Abstract
Upregulated expression of microRNA (miR)-221 is associated with downregulation of p27 and subsequent increased cell proliferation in a variety of human cancers. It is unknown whether miR-221 mimics could trigger neoplastic cellular proliferation. In vitro, we demonstrated miR-221 significantly downregulates the expression of P27 and increases proliferation of H9c2 and cardiac fibroblasts. The knockdown of PUM1 but not PUM2 abolished such effects by miR-221, as verified by RT-qPCR and western blot, direct binding of p27 3′ UTR by luciferase reporter assay and cell proliferation by Ki67. In vivo expression of P27 in the rat liver, heart, kidney, spleen, and muscle were not affected by miR-221 at 1 and 4 mg/kg and concurrently full-length (FL) PUM1 (140 kDa) was not detected. Instead, isoforms of 105 and 90 kDa were observed and generated through alternative RNA slicing verified by cDNA cloning and sequencing and cathepsin K cleavage confirmed by studies with the inhibitor odanacatib. This is the first study to address the possible pro-proliferative effects of miR-221 mimic therapeutics in cardiovascular applications. Loss of FL PUM1 expression is a key factor abrogating miR-221-mediated p27 regulation, although other concurrent mechanisms cannot be excluded. Our findings provide essential insights into the context-dependent nature of miRNA functionality.
Keywords: microRNA-221, pumilio 1, P27, alternative RNA splicing, cathepsin K cleavage, RNA binding protein, isoforms, 3' UTR, epigenetic
Graphical Abstract

miR-221 mimics deploy therapeutics for cardioprotection and prevention of cardiac fibrosis without triggering neoplastic cellular proliferation due to loss of full-length (FL) PUM1. Loss of FL PUM1 expression is a key factor abrogating miR-221-mediated p27 regulation. This finding provides essential insights into the context-dependent nature of miRNA functionality.
Introduction
RNA interference (RNAi) is a fundamental mechanism for post-transcriptional regulation of gene expression. MicroRNAs (miRNAs or miRs), small non-coding RNAs of ∼22 nucleotides (nt), are important endogenous RNAi. miRNA mimics are promising therapeutics. Base pairing between the miRNA seed region (2–8 nt) and binding site (BS) in the corresponding 3′-untranslated region (3′ UTR) of target gene mRNA initiates translational repression and/or mRNA degradation. The relatively short nt complementary binding sequence endows miRNA and miRNA mimics with multi-targeting properties.1 Synergistic multi-target effects of miRNA mimics offer unique therapeutic opportunities. Unwanted off-target effects might pose challenges in clinical applications, as observed for small interfering RNA (siRNA) prototype therapeutics. However, as an evolutionarily preserved endogenous gene regulation mechanisms with multi-layered regulations, miRNA therapeutics may be less prone to such unwanted effects.
miR-221 has been demonstrated to have cardioprotective and anti-fibrotic effects in the settings of ischemic challenges. miR-221 promotes cell survival through1 anti-apoptotic mechanisms by direct targeting of p53, Bak1, Puma, and Pten, the negative regulator of Akt signaling pathway,2, 3, 4 as well as2 anti-autophagy effects through targeting Ddit4 and Tp53inp1.5 In vivo treatment with miR-221 mimic reduces myocardial infarct size, improves cardiac function, and increases survival rate in a rat model of myocardial infarction (MI).3 Clinical studies reveal that miR-221 expression in the human failing heart is inversely correlated with the severity of cardiac fibrosis.6 We recently demonstrated the anti-fibrotic mechanism of miR-221 is mediated via direct targeting of thrombospondin-1, a key factor in latent transforming growth factor β (TGF-β) activation.7 Additionally, miR-221 has been reported to ameliorate vascular remodeling and inflammation in endothelial cells, smooth muscle cells, macrophages, and adipose tissue.8, 9, 10
Despite the promise of miR-221 mimics in the treatment of cardiac infarction and heart failure, miR-221 is a known oncomiR, raising concerns with respect to safety. Upregulation of miR-221 is associated with tumor progression and poor prognosis.11, 12, 13 Functional studies of miR-221 have been performed in cancer cell lines in vitro and in tumor xenograft model in vivo.14,15 The oncogenic mechanism of miR-221 is mediated via direct targeting of cyclin-dependent kinase inhibitor 1B (also known as Cdkn1b, p27Kip1, or p27), a cell cycle inhibitor, which normally prevents uncontrolled cell proliferation.13,16 The upregulation of miR-221 and associated downregulation of P27 have been reported in multiple cancers.16, 17, 18 This raises the question of whether miR-221 mimic therapeutics could be pro-oncogenic.
p27 is a highly conserved target of miR-221. Its 3′ UTR contains two miR-221 BSs in human and mouse and three in the rat. A seminal study in 2010 demonstrated that pumilio 1 (PUM1) is a co-factor in miR-221-induced p27 downregulation.19 p27 3′ UTR contains two conserved PUM response elements (PREs), RNA sequences specifically recognized by the PUM1 RNA-binding domain (RBD). One of the PREs is adjacent to a miR-221 BS and directly forms a hairpin by internal base pairing. This secondary structure of p27 3′ UTR blocks access of miR-221 to its BSs unless PUM1 is bound to the PRE of p27 mRNA opening up the hairpin structure. In quiescent cancer cells, co-existent high levels of miR-221 and P27 results in neither downregulation of P27 nor cell proliferation, due to lack of PUM1.19
Pumilios, RNA-binding proteins of the PUF (Pumilio/Ferm-3 mRNA-binding factor) family, large sequence-specific RNA-binding proteins, were initially discovered in Drosophila and later found to be highly conserved across species.20 They contain evolutionally conserved C-terminal RBDs that bind to a conserved 8-nt sequence UGUANAUA, called PREs, on the 3′ UTR of target mRNAs.21 Hundreds of pumilio mRNA targets have been identified in human, based on the presence of PRE and observed changes in RNA expression levels upon Pum1/Pum2 knockdown.22 PUM proteins are also involved in regulating multiple miRNA target gene interactions.23 Two classical PUMs, PUM1 and PUM2, share 76% sequence homology in Drosophila.24,25 They are believed to be ubiquitously expressed in mammals and play a pivotal role in germline development and differentiation.26 Their expression and translation in the heart are not well understood. According to the Human Protein Atlas, Pum1 mRNA is detected in heart tissue, while PUM1 protein is not. In contrast, both Pum2 mRNA and protein are expressed in the heart (https://www.proteinatlas.org/ENSG00000134644-PUM1/tissue). However, it is reported that immunohistochemical staining indicates PUM1 protein is expressed in cardiomyocytes. Whether alternative RNA splicing and protein cleavage play a role in the discrepancy of Pum1 RNA and protein expression in the heart has not been previously elucidated.
Here we report the effects of miR-221 mimics on P27 expression in vitro and in vivo in primary cell cultures and cell lines and in healthy rat heart, liver, kidney, spleen, lung, and muscle tissues versus diseased rat heart. Pum1 and Pum2 mRNA and protein expressions were measured and their functions ascertained via siRNA knockdown, luciferase reporter assay, RNA splicing, and protein cleavage studies. In summary, miR-221 downregulates p27 through binding to its 3′ UTR in vitro. This interaction is controlled by PUM1 but does not require PUM2. miR-221 does not downregulate p27 mRNA and protein in the heart, liver, kidney, spleen, and muscle due to lack of PUM1 full-length (FL) (∼140 kDa) protein expression. Instead, two novel PUM1 isoforms of 105 and 90 kDa are detected, originating from RNA splice variations involving the skipping of two exons and C-terminal cleavage respectively. The requirement of FL PUM1 for miR-221-p27 interaction and its absence in vivo may support the safe use of miR-221 mimics in therapeutic applications. The discovery of novel PUM1 isoforms and associated underlying mechanisms warrant further investigation.
Results
miR-221 downregulation of P27 in vitro but not in vivo
As a well-known gene target of miR-221, p27 mRNA and protein were readily downregulated by miR-221 mimics in H9c2 under both normal and hypoxia/re-oxygenation (H/R) conditions (Figures 1A and 1B). miR-221 mimics significantly increased H9c2 proliferation as indicated by the increases of ki67 staining, and cell count (Figure 1C i–ii). Furthermore, overexpression of P27 in H9c2 abrogated the pro-proliferative effect of miR-221 (Figure 1D i–iv). Similar experiments were carried out in primary isolated adult rat cardiac fibroblasts (cFBs), with comparable results (Figures S1A–S1D).
Figure 1.
miR-221 downregulates P27 and increases cellular proliferation in vitro
Rat ventricular myoblast H9c2 cells were transfected with 25 nM miR-221 mimics (miR-221) or control (MC) for 24 h and subjected to 20 h hypoxia/2 h re-oxygenation (H/R). The normoxia groups were incubated under normal culture conditions for an equal duration. p27 was measured by RT-qPCR (A) and western blot (B). (C) Cell proliferation was measured by ki67 staining (i) and cell count (ii). (D) p27 cDNA plasmid was co-transfected with mimics for phenotype rescue. The effects of P27 overexpression were validated by western blot (i), ki67 staining (ii), and cell count (iii). Data are expressed as mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001 versus MC, #p < 0.05 versus miR-221 by one-way ANOVA with Bonferroni post hoc analysis. Experiments performed three times in triplicate.
We then addressed whether miR-221 could downregulate P27 in vivo in the rat. Following miR-221 mimic (1 mg/kg and 4 mg/kg) intravenous (i.v.) injection, expression of miR-221 increased in the heart 6.8- and 17.3-fold; the most in the liver and spleen at 33.4- and 506.5-fold, and 86- and 223-fold, respectively; and in the kidney 2.2- and 3.5-fold, respectively, with limited increases in skeletal muscle and the lung (Figures 2A and S2A). In contrast, mimic control (MC) did not induce any changes compared with PBS control. P27 mRNA and proteins levels in these tissues did not change except for a slight decrease in the liver (Figures 2B, 2C, S2B, and S2C).
Figure 2.
miR-221 does not downregulate P27 in vivo
Healthy adult rats received PBS, 1 mg/kg MC, 1 mg/kg or 4 mg/kg miR-221 mimics through tail vein injection. The heart, liver, kidney, and skeletal muscle were harvested after 48 h for RNA and protein extraction. (A) miR-221 levels were measured by stem-loop qPCR. p27 mRNA and protein levels were measured by RT-qPCR (B) and western blot (C). Data are expressed as mean ± SD. n = 3 each group.
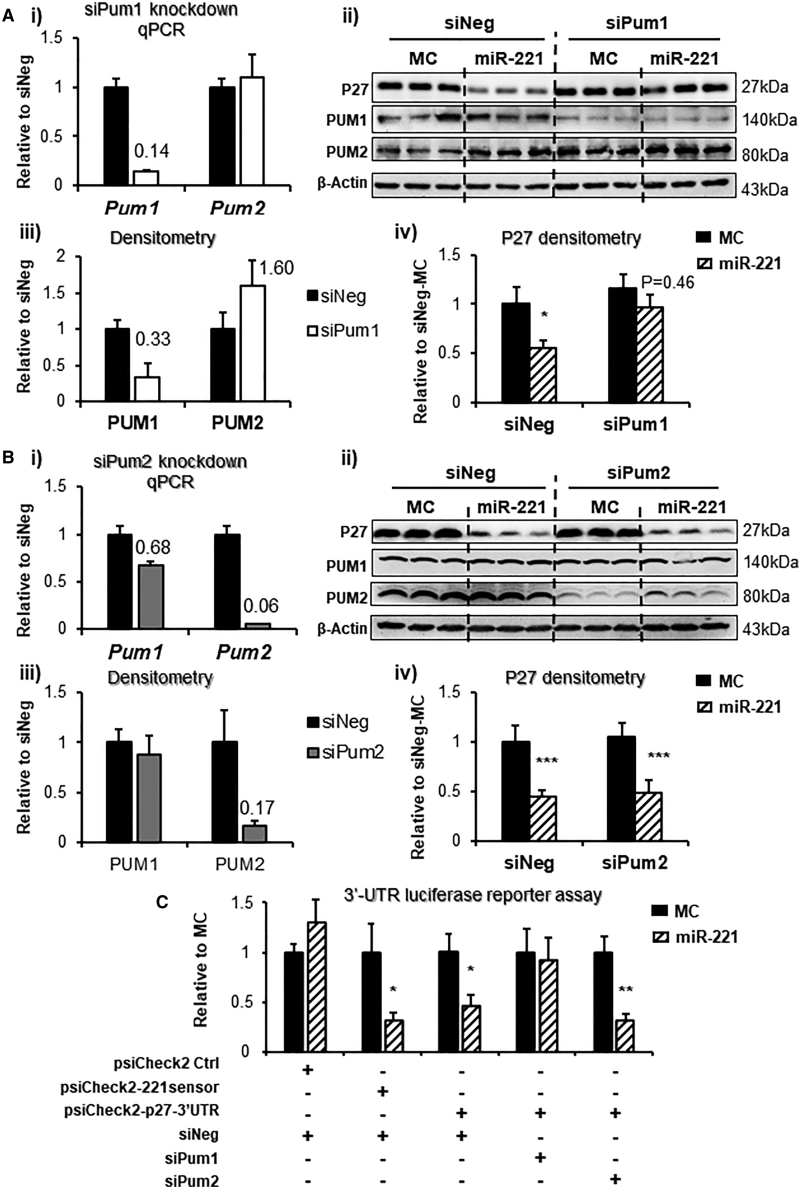
PUM1 but not PUM2 knockdown obliterates miR-221-induced P27 downregulation
RNAi knockdown of PUM1 and PUM2 was performed to demonstrate the direct regulation of P27 by miR-221. Dicer-substrate short interfering RNAs (dsiRNAs) specifically knocked down Pum1 mRNA and protein, by 86% and 67%, respectively (Figure 3A i–iii). With specific PUM1 knockdown, miR-221 no longer downregulated P27 (Figure 3A iv). In contrast, siPum2 reduced Pum2 mRNA and protein by 94% and 83% without affecting P27 regulation (Figure 3B i–iv). Moreover, 3′ UTR luciferase reporter assay showed that PUM1 knockdown abolished the binding of miR-221 to p27 3′ UTR, but PUM2 knockdown did not affect this binding (Figure 3C).
Figure 3.
miR-221 downregulation of p27 is PUM1 dependent
Rat ventricular myoblast H9c2 were transfected with 25 nM dsiRNA pumilios (siPum) or negative control (siNeg) for 48 h. In co-transfection experiment, the dsiRNA were transfected for 24 h followed by mimic transfection for 24 h. (A) Pum1 and Pum2 knockdown were validated by RT-qPCR for mRNA (i) by western blot for proteins (ii and iii). P27 downregulation by miR-221 with or without PUM1 knockdown was assessed by western blot (ii and iv). (B) Parallel experiment of Pum2 siRNA knockdown. (C) 3′ UTR luciferase reporter assay to assess p27-3′ UTR binding and suppression by miR-221 with or without Pum1 and Pum2 siRNA knockdown. 221 sensor (psiCHECK2 vector carrying perfect complementary sequence to miR-221) was used as a positive control. Data are expressed as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus MC, by unpaired two-tailed t test. Experiments performed three times in triplicate.
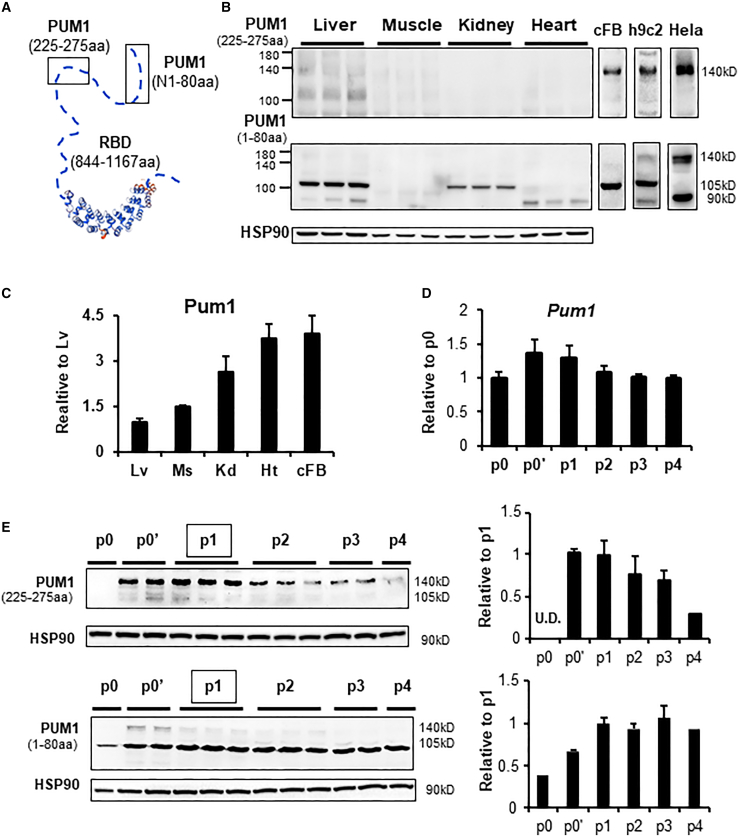
Discrepant Pum1 mRNA and protein expression in the heart, liver, kidney, and muscle
Two different antibodies (Abs) recognizing epitopes of PUM1 1–80 amino acids (aa) and 225–275 aa were used to detect PUM1 protein (Figure 4A). FL PUM1 protein was absent in heart, liver, kidney, skeletal muscle, and spleen compared with positive expression observed in cultured HeLa and H9c2 cells and cFB. FL PUM1 was detected in the lung. Interestingly, endogenous miR-221 expression is extremely high in the lung. Therefore, mimic treatment did not increase miR-221 level (Figures S2A and S2B). The observed molecular weight (MW) of 140 kDa was slightly higher than predicted MW of 126.6 kDa, which is consistent with Ab information from HeLa cell lysate (https://www.abcam.com/Pumilio-1-antibody-EPR3795-ab92545.html). Surprisingly, Pum1 mRNA was expressed in all tissues (Figure 4C). Using the Ab to PUM11–80 aa, a 105-kDa isoform was detected in the liver, kidney, and spleen, while a 90-kDa isoform was found in liver, heart, and lung (Figures 4B and S2E). Notably, PUM1, as detected by the same Ab, has been previously reported as present in the liver and kidney.27 Immunohistochemical staining does not allow differentiation of full-length and shorter isoforms of PUM1. To confirm the finding, we validated PUM1 expressions in multiple mouse tissues as well, FL PUM1 is missing in the heart, liver, kidney, muscle, bone marrow, and intestine, but expressed in the lung, spleen, and stomach (Figure S3).
Figure 4.
PUM1 expression is missing in the tissues and inducible in cultured cFB
(A) Schematic of two PUM1 Abs recognizing epitopes of 1–80 aa and 225–275 aa indicated by boxed areas. The RBD is highly conserved and is not chosen for Ab specificity concern. (B) Western blot and (C) RT-qPCR of PUM1 in adult rat tissues of liver (Lv), muscle (Ms), kidney (Kd), heart (Ht), and culture cells of cFBs; primary isolated adult rat cFBs were collected before (p0) and after plating (p0′) and after subculturing from passages 1–4. Pum1 expressions of (D) mRNA were assessed by RT-qPCR (D) and (E) proteins were assessed by western blot. Data are expressed as mean ± SD. n = 3 each group for in vivo samples. Cell culture experiments were performed three times in duplicate.
To explain the absence of FL PUM1 in the heart while it is expressed in cFB, we undertook additional investigation. Primary cFBs were isolated from adult rat heart and cultured for a few passages. Pum1 mRNA was expressed in all cell passages. Freshly isolated cFBs (p0) had no FL PUM1 and a little of the 105-kDa isoform. However, both were strongly induced after plating/attachment (p0′) and passage 1 (p1). The abundance of the FL PUM1 gradually diminished with increasing passage number, while the 105-kDa isoform remained relatively stable over successive passages (Figures 4E and 4F).
Surprisingly miR-221 downregulated P27 in neonatal rat ventricular myocytes (NRVMs) considering FL PUM1 is not expressed in the adult heart (Figure S4A). It is pertinent to ask whether FL PUM1 is expressed in the neonatal heart. We detected very low-level FL PUM1 in the neonatal heart and in freshly isolated NRVMs and nFBs (p0). FL PUM1 expression was significantly induced in nFB culture, consistent with our adult cFB results (Figure S4B). PUM1 phosphorylation was reported to be associated with functional regulation of the miR-221/P27 interaction.19 We found that p-PUM1 was not detected in the heart tissue but was present in H9c2, cFB, and NRVM (Figure S4C). The results indicate that FL PUM1 is expressed in the neonatal but not adult heart.
PUM1 expression was corroborated by western blot. Both the PUM1 (1–80 aa) and PUM1 (225–275 aa) Abs successfully pulled down FL PUM1 from HeLa cell lysate (Figure S5A). Peptide sequences were identified from PUM1 (225–275 aa) pull-down by mass spectrometry assessment. A total of 27 peptide hits aligned with human PUM1. After excluding repeated hits, 10 unique peptide sequences were identified (bold in Figure S5B). Unfortunately, immunoprecipitation (IP) of the shorter isoforms from tissue samples did not yield sufficient protein for mass spectrometry assessment. These results confirm the presence of FL PUM1 in vitro and validate the western blot data.
Pum1 alternative RNA splicing is responsible for the generation of isoform 105 kDa
The expression of PUM1 105-kDa and 90-kDa isoforms in tissues and cells has not been previously reported and the underlying mechanisms are unknown. The Pum1 mRNA exon structure open reading frame (ORF) is illustrated in Figure 5A. cDNA was cloned from heart, liver, kidney, and muscle tissue extracts, using primers flanking start and stop codons, and inserted into a mammalian expression vector, pcDNA3.1. Sequencing 10 colonies from each tissue, six mRNA splicing variants (v1–v6) were identified (Figures 5B and S6). With the exception of v2, which had GC-rich sequence at its specific exon junction and failed primer designs, all variants of v1, v3–6, were detected in four tissues with single-peak melt curves indicating specific PCR products (Figure 5C ii). BLAST with database, v2, and v4 could be mapped to predicted rat mRNA sequences and v5 and v6 were predicted mouse mRNA sequences.28 Variants v1–4 were predicted to encode smaller isoforms due to exon skipping. Among them, v1 and v3, predicting translation of proteins with MWs of 123 kDa and 105 kDa, have not been previously reported. Variants v5–6 had premature stop codons (PSCs) from an alternative splice site in exon 2 predicted to result in nonsense-mediated decay (Figure 5B).
Figure 5.
Pum1 mRNA undergoes post-transcription modification of alternative splicing
(A) Schematic of rat Pum1 exon structure with boxes depicting ORF, gray area indicating coding sequence, and arrows marking cloning primer locations. Sequences that encode Ab recognition sites and RBD are marked with brackets. ORF and PSC were predicted by translating from the original start codon. (B) Schematic of six Pum1 splice variants (v1–6) discovered from cDNA clones aligned against FL Pum1 and against human, mouse, and rat mRNA reference sequences through nucleotide (nt) BLAST. (C) RT-qPCR measurement of mRNA expressions of individual splice variants in liver, muscle, kidney, heart, cFB, and H9c2. Data are expressed as mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus Lv by unpaired two-tail t test. N = 3 for tissues and experiments performed three times in triplicate.
RNA variants v1–6-translated PUM1 isoforms were further verified. The cDNA plasmids of Pum1 variants with an N-terminal FLAG tag were overexpressed in HeLa cells. FL PUM1 overexpression was consistent with the endogenous 140 kDa observed earlier, with a slight shift due to FLAG tag. Only variants v2 and v3 generated proteins, whereas v1, v4, v5, and v6 did not produce anything detectable by anti-FLAG or PUM1 Abs (Figure 6A). PUM1 v2 was slightly smaller than PUM1-FL, consistent with predicted size reduction due to loss of two exons. However, endogenous PUM1 v2 has not been observed in any of our tissue or cell samples (Figure 4A). The 105-kDa isoform observed in liver, kidney, and cultured cells was confirmed to be PUM1 v3 as it was recognized only by PUM1 1–80-aa Ab but not by the PUM1 225–275-aa Ab due to the loss of translation of exon 6–7 (241–385 aa) (Figures 6A and 6B). Overexpression of PUM1 isoforms was repeated in H9c2 cells with similar results (Figure S7). The PUM1 v3 translated 105-kDa isoform retains RBD and may be a functional isoform. However, regulatory effect is greatly reduced. With extremely high-dose miR-221 mimic, inducing 6,297-fold increase in the liver, P27 protein was downregulated (Figure S8). We suspect that this short isoform may induce conformational change that affects its interaction with p27 3′ UTR. However, even with this high dose of miR-221 mimic, p27 mRNA and protein in the heart were not downregulated (Figure S8). We must point out that both FL PUM1 and endogenous miR-221 were highly expressed in the lung. With miR-221 mimic treatment, the increase of miR-221 was very limited and p27 was not downregulated (Figure S2B). FL PUM1 expression is a key factor in miR-221-mediated p27 regulation, although other co-contributory mechanisms cannot be excluded.
Figure 6.
The overexpression of full-length and variant PUM1
(A) The overexpression of FL and variant PUM1 in HeLa cells was detected by Abs of anti-FLAG and PUM1 1–80 aa and 225–275 aa. (B) Schematic of structure and Ab recognition sites of overexpressed PUM1 of FL and 105-kDa isoform that correspond to endogenous PUM1.
PUM1 cleavage and proteasome degradation
PUM1 90-kDa isoform was repeatedly detected in the tissues, cultured cells, and FL and v3 over-expressions. This isoform gradually accumulated after the overexpression of PUM1-FL, indicating protein cleavage (Figure S9). It was recognized by both PUM1 Abs and anti-FLAG Ab, suggesting C-terminal cleavage. In silico proteolytic cleavage prediction indicated potential cathepsin K (CatK) cleavage sites of PUM1 at amino acids 138, 776, and 919 (https://www.dmbr.ugent.be/prx/bioit2-public/SitePrediction/output/955290_955290_INPUTfasta_scorelist.html). The latter two cleavage sites could generate a ∼90-kDa isoform with complete or partial loss of RBD (Figure 7A). To verify this mechanism, the CatK inhibitor odanacatib (ODN) was incubated with HeLa and H9c2. The production of the 90-kDa isoform was significantly reduced by ODN (Figure 7B). Treatment with MG132, a proteasome inhibitor, stabilized the 90-kDa band in HeLa cells in a dose-dependent fashion, while not affecting the 140-kDa band (Figure 7C). Similar results were observed in H9c2 (Figures S10A and S10B). Altogether, these results demonstrated that the endogenous 90-kDa isomer might be a cleavage product of FL PUM1 and cleaved PUM1 might be subject to non-specific proteasome degradation.
Figure 7.
PUM1 protein cleavage and degradation
(A) Schematic of PUM1 protein structure with top three predicted cathepsin K (CatK) cleavage sites. (B) HeLa cells were treated with 5 and 10 μM CatK inhibitor ODN for 24 h. Levels of FL PUM1 and its cleavage product were assessed by western blot. (C) HeLa cells overexpressing PUM1-FL were treated with proteasome inhibitor MG132 at doses of 1, 5, 10 μM for 8 h. Levels of FL PUM1 and its cleavage products were assessed by western blot and densitometry. Data are expressed as mean ± SD. ∗p < 0.05, ∗∗∗p < 0.001 versus control by unpaired two-tail t test. Experiments were performed three times in triplicate. Ctrl, control.
Stress and repeated miR-221 treatment do not induce FL PUM1 expression or downregulate P27 in rat MI heart
We investigated the interaction of miR-221 and target P27 in the infarct heart (MI). FL PUM1 was not expressed in the infarct, border, or remote areas after MI (Figure 8A). With 1 mg/kg mimic treatment immediately after left anterior descending (LAD) coronary artery ligation, followed by a second injection at day 3, miR-221 levels increased significantly in infarct and border areas but not in the remote areas of the myocardium (Figure 8B). This treatment was cardioprotective.3 However, p27 mRNA and protein were not downregulated by miR-221 in any myocardial area with either one or two treatments (Figures 8C and 8D). Although Ki67 staining indicated a significant increase of proliferating cells in the infarct compared with the remote myocardium, miR-221 treatment did not increase Ki67+ cells, and instead showed a strong trend toward reduced cell proliferation (Figure 8E). The increase of Ki67-positive cells in the infarct was not associated with P27 but reflected injury-induced non-myocyte proliferation (e.g. cFBs, inflammatory cells, endothelial cells).29
Figure 8.
Stress and repeated miR-221 treatment do not induce FL PUM1 expression or p27 regulation in rat MI heart
(A) Rat hearts with MI were harvested at 48 h after surgery (MId2) and dissected by the infarct, border, and remote zones (Inf., Bor., Rem., respectively). PUM1 protein expression was assessed with western blot. HeLa cell lysate was used as a positive control. (B) Mimics of miR-221 were i.v. injected at 1 mg/kg immediately after surgery and tissue was harvested at MId2. A second injection was given at day 3 and tissue was harvested at MId7. miR-221 overexpression was measured by stem-loop qPCR. Results are presented as fold change over MI control. P27 expression in MI control and miR-221 mimic-treated rat hearts was measured by (C) RT-qPCR and (D) western blot. (E) Immunofluorescence staining of the heart sections (MId7) with three colors: cell proliferation marker ki67 (red), fibrosis marker α-SMA (green), nuclei stain DAPI (blue) (i). Scale bar, 200 μm. Results were analyzed as ki67% positive nuclei and normalized to MI control (ii). n = 5 animals for each group.
Discussion
In this study, we demonstrated that the pro-proliferative action of miR-221 mimics (at both 1 mg/kg and 4 mg/kg) observed in vitro is eliminated due to absence of P27 downregulation in the heart, liver, kidney, spleen, and muscle in vivo. From comprehensive in vivo and in vitro experiments by carefully following the guidelines for double-stranded RNA study,30 we have demonstrated expression of FL PUM1 is absent in these organs, which prevents such regulation. Instead, two major short PUM1 isoforms of 105 kDa and 90 kDa are found. They are generated by alternative RNA splicing and CatK cleavage respectively.
miR-221 mimics possess cardiac therapeutic potential in view of cardioprotective and anti-fibrotic effects we have previously reported in experimental MI and heart failure. In these models, miR-221 exerts anti-apoptotic and anti-autophagic effects via multi-gene targeting. Multi-targeting offers coordinated and synergistic effects but raises safety concerns. miR-221 is a known oncomiR acting via inhibition of p27 with consequent release of cancerous cellular proliferation.11 With miR-221 mimic overexpression, miR-221 downregulates P27 expression and stimulates cell proliferation in H9c2 and cFB, under normal and hypoxia conditions. The effects are blocked by p27 overexpression. These results suggest that the pro-proliferative effects of miR-221 through targeting of P27 exist in normal somatic cells as well as in cancer. To our surprise, at a therapeutic dose (1 mg/kg) of miR-221 mimic, P27 expression was unchanged in vivo. The therapeutic efficacy of this dose was retained, as confirmed by significant downregulation of key targets and sustained therapeutic effects with improved cardiac function and reduced mortality.3,7 To confirm this finding, we treated rats with high-dose mimic (4 mg/kg). P27 protein levels were not changed in the face of ∼20-, ∼223-, and ∼500-fold increases in miR-221 in the heart, liver, and spleen respectively. At this dosage, p27 mRNA was downregulated in the liver but did not affect protein protection.
Resolving the puzzling discrepancy of miR-221-induced P27 regulation in vitro versus in vivo was aided by a report from Kedde et al.19 The well-documented role of miR-221 in oncogenesis and metastasis was known to be mediated via P27 silencing, but miR-221 only regulated P27 in cycling (active proliferative) cancer cells and lost such effects in quiescent cancer cells. These investigators demonstrated that PUM1 and PUM2 specifically regulate miR-221 and p27 interaction by binding to p27 3′ UTR to open the hairpin structure, making the relevant BS accessible to miR-221.19 The loss of PUM in quiescent cancer cells or knockdown of PUM by siRNA abolished the regulation of P27 by miR-221. Through binding to mRNAs, both PUM1 and PUM2 play an important role in embryonic development, cell proliferation, and neurological function.26,31,32 The effects of PUM1 and PUM2 were not differentiated in previous reports. We showed that silencing specifically of PUM1 diminishes the direct binding of miR-221 to the p27 3′ UTR, which then blocks miR-221-induced downregulation of P27. Silencing of PUM2 does not affect direct binding or P27 expression. It has been reported that PUM2 is significantly upregulated in human hearts with end-stage dilated cardiomyopathy and regulates multiple fibrogenic genes through RNA-binding properties, while PUM1 is unchanged.33 These results indicate that PUM1 and PUM2 have different functions, warranting future investigation.
It is then necessary to know if PUM1 is expressed in vivo. Pum1 mRNA expression is readily detected in rat heart, liver, kidney, spleen, lung, and skeletal muscle, consistent with published data in human tissues34 and RNA sequencing (RNA-seq) data of major databases such as NCBI and Genecards (https://www.genecards.org/cgi-bin/carddisp.pl?gene=PUM1). PUM1 is a protein with 1,186 aa. Verified by two PUM1 Abs recognizing epitopes of 1–80 aa and 225–275 aa, FL 140-kDa PUM1 was detected in all cultured cells but missing in the tissues. Instead, two novel PUM1 isoforms were found; i.e. 105 kDa, the predominant isoform in the liver and kidney, and 90 kDa, in very low levels in the liver and heart. These isoforms are detected in cultured cells as endogenous products as well as by-products of FL PUM1 overexpression. FL PUM1 IP pull-down followed by mass spectrometry sequence detection validated western blot observations. Our finding explains why PUM1 is reported missing in the heart from the Human Protein Atlas but is detected by immunohistochemistry with Ab PUM1 1–80 aa.27 Our work expands the understanding of PUM1 in vivo expression.
Alternative splicing is a fundamental regulatory process of gene expression and increases the variety of protein production.35 A total of six Pum1 mRNA alternative splicing variants were discovered from the rat tissues by cDNA cloning and sequencing. Among them, two variants, v5 and v6, carry PSCs and do not produce proteins, as confirmed on overexpression studies. Although variants of v1–v4 contain putative ORFs, only v2 and v3 translate proteins. However, the v2 product has not been detected endogenously in any of the cell or tissue samples. V3 translates a 105-kDa isoform that is detected by Abs of anti-FLAG and PUM1 1–80 aa. It is a product of Pum1 with skipping of exons 6–7 (241–385 aa) in the N-terminal region. Therefore, it is not detected by Ab PUM1 225–275 aa. The heart exhibits predominance of v1 and v5–v6 splice variants, increased by 1.5–2-fold compared with other organs and cultured cells, which do not translate into PUM1 protein due to unknown reasons or PSC respectively.36 The 105-kDa PUM1 isoform is barely expressed, while the 90 kDa is the main isoform in the heart. PUM1 isoforms are repeatedly detected in culture cells as well. The isoform accumulation following PUM1 overexpression and recognition by Abs of N-terminal FLAG tag and both PUM1 suggest the possibility of C-terminal cleavage.
Our attempt to identify the protease for this isoform focused on cathepsin K, a cysteine protease previously studied in osteoblasts with recently recognized roles in the cardiovascular system.37,38 The top three in silico predicted cleavage sites were conserved between human and mouse: 138 aa, 776 aa, and 919 aa.39 The latter two sites could generate C-terminal cleaved proteins with predicted sizes of 80 kDa and 96 kDa, matching the ∼90 kDa observed. CatK inhibitor ODN treatment significantly inhibited the production of the 90-kDa isoform. The aspartic protease inhibitor pepstatin A had no such effects, suggesting the protease cleavage is specific. The isoforms are subjected to rapid degradation supported by a dose-dependent increase induced by the proteasome inhibitor MG132. This is a proof-of-concept study supporting involvement of selected proteases in PUM1 cleavage and degradation. We cannot exclude the involvement of other proteases, and an unbiased protease screen is needed. CatK is often thought to act upon the extracellular matrix.40 Its cleavage of an endogenous protein like pumilio is an interesting topic for further investigation.
The 105-kDa and 90-kDa isoforms of PUM1 have not been previously reported and their functions are unknown (https://blast.ncbi.nlm.nih.gov/ and https://www.proteinatlas.org/). Due to the presence of endogenous PUM1, functional miR-221 downregulation of P27 was observed in all of our cell cultures. Therefore, we were unable to directly tease out the function of individual isoforms by isoform overexpression in vitro. PUM1 105 kDa is well expressed in the liver and only with an extremely high dose of miR-221 mimic (6,297-fold increase) were the expressions of p27 mRNA and protein downregulated (Figure S6). As this isoform results from skipping of exons 6–7 at the N terminus but retains the complete RBD, we suspect that significantly reduced efficiency is due to the conformational change of 3D structure which affects the interaction of miR-221 and p27 3′ UTR. This fold change should never be achieved with therapeutic dosages. The 90-kDa isoform is a product of the C-terminal cleavage leading to the loss of RBD. It is a non-functional isoform. Our findings add important knowledge on PUM1 isoform expression and function. Previous understanding of PUM1 function was limited to its regulation of growth and development.32,41 The current study indicates potentially distinct roles of PUM1 in adult tissues, and PUM1 may not be required in the heart for healthy homeostasis.
Then we asked whether FL PUM1 is stress inducible; in other words, whether miR-221 treatment is safe under pathological stresses with respect to possible downregulation of p27. As our organ of prime interest, we observed miR-221 effects in the infarcted heart. As in the healthy heart, FL PUM1 is not expressed in the infarct, border, or remote areas in the infarcted heart. At day 2 and day 7 following one and two miR-221 treatments, P27 expression did not change. We selected this time frame because several studies have shown that miRNA mimic organ distributions remain high 1 day after injection but are back to normal within 2–4 days, with target gene silencing effects peaking at day 1.42,43 In our experience, the effects of target gene knockdown peaked in 1 day and lasted for 2–3 days. A challenge for miRNA therapeutics is maintenance of effective mimic concentrations and gene silencing effects. In our previous studies, two injections of 1 mg/kg miR-221 mimics were effective. Compared with permanent genetic upregulation or long-lasting viral miRNA delivery, short-lasting miRNA mimics have fewer safety concerns. Although PUM1 is not stress inducible, long-term miR-221 mimic treatment, the synergistic effect with another oncogenic factor, and the function of novel PUM1 isoforms warrant further investigation.
Both siRNA and miRNA mimics are ∼21-nt RNA duplexes that induce gene silencing. miRNA mimics have a potentially great therapeutic future but their development lags behind that of siRNA drugs. As endogenous products of molecular evolution, miRNAs induce coordinated gene regulation that is context dependent (e.g., healthy versus disease) and cell-type specific.44 The mechanisms are not well understood. As demonstrated in this study, FL PUM1 is missing in vivo; therefore, miR-221 did not downregulate P27, which indicates miRNA-221 mimics are unlikely to induce malignant cellular proliferation. Our results demonstrated that PUM1 is not expressed in primary cells, consistent with our in vivo findings. However, PUM1 is induced in cell culture by unknown mechanisms. We identified novel PUM1 isoforms of 105 kDa and 90 kDa generated through alternative RNA splicing and protease cleavage respectively. This is the first study to address this aspect of miR-221 mimic therapeutic safety in addressing concerns regarding possible adverse pro-proliferative effects, which theoretically may have threatened potential cardiovascular applications. Our findings illustrate the need for understanding the context-dependent nature of miRNA functionality, which is crucial for effective miRNA drug development.
Materials and methods
Animals
A total of 76 Sprague-Dawley rats were used in this study, including 28 8-week-old males for in vivo miRNA mimic delivery, 45 8-week-old males for MI, and three pregnant females for primary cell isolation of NRVMs and cFBs. All rats were kept at 21°C ± 2°C in a temperature-controlled room with 12-h light/dark cycle. Water and diet were available ad libitum. Protocols were approved by the Institutional Animal Care and Use Committee of National University of Singapore and complied with the Guide for the Care and Use of Laboratory Animals published by the National Institutes of Health (NIH publication no. 85-23, revised 1996).
Cell culture
Primary NRVM and cFB isolation from the neonatal and adult rat hearts has been described before.45 Briefly, the gentleMACS dissociator (Miltenyi Biotec, Singapore) was used following manufacturer’s instructions with modifications. NRVMs were purified by Percoll density gradient centrifugation (GE healthcare, Singapore). cFBs from the female adult rat heart were isolated with a customized enzyme solution of collagenase II (80 mg/mL) and DNase (60 U/mL) (Worthington Biochem, i-DNA Biotechnology, Singapore). Aliquots of freshly isolated cells were lysed for protein and RNA extraction (sample p0) and subcultured for transfection/treatment at p0′, p1 to p4 as needed.
Rat cardiac myoblast H9c2 cells and Hela cells were purchased from American Type Culture Collection (ATCC, Bio-REV, Singapore) and cultured according to ATCC instructions. Normal growth medium for NRVMs (DMEM and M199 at 4:1 ratio), cFBs, and h9c2 (DMEM) were all supplemented with 10% FBS and penicillin/streptomycin (Gibco, Thermo Fisher Scientific, Singapore). Hypoxia was performed by incubating in low-glucose, serum-free DMEM at 0.2% O2.46
In vivo miR-221 mimics delivery
miR-221 mimic and MC were complexed with Invivofectamine 3.0 following manufacturer’s instructions (Thermo Fisher Scientific, Singapore) and diluted in PBS to 0.8 mL volume per rat for tail vein injection. Additional equal volume of PBS control was included. Healthy rats received 1 mg/kg or 4 mg/kg mimics. Extra-high-dosage delivery was performed as three injections of 3.75 mg/kg on consecutive days. Rats were euthanized at 48 h after the last injections. Heart, liver, kidney, and skeletal muscle were snap frozen in liquid nitrogen for protein and RNA extraction.
MI
Rats were divided into sham, MI control, and MI miR-221 groups. LAD coronary artery ligation was occluded to induce MI. miR-221 mimics were injected i.v. at 1 mg/kg immediately and again at 5 days later after surgery. Hearts were harvested at day 2 (MId2) and at day 7 (MId7) respectively. Heart tissues were dissected by infarct, border, and remote zones and snap frozen for RNA and protein analysis, or formalin fixed and paraffin embedded for immunohistological analysis.
Cell proliferation
miR-221 mimics or control (MC) were transfected at 25 nM using lipofectamine RNAiMax (Thermo Fisher Scientific, Singapore). Ki67 proliferation assay was performed 24 h after transfection. Briefly, cells were trypsinized, fixed, and permeabilized (eBioscience Intracellular Fixation & Permeabilization Buffer Set, Thermo Fisher Scientific, Singapore) for anti-ki67 Ab staining (clone 8D5, Cell Signaling Technology, Research Biolabs, Singapore) and anti-mouse IgG-PE (Invitrogen, Thermo Fisher Scientific, Singapore), followed by flow cytometry on MUSE cell analyzer (Merck, Singapore). Cell number was assessed on MUSE.
In vivo cell proliferation was assessed in MId7 heart cross sections by immunofluorescence staining with anti-ki67 (clone 8D5, Cell Signaling Technology, Research Biolabs, Singapore), anti-α-SMA (clone 1A4, Sigma-Aldrich, Merck, Singapore), and DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen, Thermo Fisher Scientific, Singapore).
dsiRNA knockdown
Predesigned dsiRNAs for Pum1 and Pum2 (Integrated DNA Technologies, Singapore) were transfected at 25 nM for 48 h to verify knockdown efficiency by RT-qPCR and western blot. When combined with other transfection and/treatment, the dsiRNA transfection was performed 24 h in advance. Sequences of dsiRNA are listed in Table S1.
The 3′ UTR luciferase reporter assay
p27 3′ UTR was cloned into psiCHECK-2 dual luciferase reporter plasmid (Promega, Singapore). Sequences of cloning primers are listed in Table S2. miR-221 sensor, the complementary binding sequence to miR-221 as positive control (Integrated DNA Technologies, Singapore), was cloned into psiCHECK-2. Co-transfection of the plasmid (lipofectamine 2000) and miRNA mimics (lipofetamine RNAiMax) were performed to validate miRNA-3′ UTR binding. Renilla and Firefly luciferase activities were measured 24 h after transfection using Dual-Glo luciferase assay system (Promega, Singapore).
miRNA stem-loop qPCR and mRNA RT-qPCR
RNA was extracted with TRIzol Reagent (Thermo Fisher Scientific, Singapore) from cultured cells or mortar/pestle ground tissue powder with sonication (Qsonica XL-2000, ITS Science & Medical, Singapore). miRNA expression was determined by stem-loop qPCR (TaqMan MicroRNA Assay, QuantStudio 5 Real-Time PCR System, Thermo Fisher Scientific, Singapore) and normalized to U6B. Genomic DNA was removed by ezDNase treatment (Thermo Fisher Scientific, Singapore). Gene expressions were measured by universal reverse transcription and real-time PCR (SensiFAST cDNA Synthesis Kit, Bioline, Axil Scientific, Singapore; iTaq Universal SYBR Green Supermix, Bio-Rad Laboratories, Singapore), and normalized by β-actin or 18S. Primers are listed in Table S3.
Pum1 cDNA cloning
Rat heart, liver, kidney, and skeletal muscle RNA were treated with DNase and reverse transcribed using a mixture of random hexamer and oligo dT (ezDNase and Superscript IV, Thermo Fisher Scientific, Singapore). Pum1 cDNA was amplified using KOD-Plus-Neo DNA polymerase (Toyobo Biotech, Axil Scientific, Singapore) and cloned into pcDNA3.1 vector with 3× FLAG inserted between HindIII and EcoRI in its multiple cloning site (Thermo Fisher Scientific, Singapore). Sequences of cloning primers are listed in Table S2. Ten colonies from each tissue type were sequenced to discover Pum1 cDNA variants. Primers were designed based on sequencing results to validate alternative splice variants with specific exon junctions, as listed in Table S4.
Protease and proteasome inhibitors
In silico analysis of PUM1 protease cleavage sites were performed on the SitePrediction website (https://www.dmbr.ugent.be/prx/bioit2-public/SitePrediction/).39 The CatK inhibitor ODN was purchased from MedChemExpress (Bio.etc, Singapore) and dissolved in DMSO. H9c2 or HeLa cells were treated at 5–10 μM for either inhibitor or at matched concentration of DMSO (0.1%) for 24 h. In the proteasome inhibitor experiments, cells were treated with MG132 (Sigma-Aldrich Merck, Singapore) at 1, 5, and 10 μM for 8-h incubation.
Western blots
Protein expression in cell or tissue lysates was analyzed using SDS-PAGE and immunoblotting with different Abs: β-actin, HSP90, P27 (Santa Cruz Biotechnology, Axil Scientific, Singapore), PUM1 225–275 aa (Bethyl Laboratories Axil Scientific, Singapore), PUM1 1–80 aa, phosphorylated-PUM1 (p-PUM1, y83) and FLAG (Sigma-Aldrich, Merck, Singapore), and PUM2 (Novus Biologicals, SingLab Technologies, Singapore).
Statistics
Data were compared for differences by one-way ANOVA followed by Bonferroni post hoc analysis or unpaired two-tailed t test as appropriate (Prism; GraphPad Software, La Jolla, CA). Values were expressed as mean ± SD. Differences between groups were regarded as significant at the p < 0.05 probability level.
Acknowledgments
This work was supported by grants from the National Medical Research Council Center, Ministry of Health, Singapore (principal investigator [PI], A.M.R.) and CVRI Operating Fund, National University of Singapore, Singapore (PI, A.M.R.).
Author contributions
Conceptualization, P.W. and Y.Z.; investigation, Y.Z., D.Y.N., and P.W.; writing, P.W. and Y.Z.; review and editing, P.W. and A.M.R.; resources, A.M.R.; supervision, P.W. and A.M.R.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2021.12.012.
Supplemental information
and Tables S1–S4
information
References
- 1.Lam J.K., Chow M.Y., Zhang Y., Leung S.W. siRNA versus miRNA as therapeutics for gene silencing. Mol. Ther. Nucleic Acids. 2015;4:e252. doi: 10.1038/mtna.2015.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee T.L., Lai T.C., Lin S.R., Lin S.W., Chen Y.C., Pu C.M., Lee I.T., Tsai J.S., Lee C.W., Chen Y.L. Conditioned medium from adipose-derived stem cells attenuates ischemia/reperfusion-induced cardiac injury through the microRNA-221/222/PUMA/ETS-1 pathway. Theranostics. 2021;11:3131–3149. doi: 10.7150/thno.52677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou Y., Richards A.M., Wang P. MicroRNA-221 is cardioprotective and anti-fibrotic in a rat model of myocardial infarction. Mol. Ther. Nucleic Acids. 2019;17:185–197. doi: 10.1016/j.omtn.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun L., Zhu W., Zhao P., Zhang J., Lu Y., Zhu Y., Zhao W., Liu Y., Chen Q., Zhang F. Down-regulated exosomal MicroRNA-221 – 3p derived from senescent mesenchymal stem cells impairs heart repair. Front. Cell Dev. Biol. 2020;8:263. doi: 10.3389/fcell.2020.00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Q., Zhou Y., Richards A.M., Wang P. Up-regulation of miRNA-221 inhibits hypoxia/reoxygenation-induced autophagy through the DDIT4/mTORC1 and Tp53inp1/p62 pathways. Biochem. Biophys. Res. Commun. 2016;474:168–174. doi: 10.1016/j.bbrc.2016.04.090. [DOI] [PubMed] [Google Scholar]
- 6.Verjans R., Peters T., Beaumont F.J., van Leeuwen R., van Herwaarden T., Verhesen W., Munts C., Bijnen M., Henkens M., Diez J., et al. MicroRNA-221/222 family counteracts myocardial fibrosis in pressure overload-induced heart failure. Hypertension. 2018;71:280–288. doi: 10.1161/HYPERTENSIONAHA.117.10094. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y., Ng D.Y.E., Richards A.M., Wang P. microRNA-221 inhibits latent TGF-beta1 activation through targeting thrombospondin-1 to attenuate kidney failure-induced cardiac fibrosis. Mol. Ther. Nucleic Acids. 2020;22:803–814. doi: 10.1016/j.omtn.2020.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X., Ballantyne L.L., Yu Y., Funk C.D. Perivascular adipose tissue-derived extracellular vesicle miR-221-3p mediates vascular remodeling. FASEB J. 2019;33:12704–12722. doi: 10.1096/fj.201901548R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quero L., Tiaden A.N., Hanser E., Roux J., Laski A., Hall J., Kyburz D. miR-221-3p drives the shift of M2-macrophages to a pro-inflammatory function by suppressing JAK3/STAT3 activation. Front. Immunol. 2019;10:3087. doi: 10.3389/fimmu.2019.03087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Cheng Y., Yang J., Xu L., Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J. Mol. Cell. Cardiol. 2012;52:245–255. doi: 10.1016/j.yjmcc.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu I.M., Hengst L., Slingerland J.M. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat. Rev. Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- 12.Kneitz B., Krebs M., Kalogirou C., Schubert M., Joniau S., van Poppel H., Lerut E., Kneitz S., Scholz C.J., Strobel P., et al. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res. 2014;74:2591–2603. doi: 10.1158/0008-5472.CAN-13-1606. [DOI] [PubMed] [Google Scholar]
- 13.le Sage C., Nagel R., Egan D.A., Schrier M., Mesman E., Mangiola A., Anile C., Maira G., Mercatelli N., Ciafre S.A., et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J., Zhang Y., Jin N., Li Y., Wu S., Xu L. MicroRNA-221-3p plays an oncogenic role in gastric carcinoma by inhibiting PTEN expression. Oncol. Res. 2017;25:523–536. doi: 10.3727/096504016X14756282819385. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15.Felicetti F., Errico M.C., Bottero L., Segnalini P., Stoppacciaro A., Biffoni M., Felli N., Mattia G., Petrini M., Colombo M.P., et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Res. 2008;68:2745–2754. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 16.Fornari F., Gramantieri L., Ferracin M., Veronese A., Sabbioni S., Calin G.A., Grazi G.L., Giovannini C., Croce C.M., Bolondi L., et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 17.Yin G., Zhang B., Li J. miR2213p promotes the cell growth of nonsmall cell lung cancer by targeting p27. Mol. Med. Rep. 2019;20:604–612. doi: 10.3892/mmr.2019.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Visone R., Russo L., Pallante P., De Martino I., Ferraro A., Leone V., Borbone E., Petrocca F., Alder H., Croce C.M., et al. MicroRNAs (miR)-221 and miR-222, both overexpressed in human thyroid papillary carcinomas, regulate p27Kip1 protein levels and cell cycle. Endocr. Relat. Cancer. 2007;14:791–798. doi: 10.1677/ERC-07-0129. [DOI] [PubMed] [Google Scholar]
- 19.Kedde M., van Kouwenhove M., Zwart W., Oude Vrielink J.A., Elkon R., Agami R. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat. Cell Biol. 2010;12:1014–1020. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 20.Nusslein-Volhard C., Frohnhofer H.G., Lehmann R. Determination of anteroposterior polarity in Drosophila. Science. 1987;238:1675–1681. doi: 10.1126/science.3686007. [DOI] [PubMed] [Google Scholar]
- 21.Edwards T.A., Pyle S.E., Wharton R.P., Aggarwal A.K. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell. 2001;105:281–289. doi: 10.1016/s0092-8674(01)00318-x. [DOI] [PubMed] [Google Scholar]
- 22.Bohn J.A., Van Etten J.L., Schagat T.L., Bowman B.M., McEachin R.C., Freddolino P.L., Goldstrohm A.C. Identification of diverse target RNAs that are functionally regulated by human Pumilio proteins. Nucleic Acids Res. 2018;46:362–386. doi: 10.1093/nar/gkx1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Incarnato D., Neri F., Diamanti D., Oliviero S. MREdictor: a two-step dynamic interaction model that accounts for mRNA accessibility and Pumilio binding accurately predicts microRNA targets. Nucleic Acids Res. 2013;41:8421–8433. doi: 10.1093/nar/gkt629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parisi M., Lin H. The Drosophila pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics. 1999;153:235–250. doi: 10.1093/genetics/153.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goldstrohm A.C., Hall T.M.T., McKenney K.M. Post-transcriptional regulatory functions of mammalian pumilio proteins. Trends Genet. 2018;34:972–990. doi: 10.1016/j.tig.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin K., Qiang W., Zhu M., Ding Y., Shi Q., Chen X., Zsiros E., Wang K., Yang X., Kurita T., et al. Mammalian Pum1 and Pum2 control body size via translational regulation of the cell cycle inhibitor Cdkn1b. Cell Rep. 2019;26:2434–2450.e2436. doi: 10.1016/j.celrep.2019.01.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 28.O'Leary N.A., Wright M.W., Brister J.R., Ciufo S., Haddad D., McVeigh R., Rajput B., Robbertse B., Smith-White B., Ako-Adjei D., et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kretzschmar K., Post Y., Bannier-Helaouet M., Mattiotti A., Drost J., Basak O., Li V.S.W., van den Born M., Gunst Q.D., Versteeg D., et al. Profiling proliferative cells and their progeny in damaged murine hearts. Proc. Natl. Acad. Sci. U S A. 2018;115:E12245–E12254. doi: 10.1073/pnas.1805829115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagnon K.T., Corey D.R. Guidelines for experiments using antisense oligonucleotides and double-stranded RNAs. Nucleic Acid Ther. 2019;29:116–122. doi: 10.1089/nat.2018.0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang M., Chen D., Xia J., Han W., Cui X., Neuenkirchen N., Hermes G., Sestan N., Lin H. Post-transcriptional regulation of mouse neurogenesis by Pumilio proteins. Genes Dev. 2017;31:1354–1369. doi: 10.1101/gad.298752.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asaoka-Taguchi M., Yamada M., Nakamura A., Hanyu K., Kobayashi S. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1999;1:431–437. doi: 10.1038/15666. [DOI] [PubMed] [Google Scholar]
- 33.Chothani S., Schafer S., Adami E., Viswanathan S., Widjaja A.A., Langley S.R., Tan J., Wang M., Quaife N.M., Jian Pua C., et al. Widespread translational control of fibrosis in the human heart by RNA-binding proteins. Circulation. 2019;140:937–951. doi: 10.1161/CIRCULATIONAHA.119.039596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spassov D.S., Jurecic R. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene. 2002;299:195–204. doi: 10.1016/s0378-1119(02)01060-0. [DOI] [PubMed] [Google Scholar]
- 35.Zheng Z.M. Regulation of alternative RNA splicing by exon definition and exon sequences in viral and mammalian gene expression. J. Biomed. Sci. 2004;11:278–294. doi: 10.1159/000077096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi M., Zhang H., Wang L., Zhu C., Sheng K., Du Y., Wang K., Dias A., Chen S., Whitman M., et al. Premature termination codons are recognized in the nucleus in a reading-frame dependent manner. Cell Discov. 2015;1:15001. doi: 10.1038/celldisc.2015.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fang W., He A., Xiang M.X., Lin Y., Wang Y., Li J., Yang C., Zhang X., Liu C.L., Sukhova G.K., et al. Cathepsin K-deficiency impairs mouse cardiac function after myocardial infarction. J. Mol. Cell. Cardiol. 2019;127:44–56. doi: 10.1016/j.yjmcc.2018.11.010. [DOI] [PubMed] [Google Scholar]
- 38.Cheng X.W., Kikuchi R., Ishii H., Yoshikawa D., Hu L., Takahashi R., Shibata R., Ikeda N., Kuzuya M., Okumura K., et al. Circulating cathepsin K as a potential novel biomarker of coronary artery disease. Atherosclerosis. 2013;228:211–216. doi: 10.1016/j.atherosclerosis.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Verspurten J., Gevaert K., Declercq W., Vandenabeele P. SitePredicting the cleavage of proteinase substrates. Trends Biochem. Sci. 2009;34:319–323. doi: 10.1016/j.tibs.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 40.Lutgens S.P., Cleutjens K.B., Daemen M.J., Heeneman S. Cathepsin cysteine proteases in cardiovascular disease. FASEB J. 2007;21:3029–3041. doi: 10.1096/fj.06-7924com. [DOI] [PubMed] [Google Scholar]
- 41.Lin K., Zhang S., Shi Q., Zhu M., Gao L., Xia W., Geng B., Zheng Z., Xu E.Y. Essential requirement of mammalian Pumilio family in embryonic development. Mol. Biol. Cell. 2018;29:2922–2932. doi: 10.1091/mbc.E18-06-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang H., Chiu M., Xie Z., Chiu M., Liu Z., Chen P., Liu S., Byrd J.C., Muthusamy N., Garzon R., et al. Synthetic microRNA cassette dosing: pharmacokinetics, tissue distribution and bioactivity. Mol. Pharm. 2012;9:1638–1644. doi: 10.1021/mp2006483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Montgomery R.L., Yu G., Latimer P.A., Stack C., Robinson K., Dalby C.M., Kaminski N., van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol. Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Erhard F., Haas J., Lieber D., Malterer G., Jaskiewicz L., Zavolan M., Dolken L., Zimmer R. Widespread context dependency of microRNA-mediated regulation. Genome Res. 2014;24:906–919. doi: 10.1101/gr.166702.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou Y., Richards A.M., Wang P. Characterization and standardization of cultured cardiac fibroblasts for ex vivo models of heart fibrosis and heart ischemia. Tissue Eng. . Part C Methods. 2017;23:422–433. doi: 10.1089/ten.TEC.2017.0169. [DOI] [PubMed] [Google Scholar]
- 46.Zhou Y., Chen Q., Lew K.S., Richards A.M., Wang P. Discovery of potential therapeutic miRNA targets in cardiac ischemia-reperfusion injury. J. Cardiovasc. Pharmacol. Ther. 2016;21:296–309. doi: 10.1177/1074248415604463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
and Tables S1–S4
information