Highlights
-
•
OLFM4 is involved in development of gallbladder cancer.
-
•
Depletion of OLFM4 sensitizes gallbladder cancer cells to cisplatin by regulating apoptosis.
-
•
Low expression of OLFM4 in GBC patients indicates longer survival.
Keywords: Gallbladder cancer, OLFM4, Tumorigenesis, Chemosensitivity, Cisplatin
Abstract
Background
Gallbladder cancer (GBC) is a highly lethal malignancy that carries an extremely poor prognosis due to its chemoresistant nature. Cisplatin (CDDP) is a first-line chemotherapeutic for GBC; however, patients experienced no benefit when treated with CDDP alone. The underlying mechanisms of CDDP resistance in GBC remain largely unknown.
Methods
Agilent mRNA microarray analysis was performed between paired GBC and paracarcinoma to explore differentially expressed genes that might underlie drug resistance. Gene Set Enrichment Analysis (GSEA) was employed to identify key genes mediating CDDP resistance in GBC, and immunohistochemistry was performed to validate protein expression and test correlations with clinicopathological features. In vitro and in vivo functional assays were performed to investigate the proteins’ roles in CDDP resistance.
Results
Olfactomedin 4 (OLFM4) was differentially expressed between GBC and paracarcinoma and had the highest rank metric score in the GSEA. OLFM4 expression was increasingly upregulated from chronic cholecystitis to GBC in clinical tissue samples, and OLFM4 depletion decreased GBC cell proliferation and invasion. Interestingly, downregulation of OLFM4 reduced ARL6IP1 (antiapoptotic factor) expression and sensitized GBC cells to CDDP both in vitro and in vivo. The evidence indicated that CDDP could significantly increase Bax and Bad expression and activate caspase-3 cascade in OLFM4-depleted GBC cells through ARL6IP1. Clinically, lower OLFM4 expression was associated with good prognosis of GBC patients.
Conclusions
Our results suggest that OLFM4 is an essential gene that contributes to GBC chemoresistance and could serve as a prognostic biomarker for GBC. Importantly, OLFM4 could be a potential chemotherapeutic target.
Introduction
Gallbladder cancer (GBC) is a rare malignant neoplasm [1] and the most frequently encountered malignancy of the biliary tract [2], with a global annual incidence of 115,949 and mortality of 84,695 in 2020 [3,4]. GBC is reported to affect females two to six times more than males, and the incidence varies greatly among ethnic groups and in countries around the world [5,6]. Gallstones are recognized as a major risk factor for gallbladder carcinoma [7], but several other factors may also be important in GBC development including porcelain gallbladder, gallbladder polyps, anomalous pancreaticobiliary duct junction, carcinogens, obesity, and estrogens [8], [9], [10]. GBC is generally diagnosed at an advanced stage due to the unique anatomical position of the gallbladder and vague clinical symptoms [10,11]. According to most reports, the 5-years survival for GBC is less than 10%. The only curative therapy for this malignant disease is surgical resection [[12], [51]], and patients with unresectable and metastatic GBC have extremely poor prognosis [13,14]. No standard adjuvant chemotherapeutic regimen or targeted therapy is currently available for this disease [15,16]. The median overall survival is only 11.7 months following first-line treatment of cisplatin (CDDP)–gemcitabine chemotherapy due to GBC chemoresistance [17,18].
Chemotherapy is used to treat unresectable GBC and as an adjuvant therapy for GBC patients who underwent cholecystectomy [19], [20], [21]; however, the chemoresistant nature of GBC impedes the beneficial effects [22]. CDDP is a cytotoxic anticancer drug that is widely used to treat a variety of biliary tract cancers [19,23]. Several studies indicated that the overall clinical efficacies of CDDP containing regimens were unsatisfactory in both postoperative and unresectable GBC patients owing to the chemoresistant nature of gallbladder carcinoma [20,24,25]. Apoptosis is the dominant way that CDDP suppresses tumor growth; however, the presence of anti-apoptotic factors leads to chemoresistance, which is a major obstacle in solid tumor treatment [26]. Clarifying the drug resistance mechanism of GBC will help identify effective therapeutic targets to improve patient response to chemotherapy and determine their prognosis [27,28].
Olfactomedin 4 (OLFM4) is a multi-functional glycoprotein that belongs to the OLFM family and was first identified in myeloid cells. Substantial evidence supports the protein's anti-apoptotic role [29,30]. In addition, OLFM4 is known to regulate the innate immunity or inflammatory process [31], [32], [33]. It is upregulated in several malignant cancers, particularly those originating from the digestive system [32,34,35]. OLFM4 is involved in early-stage tumorigenesis especially in the gastrointestinal and biliary tracts [36,37]. Higher OLFM4 expression was found in colonic epithelial organoid cultures derived from patients with primary sclerosing cholangitis, and it may induce disease progression and indicate greater risk for cancer development [38]. Moreover, OLFM4 can regulate chemoresistance in non-small cell lung cancer (NSCLC) under hypoxic conditions by regulating hypoxia-inducible factor-1α [39], and it was also contributed to chemoresistance in pancreatic cancer [40]. Several studies showed OLFM4 overexpression in GBC tissues [32,41], but the biological function of strong OLFM4 expression in GBC has not been elucidated.
In this study, we sought to identify key genes mediating the chemoresistance of GBC and explore potential mechanisms. Using microarray analysis of GBC clinical specimens, we found that OLFM4 was strongly expressed in GBC and investigated the possible pathway by which OLFM4 might contributed to CDDP resistance. Our findings provide new insight into the drug-resistant nature of GBC and provide a potential GBC chemotherapy target.
Materials and methods
Patients and clinical samples
Five paired fresh-frozen GBC tissues and normal paracarcinoma tissues from patients who underwent radical surgery without preoperative chemotherapy or radiotherapy were collected for TMA analyses. Paraffin-embedded tissues from 108 cases of chronic cholecystitis, 92 cases of dysplasia, and 72 cases of GBC were obtained and cut into 4-mm sections for further analysis. Informed consent was obtained from all the patients, and sample collection complied with the approved guidelines of Ethics Committee of the Kunming Medical University. GBC TNM (Tumor-Node-Metastasis) stages were evaluated according to the 8th edition of the American Joint Committee on Cancer staging system (AJCC/UICC TNM, 8th edition).
RNA extraction
Briefly, GBC tissues and normal paracarcinoma tissues were ground into powder in liquid nitrogen, then TRIzol (Invitrogen, Carlsbad, CA, USA) was added and total RNA was extracted and purified using NucleoSpin® RNA clean-up kit (Macherey-Nagel, Düren, Germany). RNA concentration and quality were determined by NanoDrop (NanoDrop Technologies, Wilmington, DE, USA) and formaldehyde gel electrophoresis.
Microarray expression profiling
Long non-coding RNA (lncRNA) and mRNA TMA profiling of GBC and paracarcinoma samples was performed on a SurePrint G3 Human Gene Expression 8 × 60Kv2 Microarray (Agilent Technologies, Santa Clara, CA, USA) conducted by Beijing Capitalbio Technology company. Raw data were acquired and input into GeneSpring GX software (Agilent Technologies), and normalization was processed using the percentile shift method. Differentially expressed genes (DEGs) were screened out under the criterion of absolute fold change (FC) ≥ 2 and p < 0.05 (Student's t-test).
Bioinformatic analysis of differential expression levels of mRNA and lncRNA from microarrays
Gene set enrichment analysis (GSEA) was performed to identify potential biological mechanisms associated with OLFM4 expression in GBC sand normal paracarcinoma tissues. GSEA software and gene sets were downloaded from https://www.gsea-msigdb.org/. The microarray expression profiling data were submitted to the software, which output the figures. The normalized enrichment score was used to assess the results, and the p < 0.05 was considered statistically significant. Heatmap was plotted on the Omicstudio online platform.
Immunohistochemistry (IHC) and grading
The GBC TMA was purchased from Shanghai Outdo Biotechnology company (Shanghai, China). Paraffin-embedded sections from patients and TMAs were deparaffinized in dimethylbenzene and hydration was performed using the LEICAST5020 system (Letica, Wetzlar, Germany). Antigen retrieval and peroxide blocking was performed in the PT Link pretreatment system (DAKO, Glostrup, Demark). The anti-OLFM4 antibody (Cat. #14369D, Cell Signaling Technology, Danvers, MA, USA) was applied to sections at 1:1000 dilution in Antibody Diluent (DAKO), which were incubated overnight at 4 °C. An Autostainer Link 48 machine was used to apply the secondary antibody (Biotinylated Multi-Link; DAKO) for 30 min. Then the horseradish peroxidase enzyme complex was applied to complete the staining, and the slides were then counterstained in hematoxylin for 3 min. Slides were dried and covered with a coverslip. The histological score (H-score) system was used to assess the results. The staining was digitized into scores according to the intensity of the positive staining, (0 = negative, weak = 1, moderate = 2, strong = 3), then the scores were multiplied by the staining area scores, (0 = negative, 1 = 1–10%, 2 = 11–50%, 3 = 51–80%, 4 = 81–100%). H-scores ≥ 4 or staining area ≥ 60% were considered as high expressing and H-scores < 4 or staining area < 60% were considered as low expressing. The results were evaluated and recorded by two pathologists blinded to the case information.
Cell culture
GBC-SD cells were purchased from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences and cultured in Dulbecco's Modified Eagle Medium (DMEM)/F12 (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, BI, Biological Industries, Beit Haemek, Israel), and 100 U/mL penicillin and 100 μg/mL streptomycin (BI, Beit Haemek, Israel). Cell lines were authenticated by Short Tandem Repeat profiling before use in the experiments. All cells were cultured at 37 °C in a humidified incubator with 5% CO2.
Vector construction and transduction
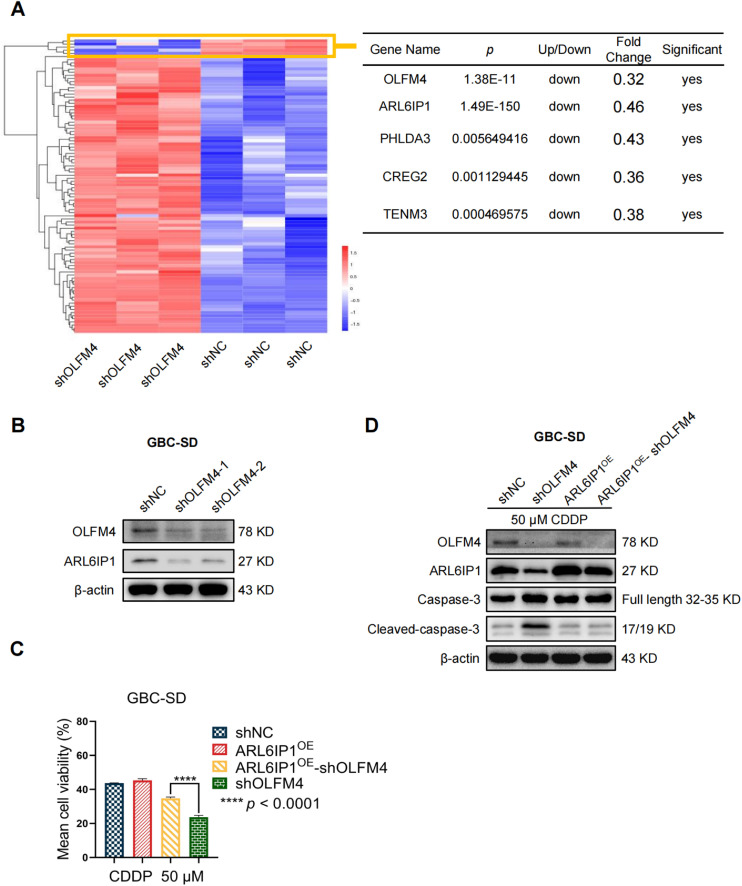
To stably knockdown OLFM4, a lentiviral short hairpin RNA (shRNA) targeting OLFM4 (puromycin-resistant) was purchased from Genechem (Shanghai, China), and ARL6IP1 overexpression lentiviral vertor was from the same company. The interference sequence targeting human OLFM4 complementary DNA is shOLFM4–1: CCGGAGTGCAGAGCATTAACTATAACTCGAGTTATAGTTAATGCTCTGCACTTTTTTG; shOLFM4–2: CCGGCCCTAATGCTGCCTATAATAACTCGAGTTATTATAGGCAGCATTAGGGTTTTTG; and a scramble shRNA was included as negative control (shNC): 5′-TTCTCCGAACGTGTCACGT-3′. An ARL6IP1 overexpression lentiviral vetor (G418 resistant) was built using NCBI Reference Sequence: NM_015161.3, and ARL6IP1 overexpression was performed on cells expressing shNC or shOLFM4 for add-back rescue experiments. Lentivirus infection was performed according to the manufacturer's instructions. Afterwards, cells were co-cultured with 2 μg/mL puromycin or 100 μg/mL G418 for 7 days to screen out cells not expressing vectors, western blot (WB) was performed to validate the protein expression level.
Cell viability and colony formation assays
After 72 h of lentivirus transduction, cells were trypsinized, resuspended, and seeded in 96-well plates (1000/well). Cell proliferation was quantified with Cell Counting Kit-8 (CCK-8, Dojindo, Kumamoto, Japan) for 5 days at the indicated time points. Optical density (OD) at 450 nm absorbance was detected using a multimode microplate reader (Varioskan, Thermo Fisher Scientific, Waltham, MA, USA). For cell clonogenic capacity assays, 1000 cells/well were seeded in 6-well plates in triplicate conditions. After 14 days of culture cells were fixed in methanol, stained with 1% crystal violet solution, air-dried, and the colonies were counted and recorded.
Cell invasion assays
For cell invasion assays, 24-well plates with Transwell chambers (8 μm pore, Corning, Corning, NY, USA) were coated with 100 μL Matrigel before use (Matrigel Basement Membrane Matrix High Concentration, lactose dehydrogenase elevating virus-free, Corning), the final concentration of Matrigel was 200 μg/mL. The cells were trypsinized and washed twice with PBS, then the cells were resuspended in 100 μL of serum free DMEM/F12 medium (Gibco) and seeded into the upper chamber. In the lower chamber, 500 μL culture medium containing 10% FBS was added. After 24 h of 37 °C, 5% CO2 incubation, the chambers were washed gently in PBS, and cotton swabs were used to wipe out the cells in upper chambers. Cells were fixed in methanol and stained with 1% crystal violet solution, and the chambers were photographed with a microscope (Eclipse 50i POL, Nikon, Tokyo, Japan) equipped with a camera. Then the cells were quantified using ImageJ software (National Institutes of Health, Bethesda, MD, USA).
Cell cycle analysis by flow cytometry
Cell cycle percentages were analyzed using Cell Cycle and Apoptosis Analysis Kits (Beyotime, Beijing, China). After 72 h of shRNA transduction, cells were trypsinized and washed in ice-cold PBS, then fixed in 70% ethanol for 24 h. After fixation was complete, cells were centrifuged for 200 g at room temperature and fixative solution was discarded. Cells were stained with propidium iodide solution for 20 min in a 37 °C incubator and then analyzed on a flow cytometer (Beckman Coulter, Brea, CA, USA), and data were analyzed and generated using Modfit software (http://www.vsh.com/products/mflt/index.asp).
CCK-8 assay
CDDP was purchased from Solarbio (Beijing, China), and was dissolved in 0.9% normal saline (NS) and sonicated. The IC50 value of GBC-SD cells to CDDP was determined using CCK-8 assays. Cells were seeded in 96-well plates (5000/well); after they adhered, different concentrations of CDDP (0, 0.1953125, 0.390525, 0.78125, 1.5625, 3.125, 6.25, 12.5, 25, 50, 100, 200 μM) were added to each well. After 24 h, CCK-8 solution was added into each well and incubated for 2 h at 37 °C, then the OD 450 nm was detected with a Varioskan microplate reader.
Cell apoptosis assessment
Intracellular caspase-3 activity as a marker of cell apoptosis after CDDP treatment (50 μM) was evaluated by colorimetric Dojindo Caspase-3 Assay Kits. According to the manufacturer's instruction, cells were seeded in 10 cm2 Petri dishes and treated with 50 μM CDDP or NS for 24 h. Next, cells were lysed and incubated with assay substrate in 37 °C for 4 h, then the p-nitroaniline absorbance (OD 405 nm) was determined on a Varioskan microplate reader. Cell apoptosis-related proteins (caspases-3 and −9, cleaved caspase-3, Bax, and Bad) were analyzed by WB after 50 μM CDDP treatment.
RNA sequencing of the OLFM4 knockdown GBC-SD cell line
To identify genes that are regulated by OLFM4, three replicates (1 × 106 cells each) of GBC-SD cells stably expressing shNC or shOLFM4 were collected. Cells were suspended in TRIzol (Invitrogen) and stored at −80 °C for further analysis. RNA-sequencing was carried out on Illumina platform (Illumina, San Diego, CA, USA), and corresponding analyses were performed by Annoroad Gene Technology Company (Beijing, China). Significantly up- and downregulated genes were screened out under the criterion of absolute FC ≥ 2, p < 0.05.
WB
Cells were lysed in radioimmunoprecipitation assay lysis buffer (Beyotime) supplemented with protease inhibitor (cOmplete Tablets, Mini, EDTA-free, Roche, Basel Switzerland). Proteins were separated by SDS-PAGE and transferred to polyvinylidene fluoride membranes. Primary antibodies were used for immunoblotting: anti-OLFM4 antibody (Cat No. 14369D, Cell Signaling Technology, Danvers, Massachusetts, USA), caspase-3/p17/p19 (Cat No. 66,470–2-Ig, Proteintech, Rosemont, IL, USA), cleaved caspase-3 (Cat No. 9664, Cell Signaling Technology), caspase-9/p35/p10 (Cat No. 66,169–1-Ig, Proteintech), Bax (Cat No. 60,267–1-Ig, Proteintech), Bad (Cat No. 10,435–1-AP, Proteintech), and ARL6IP1 (Cat No. ab24228, Abcam, Cambridge, UK). β-actin (Cat No. 66,009–1-Ig, Proteintech) was used as a loading control. The membranes were blocked with 5% skim milk diluted in TBST and incubated with primary and then secondary antibodies. Finally, the blots were incubated in enhanced chemiluminescent reagent (Affinity Biosciences, Cincinnati, OH, USA) and visualized with the ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA).
Tumor xenografts in nude mice
Four-week-old male BALB/c nude mice were purchased from the Hu Nan Animal Center. Each mouse received 1 × 106 GBC-SD cells transfected with luciferase gene suspended in 100 μl of 7 mg/mL Matrigel (Matrigel Basement Membrane Matrix High Concentration, LDEV-free, Corning) injected subcutaneously into the right side of the neck. Tumor growth was monitored by measuring the length and width every 2 days, and tumor volume was calculated as: V = π/6 × L × W2. At day 30, mice were euthanatized and the tumors were dissected for further analysis.
CDDP treatment in nude mice
To evaluate the effect of CDDP in nude mice, GBC-SD cells stably expressing shNC or shOLFM4 were subcutaneously inoculated into the right side of the neck of each nude mouse. After 15 days of tumor growth, the mice were randomly assigned into the treatment or NS group. Mice in the treatment group were given intraperitoneal CDDP according to body weight (5 mg/kg), and mice in the control group were given same amount of 0.9% NS; in total, mice were given two doses of CDDP or NS with a 3-days interval. At day 30, mice were euthanatized and the tumors were dissected for further analysis. All animal experiments were followed the guidance of National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978). Animal care and all procedures were approved and under the supervision of Yunnan Provincial Laboratory Animal Welfare Committee. The Aniview bioluminescent imaging system (Antpedia, Guang Zhou, China) was also used to detect tumor growth. Aniview software (Antpedia, Guang Zhou, China) was used to quantify bioluminescent and analyze data.
HE and TUNEL
Tissue sections were deparaffinized in xylene; dehydrated in gradient ethanol of 95%, 90%, 80%, and 70% ethanol for 5 min each; and placed in a distilled water bath. Then the sections were counterstained with hematoxylin and eosin (HE), excessive dye was washed by distilled water. The sections were then air-dried and mounted in neutral gum.
For terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL), after section dehydration, antigen retrieval was performed using proteinase K incubation at 37 °C for 25 min. Sections were washed three times with PBS (pH 7.4) in a rocker device for 5 min each. After permeabilization with 0.1% Triton-X diluted in PBS, TUNEL working solution was added to sections with DAPI counterstaining in a dark place. Sections were then coverslipped with anti-fade mounting medium and observed under a fluorescence microscope (Eclipse 50i POL, Nikon, Tokyo, Japan) for image collection (DAPI ultraviolet excitation wavelength 330–380 nm, emission wavelength 420 nm, blue light emission; CY3 excitation wavelength 510–561 nm, emission wavelength 590 nm, red light emission).
Statistical analysis
Data were acquired from experiments conducted in triplicate. Statistical analyses were performed using SPSS version 23.0 (IBM Corp., Armonk, NY, USA) or Graph Pad Prism version 8.0 (GraphPad Inc., San Diego, CA USA). Clinicopathological associations with OLFM4 H-scores were analyzed using chi-square tests. Survival curves were generated using log-rank tests. Two-groups comparisons were made with Student's t-tests, and comparison for variables in more than two groups was using one- or two-way analysis of variance (ANOVA). The data are presented as mean ± standard deviation (SD). Significance was symbolized as * p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001, n.s., non-significant.
Results
OLFM4 is a cholangiocarcinoma-related gene
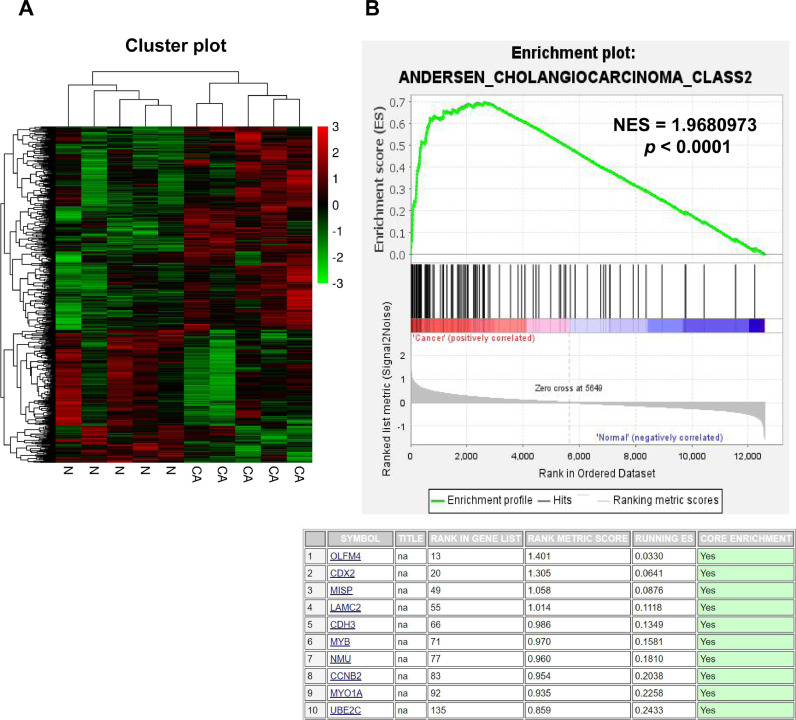
In order to identify DEGs in GBC, we performed mRNA microarray for five pairs of GBC and normal paracarcinoma tissues using Agilent Sure Print G3 Human Gene Expression 8 × 60 K v2 Microarray. In total, there were 252 significantly upregulated and 379 downregulated genes identified through microarray (p < 0.05, FC ≥ 2), cluster analysis shows the DEGs, (Fig. 1A). GSEA was then performed to screen out genes that play key roles in GBC. Among the upregulated DEGs, OLFM4 was enriched in GBC tissues in related to cholangiocarcinoma with highest rank metric score of 1.401 (Fig. 1B, p < 0.0001).
Fig. 1.
OLFM4 was identified as a cholangiocarcinoma-related gene. (A) Heatmap showing the 379 upregulated (red) and 252 downregulated (green) genes in GBC and normal paracarcinoma tissues. (B) The upper panel shows the GSEA plot of the DEGs related to cholangiocarcinoma, and the lower panel shows top 10 enriched cholangiocarcinoma-related genes in GSEA (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
OLFM4 expression progressively increased from precancerous lesions to carcinoma
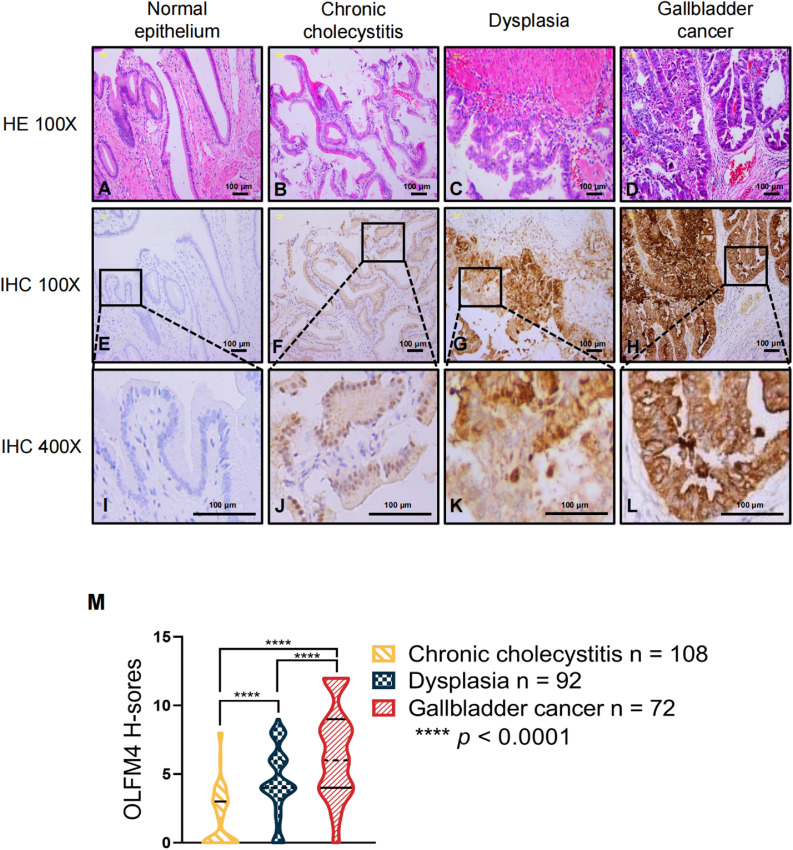
Previous studies have found that OLFM4 is responsible for initiation of gastric cancer and tumorigenesis in extrahepatic bile duct carcinomas [36,37]. Other studies described the malignant transformation from chronic cholecystitis to dysplasia and eventually GBC [42,43]. Considering this evidence, we investigated the role that OLFM4 might play in tumorigenesis of GBC in tissue from 108 cases of chronic cholecystitis, 92 cases of precancerous lesion (dysplasia), and 72 cases of GBC. Histological features of chronic cholecystitis, dysplasia, and GBC were confirmed by HE staining (Fig. 2A-D). In normal epithelium (not affected by inflammation) of chronic cholecystitis cases, the simple columnar epithelium cells and sub-mucosa were intact (Fig. 2A). In chronic cholecystitis tissues, inflammatory infiltrating mononuclear cells were observed (Fig. 2B). Dysplasia tissues showed atypical surface epithelium with obvious cytologic atypia (Fig. 2C). Finally, GBC tissues had pleomorphic, irregularly distributed nuclei (Fig. 2D).
Fig. 2.
OLFM4 expression progressively increased from precancerous lesions to carcinoma. (A–D) HE staining of the gallbladder normal epithelium, chronic cholecystitis, dysplasia, and GBC, respectively. (E,I) IHC staining of OLFM4 in normal epithelium. (F,J) IHC staining of OLFM4 in chronic cholecystitis tissue (n = 108). (G,K) IHC staining of OLFM4 in dysplasia tissue (n = 92). (H,L) IHC staining of OLFM4 in gall bladder cancer tissue (n = 72). (M) Violin plot of OLFM4 H-scores in three groups; the solid and dotted lines show the quartile and median value, respectively. H-scores of the three groups were compared with one-way ANOVA, **** p < 0.0001. Scale bars, 100 μm.
We then evaluated OLFM4 expression with tissue IHC H-scores were calculated to represent different OLFM4 expression levels in the above three groups. They ranged from 0 to 8 in the chronic cholecystitis and dysplasia groups and from 0 to 12 in the GBC group. OLFM4 expression was markedly increased in GBC tissues compared to cholecystitis and dysplasia tissues, with a gradual increase during the tumourigenesis process from precancerous lesions to GBC (Fig. 2E-L, M, p < 0.0001). To further examine the correlations between clinicopathological characteristics with OLFM4 expression in GBC patients, clinical data were collected and analyzed with OLFM4 expression using chi-square tests. The results showed that there were no correlations of OLFM4 expression with patients’ age, sex, or TNM stage; the clinical features are summarized in supplementary Table S1. These results suggest that OLFM4 is an important carcinogenic factor in GBC.
OLFM4 depletion suppressed GBC progression
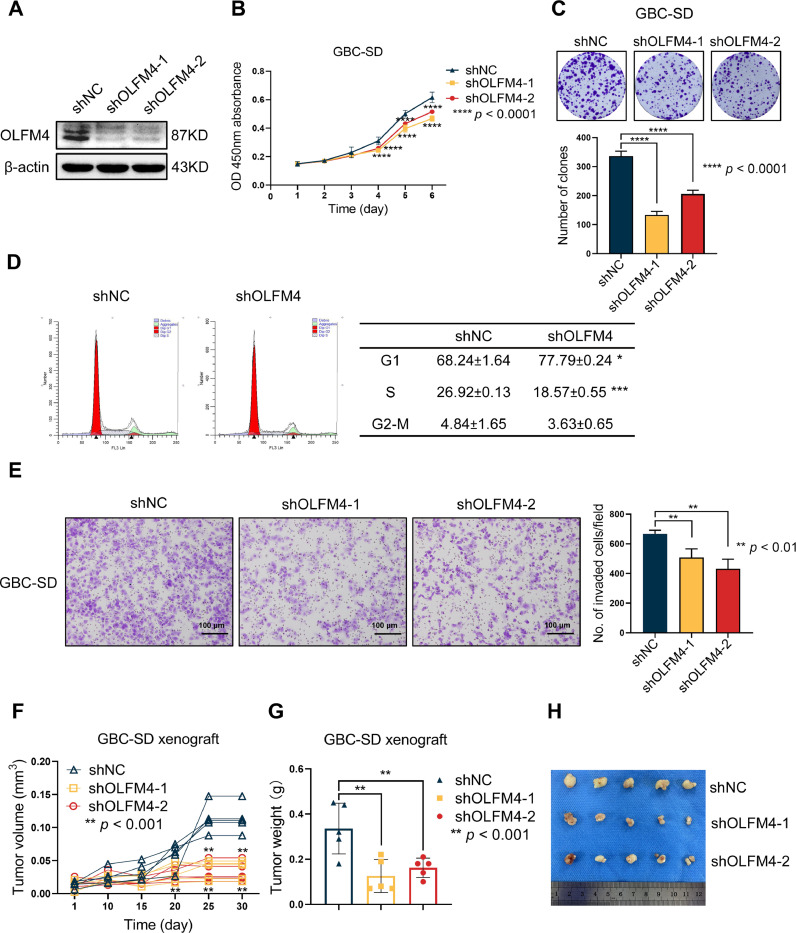
Given that OLFM4 played a role in gallbladder tumorigenesis [36–38], we explored the protein's biological functions in GBC with loss-of-function assays. After validating the knockdown effect of shRNA by WB (Fig. 3A), cell proliferation was evaluated with CCK-8 assays. Proliferation was remarkably hindered after OLFM4 knockdown (Fig. 3B). Further experiments showed impaired clone formation ability in OLFM4 knockdown cells (Fig. 3C). The cell cycle analysis showed that the G1 phase percentage was increased in shOLFM4-expressing cells with a significant reduction in S phase, suggesting G1 arrest (Fig. 3D). To assess cell invasion ability, Matrigel chamber assays were performed after OLFM4 knockdown. Low OLFM4 expression cells showed obviously reduced invasion tendency (Fig. 3E). We then validated the proliferation-inhibiting effect with a tumor xenograft model in nude mice. Tumor growth was significantly inhibited in the shOLFM4 group compared with the shNC group (Fig. 3F), and the final tumor size was significantly smaller (Fig. 3G,H). These results indicate that OLFM4 is crucial for cancer development by affecting proliferation and invasion abilities.
Fig. 3.
OLFM4 depletion suppressed GBC progression. Cell biological function experiments were performed 72 h after lentivirus transfection. (A) WB was used to verify OLFM4 expression, which was only significantly reduced in shOLFM4-expressing cells. (B) CCK-8 assays were performed to measure cell proliferation, (Data shown as mean ± SD, two-way ANOVA comparing shNC, shOLFM4–1, shOLFM4–2 cell growth over time, **** p < 0.0001). (C) Clone formation assay. And bar graph shows the clones counted in three groups. (n = 3, one-way ANOVA comparing shNC, shOLFM4–1, and shOLFM4–2, **** p < 0.0001). (D) Representative images of cell cycle analyses. Flow cytometry was performed to indicate cell cycle. The cell cycle distributions of each phase of shNC and shOLFM4 cells are summarized in the table. In shOLFM4 cells, the percentages of G1 and S phase cells were increased and decreased, respectively (n = 3, t-test comparing shNC and shOLFM4). The results are shown as mean ± SD, * p < 0.05, *** p < 0.001. (E) Knocking down OLFM4 in GBC-SD cells decreased cellular invasion in Matrigel chamber assays. After 24 h incubation, invaded cells were fixed with methyl alcohol and dyed with 0.1% crystal violet, and counted. Original magnification, 200 × . Scale bars, 100 μm. Data of invaded cells shown as mean with SD (n = 3, one-way ANOVA comparing shNC, shOLFM4–1, and sh-OLFM4–2, ** p < 0.01). (F) OLFM4 depletion in GBC-SD cells inhibited tumor growth in vivo. Spider plot showing individual tumor growth curves of the shOLFM4–1, shOLFM4–2, and shNC groups (all n = 5), one-way ANOVA comparing shNC, shOLFM4–1, and sh-OLFM4–2, ** p < 0.01. (G) Each tumor was weighed and recorded. Data are shown as individual values and were compared with one-way ANOVA, ** p < 0.01. (H) Tumors were excised from euthanized mice at day 30. Tumors were smaller in the shOLFM4 group than the shNC group.
OLFM4 depletion increased CDDP chemosensitivity in vitro
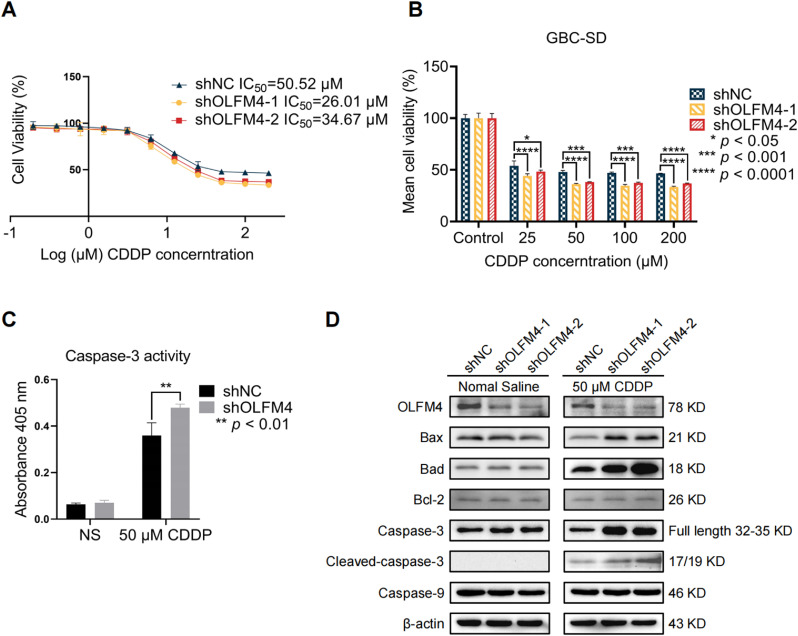
Given that elevated OLFM4 contributed to chemoresistance in pancreatic cancer and NSCLC [39,40], the potential of knocking down OLFM4 in CDDP treatment was assessed. We first measured IC50 value of cells to CDDP in shNC and shOLFM4 cells, the IC50 value of shNC, shOLFM4–1 cells, and shOLFM4–2 was 50.52, 26.01, and 34.67 μM, respectively, which indicate that knock-down of OLFM4 increased cell chemosensitivity to CDDP (Fig. 4A). Cell viability was significantly reduced in shOLFM4 groups under 25, 50, 100, and 200 μM treatment compared to the shNC group (Fig. 4B). Since previous studies have suggested that OLFM4 is an anti-apoptotic factor [30,35], we hypothesized that OLFM4 depletion might promote apoptosis as measured by caspase-3 activity. The results showed that caspase-3 activity was higher in OLFM4-depleted cells after CDDP treatment compared to shNC group (Fig. 4C). We further measured other apoptosis-related proteins by WB and found increased level of caspase-3 and cleaved caspase-3. Expression of other apoptotic molecules including Bax and Bad were also significantly enhanced in the shOLFM4 group after CDDP treatment, but their levels were unchanged in NS treatment with the exception of Bcl-2 and caspase-9, which had similar levels in both two groups with or without CDDP treatment (Fig. 4D). These results showed CDDP treatment notably activated cell apoptosis in OLFM4-deficient cells.
Fig. 4.
OLFM4 knockdown enhanced cell sensitivity to CDDP by regulating caspase-3. (A) The IC50 values of GBC-SD cells exposed to CDDP were evaluated with CCK-8 assays. The maximum concentration of CDDP was 200 μM, and NS was used as blank control. (B) The cell viabilities of three groups are shown in the bar graph, one-way ANOVA comparing shNC, shOLFM4–1, and sh-OLFM4–2, * p < 0.05, *** p < 0.001, **** p < 0.0001. (C) Caspase-3 activity in shNC and shOLFM4 cells was measured after 24 h 50 μM CDDP treatment. Caspase-3 activity was significantly increased in shOLFM4 cells, ** p < 0.01. (D) OLFM4, and apoptosis-related proteins were detected by WB in OLFM4 knockdown cells and control cells treated with NS or 50 μM CDDP. The levels of apoptotic proteins (Bax, Bad, Caspase-3, Cleaved-caspase-3) were elevated in shOLFM4 cells treated with CDDP compared with shNC cells, while Bcl-2 and Caspase-9 were not changed after NS of CDDP treatment in both groups.
ARL6IP1 (ARMER) is downregulated after OLFM4 knockdown
To identify the apoptosis-promoting mechanisms regulated by OLFM4, we performed RNA-sequencing in OLFM4 knocked-down GBC-SD cells. Fig. 5A shows the up- and downregulated genes in OLFM4-depleted cells under the criteria of FC ≥ 2, p < 0.05. Four genes were significantly downregulated: ARL6IP1 (ARMER), PHLDA3, CREG2, and TENM3. Previous research suggested that ARL6IP1 is an apoptosis regulator and could prevent CDDP-induced apoptosis [44], [45], [46], [47]. Based on this, we hypothesized that ARL6IP1 might be an important gene regulated by OLFM4 and that apoptosis might be increased when the levels of these two anti-apoptosis factors (OLFM4, ARL6IP1) are low. WB for ARL6IP1 showed its downregulated expression in OLFM4-deficient cells (Fig. 5B). We then performed add-back rescue experiments to verify functional changes regulated by OLFM4/ARL6IP1/Capase-3 axis, and the results showed that ARL6IP1 overexpression in OLFM4-depleted cells could reverse CDDP-induced apoptosis (Fig. 5C,D), but it did not affect cell proliferation (Fig. S1). Collectively, these findings demonstrate that apoptosis can be regulated by downregulating ARL6IP1 in OLFM4 knockdown cells.
Fig. 5.
ARL6IP1 is a downstream effector of OLFM4. (A) Left panel: the heatmap shows the significantly up- and downregulated genes after OLFM4 knockdown detected by RNAseq; criteria: FC ≥ 2, p < 0.05. Right panel: the table shows the five most significantly downregulated genes. (B) WB showed that OLFM4 knockdown could downregulate ARL6IP1. (C) Add-back rescue experiments showed that ARL6IP1 overexpression in shOLFM4 cells increased viability when cells were treated with 50 μM CDDP. (Data shown as mean ± SD, * Student's t-test comparing the shNC and shOLFM4 groups, * p < 0.05). (D) Add-back rescue experiments showed ARL6IP1 overexpression in shOLFM4 cells attenuated caspase-3 activation and reduced cleaved-caspase-3 levels after CDDP treatment.
OLFM4 depletion enhanced CDDP chemotherapeutic effects in vivo
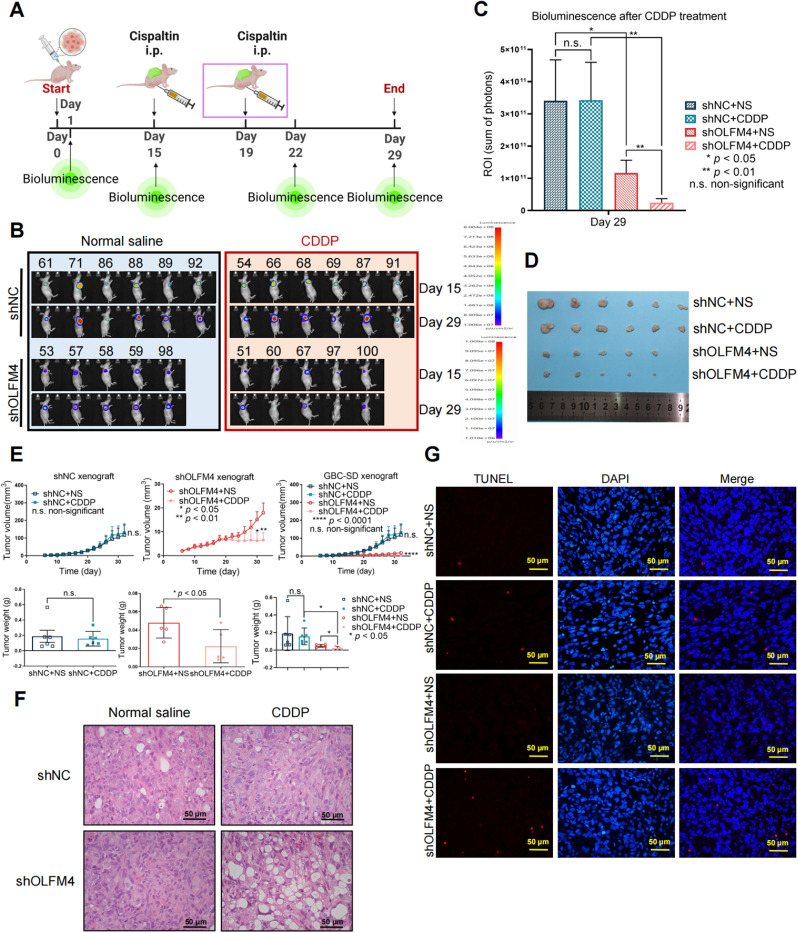
We further evaluated the tumor-suppressive effect of CDDP in OLFM4-depleted tumors. Remarkably, OLFM4 knockdown increased CDDP's tumor-suppressing effect in nude mice. The animals were inoculated with shOLFM4- or shNC-expressing cells, followed by two doses of CDDP (5 mg/kg, experimental timeline displayed in Fig. 6A). CDDP significantly reduced shOLFM4 tumor load as measured by bioluminescence (Figs. 6B, C and S2), tumor growth (Fig. 6E, upper panels), and final tumor weight (Fig. 6E, lower panels). Tumor growth in the shOLFM4 + CDDP group was significantly inhibited and tumors drastically shrunk in response to CDDP treatment (Fig. 6D,E). In the shNC group, after the treatment completion there was no difference between the CDDP and NS groups (Fig. 6C–E). HE staining of tumor tissue from the shNC + CDDP group showed no significant changes in histological features compared to the shNC + NS group (Fig. 6F), and no cell apoptosis was detected by TUNEL (Fig. 6G). Conversely, there was a obvious tissue reduction in the shOLFM4 + CDDP group compared to shOLFM4 + NS group which had compact tumor cell arrangement; and the tumor tissue became loose in response to CDDP treatment (Fig. 6F). Significant cell apoptosis was detected in shOLFM4 + CDDP group while no significant apoptosis was detected in shOLFM4 + NS group (Fig. 6G). These results suggest that OLFM4 depletion enhanced the CDDP treatment response in vivo.
Fig. 6.
OLFM4 depletion increased the CDDP tumor-suppressing effect in vivo. (A) Experiment timeline of CDDP treatment in nude mice. At 15 days after tumor cell inoculation, mice bearing shNC (n = 12) or shOLFM4 (n = 10) tumors were randomly assigned into the NS or CDDP group. The first dose of CDDP (5 mg/kg) was given intraperitoneally at day 15, and the second dose was given intraperitoneally at day 19. Bioluminescence was measured before and after the two-dose CDDP treatment. (B) Bioluminescence images of tumor shNC and shOLFM4 after CDDP treatment. (C) On day 29 (7 days after the two-dose CDDP treatment) shOLFM4 + CDDP bioluminescence was significantly reduced compared to the shOLFM4 + NS group, (** p < 0.01, two-way ANOVA), while shNC + CDDP bioluminescence remained the same level with shNC + NS group, (n.s., non-significant). Bioluminescence was lower in the shOLFM4 + NS group compared to the shNC + NS group, (* p < 0.05, two-way ANOVA). (D) Tumor size was significantly reduced in the shOLFM4 + CDDP group compared to the shOLFM4 + NS group, but there was no significant change in the shNC group after CDDP treatment. (E) The left upper and lower panels show the tumor growth curve and final tumor weight of shNC xenografts; the middle upper and lower panels show the tumor growth curve and final tumor weight of shOLFM4 xenografts; the left upper and lower panels show the tumor growth curve and final tumor weight of shNC and shOLFM4 xenografts. Tumor growth and weight were not reduced in the shNC + CDDP group compared to the shNC + NS group, while CDDP significantly reduced tumor weight and growth in the shOLFM4 group. Data are shown as mean ± SEM, * p < 0.05, ** p < 0.01, n.s., non-significant. (F) HE-stained tumor tissues. In the upper two figures, shNC + NS and shNC + CDDP xenograft histology show densely arranged tumor cells, there was no marked tissue loss in shNC + CDDP group. The lower two figures show the shOLFM4 + CDDP xenografts with obvious reduction of tumor cells and the tissues became loose, while shOLFM4 + NS tissues had compact cancer cell arrangement. Original magnification 400 × . Scale bars, 50 μm. (G) TUNEL (red fluorescence) showed scarce cell apoptosis in the shNC + CDDP group, and there was a remarkable increase in apoptotic cells in the shOLFM4 group after CDDP treatment. Original magnification 400 × . Scale bars, 50 μm (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Low OLFM4 expression is associated with longer survival in patients with GBC
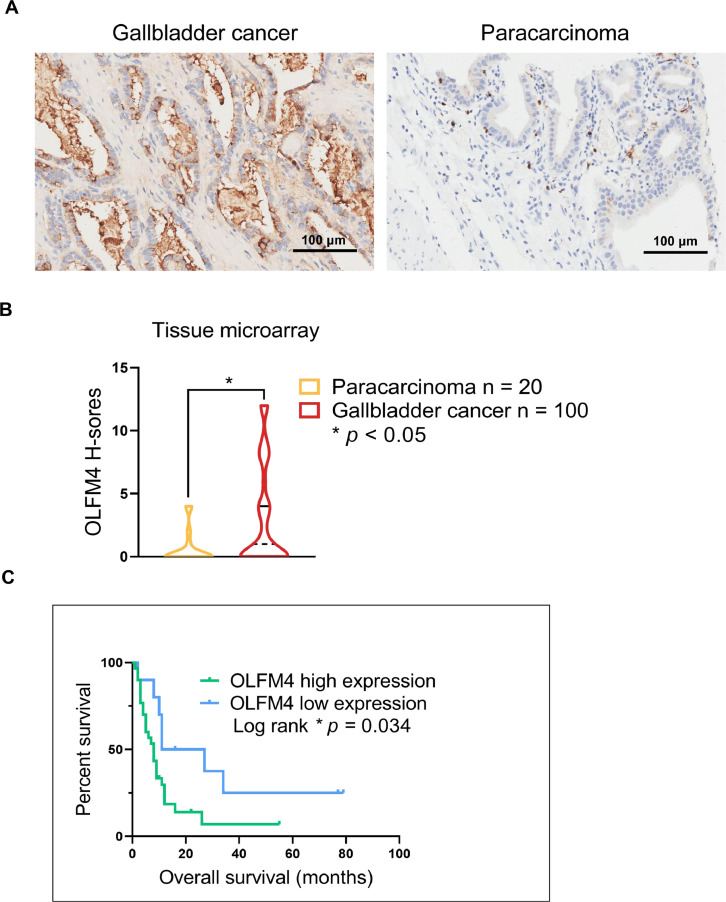
The prognostic value of OLFM4 in GBC has not been described, so we performed IHC labeling of OLFM4 in GBC TMAs to determine its prognostic value. There were 100 cases of GBC and 20 cases of normal paracarcinoma tissues included in the TMA, and 40 GBC cases had survival data. OLFM4 expression was detected in cancer and normal paracarcinoma tissues (Fig. 7A) and given H-scores. OLFM4 was overexpressed in GBC tissues compared to paracarcinoma tissues (Fig. 7B). Among 40 cases with survival data, 30 were high expressing and 10 were low expressing. The longest and shortest survival periods in the high/low OLFM4 groups were 55/79 and 1/2 months, respectively. Log-rank testing showed that the survival difference was significantly (log-rank p = 0.034, Fig. 7C). These results demonstrate that low OLFM4 expression is associated with improved survival of GBC patients.
Fig. 7.
Low OLFM4 expression is associated with longer survival in patients with GBC. (A) IHC staining of OLFM4 in GBC (n = 100) and normal paracarcinoma tissues (N = 20) in TMAs. Scale bars on right bottom, 100 μm. (B) H-scores were used to evaluate positive staining and staining areas. Violin plot of OLFM4 H-scores in GBC and paracarcinoma tissues; the solid and dotted lines show the quartile and median value, respectively (Student's t-test, * p < 0.05). (C) Kaplan-Meier curves of overall survival of GBC patients (n = 40) with high (n = 30) and low (n = 10) OLFM4 expression. * Log-rank p = 0.034.
Discussion
The mechanisms of GBC onset and progression remain unclear, and patients are often diagnosed too late to undergo surgery, which is the only curative method. Unfortunately, due to GBC chemoresistance, the chemotherapy can not be applied earlier, and the palliative chemotherapy is not efficient. Understanding GBC pathogenesis, discovering new targets to sensitize GBC to chemotherapy, and excavating prognostic biomarkers are therefore important to prolong patient survival.
Aberrant OLFM4 expression has been detected in different types of cancers, including colon cancer, prostate cancer, and GBC [34,48,49]. OLFM4 is also involved in multiple biological processes in various cancers, including proliferation, metastasis, apoptosis regulation, and cell differentiation, especially in early stages of gastric cancer and cholangiocarcinoma [36], [37], [38]. Here, in our study, the findings of OLFM4 in GBC may provide a multiple-role playing maker that can be a therapeutic target and prognostic indicator. Unlike other biomarkers in cancer, the gradually expression pattern from precancerous lesions to cancer of OLFM4 endowed it the capability of predicting both the tumorigenesis and survival.
There is a mainstream hypothesis is that chronic inflammation contributes to tumorigenesis in GBC [50]. Notably, OLFM4 is regulated by a variety of factors correlated with inflammation and tumorigenesis [35], which indicates that a similar process can happen in GBC. Presumably, long-term chronic inflammation may stimulate excessive OLFM4 expression, leading to the initiation of cancer regulated by OLFM4, followed by progression due to autogenic regulating actions in cancer promotion. In our study, through microarray and bioinformatic analyses, we found that OLFM4 is highly correlated with cholangiocarcinoma. More importantly, OLFM4 expression gradually increased from chronic cholecystitis tissues to dysplasia (i.e., precancerous lesions) and GBC, which is consistent with previous studies [32,41]. These findings suggest that OLFM4 is vital in the development of GBC, especially cancer that originated from chronically inflamed tissues [33], and then we found that knocking down OLFM4 inhibited cell proliferation and invasion, supporting the hypothesis that OLFM4 is involved in tumor proliferation and metastasis. To predict patient outcome, the IHC of OLFM4 in TMA supported that low OLFM4 expression predicted longer patient survival. However, more work is needed to verify a direct connection of chronic inflammation and the effects OLFM4, and larger clinical sample sizes are needed to be more representative of the entire GBC patient population.
As it is known that OLFM4 is an anti-apoptotic factor, we evaluated the value of OLFM4 as a chemotherapeutic target. According to its strong anti-apoptotic regulating function reported in preliminary research, which can hinder the chemotherapy effects [29,30,35], our study validated that OLFM4 depletion could sensitize GBC cells to CDDP treatment by regulating ARL6IP1/caspase-3, indicating that OLFM4 or ARL6IP1 can be potentially used as a chemotherapeutic targets if OLFM4 or ARL6IP1 inhibitors are available. However, it requires more proof from further research to verify if the low expression of OLFM4 indicate a better response to chemotherapy, and to support whether the anti-apoptotic effect of OLFM4 is responsible for GBC chemoresistance. Future studies are needed to elucidate how OLFM4 might regulate chemoresistance of GBC to identify more targets for precise treatment.
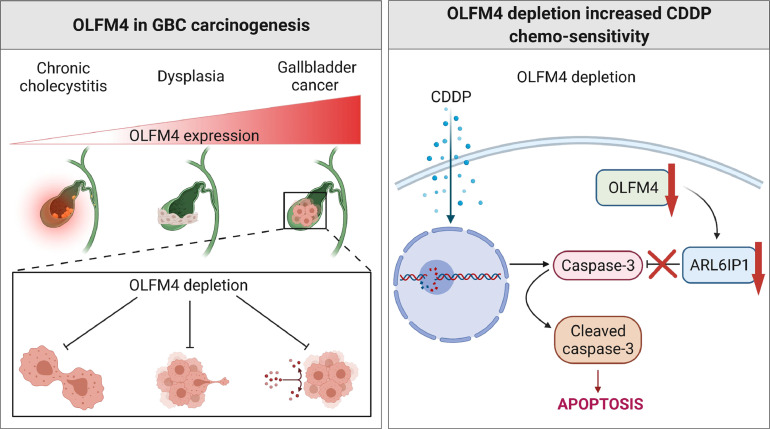
In summary, our study suggests that OLFM4 is being critical in GBC tumorigenesis and chemoresistance. Thus, OLFM4 can be a useful prognostic biomarker in tumorigenesis and progression. Most importantly, our study offers a novel potential treatment target in GBC and a chemosensitivity marker. A summarizing diagram for the whole content of this study is shown in Fig. 8 (generated using https://BioRender.com, agreement number: SN22SZSL8P).
Fig. 8.
Diagram representing the whole content of the study. The left upper panel show the expression of OLFM4 gradually increased from cholecystitis to dysplasia and gallbladder cancer. The left lower panel show the biological role of OLFM4 in gallbladder cancer is to participate in proliferation, invasion, and chemoresistance. And the right panel show the mechanism of the enhanced chemosensitivity induced by OLFM4 depletion in gallbladder cancer.
Consent
Written informed consent was obtained from the patient for publication of this case report and accompanying images. A copy of the written consent is available for review by the Editor-in-Chief of this journal on request.
Funding resources
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81960499, 81360245, 81660477); the Yunnan Province Science and Technology Department of Kunming Medical University (Grant No. 202001AY070001–168); the Science and Technology Fund of Yunnan Province (Grant No. 2017IB020); and the Science and Technology Fund of Kunming City (Grant No. 2019–1-N-25318000002027).
CRediT authorship contribution statement
Zhuying Lin: Conceptualization, Writing – original draft. Songlin Yang: Investigation. Yong Zhou: Investigation. Zongliu Hou: Supervision, Resources. Lin Li: Writing – review & editing. Mingyao Meng: Project administration. Chunlei Ge: Data curation. Baozhen Zeng: Visualization. Jinbao Lai: Data curation. Hui Gao: Validation. Yiyi Zhao: Validation. Yanhua Xie: Resources. Shan He: Formal analysis. Weiwei Tang: Investigation. Ruhong Li: Supervision, Funding acquisition. Jing Tan: Funding acquisition, Project administration. Wenju Wang: Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Prof. Xin Song for important advisory for this research, and his kind encouragement to us. He will be long remembered for his devotion into the scientific research.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101331.
Contributor Information
Ruhong Li, Email: lrh272@126.com.
Jing Tan, Email: kmtjing@sina.com.
Wenju Wang, Email: wangwenju_vip@hotmail.com.
Appendix. Supplementary materials
References
- 1.Misra S., Chaturvedi A., Misra N.C., Sharma I.D. Carcinoma of the gallbladder. Lancet Oncol. 2003;4:167–176. doi: 10.1016/s1470-2045(03)01021-0. [DOI] [PubMed] [Google Scholar]
- 2.Wistuba I.I., Gazdar A.F. Gallbladder cancer: lessons from a rare tumour. Nat. Rev. Cancer. 2004;4:695–706. doi: 10.1038/nrc1429. [DOI] [PubMed] [Google Scholar]
- 3.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21590. [DOI] [PubMed] [Google Scholar]
- 4.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt M.A., Marcano-Bonilla L., Roberts L.R. Gallbladder cancer: epidemiology and genetic risk associations. Chin. Clin. Oncol. 2019;8:31. doi: 10.21037/cco.2019.08.13. [DOI] [PubMed] [Google Scholar]
- 6.Ertel A.E., Bentrem D., Abbott D.E. Gall bladder cancer. Cancer Treat. Res. 2016;168:101–120. doi: 10.1007/978-3-319-34244-3_6. [DOI] [PubMed] [Google Scholar]
- 7.Ryu S., Chang Y., Yun K.E., Jung H.S., Shin J.H., Shin H. Gallstones and the risk of gallbladder cancer mortality: a cohort study. Am. J. Gastroenterol. 2016;111:1476–1487. doi: 10.1038/ajg.2016.345. [DOI] [PubMed] [Google Scholar]
- 8.Lazcano-Ponce E.C., Miquel J.F., Muñoz N., Herrero R., Ferrecio C., Wistuba, Alonso de Ruiz P., Aristi Urista G., Nervi F. Epidemiology and molecular pathology of gallbladder cancer. CA Cancer J. Clin. 2001;51:349–364. doi: 10.3322/canjclin.51.6.349. [DOI] [PubMed] [Google Scholar]
- 9.Barahona Ponce C., Scherer D., Brinster R., Boekstegers F., Marcelain K., Gárate-Calderón V., Müller B., de Toro G., Retamales J., Barajas O., Ahumada M., Morales E., Rojas A., Sanhueza V., Loader D., Rivera M.T., Gutiérrez L., Bernal G., Ortega A., Montalvo D., Portiño S., Bertrán M.E., Gabler F., Spencer L., Olloquequi J., Fischer C., Jenab M., Aleksandrova K., Katzke V., Weiderpass E., Bonet C., Moradi T., Fischer K., Bossers W., Brenner H., Hveem K., Eklund N., Völker U., Waldenberger M., Fuentes Guajardo M., Gonzalez-Jose R., Bedoya G., Bortolini M.C., Canizales-Quinteros S., Gallo C., Ruiz-Linares A., Rothhammer F., Lorenzo Bermejo J. Gallstones, body mass index, C-reactive protein, and gallbladder cancer: mendelian randomization analysis of chilean and european genotype data. Hepatology. 2021;73:1783–1796. doi: 10.1002/hep.31537. [DOI] [PubMed] [Google Scholar]
- 10.Giannis D., Cerullo M., Moris D., Shah K.N., Herbert G., Zani S., Blazer D.G., Allen P.J., Lidsky M.E. Validation of the 8th edition American joint commission on cancer (AJCC) gallbladder cancer staging system: prognostic discrimination and identification of key predictive factors. Cancers. 2021;13:547. doi: 10.3390/cancers13030547. (Basel) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Javle M., Zhao H., Abou-Alfa G.K. Systemic therapy for gallbladder cancer. Chin. Clin. Oncol. 2019;8:44. doi: 10.21037/cco.2019.08.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mao W., Deng F., Wang D., Gao L., Shi X. Treatment of advanced gallbladder cancer: a SEER-based study. Cancer Med. 2020;9:141–150. doi: 10.1002/cam4.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jaruvongvanich V., Yang J.D., Peeraphatdit T., Roberts L.R. The incidence rates and survival of gallbladder cancer in the USA. Eur. J. Cancer Prev. 2019;28:1–9. doi: 10.1097/CEJ.0000000000000402. [DOI] [PubMed] [Google Scholar]
- 14.Eslick G.D. Epidemiology of gallbladder cancer. Gastroenterol. Clin. N. Am. 2010;39:307–330. doi: 10.1016/j.gtc.2010.02.011. ix. [DOI] [PubMed] [Google Scholar]
- 15.Soreide K., Guest R.V., Harrison E.M., Kendall T.J., Garden O.J., Wigmore S.J. Systematic review of management of incidental gallbladder cancer after cholecystectomy. Br. J. Surg. 2019;106:32–45. doi: 10.1002/bjs.11035. [DOI] [PubMed] [Google Scholar]
- 16.Hickman L., Contreras C. Gallbladder cancer: diagnosis, surgical management, and adjuvant therapies. Surg. Clin. N. Am. 2019;99:337–355. doi: 10.1016/j.suc.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Valle J., Wasan H., Palmer D.H., Cunningham D., Anthoney A., Maraveyas A., Madhusudan S., Iveson T., Hughes S., Pereira S.P., Roughton M., Bridgewater J. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010;362:1273–1281. doi: 10.1056/NEJMoa0908721. [DOI] [PubMed] [Google Scholar]
- 18.Wang S.J., Lemieux A., Kalpathy-Cramer J., Ord C.B., Walker G.V., Fuller C.D., Kim J.S., Thomas C.R. Nomogram for predicting the benefit of adjuvant chemoradiotherapy for resected gallbladder cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2011;29:4627–4632. doi: 10.1200/JCO.2010.33.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McNamara M.G., Goyal L., Doherty M., Springfeld C., Cosgrove D., Sjoquist K.M., Park J.O., Verdaguer H., Braconi C., Ross P.J., Gramont A., Zalcberg J.R., Palmer D.H., Valle J.W., Knox J.J. NUC-1031/cisplatin versus gemcitabine/cisplatin in untreated locally advanced/metastatic biliary tract cancer (NuTide:121) Future Oncol. 2020;16:1069–1081. doi: 10.2217/fon-2020-0247. [DOI] [PubMed] [Google Scholar]
- 20.Kim B.J., Hyung J., Yoo C., Kim K.P., Park S.J., Lee S.S., Park D.H., Song T.J., Seo D.W., Lee S.K., Kim M.H., Park J.H., Cho H., Ryoo B.Y., Chang H.M. Prognostic factors in patients with advanced biliary tract cancer treated with first-line gemcitabine plus cisplatin: retrospective analysis of 740 patients. Cancer Chemother. Pharmacol. 2017;80:209–215. doi: 10.1007/s00280-017-3353-2. [DOI] [PubMed] [Google Scholar]
- 21.Zaidi M.Y., Maithel S.K. Updates on gallbladder cancer management. Curr. Oncol. Rep. 2018;20:21. doi: 10.1007/s11912-018-0664-3. [DOI] [PubMed] [Google Scholar]
- 22.Azizi A.A., Lamarca A., Valle J.W. Systemic therapy of gallbladder cancer: review of first line, maintenance, neoadjuvant and second line therapy specific to gallbladder cancer. Chin. Clin. Oncol. 2019;8:43. doi: 10.21037/cco.2019.07.05. [DOI] [PubMed] [Google Scholar]
- 23.Goetze T.O., Bechstein W.O., Bankstahl U.S., Keck T., Königsrainer A., Lang S.A., Pauligk C., Piso P., Vogel A., Al-Batran S.E. Neoadjuvant chemotherapy with gemcitabine plus cisplatin followed by radical liver resection versus immediate radical liver resection alone with or without adjuvant chemotherapy in incidentally detected gallbladder carcinoma after simple cholecystectomy or in front of radical resection of BTC (ICC/ECC)-a phase III study of the German registry of incidental gallbladder carcinoma platform (GR)-the AIO/CALGP/ ACO-GAIN-trial. BMC Cancer. 2020;20:122. doi: 10.1186/s12885-020-6610-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma A., Kalyan Mohanti B., Pal Chaudhary S., Sreenivas V., Kumar Sahoo R., Kumar Shukla N., Thulkar S., Pal S., Deo S.V., Pathy S., Ranjan Dash N., Kumar S., Bhatnagar S., Kumar R., Mishra S., Sahni P., Iyer V.K., Raina V. Modified gemcitabine and oxaliplatin or gemcitabine + cisplatin in unresectable gallbladder cancer: results of a phase III randomised controlled trial. Eur. J. Cancer. 2019;123:162–170. doi: 10.1016/j.ejca.2019.10.004. [DOI] [PubMed] [Google Scholar]
- 25.You M.S., Ryu J.K., Choi Y.H., Choi J.H., Huh G., Paik W.H., Lee S.H., Kim Y.T. Therapeutic outcomes and prognostic factors in unresectable gallbladder cancer treated with gemcitabine plus cisplatin. BMC Cancer. 2019;19:10. doi: 10.1186/s12885-018-5211-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiao L., Lan X., Shi X., Zhao K., Wang D., Wang X., Li F., Huang H., Liu J. Cytoplasmic RAP1 mediates cisplatin resistance of non-small cell lung cancer. Cell Death Dis. 2017;8:e2803. doi: 10.1038/cddis.2017.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galluzzi L., Senovilla L., Vitale I., Michels J., Martins I., Kepp O., Castedo M., Kroemer G. Molecular mechanisms of cisplatin resistance. Oncogene. 2011;31:1869–1883. doi: 10.1038/onc.2011.384. [DOI] [PubMed] [Google Scholar]
- 28.Zhang H., Deng T., Liu R., Ning T., Yang H., Liu D., Zhang Q., Lin D., Ge S., Bai M., Wang X., Zhang L., Li H., Yang Y., Ji Z., Wang H., Ying G., Ba Y. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol. Cancer. 2020;19:43. doi: 10.1186/s12943-020-01168-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang X., Huang Q., Yang Z., Li Y., Li C.Y. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 2004;64:2474–2481. doi: 10.1158/0008-5472.can-03-3443. [DOI] [PubMed] [Google Scholar]
- 30.Liu W., Liu Y., Li H., Rodgers G.P. Olfactomedin 4 contributes to hydrogen peroxide-induced NADPH oxidase activation and apoptosis in mouse neutrophils. Am. J. Physiol. Cell Physiol. 2018;315:C494–c501. doi: 10.1152/ajpcell.00336.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu W., Yan M., Liu Y., Wang R., Li C., Deng C., Singh A., Coleman W.G., Rodgers G.P. Olfactomedin 4 down-regulates innate immunity against helicobacter pylori infection. Proc. Natl. Acad. Sci. U.S.A. 2010;107:11056–11061. doi: 10.1073/pnas.1001269107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang X.Y., Chen S.H., Zhang Y.N., Xu C.F. Olfactomedin-4 in digestive diseases: a mini-review. World J. Gastroenterol. 2018;24:1881–1887. doi: 10.3748/wjg.v24.i17.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W., Rodgers G.P. Olfactomedin 4 expression and functions in innate immunity, inflammation, and cancer. Cancer Metastasis Rev. 2016;35:201–212. doi: 10.1007/s10555-016-9624-2. [DOI] [PubMed] [Google Scholar]
- 34.Luo Z., Zhang Q., Zhao Z., Li B., Chen J., Wang Y. OLFM4 is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. J. Cancer Res. Clin. Oncol. 2011;137:1713–1720. doi: 10.1007/s00432-011-1042-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R.H., Yang M.H., Xiang H., Bao L.M., Yang H.A., Yue L.W., Jiang X., Ang N., Wu L.Y., Huang Y. Depletion of OLFM4 gene inhibits cell growth and increases sensitization to hydrogen peroxide and tumor necrosis factor-alpha induced-apoptosis in gastric cancer cells. J. Biomed. Sci. 2012;19:38. doi: 10.1186/1423-0127-19-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jun S.Y., An S., Huh T., Chung J.Y., Hong S.M. Clinicopathological significance of olfactomedin-4 in extrahepatic bile duct carcinoma. Pathol. Res. Pract. 2020;216 doi: 10.1016/j.prp.2020.152940. [DOI] [PubMed] [Google Scholar]
- 37.Jang B.G., Lee B.L., Kim W.H. Olfactomedin-related proteins 4 (OLFM4) expression is involved in early gastric tumorigenesis and of prognostic significance in advanced gastric cancer. Virchows Arch. Int. J. Pathol. 2015;467:285–294. doi: 10.1007/s00428-015-1793-9. [DOI] [PubMed] [Google Scholar]
- 38.Neyazi M., Bharadwaj S.S., Bullers S., Varenyiova Z., Travis S., Arancibia-Cárcamo C.V., Powrie F., Geremia A. Overexpression of cancer-associated stem cell gene OLFM4 in the colonic epithelium of patients with primary sclerosing cholangitis. Inflamm. Bowel Dis. 2021;27:1316–1327. doi: 10.1093/ibd/izab025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao X.Z., Wang G.N., Zhao W.G., Han J., Diao C.Y., Wang X.H., Li S.L., Li W.C. Blocking OLFM4/HIF-1α axis alleviates hypoxia-induced invasion, epithelial-mesenchymal transition, and chemotherapy resistance in non-small-cell lung cancer. J. Cell. Physiol. 2019 doi: 10.1002/jcp.28144. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 40.Ohkuma R., Yada E., Ishikawa S., Komura D., Ishizaki H., Tamada K., Kubota Y., Hamada K., Ishida H., Hirasawa Y., Ariizumi H., Satoh E., Shida M., Watanabe M., Onoue R., Ando K., Tsurutani J., Yoshimura K., Yokobori T., Sasada T., Aoki T., Murakami M., Norose T., Ohike N., Takimoto M., Izumizaki M., Kobayashi S., Tsunoda T., Wada S. High expression of olfactomedin-4 is correlated with chemoresistance and poor prognosis in pancreatic cancer. PLoS One. 2020;15 doi: 10.1371/journal.pone.0226707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ge C., Zhu X., Niu X., Zhang B., Chen L. A transcriptome profile in gallbladder cancer based on annotation analysis of microarray studies. Mol. Med. Rep. 2021;23:25. doi: 10.3892/mmr.2020.11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castillo J., García P., Roa J.C. [Genetic alterations in preneoplastic and neoplastic injuries of the gallbladder] Rev. Med. Chil. 2010;138:595–604. [PubMed] [Google Scholar]
- 43.Rosa L., Lobos-González L., Muñoz-Durango N., García P., Bizama C., Gómez N., González X., Wichmann I.A., Saavedra N., Guevara F., Villegas J., Arrese M., Ferreccio C., Kalergis A.M., Miquel J.F., Espinoza J.A., Roa J.C. Evaluation of the chemopreventive potentials of ezetimibe and aspirin in a novel mouse model of gallbladder preneoplasia. Mol. Oncol. 2020;14:2834–2852. doi: 10.1002/1878-0261.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G.L., Shi X., Salisbury E., Timchenko N.A. Regulation of apoptotic and growth inhibitory activities of C/EBPalpha in different cell lines. Exp. Cell Res. 2008;314:1626–1639. doi: 10.1016/j.yexcr.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo F., Liu Y., Li Y., Li G. Inhibition of ADP-ribosylation factor-like 6 interacting protein 1 suppresses proliferation and reduces tumor cell invasion in CaSki human cervical cancer cells. Mol. Biol. Rep. 2010;37:3819–3825. doi: 10.1007/s11033-010-0037-y. [DOI] [PubMed] [Google Scholar]
- 46.Lui H.M., Chen J., Wang L., Naumovski L. ARMER, apoptotic regulator in the membrane of the endoplasmic reticulum, a novel inhibitor of apoptosis. Mol. Cancer Res. MCR. 2003;1:508–518. [PubMed] [Google Scholar]
- 47.Guo F., Li Y., Liu Y., Wang J., Li G. ARL6IP1 mediates cisplatin-induced apoptosis in CaSki cervical cancer cells. Oncol. Rep. 2010;23:1449–1455. doi: 10.3892/or_00000783. [DOI] [PubMed] [Google Scholar]
- 48.Quesada-Calvo F., Massot C., Bertrand V., Longuespée R., Blétard N., Somja J., Mazzucchelli G., Smargiasso N., Baiwir D., De Pauw-Gillet M.C., Delvenne P., Malaise M., Coimbra Marques C., Polus M., De Pauw E., Meuwis M.A., Louis E. OLFM4, KNG1 and Sec24C identified by proteomics and immunohistochemistry as potential markers of early colorectal cancer stages. Clin. Proteom. 2017;14:9. doi: 10.1186/s12014-017-9143-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Valo I., Raro P., Boissard A., Maarouf A., Jézéquel P., Verriele V., Campone M., Coqueret O., Guette C. OLFM4 expression in ductal carcinoma in situ and in invasive breast cancer cohorts by a SWATH-based proteomic approach. Proteomics. 2019;19 doi: 10.1002/pmic.201800446. [DOI] [PubMed] [Google Scholar]
- 50.Goetze T.O. Gallbladder carcinoma: prognostic factors and therapeutic options. World J. Gastroenterol. 2015;21:12211–12217. doi: 10.3748/wjg.v21.i43.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rizzo A., Ricci AD., Brandi G. Recent advances of immunotherapy for biliary tract cancer. Expert Rev Gastroenterol Hepatol. 2021;15:527–536. doi: 10.1080/17474124.2021.1853527. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.