Abstract
Cardiac hypertrophy is a physiological adaptation to pressure stress that augments or preserves cardiac function. Prolonged hypertrophy can, however, eventually lead to heart failure. Although some effector molecules and signaling pathways have been associated with myocardial hypertrophy, progress has been limited, and further studies are needed to thoroughly explore the underlying mechanisms and to discover novel and effective therapeutic targets. Recently, non-coding RNAs, which are well-known physiological regulators, have attracted much attention in the field of cardiovascular research. Circular RNA, in particular, has emerged as a key player in cardiac hypertrophy, and increasing numbers of papers are now being devoted to this topic. In this review, we will give a brief introduction to circular RNA and then focus on its role as a potential therapeutic target in cardiac hypertrophy.
Keywords: cardiac hypertrophy, circular RNA, sponge, regulation, therapeutic target
Graphical abstract
Cardiac hypertrophy is an adaptive response to pressure overload and can eventually lead to heart failure. circRNA has recently emerged as a key player in myocardial hypertrophy. In this review, we mainly summarize the crucial roles of circRNA in the regulation of cardiac hypertrophy.
Introduction
Cardiac hypertrophy, which is initially a compensatory mechanism to sustained ventricular wall stress caused by pressure or volume overload, acts by diminishing wall stress and reducing oxygen consumption.1,2 Chronic hypertrophy, however, undermines the structure and pumping capacity of the heart.3 Pathological hypertrophic growth, accompanied by fibrosis and maladaptive cardiac remodeling, is a potential contributor to heart failure, malignant arrhythmia, and even sudden death.4 Myocardial hypertrophy can thus be used as a predictor of cardiovascular morbidity and mortality and could signal the need for early intervention. A better and more comprehensive understanding of the molecular mechanisms that regulate cardiac hypertrophy would be invaluable in developing more effective diagnostics and therapies.
Non-coding RNAs (ncRNAs), which account for around 98% of the human genome, are a type of RNA that cannot be used as a template for the translation of proteins. Instead, ncRNAs regulate a variety of physiological and pathophysiological processes, such as mRNA translation, RNA splicing, DNA replication and repair, gene transcription, and cell differentiation.5,6 ncRNAs, which consist mainly of microRNA (miRNA), circular RNA (circRNA), long ncRNA (lncRNA), and small nucleolar RNA (snoRNA), are classified by structure, length, and function. Unlike traditional linear RNA molecules, circRNA molecules are covalently closed and do not have free termini.7 Endogenous circRNAs are abundant and highly conserved and, in eukaryotic cells, display expression patterns that are specific to cell type, tissue type, and developmental stage.8, 9, 10 In the past few years, rapid progress in biochemical techniques and the use of high-throughput sequencing have enabled further understanding and identification of circRNAs.11 In this review, we will give a brief introduction to circRNA and then focus on its role in the regulation of cardiac hypertrophy.
Discovery, biogenesis, and functions of circRNA
The existence of circRNA was first reported by the Sanger group in 1976 when they showed that plant viroids are single-stranded covalently closed RNA loops.12 In 1979, Hsu et al. discovered the first mammalian circRNAs in the cytoplasm of HeLa (immortalized human cervical carcinoma) and other eukaryotic cells by electron microscopy.13 In the early years, however, circRNAs were considered to be merely byproducts of aberrant RNA splicing and received little attention. The nascent field of circRNA research only really took off in 2013 with the striking discovery by the Memczak group of a human antisense circRNA to the cerebellar degeneration-related protein 1 transcript (CDR1as), which was shown to play a role in neural development.14 There are now a number of systematic databases, including CSCD, Circ2Disease, exoRBase, and circNet, that were established to catalog and classify circRNAs from different species and to provide access to associations between human diseases and ncRNAs.15, 16, 17, 18 Such databases have greatly facilitated further exploration of the functions of circRNA and elucidation of underlying regulatory mechanisms. The number of publications on circRNAs will undoubtedly continue to grow exponentially.
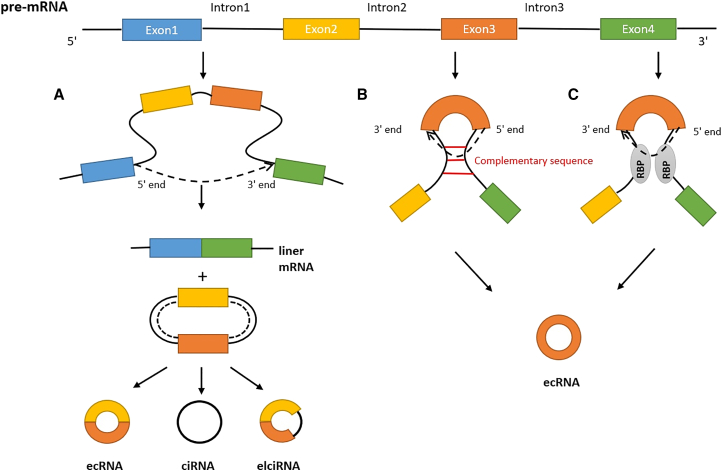
circRNAs are generated by a unique splicing mechanism originally proposed by Jeck et al. and termed “back-splicing.” 19 In back-splicing, the 5′ terminal of a pre-mRNA upstream exon is ligated in a non-colinear fashion to the 3′ terminal of a downstream exon. Unlike canonical splicing, back-splicing leads to the disordered reading of exon sequences because of the circular structure of circRNA and thus gives rise to totally different transcripts. Besides, base pairing of flanking reverse complementary sequences (i.e., Alu repeats) of introns promotes the biogenesis. Jeck et al. also proposed two separate models of circRNA biogenesis through back-splicing: lariat-driven circularization and intron-pairing-driven circulation (Figures 1A and 1B).19 The final products can be classified into three categories: exonic circRNA (ecRNA), circular intronic RNA (ciRNA), and exon-intron circRNA (EIciRNA). It is noteworthy that a number of RNA-binding proteins (RBPs), such as the RBP quaking 1, RBM20, and muscleblind, have recently been found to regulate circRNA biogenesis by increasing circRNA expression and formation.20,21 They connect to flanking introns to make the donor site and the acceptor site closer, thereby promoting the exon cyclization (Figure 1C).22, 23, 24 On the other hand, the RNA-editing enzyme ADAR1 has a negative effect on circRNA biogenesis, although the underlying mechanism is uncertain.9
Figure 1.
The biogenesis of circRNA
(A) In lariat-driven circularization, the pre-mRNA is partially folded during transcription, and the 5′ donor site of the upstream intron comes close to and attacks the 3′ acceptor site of the downstream intron. A linear mRNA and an exon-containing lariat with a 2′–5′ phosphodiester bond are then formed. After a second splicing reaction, the lariat intermediate is finally transformed into a circular RNA. The final products can be divided into three categories: ecRNA, ciRNA, and EIciRNA. (B) In intron-pairing-driven circularization, complementary flanking intronic sequencing of pre-mRNA leads to the formation of secondary structure, which allows the 5′ end of the downstream intron to attack the 3′ end of the upstream intron to form a circular RNA and a terminated fork product. (C) RNA-binding proteins could promote splicing and circRNA formation by binding flanking introns to make the donor site and the acceptor site closer.
A growing body of evidence points to the involvement of circRNAs in different physiological and pathological processes and indicates that circRNAs could act as novel biological targets for disease diagnosis and therapy. Groundbreaking studies by the Memczak14 and Hansen25 groups showed that some circRNAs contain numerous miRNA-binding sites and act as sponges that sequester miRNA. This reduces levels of specific miRNAs that could otherwise inhibit the translation or promote the degradation of target mRNAs and thus post-transcriptionally regulates gene expression. Memczak et al. found that CDR1as (mentioned above) contains 63 conserved binding sites for miR-7 and has a miRNA-binding capacity much higher than that of any other known transcript. CDR1as strongly suppressed miR-7 activity in human and mouse neural tissues and knock down of CDR1as led to miR-7 dysregulation and subsequent nervous system diseases. The Hansen group similarly found that the transcript, which they termed ciRS-7, harbors over 70 selectively conserved miRNA target sites. ciRS-7 was discovered to be highly expressed in cerebral tissue and to strongly suppress miR-7 activity, facilitating increased levels of miR-7 targets. Hansen et al. also described a testis-specific circRNA, sex-determining region Y (Sry)9, which acts as a miR-138 sponge, implying that the phenomenon of miRNA sponging by circRNA should not be a coincidence.
Since miRNAs play essential roles in cellular homeostasis, it is likely that some circRNAs may regulate stress response pathways by inhibiting miRNA activity. In addition to neurological diseases, circRNAs have been associated with cell proliferation, tumor progression, insulin secretion, and myocardial infarction (MI).26, 27, 28, 29, 30, 31, 32, 33, 34, 35 Interestingly, circRNAs can exert opposing effects on tumor progression through binding to different miRNAs. For example, CDR1as is reported to be involved in the pathogenesis of esophageal squamous cell carcinoma by impairing the tumor-suppressive effects of miR-7 and activating the downstream HOXB13-mediated NF-κB/p65 pathway, whereas it plays an anti-oncogenic role in bladder cancer by sponging miR-135a and modulating P21 expression levels.31,32 circRNAs can also have opposing effects in cardiovascular disease. In myocardial ischemia and infarction, Geng et al. showed that CDR1as/ciRS-7 aggravated cardiomyocyte apoptosis and increased the size of myocardial infarcts by binding to miR-7a.33 Wang et al. suggested that mitochondrial fission and apoptosis-related circRNA (MFACR), also referred to as mm9-circ-016597, promotes cardiomyocyte apoptosis by directly restraining the beneficial activity of miR-652-3p, which downregulates the expression of mitochondrial 18 kDa protein (MTP18) and attenuates MI during myocardial ischemia-reperfusion.34 On the other hand, the Cai group reported that the circRNA circ-Ttc3, which acts as a sponge for miR-15b-5p, plays a protective role in MI by upregulating Arl2 gene expression and helping to overcome hypoxia-induced ATP depletion and apoptotic death in cardiomyocytes.35
As well as binding to miRNAs, circRNAs can exert biological effects by interacting with specific proteins. One of the most prominent examples here is circ-Foxo3. Du et al. found that dysregulation of circ-Foxo3, which sponges cyclin-dependent kinase 2 (CDK2) and CDK2 inhibitor 1 (or P21) to form a ternary complex, disrupted cell-cycle progression and led to the development of cancer.36 The same group later demonstrated that circ-Foxo3 sequesters the anti-senescence protein ID-1, the anti-stress proteins FAK and HIF1α, and the transcription factor E2F1 from the cytoplasm into the nucleus and mitochondria, thus repressing their normal functions and inducing cellular senescence.37 There are, of course, other protein-sponging circRNAs. In doxorubicin-induced cardiomyopathy, circ-Amotl1 alleviates myocardial injury and enhances cardiac repair in vivo by interacting with serine/threonine-protein kinase 1 (AKT1) and phosphoinositide-dependent kinase1 (PDK1), leading to AKT1 phosphorylation and cardio-protective nuclear translocation.38 Additionally, circPABPN1 has been suggested to have a strong affinity for HuR (an RBP) and to reduce PABPN1 translation in HeLa cells. It is noteworthy that this is the first example of competition between a circRNA and its cognate mRNA for the same RBP.39
Although most circRNAs can barely encode proteins, those that contain an internal ribosome entry site (IRES) or N6-methyladenosine modification (m6A) have the potential to be translated into small peptides or proteins.40, 41, 42 For instance, Legnini et al. recently proposed that human circ-ZNF609 serves as a functional part in modulating myoblast proliferation and can be translated into peptides in a splicing-dependent and cap-independent manner.43 Other circRNAs, such as circPINT87aa, circMbl, circ-FBXW7, and circSHPRH, have also been proven to have coding potential,44, 45, 46 but the details are beyond the scope of this review.
circRNA and cardiac hypertrophy
Pathological myocardial hypertrophy is now a well-acknowledged risk factor for heart failure, arrhythmia, and even sudden cardiac death, which are some of the main causes of morbidity and mortality worldwide.4 Attempts to explore the molecular mechanisms underlying cardiac hypertrophy and to identify therapeutic targets have thus recently accelerated. Approaches to avoid excessive hypertrophy and reverse maladaptive cardiac remodeling and even heart failure would be particularly attractive. In addition to known regulators, such as AKT and Gαq-coupled receptors,47,48 circRNA research has identified several attractive targets that have the potential to regulate the pathological processes involved in myocardial hypertrophy and that show promise as new diagnostic or therapeutic targets.
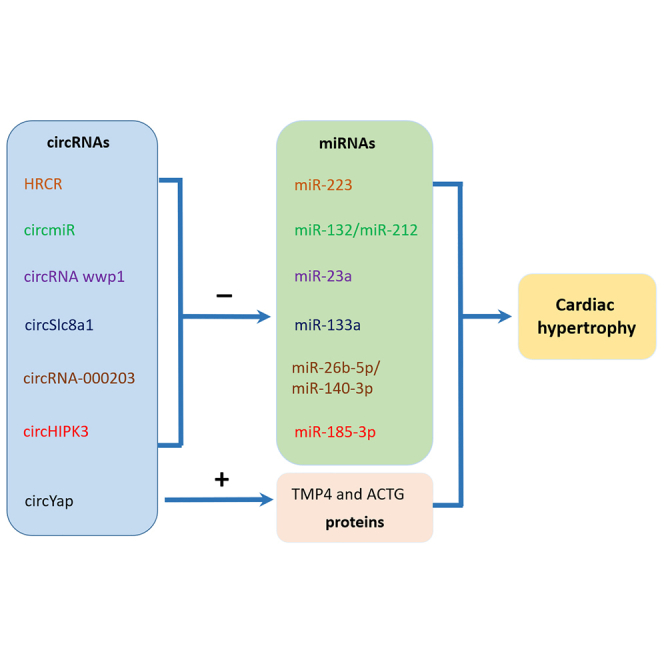
Early research by the Werfel group included the RNA sequencing (RNA-seq) analysis of ribosome-depleted libraries from rats (neonatal and adult), mice (sham or after transverse aortic constriction [TAC]), and humans (failing and non-failing). What is more, Werfel et al. noted a tendency toward increased cardiac expression of circRNA in a TAC-induced mouse model of cardiac hypertrophy and provided the first hint that circRNA may be associated with cardiac hypertrophy.49 In this review, we will summarize several specific circRNAs involved in cardiac hypertrophy (Table 1).
Table 1.
circRNAs involved in the regulation of cardiac hypertrophy
| circRNAs | Mechanisms | Effect on cardiac hypertrophy | References |
|---|---|---|---|
| HRCR | acting as an endogenous miR-223 sponge to repress its activity and aggravate the influence of ARC | negative | Wang et al.50 |
| circSlc8a1 | serving as a sponge for miR-133a and further affecting its targets: Srf, Ctgf, Adrb1, and Adcy6 | positive | Lim et al.51 |
| circRNA-000203 | sponging miR-26b-5p and miR-140-3p to abolish the suppression of target gene Gata4 and then upregulate GATA4 expression | positive | Li et al.52 |
| circmiR | synthesizing a circRNA with certain miRNA binding sites artificially to target miR-132 and miR-212 | negative | Lavenniah et al.53 |
| circRNA wwp1 | exerting functions by targeting miR-23a and ANF on isoproterenol-induced cardiac hypertrophy | negative | Yang et al.54 |
| circHIPK3 | sponging miR-185-3p and subsequently modulating its target, CaSR | positive | Xu et al.55 |
| circYap | binding directly with TMP4 and ACTG and facilitating the interaction between the two to suppress actin polymerization and the following fibrosis | negative | Wu et al.56 |
circmiR, custom circRNA sponge; ANF, atrial natriuretic factor; CaSR, calcium-sensing receptor; TMP4, tropomyosin-4; ACTG, gamma-actin.
Heart-related circRNA
In a study to investigate the molecular mechanisms that regulate cardiac hypertrophy, Wang et al.50 found that heart-related circRNA (HRCR), which serves as an endogenous miR-223 sponge, inhibited hypertrophy induced by both isoproterenol and TAC. Using cardiac-specific transgenic mice, miR-223 was shown to contribute to cardiac hypertrophy in failing hearts, whereas its downstream target—apoptosis repressor with CARD domain (ARC)—reduced hypertrophy. HRCR can thus attenuate hypertrophic responses in vivo by repressing the activity of miR-223 and enhancing the effect of ARC.
circSlc8a1
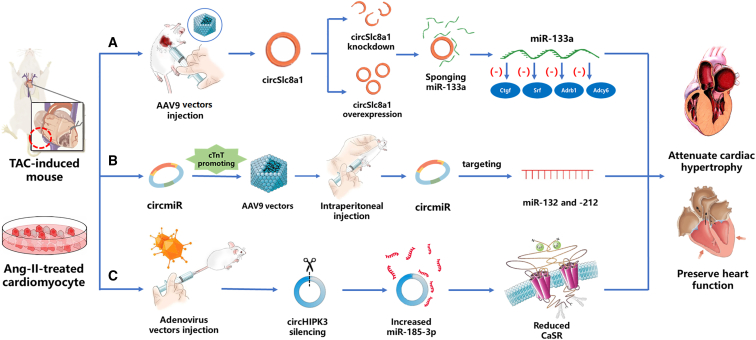
Due to its high abundance in cardiomyocytes, circSlc8a1 has attracted much attention from researchers investigating myocardial hypertrophy. The leading investigators, Lim et al.,51 revealed that circSlc8a1 acts as a sponge for miR-133a, which inhibits cardiac hypertrophy,57 through locked-nucleic-acid-based miRNA qPCR, luciferase assays, and reciprocal pull-down assays. In mice, RNA-interference knock down of circSlc8a1 using adeno-associated virus serotype 9 (AAV9) vectors alleviated cardiac hypertrophy arising from TAC-induced pressure overload, whereas forced cardiomyocyte-specific overexpression of circSlc8a1 using the AAV9-based vector system resulted in heart failure (Figure 2A). miR-133a targets, including serum response factor (Srf), connective tissue growth factor (Ctgf), adrenoceptor beta 1 (Adrb1), and adenylate cyclase 6 (Adcy6), as well as associated transcription factors, were also shown to be regulated by circSlc8a1. This study thus provided compelling evidence that circSlc8a1-mediated inhibition of miR-133a could potentially be used to treat cardiac hypertrophy.
Figure 2.
Schematic representation of mechanisms of selected circRNAs
(A) Knock down of circSlc8a1 alleviated cardiac hypertrophy through sponging miR-133a, whereas circSlc8a1 overexpression promoted cardiac hypertrophy. (B) When delivered to hypertrophic myocardium using AAV9 vectors, artificial circRNA (also termed circmiR), which targets miR-132 and miR-212, could reduce cardiac hypertrophy and improve heart function. (C) circHIPK3 silencing by RNA interference inhibited cardiomyocyte hypertrophy by reducing CaSR expression and consequently preserved cardiac function.
circRNA-000203/hsa-circ-0036167
Li et al.52 found that circRNA-000203 aggravated cardiac hypertrophy in angiotensin-II (Ang-II)-infused mice. circRNA-000203 was upregulated in both the mouse myocardium and the cytoplasm of neonatal mouse ventricular cardiomyocytes, probably as a result of increased NF-κB signaling. Mechanistically, circRNA-000203 was shown to modulate cardiac hypertrophic growth by specifically sponging miR-26b-5p and miR-140-3p and thus abolishing suppression of the target gene Gata4 by these two miRNAs. Upregulation of GATA4 expression has previously been shown to play a pro-hypertrophic role both in vivo and in vitro.58 Importantly, human hsa-circ-0036167, the homolog of circRNA-000203, was also shown to be significantly increased in the hypertrophic myocardium of heart failure patients, suggesting that hsa-circ-0036167 may play a similar role in humans and providing hope that reducing levels of hsa-circ-0036167 could reverse myocardial hypertrophy.
Artificial circRNA sponge for miR-132 and miR-212
Since endogenous circRNAs are known to prevent cardiac hypertrophy, the Lavenniah group53 proposed that artificial circRNAs could be used therapeutically to sponge target miRNAs. In support of this hypothesis, they showed that custom-built circRNA sponges (circmiRs) targeting miR-132 and miR-212 were able to drive the hypertrophic growth of cardiomyocytes.59 circmiRs containing 12 miRNA binding sites were found to be optimal, and, when delivered to cardiomyocytes in vivo using AAV9 vectors, they attenuated left ventricular hypertrophy and preserved cardiac function in a TAC-induced mouse model of hypertrophy (Figure 2B). Compared with natural circRNAs, circmiRs could have the advantages of low dosage requirements and extended half-lives. The length of their flanking introns can also be adjusted to meet the requirements of circularization efficiency or spacer size. Apparently, this study shows the potential of engineered circRNAs as new treatments for cardiac hypertrophy that warrant future clinical investigation.
circRNA wwp1
The Yang and Wang group54 identified the expression profiles of circRNAs in a mouse model of cardiac hypertrophy in which 8-week-old mice were infused with isoproterenol for 14 days. By circRNA sequencing, they found that 401 out of 3,323 total circRNAs were dysregulated in the hearts of mice treated with isoproterenol—303 circRNAs were upregulated and 98 were downregulated. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses further showed that the parental genes of differentially expressed circRNAs were associated with pathways involved in the circulatory system and in cardiovascular diseases. Out of the possible mRNA targets of the many miRNAs predicted to bind to the differentially expressed circRNAs, atrial natriuretic factor (ANF) and miR-23a were identified as downstream targets of circRNA wwp1, suggesting that circRNA wwp1 reduces cardiac hypertrophy by downregulating ANF and miR-23a. This comprehensive analysis of circRNA expression in isoproterenol-induced myocardial hypertrophy confirms the value of circRNAs as novel therapeutic targets.
circHIPK3
Using both TAC- and Ang-II-induced hypertrophy models in mice, Xu et al.55 discovered that another specific circRNA, circHIPK3, regulates cardiac hypertrophy. Real-time PCR analysis showed that circHIPK3 expression was significantly increased in cardiac tissues from mice with TAC-induced cardiac hypertrophy. Knock down of circHIPK3 using an adenovirus vector was shown to reduce both left ventricular end-diastolic and end-systolic diameters and to increase both left ventricular fraction shortening and ejection fraction. Expression of circHIPK3 was also increased in mouse cardiomyocytes treated with Ang-II in vitro. Silencing circHIPK3 markedly reduced the size of the cardiomyocytes and reversed upregulation of mRNA for atrial natriuretic peptide, brain natriuretic peptide, and β-myosin heavy chain. The mechanism underlying regulation of hypertrophy by circHIPK3 was determined to be miR-185-3p sponging and the subsequent modulation of its target, calcium-sensing receptor (CaSR) (Figure 2C). Knock down of circHIPK3 thus clearly suppressed cardiac TAC-induced hypertrophy in vivo and reduced markers of hypertrophy induced by Ang-II in vitro. The circHIPK3/miR-185-3p/CaSR axis is thus expected to provide a therapeutic target for overload-driven cardiac hypertrophy.
YAP circRNA (circYap)
Cardiac fibrosis is a common pathological feature of cardiac hypertrophy, and Wu et al.56 investigated a novel function of circYap, whose parental gene, Yes-associated protein (YAP), is known to mediate crucial pathways in heart development, injury, and regeneration60, 61, 62 and in cardiac fibrosis and hypertrophy. In mice with TAC-induced cardiac hypertrophy, increasing cardiac circYap levels by injecting the circYap plasmid repressed cardiac fibrosis and protected the heart-pumping function. RNA pull-down assays in mouse cardiac fibroblasts and cardiomyocytes showed that tropomyosin-4 (TMP4) and gamma-actin (ACTG) bind directly to circYap. This facilitates the interaction of TPM4 with ACTG, thereby increasing the suppression of actin polymerization and ensuing fibrosis. Wu et al. have thus uncovered a new molecule, circYap, that regulates cardiac fibrosis. Since cardiac fibrosis is associated with cardiac hypertrophy, this study suggests that circYap may have a future role in cardiovascular medicine.
Other circRNAs
Through culturing hypertrophic cardiomyocytes induced by high glucose, Meng et al.63 investigated differentially expressed circRNAs and found three downregulated circRNAs, ciRNA26, circRNA1191, and circRNA4251, and two upregulated circRNAs, ciRNA261 and circRNA6913. The circRNAs, DNAJC6, TMEM56, and MBOAT2 were found to be downregulated in serum from patients with hypertrophic obstructive cardiomyopathy. In these patients, circTMEM56 and circDNAJC6 showed significant inverse correlations with echocardiographic parameters, such as left ventricular outflow tract gradient and thickness of interventricular septum.64 circRNAs could thus be used as non-invasive diagnostic indicators of disease severity and could facilitate timely clinical decisions.
Conclusions and prospectives
In this review, we first briefly introduced the discovery, biogenesis, and general functions of circRNAs. We then highlighted the essential role of circRNAs in cardiac hypertrophy and their potential as therapeutic targets and also explained the presumed mechanisms underlying their biological actions. We have described relevant studies that shed light on the therapeutic and diagnostic potential of circRNAs with an emphasis on cardiovascular clinical practice.
Although the field of circRNAs is gradually maturing and attracting increased attention, there are still some barriers to overcome. So far, only a handful of circRNAs have been definitely proven to be involved in cardiac hypertrophy, and more in-depth studies are needed to identify whether other circRNAs contribute to myocardial hypertrophy. The majority of studies have identified the sponging of miRNAs or RBPs as the mechanism underlying the effect of circRNAs in cardiac hypertrophy, and whether other mechanisms are at play still needs to be explored. Large-scale discovery and validation studies must be undertaken before circRNAs are widely acknowledged as clinically valuable molecules for diagnosis and treatment purposes. Another shortcoming is that there is currently no standard nomenclature or a comprehensive classification system for circRNAs, both of which would facilitate future research. The consistency of computational tools used in studies on circRNAs also seems to be relatively low, and a systematic approach for identifying circRNAs in the human transcriptome is not available. Improvements in standardization of the assessment of circRNAs and the development of clinically applicable diagnostic and therapeutic assays would undoubtedly provide a major impetus for future work. Moreover, most of the studies that we have described were conducted using animal models, and the applicability of the results to humans needs to be verified.
In summary, we have reviewed recent findings that show an important role for circRNAs as therapeutic targets in cardiac hypertrophy. These studies provide novel insights into future personalized healthcare for patients with cardiac hypertrophy. With continuing research efforts, circRNA-based targeted drugs or technologies are sure to achieve great breakthroughs and become a major emerging trend in cardiovascular clinical applications in near future.
Acknowledgments
Author contributions
S.W. and L.C. wrote the manuscript, and X.Z. revised the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Grossman W., Jones D., McLaurin L.P. Wall stress and patterns of hypertrophy in the human left ventricle. J. Clin. Invest. 1975;56:56–64. doi: 10.1172/JCI108079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sandler H., Dodge H.T. Left ventricular tension and stress in man. Circ. Res. 1963;13:91–104. doi: 10.1161/01.res.13.2.91. [DOI] [PubMed] [Google Scholar]
- 3.Mozaffarian D., Benjamin E.J., Go A.S., Arnett D.K., Blaha M.J., Cushman M., Das S.R., de Ferranti S., Després J.P., Fullerton H.J., et al. Executive summary:heart disease and stroke statistics –2016update :areport from the American Heart Association. Circulation. 2016;133:447–454. doi: 10.1161/CIR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 4.Levy D., Garrison R.J., Savage D.D., Kannel W.B., Castelli W.P. Prognostic implications of echocardiographically determined left ventricular mass in the Framingham Heart Study. N. Engl. J. Med. 1990;322:1561–1566. doi: 10.1056/NEJM199005313222203. [DOI] [PubMed] [Google Scholar]
- 5.Chi K.R. The dark side of the human genome. Nature. 2016;538:275–277. doi: 10.1038/538275a. [DOI] [PubMed] [Google Scholar]
- 6.Djebali S., Davis C.A., Merkel A., Dobin A., Lassmann T., Mortazavi A., Tanzer A., Lagarde J., Lin W., Schlesinger F., et al. Landscape of transcription in human cells. Nature. 2012;489:101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 8.Yu C.-Y., Kuo H.-C. The emerging roles and functions of circular RNAs and their generation. J. Biomed. Sci. 2019;26:29. doi: 10.1186/s12929-019-0523-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rybak-Wolf A., Stottmeister C., Glazar P., Jens M., Pino N., Giusti S., Hanan M., Behm M., Bartok O., Ashwal-Fluss R., et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 2015;58:870–885. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 10.Salzman J., Chen R.E., Olsen M.N., Wang P.L., Brown P.O. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansen T.B., Venø M.T., Damgaard C.K., Kjems J. Comparison of circular RNA prediction tools. Nucleic Acids Res. 2016;44:e58. doi: 10.1093/nar/gkv1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanger H.L., Klotz G., Riesner D., Gross H.J., Kleinschmidt A.K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl. Acad. Sci. U S A. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu M.T., Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280:339–340. doi: 10.1038/280339a0. [DOI] [PubMed] [Google Scholar]
- 14.Memczak S., Jens M., Elefsinioti A., Torti F., Krueger J., Rybak A., Maier L., Mackowiak S.D., Gregersen L.H., Munschauer M., et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 15.Xia S., Feng J., Chen K., Ma Y., Gong J., Cai F., Jin Y., Gao Y., Xia L., Chang H., et al. CSCD: a database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46:D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao D., Zhang L., Zheng M., Sun X., Lu Y., Liu P. Circ2Disease: a manually curated database of experimentally validated circRNAs in human disease. Sci. Rep. 2018;8:11018. doi: 10.1038/s41598-018-29360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y.C., Li J.R., Sun C.H., Andrews E., Chao R.F., Lin F.M., Weng S.L., Hsu S.D., Huang C.C., Cheng C., et al. CircNet: a database of circular RNAs derived from transcriptome sequencing data. Nucleic Acids Res. 2016;44:D209–D215. doi: 10.1093/nar/gkv940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S., Li Y., Chen B., Zhao J., Yu S., Tang Y., Zheng Q., Li Y., Wang P., He X., et al. exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018;46:D106–D112. doi: 10.1093/nar/gkx891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jeck W.R., Sorrentino J.A., Wang K., Slevin M.K., Burd C.E., Liu J., Marzluff W.F., Sharpless N.E. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L., Chen L.L. Circular intronic long noncoding RNAs. Mol. Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Li Z., Huang C., Bao C., Chen L., Lin M., Wang X., Zhong G., Yu B., Hu W., Dai L., et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 22.Conn S.J., Pillman K.A., Toubia J., Conn V.M., Salmanidis M., Phillips C.A., Roslan S., Schreiber A.W., Gregory P.A., Goodall G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–1134. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Khan M.A., Reckman Y.J., Aufiero S., van den Hoogenhof M.M., van der Made I., Beqqali A., Koolbergen D.R., Rasmussen T.B., van der Velden J., Creemers E.E., et al. RBM20 regulates circular RNA production from the Titin gene. Circ. Res. 2016;119:996–1003. doi: 10.1161/CIRCRESAHA.116.309568. [DOI] [PubMed] [Google Scholar]
- 24.Ashwal-Fluss R., Meyer M., Pamudurti N.R., Ivanov A., Bartok O., Hanan M., Evantal N., Memczak S., Rajewsky N., Kadener S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Hansen T.B., Jensen T.I., Clausen B.H., Bramsen J.B., Finsen B., Damgaard C.K., Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 26.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B., Luo Y., Lyu D., Li Y., Shi G., et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G., Shi Y., Liu M., Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding K., Liao Y., Chen Y., Gong D., Zhao X., Ji W. Circular RNA HIPK3 promotes gallbladder cancer cell growth by sponging microRNA-124. Biochem. Biophys. Res. Commun. 2018;503:863–869. doi: 10.1016/j.bbrc.2018.06.088. [DOI] [PubMed] [Google Scholar]
- 29.Xu H., Guo S., Li W., Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stoll L., Sobel J., Rodriguez-Trejo A., Guay C., Lee K., Venø M.T., Kjems J., Laybutt D.R., Regazzi R. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol. Metab. 2018;9:69–83. doi: 10.1016/j.molmet.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li R.C., Ke S., Meng F.K., Lu J., Zou X.J., He Z.G., Wang W.F., Fang M.H. CiRS-7 promotes growth and metastasis of esophageal squamous cell carcinoma via regulation of miR-7/HOXB13. Cell Death Dis. 2018;9:838. doi: 10.1038/s41419-018-0852-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li P., Yang X., Yuan W., Yang C., Zhang X., Han J., Wang J., Deng X., Yang H., Li P., et al. CircRNA-Cdr1as exerts anti-oncogenic functions in bladder Cancer by sponging MicroRNA-135a. Cell. Physiol. Biochem. 2018;46:1606–1616. doi: 10.1159/000489208. [DOI] [PubMed] [Google Scholar]
- 33.Geng H.H., Li R., Su Y.M., Xiao J., Pan M., Cai X.X., Ji X.P. The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS One. 2016;11:e0151753. doi: 10.1371/journal.pone.0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K., Gan T.Y., Li N., Liu C.Y., Zhou L.Y., Gao J.N., Chen C., Yan K.W., Ponnusamy M., Zhang Y.H., et al. Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ. 2017;24:1111–1120. doi: 10.1038/cdd.2017.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai L., Qi B., Wu X., Peng S., Zhou G., Wei Y., Xu J., Chen S., Liu S. Circular RNA Ttc3 regulates cardial function after myocardial infarction by sponging miR-15b. J. Mol. Cell. Cardiol. 2019;130:10–22. doi: 10.1016/j.yjmcc.2019.03.007. [DOI] [PubMed] [Google Scholar]
- 36.Du W.W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Du W.W., Fang L., Yang W., Wu N., Awan F.M., Yang Z., Yang B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24:357–370. doi: 10.1038/cdd.2016.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Y., Du W.W., Wu Y., Yang Z., Awan F.M., Li X., Yang W., Zhang C., Yang Q., Yee A., et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics. 2017;7:3842–3855. doi: 10.7150/thno.19764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abdelmohsen K., Panda A.C., Munk R., Grammatikakis I., Dudekula D.B., De S., Kim J., Noh J.H., Kim K.M., Martindale J.L., et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. doi: 10.1080/15476286.2017.1279788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21:172–179. doi: 10.1261/rna.048272.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abe N., Matsumoto K., Nishihara M., Nakano Y., Shibata A., Maruyama H., Shuto S., Matsuda A., Yoshida M., Ito Y., et al. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 2015;5:16435. doi: 10.1038/srep16435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang Y., Fan X., Mao M., Song X., Wu P., Zhang Y., Jin Y., Yang Y., Chen L.L., Wang Y., et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Legnini I., Di Timoteo G., Rossi F., Morlando M., Briganti F., Sthandier O., Fatica A., Santini T., Andronache A., Wade M., et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell. 2017;66:22–37.e9. doi: 10.1016/j.molcel.2017.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pamudurti N.R., Bartok O., Jens M., Ashwal-Fluss R., Stottmeister C., Ruhe L., Hanan M., Wyler E., Perez-Hernandez D., Ramberger E., et al. Translation of CircRNAs. Mol. Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Y., Gao X., Zhang M., Yan S., Sun C., Xiao F., Huang N., Yang X., Zhao K., Zhou H., et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl. Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang M., Huang N., Yang X., Luo J., Yan S., Xiao F., Chen W., Gao X., Zhao K., Zhou H., et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–1814. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 47.Matsui T., Li L., Wu J.C., Cook S.A., Nagoshi T., Picard M.H., Liao R., Rosenzweig A. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J. Biol. Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- 48.Adams J.W., Sakata Y., Davis M.G., Sah V.P., Wang Y., Liggett S.B., Chien K.R., Brown J.H., Dorn G.W. Enhanced G alpha q signaling:a common pathway mediates cardiac hypertrophy and apoptotic heart failure. Proc. Natl. Acad. Sci. U S A. 1998;95:10140–10145. doi: 10.1073/pnas.95.17.10140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werfel S., Nothjunge S., Schwarzmayr T., Strom T.M., Meitinger T., Engelhardt S. Characterization of circular RNAs in human, mouse and rat hearts. J. Mol. Cell. Cardiol. 2016;98:103–107. doi: 10.1016/j.yjmcc.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Wang K., Long B., Liu F., Wang J.X., Liu C.Y., Zhao B., Zhou L.Y., Sun T., Wang M., Yu T., et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 51.Lim T.B., Aliwarga E., Luu T.D.A., Li Y.P., Ng S.L., Annadoray L., Sian S., Ackers-Johnson M.A., Foo R.S. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 2019;115:1998–2007. doi: 10.1093/cvr/cvz130. [DOI] [PubMed] [Google Scholar]
- 52.Li H., Xu J.D., Fang X.H., Zhu J.N., Yang J., Pan R., Yuan S.J., Zeng N., Yang Z.Z., Yang H., et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc. Res. 2020;116:1323–1334. doi: 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lavenniah A., Luu T.D.A., Li Y.P., Lim T.B., Jiang J., Ackers-Johnson M., Foo R.S. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 2020;28:1506–1517. doi: 10.1016/j.ymthe.2020.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang M.H., Wang H., Han S.N., Jia X., Zhang S., Dai F.F., Zhou M.J., Yin Z., Wang T.Q., Zang M.X., et al. Circular RNA expression in isoproterenol hydrochloride-induced cardiac hypertrophy. Aging. 2020;12:2530–2544. doi: 10.18632/aging.102761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu X., Wang J., Wang X. Silencing of circHIPK3 inhibits pressure overload-induced cardiac hypertrophy and dysfunction by sponging miR-185-3p. Drug Des. Devel. Ther. 2020;14:5699–5710. doi: 10.2147/DDDT.S245199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu N., Xu J., Du W.W., Li X., Awan F.M., Li F., Misir S., Eshaghi E., Lyu J., Zhou L., et al. YAP circular RNA, circYap, attenuates cardiac fibrosis via binding with tropomyosin-4 and gamma-actin decreasing actin polymerization. Mol. Ther. 2020;29:1138–1150. doi: 10.1016/j.ymthe.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Care A., Catalucci D., Felicetti F., Bonci D., Addario A., Gallo P., Bang M.L., Segnalini P., Gu Y., Dalton N.D., et al. MicroRNA-133 controls cardiac hypertrophy. Nat. Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 58.Pikkarainen S., Tokola H., Kerkelä R., Ruskoaho H. GATA transcription factors in the developing and adult heart. Cardiovasc. Res. 2004;63:196–207. doi: 10.1016/j.cardiores.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 59.Ucar A., Gupta S.K., Fiedler J., Erikci E., Kardasinski M., Batkai S., Dangwal S., Kumarswamy R., Bang C., Holzmann A., et al. The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun. 2012;3:1078. doi: 10.1038/ncomms2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Morikawa Y., Heallen T., Leach J., Xiao Y., Martin J.F. Dystrophin-glycoprotein complex sequesters Yap to inhibit cardiomyocyte proliferation. Nature. 2017;547:227–231. doi: 10.1038/nature22979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Del Re D.P., Yang Y., Nakano N., Cho J., Zhai P., Yamamoto T., Zhang N., Yabuta N., Nojima H., Pan D., et al. Yes-associated protein isoform 1 (Yap1) promotes cardiomyocyte survival and growth to protect against myocardial ischemic injury. J. Biol. Chem. 2013;288:3977–3988. doi: 10.1074/jbc.M112.436311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xin M., Kim Y., Sutherland L.B., Murakami M., Qi X., McAnally J., Porrello E.R., Mahmoud A.I., Tan W., Shelton J.M., et al. Hippo pathway effector Yap promotes cardiac regeneration. Proc. Natl. Acad. Sci. U S A. 2013;110:13839–13844. doi: 10.1073/pnas.1313192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng Z., Chen C., Cao H., Wang J., Shen E. Whole transcriptome sequencing reveals biologically significant RNA markers and related regulating biological pathways in cardiomyocyte hypertrophy induced by high glucose. J. Cell. Biochem. 2019;120:1018–1027. doi: 10.1002/jcb.27546. [DOI] [PubMed] [Google Scholar]
- 64.Sonnenschein K., Wilczek A.L., de Gonzalo-Calvo D., Pfanne A., Derda A.A., Zwadlo C., Bavendiek U., Bauersachs J., Fiedler J., Thum T. Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci. Rep. 2019;9:20350. doi: 10.1038/s41598-019-56617-2. [DOI] [PMC free article] [PubMed] [Google Scholar]