Abstract
EFG1 is a central transcriptional regulator of filamentation that is an important virulence factor of Candida albicans. This study serves to assess in vivo the applicability of the anti-EFG1 2′-OMethylRNA oligomer for inhibiting C.albicans filamentation and to attenuate candidiasis, using the Galleria mellonella model. For that, larvae infected with a lethal concentration of C. albicans cells were treated with a single dose and with a double dose of the anti-EFG1 2′OMe oligomer (at 40 and 100 nM). The anti-EFG1 2′OMe oligomer toxicity and effect on larvae survival was evaluated. No evidence of anti-EFG1 2′OMe oligomer toxicity was observed and the treatment with double dose of 2′OMe oligomer empowered larvae survival over 24 h (by 90%–100%) and prolonged its efficacy until 72 h of infection (by 30%). Undoubtedly, this work validates the in vivo therapeutic potential of anti-EFG1 2′OMe oligomer for controlling C. albicans infections.
Keywords: antisense oligonucleotides, 2′-OMethyl chemical modification, candidiasis, Galleria mellonella, virulence
Graphical abstract
Introduction
Candida albicans remains the most common human fungal pathogen,1 and the most prevalent of all Candida species over the world.1 The pathogenicity of C. albicans is dependent of certain virulence factors in which the morphological transition from yeast to filamentous forms is recognized as one of the most alarming.2, 3, 4, 5 The EFG1 gene is one of the most important and well-studied regulators of C. albicans filamentation.6, 7, 8, 9, 10, 11 Recently, we applied antisense technology to project the anti-EFG1 2′-OMethylRNA (2′OMe) oligomer, to control EFG1 gene expression, and to prevent C. albicans filamentation.12 The anti-EFG1 2′OMe oligomer was designed based on the second generation of chemical modifications (2′-OMethyl) to guarantee nuclease resistance, improve RNA affinity and potency, and to reduce its toxicity.13 Our in vitro work revealed the anti-EFG1 2′OMe oligomer’s ability to reduce C. albicans cell filamentation (by 80%). Moreover, it was verified that the anti-EFG1 2′OMe oligomer maintains efficacy in different human body fluids.12 Given these findings, anti-EFG1 2′OMe oligomer’s in vivo validation is crucial. Among the in vivo models available, invertebrate models, such as Galleria mellonella, have emerged at the forefront ion the study of fungal pathogenesis.14,15 The possibilities of pathogen delivery into larvae, by topical, oral, and injection application is suited to study pathogens at human body temperature makes it a desirable model for the study of fungal pathogenesis.15,16
Based on promising in vitro results,12 the main goal of this work is to validate in vivo the applicability of the anti-EFG1 2′OMe oligomer for inhibiting C. albicans filamentation and to attenuate candidiasis.
Results
Anti-EFG1 2′OMe oligomer toxicity
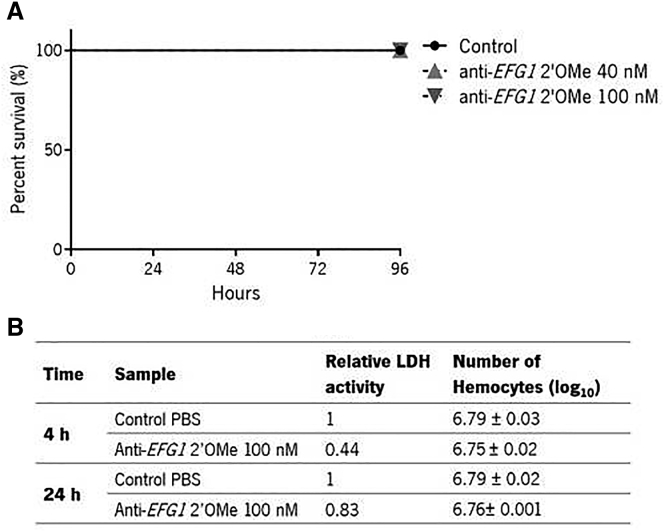
To assess the anti-EFG1 2′OMe oligomer toxicity, the G. mellonella survival rate was determined through the lactate dehydrogenase (LDH) released, and the number of hemocytes was also quantified. For that, G. mellonella larvae were injected with two different concentrations of oligomer (40 and 100 nM) and the survival evaluated during 96 h. As shown in Figure 1A, no death was observed for both tested concentrations over 96 h. Moreover, the injection of anti-EFG1 2′OMe oligomer did not increase the levels of LDH released on the hemolymphs of larvae after 4 and 24 h, since the levels of LDH are lower compared with levels released from untreated larvae (injected only with phosphate-buffered saline [PBS]) (Figure 1B). In terms of the total number of hemocytes, there is no evidence of differences between larvae injected with oligomer and the control larvae (Figure 1B). Thus, the anti-EFG1 2′OMe oligomer did not reveal toxic effects on G. mellonella.
Figure 1.
Anti-EFG1 2′OMe oligomer toxicity evaluation in a G. mellonella model
(A) Survival curves of larvae injected with 40 and 100 nM of anti-EFG1 2′OMe oligomer. For each condition, 10 larvae were injected with 40 and 100 nM of oligomer and their survival was monitored over 96 h. (B) Relative LDH activity released and total number of hemocytes counted after 4 and 24 h after injection with 100 nM of anti-EFG1 2′OMe oligomer. As controls, larvae were injected only with PBS.
G. mellonella survival
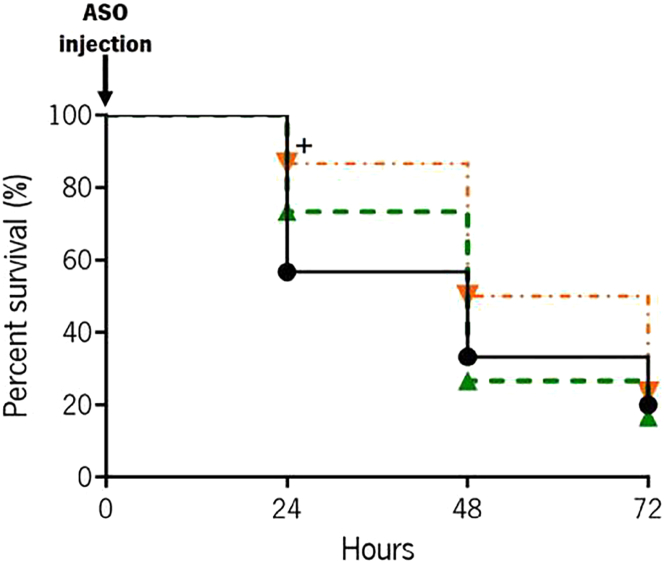
To investigate the in vivo effects of the anti-EFG1 2′OMe oligomer on attenuation of C. albicans infections, a G. mellonella larvae model was used, infected with a lethal dose of yeast cells (7 × 107 cells mL−1). A first set of larvae was treated with a single dose (0 h post infection) of anti-EFG1 2′OMe oligomer at 40 and 100 nM (Figure 2). It is noteworthy that the treatment of infected G. mellonella with a single dose of anti-EFG1 2′OMe oligomer enhances the survival of larvae over 24 h by 16% with 40 nM (p > 0.05) and by 30% with 100 nM (p < 0.05). Although, no effect was observed in larvae treated with 40 nM of anti-EFG1 2′OMe oligomer at 48 h (p > 0.05), the treatment with 100 nM intensified the larvae survival into 17% (p > 0.05). No significant effects were observed with a single dose after 72 h of infection for both concentrations tested (p > 0.05).
Figure 2.
Single-dose effect of anti-EFG1 2′OMe oligomer on the survival of G. mellonella infected with C. albicans
Survival curves of infected larvae were treated with a single dose of anti-EFG1 2′OMe oligomer (0 h post infection). Larvae infected with C. albicans cells were treated with 40 and 100 nM of anti-EFG1 2′OMe oligomer. As controls, larvae infected were injected only with PBS. +Significant difference among control and a single dose of 100 nM of anti-EFG1 2′OMe oligomer at 24 h (p < 0.05).
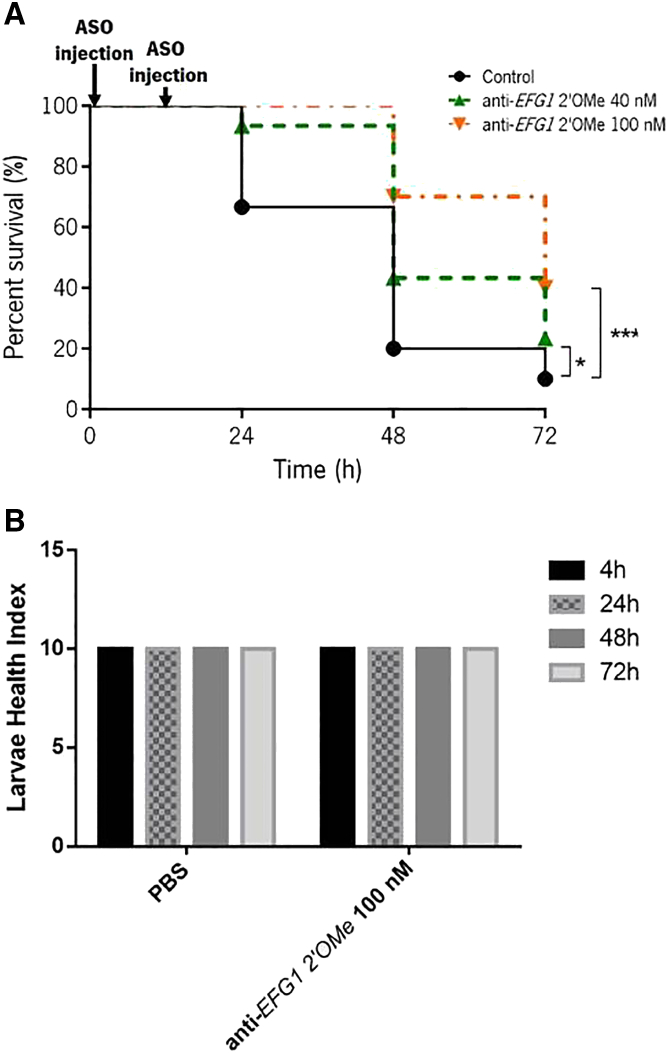
A second set of infected larvae was treated with a double dose of anti-EFG1 2′OMe oligomer (0 and 12 h post infection), since the treatments are usually not carried out only with a unique dose (Figure 3A). The results showed that a double dose of anti-EFG1 2′OMe oligomer significantly enhances the G. mellonella survival. Of note, 90% and 100% of the larvae treated with 40 nM (p < 0.05) and 100 nM (p < 0.001) survived over the first 24 h of infection. An increase in G. mellonella survival was also evident at 48 h with a rate of 23% for 40 nM (p < 0.05) and of 50% for 100 nM (p < 0.001). Note that, the administration of a double dose of anti-EFG1 2′OMe oligomer not only was responsible by enhancing the larvae survival but also for prolonging the anti-EFG1 2′OMe oligomer effects over 72 h, achieving 30% more on the survival rate with 100 nM of oligomer (p < 0.001). To infer about larvae health, the health index scores were also determined for larvae treated with 100 nM of oligomer. The larvae activity, cocoon formation, melanization, and survival were scored (Figure 3B). As can be seen, the injection of the larvae with anti-EFG1 2′OMe oligomer resulted in high health index scores even after 72 h, with a higher activity and cocoon formation.
Figure 3.
Double-dose effect of anti-EFG1 2′OMe oligomer on G. mellonella infected with C. albicans
(A) Survival curves of infected larvae treated with a double dose of anti-EFG1 2′OMe oligomer (0 and 12 h post infection). Larvae infected with C. albicans cells were treated with 40 and 100 nM of anti-EFG1 2′OMe oligomer. As controls, larvae infected were injected only with PBS. (B) The health index scores of larvae treated with a double dose of 100 nM of anti-EFG1 2′OMe oligomer. Control represents the infected larvae treated only with PBS after 12 h post infection. ∗Significant difference among control and a double dose of 40 nM of anti-EFG1 2′OMe oligomer for all times (p < 0.05). ∗∗∗Significant difference among control and a double dose of 100 nM of anti-EFG1 2′OMe ASO for all times (p < 0.001).
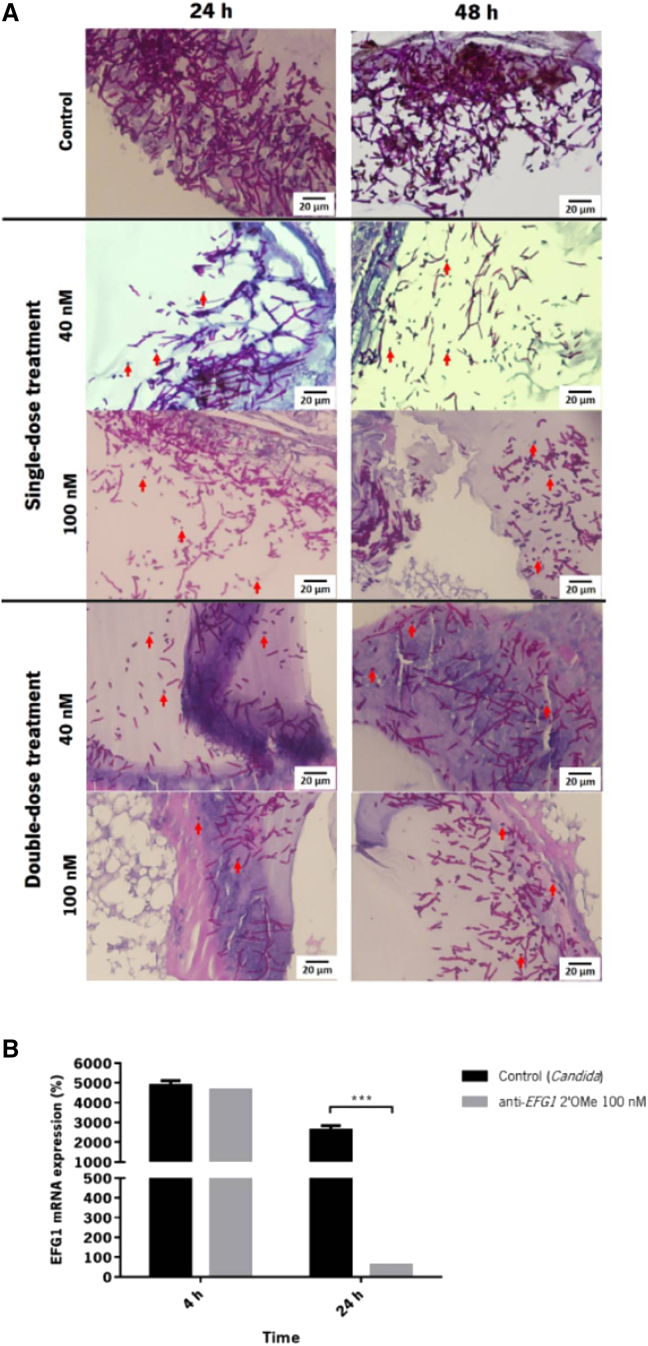
To assess the effect of the anti-EFG1 2′OMe oligomer on candidiasis progression and C. albicans morphology, the fat body of larvae was fixed, sectioned, stained, and evaluated. Figure 4A reveals the quantity and invasiveness progression of C. albicans without treatment after 24 and 48 h of infection. It is evident, that C. albicans cells are located mainly in digestive system, around the fat body and tend to organize into clusters with an extensive progression on quantity over the time. Candida albicans exhibits predominantly filamentous growth. The images highlight the contrast among the single- and double-dose treatments with the control, exhibiting both an expressive lower quantity of filaments with a significant decrease on fat body area occupied by C. albicans cells, with a more pronounced effect on sections of larvae treated with 100 nM of anti-EFG1 2′OMe oligomer. The effect of anti-EFG1 2′OMe oligomer on EFG1 gene expression was also determined at 4 and 24 h post infection (Figure 4B). The results revealed no significant differences after 4 h despite a huge reduction in the levels of EFG1 expression after 24 h post infection comparatively to the levels on untreated larvae (p < 0.001).
Figure 4.
Anti-EFG1 2′OMe oligomer effect on C. albicans cell morphology and progression into the fat body of G. mellonella
(A) Histological images of larvae infected with C. albicans (at 24 and 48 h) and treated with a single dose (0 h post infection) and with a double dose (0 and 12 h post infection) of 40 and 100 nM of anti-EFG1 2′OMe oligomer. The larvae sections were labeled with periodic acid Schiff coloration. The magnification images were at 400×. (B) Levels of EFG1 gene expression of larvae treated with a double dose of 100 nM of anti-EFG1 2′OMe oligomer evaluated by quantitative real-time PCR and analyzed by the ΔCt method and normalized to the CaACT1 mRNA levels after 4 and 24 h post infection. Control represents the infected larvae treated only with PBS after 12 h post infection. Error bars represent standard deviation. ∗∗∗Significant difference among control and a double dose of 100 nM of anti-EFG1 2′OMe oligomer at 24 h post infection (p < 0.001).
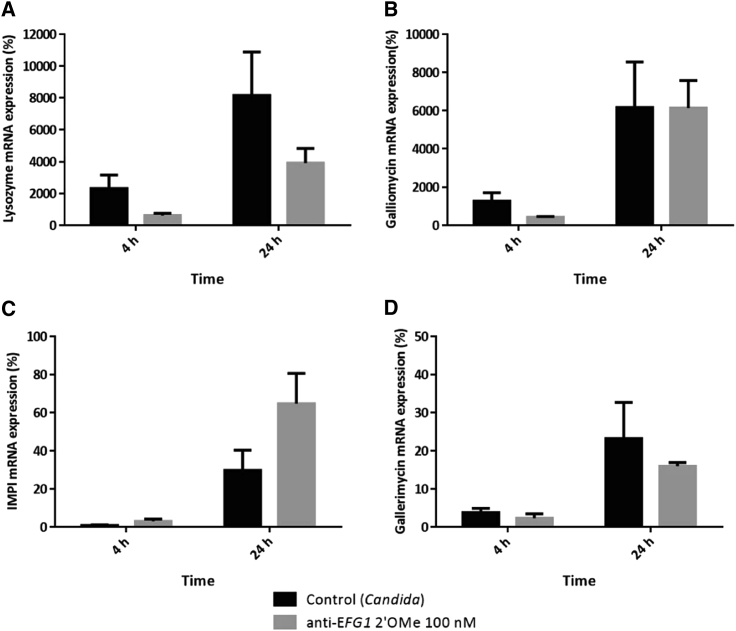
G. mellonella immune response
G. mellonella has an immune system with a high similarity to mammalians in terms of its ability to produce antimicrobial peptides, and with the ability to eliminate microorganisms.17, 18, 19 For that, the transcript levels of four encoding peptides with antimicrobial peptides, namely lysozyme, gallerimycin, galliomycin, and inducible metalloproteinase inhibitor (IMPI), were quantified by quantitative real-time PCR. The expression levels of AMPs vary according to the peptide, in which lysozyme (Figure 5A) and galliomycin (Figure 5B) presented higher levels of expression at 4 and 24 h post infection compared with the IMPI (Figure 5C) and gallerimycin (Figure 5D), indicating that these AMPs are expressed in a latter response to fight the infection. In the presence of anti-EFG1 2′OMe, in general, the levels of AMPs decreased both at 4 and 24 h post infection (p > 0.05), with the exception of IMPI, which interestingly resulted in an increase in gene expression. No change in gene expression levels of galliomycin was observed after 24 h (Figure 5B).
Figure 5.
Anti-EFG1 2′OMe oligomer effect on G. mellonella immune response
Levels of gene expression on G. mellonella treated with a double dose of 100 nM of anti-EFG1 2′OMe oligomer (0 and 12 h post infection) of (A) lysozyme, (B) galliomycin, (C) inducible metalloproteinase inhibitor, and (D) gallerimycin, after 4 and 24 h of infection by C. albicans SC5314. These results were obtained by quantitative real-time PCR and analyzed by ΔCt method and normalized to the GmACT1 mRNA levels. As a control, G. mellonella injected only with PBS after 12 h post infection was used. Error bars represent standard deviation.
Discussion
Candidiasis is supported by a series of virulence factors, and one of the most important is the ability of C. albicans cells to switch from yeast to filamentous forms. The filamentation is essential for C. albicans pathogenicity,2, 3, 4, 5 and it is regulated by a complex network of genes in which EFG1 is one of the most important virulence determinants.6,9, 10, 11 Anti-EFG1 2′OMe was suggested to degrade EFG1 mRNA by RNase activation. The in vitro results demonstrated the ability of anti-EFG1 2′OMe to reduce C. albicans cell filamentation (by 80%) and EFG1 gene expression (by 60%).12 Taking into account the promising in vitro results, the aim of this work was to validate in vivo its applicability for inhibiting C. albicans filamentation and to attenuate candidiasis using the G. mellonella model. As in other microbiological relevant studies,17, 18, 19 we opted to use the G. mellonella model to validate the in vivo performance of the anti-EFG1 2′OMe oligomer, since it is a model that provides a rapid, inexpensive, and reliable way to evaluate the effects and toxicity in vivo of nano-drugs.
As in our in vitro results,12 no evidence of in vivo toxicity was observed over 96 h (Figure 1). In fact, all larvae stayed alive over 96 h (Figure 1A) with no significant differences in terms of LDH released and in the total number of hemocytes (Figure 1B) on the hemolymphs of larvae compared with larvae injected only with PBS.
The infected G. mellonella larvae with 7 × 107 cells mL−1 of C. albicans cells were treated with a single dose of anti-EFG1 2′OMe oligomer (0 h post infection). It was clear that the anti-EFG1 2′OMe oligomer maintains its performance in vivo, as an increase in larvae survival compared with untreated larvae was observed. Moreover, with these results it is also clear that the in vivo anti-EFG1 2′OMe oligomer efficacy is concentration dependent. In fact, the treatment of infected G. mellonella with a single dose of anti-EFG1 2′OMe oligomer enhances the survival of larvae over 24 h (16%), being more pronounced with 100 nM of oligomer (30%) (Figure 2). However, after 48 h of infection the anti-EFG1 2′OMe oligomer loses its effectiveness. This result was expected, as once in a clinical context, an infection is rarely controlled with a single dose of antimicrobial and the treatments are not usually carried out over a precise time.20, 21, 22, 23 To mimic that, a double dose of anti-EFG1 2′OMe oligomer was administered (0 and 12 h post infection) on G. mellonella larvae infected with C. albicans cells (Figure 3). The results indicate that, with a double-dose administration of anti-EFG1 2′OMe oligomer, it is possible to intensify the molecule efficacy and prolong its effect over time. In fact, larvae treated with the double dose of oligomer survived around 90% (with 40 nM) and 100% (with 100 nM) over the first 24 h (Figure 3A). Moreover, an increase on larvae survival was also evident at 48 h (by 50%) and 72 h (by 30%), with more pronounced effect in case of 100 nM of oligomer, with a high health index score. These findings corroborate with the observed histological images of G. mellonella fat body, which demonstrates a strong decrease in the number of C. albicans as filaments, and an evident reduction on the extension of area occupied by Candida cells in tissues from larvae treated with anti-EFG1 2′OMe oligomer (Figure 4A). The quantitative real-time PCR assays confirm a huge reduction in the levels of EFG1 transcripts after 24 h of post infection and treated with the oligomer (Figure 4B), which is in accordance with the decrease in the number of C. albicans filaments.
The G. mellonella system presents an immune system with a highly similarity to the mammalian immune system, and the ability to release AMPs is important to fight infection.19,24 In general, the expression of AMPs was lower in the presence of 100 nM of anti-EFG1 2′OMe oligomer, indicating a possible reduction in C. albicans infection (Figure 5) when larvae are treated with the oligomer.
Numerous studies have documented the use of antisense therapy (AST) as a biochemical tool for studying human target diseases; as of now, there are ten antisense drugs in the market. However, the application of AST as anti-Candida agents is still rare, and there is one study using AST to interrupt and efficiently inhibit C. albicans by in vivo splicing using a PS-modified antisense oligonucleotide (ASO).25 Our results reveal that it is possible to synthesize an ASO modified by 2′-OMethyl chemical modification to control the virulence factor of C. albicans. Moreover, this study suggests that systemic delivery of anti-EFG1 2′OMe oligomer is feasible, devoid of toxicity, and could be a promising treatment strategy for C. albicans infections. Therefore, it warrants further studies in other animal models.
Conclusions
Hereby, the present work confirms that the anti-EFG1 2′OMe oligomer is able to inhibit C. albicans filamentation and attenuates the candidiasis on G. mellonella model. Undoubtedly, this work reveals the in vivo therapeutic potential of anti-EFG1 2′OMe oligomer for controlling C. albicans infections.
Materials and methods
Anti-EFG1 2′OMe oligomer preparation
The anti-EFG1 2′OMe oligomer was designed and synthesized based on the second generation of chemical modifications of nucleic acid mimics as described in our recent published works.12,26 Aliquots of anti-EFG1 2′OMe oligomer were prepared in sterile ultrapure water to 4 μM and stored at −20°C for later use. Whenever necessary, oligomer molecules were diluted in PBS to final concentrations of 40 and 100 nM. The lower concentration was selected according to our previous results in vitro12 and 100 nM was used to be tested as a higher concentration.
C. albicans cells and growth conditions
The C. albicans SC5314, belonging to the Candida strain collection of the Biofilm group of the Centre of Biological Engineering, was used during these studies. For all experiments, the yeast strain was subcultured on sabouraud dextrose agar (Merck, Germany) and incubated for 24 h at 37°C. Cells were then inoculated in sabouraud dextrose broth (Merck) and incubated overnight at 37°C, 120 rpm. After incubation, the cell suspensions were centrifuged for 10 min at 3,000 × g and 4°C, and washed twice with PBS (pH 7, 0.1 M). Pellets were suspended in 5 mL of PBS, and the cellular density was adjusted using a Neubauer chamber (Marienfild, Land-Konicshofem, Germany) to 7 × 107 cells mL−1.
G. mellonella larvae
G. mellonella larvae were reared on a pollen grain and bee wax diet at 25°C in darkness and used in a final stage of development with a weight of approximately 250 mg. The larvae were injected into hemolymph via the hindmost left proleg, previously sanitized with 70% (v/v) ethanol, using a micro syringe adapted in micrometres to control the volume of injection.20 All experiments were performed in triplicate and in a minimum of three independent assays.
Toxicity assays
To test the in vivo toxicity of the anti-EFG1 2′OMe oligomer, 10 larvae of G. mellonella were injected with 5 μL of 40 and 100 nM of oligomer prepared in PBS. As control, a set of larvae were injected with the same volume but only with PBS. Larvae were placed in Petri dishes and stored in the dark at 37°C. Larvae morphology and survival were followed over 4 days and the survival curves were constructed.
The LDH activity released from larvae tissues to hemolymph was also evaluated. For that, larvae were sacrificed at 4 and 24 h after injection and the hemolymphs of five larvae were collected in an Eppendorf tube. This assay was performed using the CytoTox-ONE Homogeneous Membrane Integrity Assay Kit (Promega) according to the manufacturer’s instructions. LDH activity was quantified by fluorescence spectrometer evaluation (Cytation 3 Cell Imaging Multi-Rode Reader, BioTek) by measuring the NADH disappearance rate at 560 nm excitation and 590 nm emission during the LDH-catalyzed conversion of pyruvate to lactate. The value of LDH activity of the larvae injected only with PBS used as a control was subtracted from the LDH activity of the larvae injected with 100 nM of anti-EFG1 2′OMe. The levels of LDH released were expressed as relative LDH activity.
The total numbers of hemocytes present in the hemolymphs of larvae were also evaluated. For that, three larvae previously sanitized with 70% (v/v) ethanol were punctured in the abdomen with a sterile needle and the hemolymphs were recovered in a sterile microtube. The hemolymph mixture was diluted 10-fold in sterile PBS and hemocytes were counted using a hemocytometer. The results are presented as a logarithm of the concentration (Log10).
G. mellonella survival assays
To study the effect of the anti-EFG1 2′OMe oligomer on the survival rate of G. mellonella, larvae were infected with 5 μL of a lethal dose of C. albicans cells (7 × 107 cells mL−1) and randomly allocated to five different experimental groups (with a set of ten larvae). The concentration of C. albicans to be injected (7 × 107 cells mL−1) was selected on the basis of the G. mellonella lethality results after injection with different concentrations of yeast cells (between 7 × 107 and 2 × 108 cells mL−1) (Figure S1). Two sets of larvae were treated with a single dose of 40 and 100 nM of oligomer (0 h of post infection); two sets of larvae with a double dose of 40 and 100 nM of oligomer (0 and 12 h of post infection); and a set only with PBS. As control, a set of larvae were injected only with the same volume of PBS. After injections, the larvae were placed in Petri dishes and stored in the dark at 37°C, over 72 h, and survival curves were subsequently constructed. The larvae were considered dead when they displayed no movement in response to touch. The G. mellonella health index was also determined for the larvae treated with a double dose of 100 nM of oligomer, which scores four main parameters: larvae activity, cocoon formation, melanization, and survival.27
Gene expression analysis
Quantitative real-time PCR was used to determine the EFG1 gene expression on C. albicans after the treatment with 100 nM of anti-EFG1 oligomer. The transcript levels of genes encoding the G. mellonella antimicrobial peptides, gallerimycin, galliomycin, IMPI, and lysozyme, were also determined to infer the G. mellonella immune response. For that, three larvae treated with oligomers at 4 and 24 h post infection and three larvae untreated were cryopreserved, sliced, and homogenized in lysis buffer reagent.
RNA extraction was performed using the PureLink RNA Mini Kit (Invitrogen, Carlsbad, CA, USA).12 To avoid potential DNA contamination, samples were treated with DNase I (Deoxyrybonuclease I, Amplification Grade, Invitrogen), and the RNA concentration was determined by optical density measurement (NanoDrop 1000 Spectrophotometer, Thermo Scientific). The cDNA was synthesized using the iScript Reverse Transcriptase (Bio-Rad, Berkeley, USA) in accordance with the manufacturer’s instructions, and real-time qPCR (CFX96, Bio-Rad) was performed on a 96-well microtiter plate using Eva Green Supermix (Bio-Rad). Each reaction was performed in triplicate and mean values of expression were determined by the ΔCt method. Non-transcriptase reverse controls were included in each run. The primers used are presented in Table 1.
Table 1.
Primers used for quantitative real-time PCR
| Gene name | Sequence (5′–3′) | Primer | |
|---|---|---|---|
| Candida albicans | EFG1 | TTCTGGTGCAGGTTCCAC | forward |
| CCTGGTTGTGATGCAGGT | reverse | ||
| ACT1 | AATGGGTAGGGTGGGAAAAC | forward | |
| AGCCATTTCCATTGATCGTC | reverse | ||
| Galleria mellonella | actin | ATCCTCACCCTGAAGTACCC28 | P1RT |
| CCACACGCAGCTCATTGTA28 | P2RT | ||
| lysozyme | TCCCAACTCTTGACCGACGA28 | P1RT | |
| AGTGGTTGCGCCATCCATAC28 | P2RT | ||
| galliomycin | TCGTATCGTCACCGCAAAATG29 | P1RT | |
| GCCGCAATGACCACCTTTATA29 | P2RT | ||
| IMPI | AGATGGCTATGCAAGGGATG28 | P1RT | |
| AGGACCTGTGCAGCATTTCT28 | P2RT | ||
| gallerimycin | CGCAATATCATTGGCCTTCT28 | P1RT | |
| CCTGCAGTTAGCAATGCAC28 | P2RT |
G. mellonella histological fat body analysis
Histological analysis of G. mellonella was performed to study the effect of the anti-EFG1 2′OMe oligomer on candidiasis progression and C. albicans morphology into fat body of larvae. For that, one larva from each group of study were recovered at 24 and 48 h, to be processed histologically. The fat body was removed, from each larva, through an incision in the midline of the ventral with a scalpel blade. The fat body was placed in 4% (v/v) of paraformaldehyde and stored for 24 h at 4°C to preserve the structures. The paraffin blocks were cut on sections of 4–5 μm, and the sections were stained with periodic acid Schiff and hematoxylin and eosin. Tissue sections were viewed and photographed with an Olympus BX51 microscope coupled with a DP71 digital camera (Olympus Portugal SA, Porto, Portugal).
Statistical analysis
Data are expressed as the mean ± standard deviation of a least three independent experiments. Results were compared using two-way analysis of variance using GraphPad Prism 6 (GraphPad Software, CA, USA). All tests were performed with a confidence level of 95%. Kaplan-Meier survival curves were plotted and differences in survival were calculated by using log rank Mantel-Cox statistical test, all performed with GraphPad Prism 6.
Acknowledgments
This study was supported by the Portuguese Foundation for Science and Technology (FCT) under the scope of the strategic funding of UIDB/04469/2020 unit and BioTecNorte operation (NORTE-01-0145-FEDER-000004) funded by the European Regional Development Fund under the scope of Norte2–20 – Programa Operacional Regional do Norte and Daniela Eira Araújo [SFRH/BD/121417/2016] PhD grant. The authors also acknowledge the project funding by the “02/SAICT/2017 – Projetos de Investigação Científica e Desenvolvimento Tecnológico (IC&DT) – POCI-01-0145-FEDER-028893.” Funding received by IBB – Institute for Bioengineering and Biosciences from FCT (UID/BIO/04565/2020) and Programa Operacional Regional de Lisboa 2020 (project no. 007317) is also acknowledged. We acknowledge Dr Lucília Goreti Pinto, Life and Health Sciences Research Institute (ICVS), School of Medicine, University of Minho, for processing and sectioning tissue samples of G. mellonella.
Author contributions
D.A. and S.S. conceived and designed the study. D.A. and D.M.-H. conducted the experiments. D.A. wrote the manuscript. M.H. and S.S. performed the analysis and read the paper. All authors read and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.018.
Supplemental information
References
- 1.Koehler P., Stecher M., Cornely O.A., Koehler D., Vehreschild M.J.G.T., Bohlius J., Wisplinghoff H., Vehreschild J.J. Morbidity and mortality of candidaemia in Europe: an epidemiologic meta-analysis. Clin. Microbiol. Infect. 2019;25:1200–1212. doi: 10.1016/j.cmi.2019.04.024. [DOI] [PubMed] [Google Scholar]
- 2.Silva S., Negri M., Henriques M., Oliveira R., Williams D.W., Azeredo J. Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 2012;36:288–305. doi: 10.1111/j.1574-6976.2011.00278.x. [DOI] [PubMed] [Google Scholar]
- 3.Silva S., Rodrigues C., Araújo D., Rodrigues M., Henriques M. Candida species biofilms’ antifungal resistance. J. Fungi. 2017;3:8. doi: 10.3390/jof3010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer F.L., Wilson D., Hube B. Candida albicans pathogenicity mechanisms. Virulence. 2013;4:119–128. doi: 10.4161/viru.22913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araújo D., Henriques M., Silva S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017;1:62–75. doi: 10.1016/j.tim.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Stoldt V.R., Sonneborn A., Leuker C.E., Ernst J.F. Efg1p, an essential regulator of morphogenesis of the human pathogen Candida albicans, is a member of a conserved class of bHLH proteins regulating morphogenetic processes in fungi. EMBO J. 1997;16:1982–1991. doi: 10.1093/emboj/16.8.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ernst J.F. Transcription factors in Candida albicans—environmental control of morphogenesis. Microbiology. 2000;146:763–774. doi: 10.1099/00221287-146-8-1763. [DOI] [PubMed] [Google Scholar]
- 8.Ramage G., Vandewalle K., López-Ribot J.L., Wickes B.L. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS Microbiol. Lett. 2002;214:95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 9.Nobile C.J., Mitchell A.P. Regulation of cell-surface genes and biofilm formation by the C. albicans transcription factor Bcr1p. Curr. Biol. 2005;15:1150–1155. doi: 10.1016/j.cub.2005.05.047. [DOI] [PubMed] [Google Scholar]
- 10.Nobile C.J., Fox E.P., Nett J.E., Sorrells T.R., Mitrovich Q.M., Hernday A.D., Tuch B.N., Andes D.R., Johnson A.D. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell. 2011;148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Connolly L., Riccombeni A., Grózer Z., Holland L.M., Lynch D.B., Andes D.R., Gácser A., Butler G. The APSES transcription factor Efg1 is a global regulator that controls morphogenesis and biofilm formation in Candida parapsilosis. Mol. Microbiol. 2013;90:36–53. doi: 10.1111/mmi.12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araújo D., Azevedo N.M., Barbosa A., Almeida C., Rodrigues M.E., Henriques M., Silva S. Application of 2′-OMethylRNA antisense oligomer to control Candida albicans EFG1 virulence determinant. Mol. Ther. Nucleic Acids. 2019;18:508–517. doi: 10.1016/j.omtn.2019.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khvorova A., Watts J.K. The chemical evolution of oligonucleotide therapies of clinical utility. Nat. Biotechnol. 2017;35:238–248. doi: 10.1038/nbt.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fedhila S., Buisson C., Dussurget O., Serror P., Glomski I.J., Liehl P., Lereclus D., Nielsen-LeRoux C. Comparative analysis of the virulence of invertebrate and mammalian pathogenic bacteria in the oral insect infection model Galleria mellonella. J. Invertebr. Pathol. 2010;103:24–29. doi: 10.1016/j.jip.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Junqueira J.C. Galleria mellonella as a model host for human pathogens. Virulence. 2012;3:474–476. doi: 10.4161/viru.22493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs B.B., O’Brien E., Khoury J.B.E., Mylonakis E. Methods for using Galleria mellonella as a model host to study fungal pathogenesis. Virulence. 2010;1:475–482. doi: 10.4161/viru.1.6.12985. [DOI] [PubMed] [Google Scholar]
- 17.Mak P., Zdybicka-barabas A. A different repertoire of Galleria mellonella antimicrobial peptides in larvae challenged with bacteria and fungi. Dev. Comp. Immunol. 2010;34:1129–1136. doi: 10.1016/j.dci.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Moghaddam M., Tonk M., Schreiber C., Salzig D., Czermak P., Vilcinskas A., Rahnamaeian M. The potential of the Galleria mellonella innate immune system is maximized by the co-presentation of diverse antimicrobial peptides. Biol. Chem. 2011;397:939–945. doi: 10.1515/hsz-2016-0157. [DOI] [PubMed] [Google Scholar]
- 19.Tsai C.J., Mei J., Loh S., Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7:214–229. doi: 10.1080/21505594.2015.1135289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mil-Homens D., Ferreira-Dias S., Fialho A.M. Fish oils against Burkholderia and Pseudomonas aeruginosa: in vitro efficacy and their therapeutic and prophylactic effects on infected Galleria mellonella larvae. J. Appl. Microbiol. 2016;120:1509–1519. doi: 10.1111/jam.13145. [DOI] [PubMed] [Google Scholar]
- 21.Vilela S.F.G., Barbosa J.O., Rossoni R.D., Santos J.D., Prata M.C.A., Anbinder A.L., Jorge A.O.C., Junqueira J.C. Lactobacillus acidophilus ATCC 4356 inhibits biofilm formation by C. albicans and attenuates the experimental candidiasis in Galleria mellonella. Virulence. 2015;6:29–39. doi: 10.4161/21505594.2014.981486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rossoni R.D., dos Santos Velloso M., Figueiredo L.M.A., Martins C.P., Jorge A.O.C., Junqueira J.C. Clinical strains of Lactobacillus reduce the filamentation of Candida albicans and protect Galleria mellonella against experimental candidiasis. Folia Microbiol. 2018;63:307–314. doi: 10.1007/s12223-017-0569-9. [DOI] [PubMed] [Google Scholar]
- 23.Straarup E.M., Fisker N., Hedtjärn M., Lindholm M.W., Rosenbohm C., Aarup V., Hansen H.F., Ørum H., Hansen J.B.R., Koch T. Short locked nucleic acid antisense oligonucleotides potently reduce apolipoprotein B mRNA and serum cholesterol in mice and non-human primates. Nucleic Acids Res. 2010;38:7100–7111. doi: 10.1093/nar/gkq457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jemel S., Guillot J., Kallel K., Botterel F., Dannaoui E. Galleria mellonella for the evaluation of antifungal efficacy against medically important fungi, a narrative review. Microorganisms. 2020;8:1–19. doi: 10.3390/microorganisms8030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Testa, S., Disney, M., Gryaznov, S., and Turner, D. Methods and compositions for inhibition of RNA splicing. U.S. patent WO/2000/055374, 2000.
- 26.Silva, S., Araújo, D., Azevedo, N.M., Almeida, C., Henriques, M. Antisense oligomers for controlling Candida albicans infections. International patent WO 2020/174366 A1, 2020.
- 27.Loh J.M.S., Adenwalla N., Wiles S., Proft T. Galleria mellonella larvae as an infection model for group A streptococcus. Virulence. 2013;4:419–428. doi: 10.4161/viru.24930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altincicek B., Vilcinskas A. Metamorphosis and collagen-IV-fragments stimulate innate immune response in the greater wax moth, Galleria mellonella. Dev. Comp. Immunol. 2006;30:1108–1118. doi: 10.1016/j.dci.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 29.Wojda I., Kowalski P., Jakubowicz T. Humoral immune response of Galleria mellonella larvae after infection by Beauveria bassiana under optimal and heat-shock conditions. J. Insect Physiol. 2009;55:525–531. doi: 10.1016/j.jinsphys.2009.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.