tianjin 300070, China; Department of Epidemiology and Biostatistics, Tianjin Key Laboratory of Molecular Cancer Epidemiology, Tianjin Medical University Cancer Institute and Hospital, Tianjin Medical University, Tianjin 300060, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese National Key Research and Development Project [2018YFC1315600]; National Natural Science Foundation of China [32070675, 31871327]; Natural Science Foundation of Tianjin [19JCJQJC63600]. Funding for open access charge: National Natural Science Foundation of China [31871327].
Conflict of interest statement. None declared.
REFERENCES
- 1. Loos R.J.F. 15 years of genome-wide association studies and no signs of slowing down. Nat. Commun. 2020; 11:5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium Pan-cancer analysis of whole genomes. Nature. 2020; 578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. ENCODE Project Consortium Moore J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A.et al.. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020; 583:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Roadmap Epigenomics Consortium Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J.et al.. Integrative analysis of 111 reference human epigenomes. Nature. 2015; 518:317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stunnenberg H.G., International Human Epigenome C., Hirst M.. The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. 2016; 167:1145–1149. [DOI] [PubMed] [Google Scholar]
- 6. Rentzsch P., Witten D., Cooper G.M., Shendure J., Kircher M.. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019; 47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang S., He Y., Liu H., Zhai H., Huang D., Yi X., Dong X., Wang Z., Zhao K., Zhou Y.et al.. regBase: whole genome base-wise aggregation and functional prediction for human non-coding regulatory variants. Nucleic Acids Res. 2019; 47:e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ritchie G.R., Dunham I., Zeggini E., Flicek P.. Functional annotation of noncoding sequence variants. Nat. Methods. 2014; 11:294–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen L., Jin P., Qin Z.S.. DIVAN: accurate identification of non-coding disease-specific risk variants using multi-omics profiles. Genome Biol. 2016; 17:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hinrichs A.S., Raney B.J., Speir M.L., Rhead B., Casper J., Karolchik D., Kuhn R.M., Rosenbloom K.R., Zweig A.S., Haussler D.et al.. UCSC data integrator and variant annotation integrator. Bioinformatics. 2016; 32:1430–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hunt S.E., McLaren W., Gil L., Thormann A., Schuilenburg H., Sheppard D., Parton A., Armean I.M., Trevanion S.J., Flicek P.et al.. Ensembl variation resources. Database. 2018; 2018:bay119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kopanos C., Tsiolkas V., Kouris A., Chapple C.E., Albarca Aguilera M., Meyer R., Massouras A.. VarSome: the human genomic variant search engine. Bioinformatics. 2019; 35:1978–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kotlar A.V., Trevino C.E., Zwick M.E., Cutler D.J., Wingo T.S.. Bystro: rapid online variant annotation and natural-language filtering at whole-genome scale. Genome Biol. 2018; 19:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyle A.P., Hong E.L., Hariharan M., Cheng Y., Schaub M.A., Kasowski M., Karczewski K.J., Park J., Hitz B.C., Weng S.et al.. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012; 22:1790–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ward L.D., Kellis M.. HaploReg v4: systematic mining of putative causal variants, cell types, regulators and target genes for human complex traits and disease. Nucleic Acids Res. 2016; 44:D877–D881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karczewski K.J., Francioli L.C., Tiao G., Cummings B.B., Alfoldi J., Wang Q., Collins R.L., Laricchia K.M., Ganna A., Birnbaum D.P.et al.. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020; 581:434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. 1000 Genomes Project Consortium Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A.et al.. A global reference for human genetic variation. Nature. 2015; 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K.. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001; 29:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jager M., Wang K., Bauer S., Smedley D., Krawitz P., Robinson P.N.. Jannovar: a java library for exome annotation. Hum. Mutat. 2014; 35:548–555. [DOI] [PubMed] [Google Scholar]
- 20. GenomeAsia 100K Consortium The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature. 2019; 576:106–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tadaka S., Hishinuma E., Komaki S., Motoike I.N., Kawashima J., Saigusa D., Inoue J., Takayama J., Okamura Y., Aoki Y.et al.. jMorp updates in 2020: large enhancement of multi-omics data resources on the general Japanese population. Nucleic Acids Res. 2021; 49:D536–D544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Naslavsky M.S., Yamamoto G.L., de Almeida T.F., Ezquina S.A.M., Sunaga D.Y., Pho N., Bozoklian D., Sandberg T.O.M., Brito L.A., Lazar M.et al.. Exomic variants of an elderly cohort of Brazilians in the ABraOM database. Hum. Mutat. 2017; 38:751–763. [DOI] [PubMed] [Google Scholar]
- 23. UK10K Consortium Walter K., Min J.L., Huang J., Crooks L., Memari Y., McCarthy S., Perry J.R., Xu C., Futema M.et al.. The UK10K project identifies rare variants in health and disease. Nature. 2015; 526:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sudlow C., Gallacher J., Allen N., Beral V., Burton P., Danesh J., Downey P., Elliott P., Green J., Landray M.et al.. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015; 12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhao H., Sun Z., Wang J., Huang H., Kocher J.P., Wang L.. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014; 30:1006–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kircher M., Witten D.M., Jain P., O’Roak B.J., Cooper G.M., Shendure J.. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014; 46:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pollard K.S., Hubisz M.J., Rosenbloom K.R., Siepel A.. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010; 20:110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S.et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Davydov E.V., Goode D.L., Sirota M., Cooper G.M., Sidow A., Batzoglou S.. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010; 6:e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gulko B., Hubisz M.J., Gronau I., Siepel A.. A method for calculating probabilities of fitness consequences for point mutations across the human genome. Nat. Genet. 2015; 47:276–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McVicker G., Gordon D., Davis C., Green P.. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009; 5:e1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Garber M., Guttman M., Clamp M., Zody M.C., Friedman N., Xie X.. Identifying novel constrained elements by exploiting biased substitution patterns. Bioinformatics. 2009; 25:i54–i62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li M.J., Wang L.Y., Xia Z., Wong M.P., Sham P.C., Wang J.. dbPSHP: a database of recent positive selection across human populations. Nucleic Acids Res. 2014; 42:D910–D916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pybus M., Dall’Olio G.M., Luisi P., Uzkudun M., Carreno-Torres A., Pavlidis P., Laayouni H., Bertranpetit J., Engelken J.. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 2014; 42:D903–D909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Buniello A., MacArthur J.A.L., Cerezo M., Harris L.W., Hayhurst J., Malangone C., McMahon A., Morales J., Mountjoy E., Sollis E.et al.. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 2019; 47:D1005–D1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang J., Huang D., Zhou Y., Yao H., Liu H., Zhai S., Wu C., Zheng Z., Zhao K., Wang Z.et al.. CAUSALdb: a database for disease/trait causal variants identified using summary statistics of genome-wide association studies. Nucleic Acids Res. 2020; 48:D807–D816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020; 369:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng Z., Huang D., Wang J., Zhao K., Zhou Y., Guo Z., Zhai S., Xu H., Cui H., Yao H.et al.. QTLbase: an integrative resource for quantitative trait loci across multiple human molecular phenotypes. Nucleic Acids Res. 2020; 48:D983–D991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Huang D., Yi X., Zhou Y., Yao H., Xu H., Wang J., Zhang S., Nong W., Wang P., Shi L.et al.. Ultrafast and scalable variant annotation and prioritization with big functional genomics data. Genome Res. 2020; 30:1789–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li M.J., Li M., Liu Z., Yan B., Pan Z., Huang D., Liang Q., Ying D., Xu F., Yao H.et al.. cepip: context-dependent epigenomic weighting for prioritization of regulatory variants and disease-associated genes. Genome Biol. 2017; 18:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu Q., Powles R.L., Abdallah S., Ou D., Wang Q., Hu Y., Lu Y., Liu W., Li B., Mukherjee S.et al.. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer's disease. PLos Genet. 2017; 13:e1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Backenroth D., He Z., Kiryluk K., Boeva V., Pethukova L., Khurana E., Christiano A., Buxbaum J.D., Ionita-Laza I.. FUN-LDA: A latent dirichlet allocation model for predicting Tissue-Specific functional effects of noncoding variation: methods and applications. Am. J. Hum. Genet. 2018; 102:920–942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. He Z., Liu L., Wang K., Ionita-Laza I.. A semi-supervised approach for predicting cell-type specific functional consequences of non-coding variation using MPRAs. Nat. Commun. 2018; 9:5199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gulko B., Siepel A.. An evolutionary framework for measuring epigenomic information and estimating cell-type-specific fitness consequences. Nat. Genet. 2019; 51:335–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Boix C.A., James B.T., Park Y.P., Meuleman W., Kellis M.. Regulatory genomic circuitry of human disease loci by integrative epigenomics. Nature. 2021; 590:300–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Huang D., Yi X., Zhang S., Zheng Z., Wang P., Xuan C., Sham P.C., Wang J., Li M.J.. GWAS4D: multidimensional analysis of context-specific regulatory variant for human complex diseases and traits. Nucleic Acids Res. 2018; 46:W114–W120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schofield E.C., Carver T., Achuthan P., Freire-Pritchett P., Spivakov M., Todd J.A., Burren O.S.. CHiCP: a web-based tool for the integrative and interactive visualization of promoter capture Hi-C datasets. Bioinformatics. 2016; 32:2511–2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Weirauch M.T., Yang A., Albu M., Cote A.G., Montenegro-Montero A., Drewe P., Najafabadi H.S., Lambert S.A., Mann I., Cook K.et al.. Determination and inference of eukaryotic transcription factor sequence specificity. Cell. 2014; 158:1431–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B.. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004; 32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kheradpour P., Kellis M.. Systematic discovery and characterization of regulatory motifs in ENCODE TF binding experiments. Nucleic Acids Res. 2014; 42:2976–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li M.J., Wang L.Y., Xia Z., Sham P.C., Wang J.. GWAS3D: detecting human regulatory variants by integrative analysis of genome-wide associations, chromosome interactions and histone modifications. Nucleic Acids Res. 2013; 41:W150–W158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zheng R., Wan C., Mei S., Qin Q., Wu Q., Sun H., Chen C.H., Brown M., Zhang X., Meyer C.A.et al.. Cistrome Data Browser: expanded datasets and new tools for gene regulatory analysis. Nucleic Acids Res. 2019; 47:D729–D735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Albrecht F., List M., Bock C., Lengauer T.. DeepBlueR: large-scale epigenomic analysis in R. Bioinformatics. 2017; 33:2063–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kolmykov S., Yevshin I., Kulyashov M., Sharipov R., Kondrakhin Y., Makeev V.J., Kulakovskiy I.V., Kel A., Kolpakov F.. GTRD: an integrated view of transcription regulation. Nucleic Acids Res. 2021; 49:D104–D111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vierstra J., Lazar J., Sandstrom R., Halow J., Lee K., Bates D., Diegel M., Dunn D., Neri F., Haugen E.et al.. Global reference mapping of human transcription factor footprints. Nature. 2020; 583:729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Abramov S., Boytsov A., Bykova D., Penzar D.D., Yevshin I., Kolmykov S.K., Fridman M.V., Favorov A.V., Vorontsov I.E., Baulin E.et al.. Landscape of allele-specific transcription factor binding in the human genome. Nat. Commun. 2021; 12:2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liu X., Li C., Mou C., Dong Y., Tu Y.. dbNSFP v4: a comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome medicine. 2020; 12:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Jian X., Boerwinkle E., Liu X.. In silico prediction of splice-altering single nucleotide variants in the human genome. Nucleic Acids Res. 2014; 42:13534–13544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jagadeesh K.A., Paggi J.M., Ye J.S., Stenson P.D., Cooper D.N., Bernstein J.A., Bejerano G.. S-CAP extends pathogenicity prediction to genetic variants that affect RNA splicing. Nat. Genet. 2019; 51:755–763. [DOI] [PubMed] [Google Scholar]
- 60. Jaganathan K., Kyriazopoulou Panagiotopoulou S., McRae J.F., Darbandi S.F., Knowles D., Li Y.I., Kosmicki J.A., Arbelaez J., Cui W., Schwartz G.B.et al.. Predicting splicing from primary sequence with deep learning. Cell. 2019; 176:535–548. [DOI] [PubMed] [Google Scholar]
- 61. Landrum M.J., Chitipiralla S., Brown G.R., Chen C., Gu B., Hart J., Hoffman D., Jang W., Kaur K., Liu C.et al.. ClinVar: improvements to accessing data. Nucleic Acids Res. 2020; 48:D835–D844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M., Bindal N., Boutselakis H., Cole C.G., Creatore C., Dawson E.et al.. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019; 47:D941–D947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang J., Bajari R., Andric D., Gerthoffert F., Lepsa A., Nahal-Bose H., Stein L.D., Ferretti V.. The international cancer genome consortium data portal. Nat. Biotechnol. 2019; 37:367–369. [DOI] [PubMed] [Google Scholar]
- 64. Griffith M., Spies N.C., Krysiak K., McMichael J.F., Coffman A.C., Danos A.M., Ainscough B.J., Ramirez C.A., Rieke D.T., Kujan L.et al.. CIViC is a community knowledgebase for expert crowdsourcing the clinical interpretation of variants in cancer. Nat. Genet. 2017; 49:170–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V., Li X., Li H., Kuperwasser N., Ruda V.M.et al.. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010; 466:714–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Pairo-Castineira E., Clohisey S., Klaric L., Bretherick A.D., Rawlik K., Pasko D., Walker S., Parkinson N., Fourman M.H., Russell C.D.et al.. Genetic mechanisms of critical illness in COVID-19. Nature. 2021; 591:92–98. [DOI] [PubMed] [Google Scholar]
- 67. COVID-19 Host Genetics Initiative Mapping the human genetic architecture of COVID-19. Nature. 2021; 10.1038/s41586-021-03767-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zeberg H., Paabo S.. A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. PNAS. 2021; 118:e2026309118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Johnson R., Richter N., Bogu G.K., Bhinge A., Teng S.W., Choo S.H., Andrieux L.O., de Benedictis C., Jauch R., Stanton L.W.. A genome-wide screen for genetic variants that modify the recruitment of REST to its target genes. PLoS Genet. 2012; 8:e1002624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yuan X., Larsson C., Xu D.. Mechanisms underlying the activation of TERT transcription and telomerase activity in human cancer: old actors and new players. Oncogene. 2019; 38:6172–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cano-Gamez E., Trynka G.. From GWAS to function: using functional genomics to identify the mechanisms underlying complex diseases. Frontiers in genetics. 2020; 11:424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van der Wijst M., de Vries D.H., Groot H.E., Trynka G., Hon C.C., Bonder M.J., Stegle O., Nawijn M.C., Idaghdour Y., van der Harst P.et al.. The single-cell eQTLGen consortium. eLife. 2020; 9:e52155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Nelson M.R., Tipney H., Painter J.L., Shen J., Nicoletti P., Shen Y., Floratos A., Sham P.C., Li M.J., Wang J.et al.. The support of human genetic evidence for approved drug indications. Nat. Genet. 2015; 47:856–860. [DOI] [PubMed] [Google Scholar]
- 74. Cui H., Zuo S., Liu Z., Liu H., Wang J., You T., Zheng Z., Zhou Y., Qian X., Yao H.et al.. The support of genetic evidence for cardiovascular risk induced by antineoplastic drugs. Sci. Adv. 2020; 6:eabb8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Ghandi M., Lee D., Mohammad-Noori M., Beer M.A.. Enhanced regulatory sequence prediction using gapped k-mer features. PLoS Comput. Biol. 2014; 10:e1003711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Guo Y., Tian K., Zeng H., Guo X., Gifford D.K.. A novel k-mer set memory (KSM) motif representation improves regulatory variant prediction. Genome Res. 2018; 28:891–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.