Abstract
Multiple sclerosis (MS) is an autoimmune disease of the central nervous system in which the interaction between genetic and environmental factors plays an important role in disease pathogenesis. Although environmental factors account for 70% of disease risk, the exact environmental factors associated with MS are unknown. Recently, gut microbiota has emerged as a potential missing environmental factor linked with the pathobiology of MS. Yet, how genetic factors, such as HLA class-II gene(s), interact with gut microbiota and influence MS is unclear. In the present study, we investigated whether HLA class-II genes that regulate EAE and MS susceptibility also influence gut microbiota. Previously, we have shown that HLA-DR3 transgenic mice lacking endogenous mouse class II genes (AE-KO) were susceptible to myelin proteolipid protein (PLP91-110)-induced experimental autoimmune encephalomyelitis (EAE), an animal model of MS, while AE-KO.HLA-DQ8 transgenic mice were resistant. Surprisingly, HLA-DR3.DQ8 double transgenic mice showed higher disease incidence and severity compared to HLA-DR3 mice. Gut microbiota analysis showed that HLA-DR3, HLA-DQ8, and HLA-DR3.DQ8 double transgenic mice microbiota are compositionally different from AE-KO mice. Within HLA-class-II transgenic mice, the microbiota of HLA-DQ8 mice were more similar to HLA-DR3.DQ8 than HLA-DR3. As the presence of DQ8 on an HLA-DR3 background increases disease severity, our data suggests that HLA-DQ8-specific microbiota may contribute to disease severity in HLA-DR3.DQ8 mice. Altogether, our study provides evidence that the HLA-DR and -DQ genes linked to specific gut microbiota contribute to EAE susceptibility or resistance in a transgenic animal model of MS.
Keywords: Experimental autoimmune encephalomyelitis (EAE), HLA transgenic mice, multiple sclerosis, gut microbiota, HLA polymorphisms
INTRODUCTION:
Multiple Sclerosis (MS) is a chronic, autoimmune inflammatory demyelinating disease of the central nervous system (CNS) resulting from aberrant CD4 T cell immune response to a number of myelin antigens, including proteolipid protein (PLP), and myelin oligodendrocyte glycoprotein (MOG) (1, 2). The etiology of MS is poorly understood but collective evidence suggests that the interaction of both genetic and environmental factors plays an important role (3). Furthermore, recent studies showed a greater consensus on the contribution of gene-environment (GxE) interactions in the pathogenesis of MS (4-6). Genetic factors account for approximately 30% of disease risk as determined from studies of identical twins (7). In addition, environmental factors account for 70% of disease risk (8). However, how genetic and environmental factors are linked with a predisposition to induce MS or protect from it is unknown.
Among all the genetic factors associated with MS susceptibility, the strongest association has been found with major histocompatibility complex (MHC) genes. MHC genes are among the most polymorphic genes in vertebrates (9), also known in humans as the human leukocyte antigen (HLA) genes. Previous studies have reported that individuals with HLA-DR2/HLA-DQ6, HLA-DR3/HLA-DQ2, and HLA-DR4/HLA-DQ8 class II haplotypes have increased frequency of MS (10, 11). Although a direct role of HLA-DR alleles in MS has been elucidated, the contribution of HLA-DQ alleles in disease pathogenesis is not known. Human studies have shown that HLA-DQ alleles may play a modulatory role in MS progression (12, 13). Utilizing single transgenic mice expressing HLA class II genes, we showed that while both HLA-DR3, and HLA-DQ8 were able to recognize and mount a CD4 T cell response to PLP91-110 only HLA-DR3 transgenic mice were susceptible to myelin proteolipid protein-PLP91-110 induced EAE, while HLA-DQ8 (DQB1*0302) transgenic mice were resistant (14, 15). The CD4 T cells from HLA-DR3/HLA-DQ8 double transgenic mice also recognized and proliferated to PLP91-110 peptide. However, the HLA-DR3/HLA-DQ8 double transgenic mice showed higher disease incidence and severity (14, 15) suggesting that the disease resistant HLA-DQ8 allele may synergize with HLA-DR3 to modulate the disease severity and progression. We have also shown that HLA-DQ8 induced IL-17 plays a role in modulating disease HLA-DR3.DQ8 transgenic mice. As gut microbiota has been shown to play a central role in development of IL-17 producing CD4 T cells (16), it is possible that HLA-DQ8 modulate disease through its influence on gut microbiota.
Although environmental factors account for 70% of MS risk, the identification of an exact environmental factor associated with MS remains unidentified. We, and others, have shown that the gut microbiota are an important enviromental factor linked with the etiopathogenesis of MS (17-22). The consensus of these studies is that MS patients exhibit gut dysbiosis, i.e., a change in the composition of the gut microbiota community characterized by an increase in harmful bacteria (pathobionts) and a decrease in beneficial bacteria. A healthy gut microbiota helps to maintain healthy state of the host by multiple ways, including food metabolism, maintenance of intestinal barrier, energy homeostasis, inhibition of colonization by pathogenic organisms, regulation of host physiology, and shaping of immune responses (23-26). Change in gut microbiota due to various important endogenous (genetic factors) and exogenous factors (diets, antibiotics other drugs, lifestyle, and smoking) result in alterations in its metabolites’ synthesis and perturbation of normal homeostasis, even leading to intestinal and systemic disorders (27-29).
Although prior studies have reported a role of MHC genes in gut microbiota dysbiosis (30-32), the importance of HLA-DQ8 gene in modulating HLA-DR3 microbiome has not been studied previously. In the present study, we investigated whether changes in HLA Class-II molecules from HLA-DQ8 to HLA-DR3 influences gut microbiota. Additionally, we ask whether the HLA-DQ8 gene can modulate gut microbiota of HLA-DR3 transgenic mice and modulate disease severity. We found that the gut microbiota of AE-KO mice differ from HLA-DR3, HLA-DQ8, and HLA-DR3.DQ8 double transgenic mice. We also further observedthat the gut microbiota of HLA-DR3 mice are different from that of HLA-DQ8. Furthermore, our results showed that though HLA-DQ8 confers resistance to EAE, its shared microbiota with HLA-DR3/HLA-DQ8 double transgenic mice increase disease severity. Therefore, our study, for the first time, demonstrates an important role of the HLA-DQ8 and HLA-DR3 genes in shaping gut microbiota and the HLA-DQ8 gene’s influence on the gut microbiota of HLA-DR3 transgenic mice.
Materials and Methods
Generation of Single and Double Transgenic Mice and Breeding Scheme
Major Histocompatibility Complex (MHC) Class II Genes Knockout (AE-KO) Mice
MHC class II genes knockout (AE-KO) mice were originally generated by Mathis and Benoist (33). Briefly, AE-KO mice lack all four conventional MHC class II genes (Aα, Aβ, Eα and Eβ) due to a large (80 kb) deletion of the entire mouse class II region. Embryonic stem cells with deleted MHC-II locus were injected into C57BL/6 blastocysts following standard procedures (34). Chimeras were then crossed with C57BL/6 mice to generate AE-KO mice which were then interbred to generate homozygous AE-KO mice and maintained in homozygous condition for prolonged period by interbreeding.
HLA-DQ8. AE-KO Transgenic Mice
Generation of DQ8 transgenic mice on B10 background was achieved as follows: Briefly, cosmids H11A (30-kb DNA) and X10A (38-kb DNA fragment), which contain the DQA*0301 and DQB*0302 genes, respectively microinjected into (CBA/J. B10.M) F 2 embryos, as previously described (35). Transgene-positive founders were identified by Southern blot analysis of tail DNA and subsequently mated to B10.M mice. The HLA-DQ8 transgenes were crossed with AE-KO mice to generate HLA-DQ8.AE-KO transgenic mice (36). Animals from F1 cross were genotyped by PCR to select mice positive for DQ8 genes and negative for endogenous MHC-class-II genes. Mice with correct genotype were inter-crossed and maintained in homozygous condition by prolonged interbreeding.
AE-KO.HLA-DR3 Transgenic Mice:
Generation of transgenic mice expressing HLA-DR3 (DRB1*0301) (gift from Dr. Gunter Hammerling, Germany) has been described previously (37). Briefly, 6 kilobase (kb) Nde I fragment of a HLA-DRA genomic construct and a 24 kb CLa I x Sal I fragment containing the HLA-DRB gene of DRBI*0301 were co-injection into fertilized eggs from (C57B1/6 x DBA/2) F1 donors mated with C57BL/6j males, as described previously (37). Transgene-positive founders were identified by Southern blot analysis of tail DNA and subsequently mated to B10.M mice. The HLA-DQ8 transgenes were crossed with AE-KO mice to generate HLA-DR3.AE-KO transgenic mice. Mice from F1 cross were genotyped by PCR to select for mice positive for DR3 genes and negative for endogenous MHC-class-II genes. Mice with correct genotype were inter-crossed and maintained in homozygous condition by interbreeding.
DR3.DQ8 transgenic mice
The homozygous HLA-DQ8.AE-KO mice were mated with the homozygous HLA.DR3.AE-KO mice to obtain the HLA-DR3.DQ8.AE-KO double transgenic mice lines (14). All the HLA-DR3.DQ8.AE-KO mice used in the study were from F1 cross between HLA-DR3 and HLA-DQ8 transgenic mice. Double transgenic mice were genotyped to select pups positive for both HLA-DR3 and HLA-DQ8 genes.
HLA- transgenic as well as control AE-KO mice strains were maintained in the animal facility at the University of Iowa. Eight to 12-week-old male and female mice from HLA class-II trasnegnic strains or AE-KO control were utilized in this study. For the simplicity these mice strains were referred as HLA-DR3, HLA-DQ8, HLA-DR3.DQ8, and AE-KO mice througout the manuscript. All experiments were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Iowa and animals were maintained in accordance with NIH and institutional guidelines.
Microbiome Analysis
Mouse fecal samples were collected from naïve HLA-DR3, HLA-DQ8, HLA-DR3.DQ8 transgenic mice and AE-KO mice groups. Microbial DNA extraction, 16S amplicon (V3-V4 region), and sequencing were done as described previously (38). Raw 16S sequence data was processed by R script dada2 to generate amplicon sequence variants (ASVs) which were then assigned taxonomies using a naive Bayesian classifier with the Silva database as a reference. Functional profiling of these bacterial communities was then performed using PICRUSt2 (39) to generate pathway abundance tables for the samples.
Analysis of these data were then conducted using online microbiome tools METAGENassist (40) and MicrobiomeAnalyst (41), as well as in-house scripts. MicrobiomeAnalyst was used to generate relative abundance bar graphs as well as to perform LEfSe analysis. METAGENassist was used for ordination of the data and generation of principal components for analysis (PCA). Differential abundance analysis for taxonomic and pathway data was performed using non-parametric tests (Wilcoxon signed-rank test for 2 groups and Kruskal-Wallis test for ≥3 groups) and FDR-adjusted for multiple comparisons using the Benjamini-Hochberg algorithm.
Disease Induction and Scoring
HLA-DR3, and HLA-DR3.DQ8 transgenic mice (8 to 12 weeks old) were immunized subcutaneously in both flanks using 25 μg of PLP91-110 peptides (GenScript, NJ, USA) that was emulsified in CFA containing Mycobacterium tuberculosis H37Ra (100 μg/mouse; Becton, Dickinson and Company, Sparks, MD, USA). Pertussis toxin (PTX) (Sigma Chemicals, St. Louis, MO; 80 ng) was administered intraperitoneally (i.p.) at days 0 and 2 post immunization. Mice were observed daily for clinical disease up to day 19. Disease severity was scored according to the standard 0–5 scoring system described previously (42). Briefly, 0 score for normal; 1, loss of tail tonicity; 2, hind limb weakness; 3, hind limb paralysis; 4, complete hind limb paralysis and forelimb paralysis or weakness; 5, moribund/death. Disease resistance phenotype in AE-KO and HLA-DQ8 transgenic mice have been previously characterized (14, 15).
Results
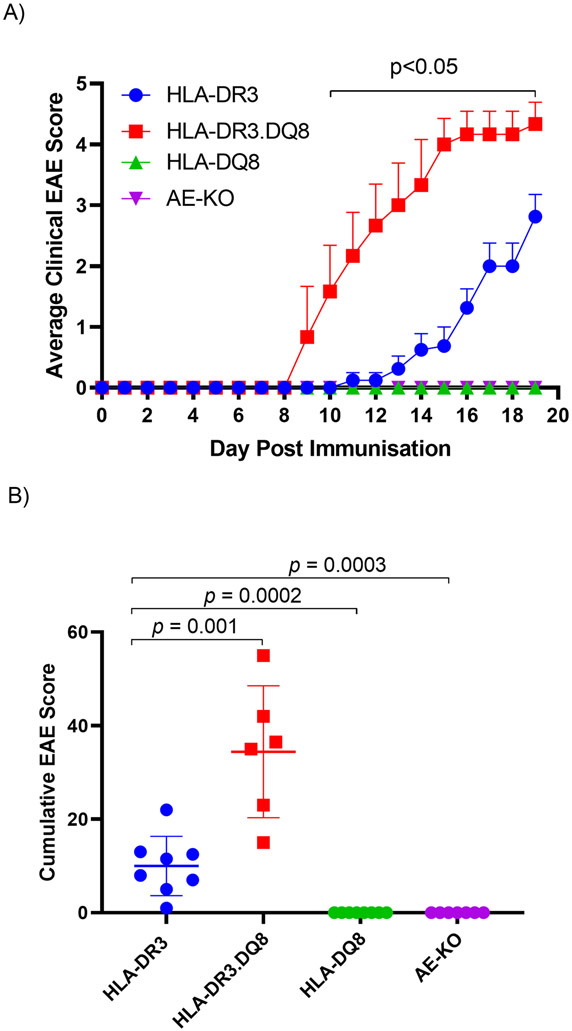
To analyze the effect of HLA polymorphism on EAE, we immunized AE-KO, HLA-DR3, HLA-DQ8 and HLA-DR3.DQ8 mice with PLP91-110 peptide (43). The HLA-DR3 mice began showing clinical signs of EAE disease on day 11 post-immunization, with a steadily increasing average EAE score that reached around 2.8±0.4 by day 19 (Fig 1A). In contrast, no disease was observed in mice either lacking endogenous MHC class-II (AE-KO) or expressing HLA-DQ8 (HLA-DQ8). However, the HLA-DR3.DQ8 double transgenic mice began showing symptoms at day 9 post-immunization and disease severity worsened quickly, reaching around 4.3±0.4 by day 19 (Fig 1A). Figure 1B further demonstrates the significant severity of disease, showing a mean cumulative EAE score over three times higher in the HLA-DR3.DQ8 group compared with the HLA-DR3 group.
Figure 1. PLP91-110-induced EAE in AE-KO, HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 transgenic mice.
A) Daily average EAE scores of AE-KO, HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 mice 19 days after immunization. P-values determined by multiple t tests B) Averaged cumulative EAE score for both groups of mice across all 19 days. P-value determined by Mann-Whitney unpaired U test (B).
To characterize the relationship between the HLA polymorphisms and microbiome composition, DNA was extracted from fecal samples of AE-KO (n=16; 8 male, 8 female), HLA-DQ8 (n=12; 5 male, 7 female), HLA-DR3 (n=18; 9 male, 9 female), and HLA-DR3.DQ8 (n=15; 7 male, 8 female) mice. The V3-V4 region of 16S ribosomal RNA was then amplified, sequenced, and the sequence data was processed through the open-access, R-based dada2 pipeline to perform sequence quality checks, trimming, merging of paired reads, alignment, and taxonomy assignment of the aligned sequences. The resultant median read depth was 38085, ranging from 5,146 to 99,124.
Distinct Gut Microbiota among AE-KO and HLA-class II Transgenic Mice
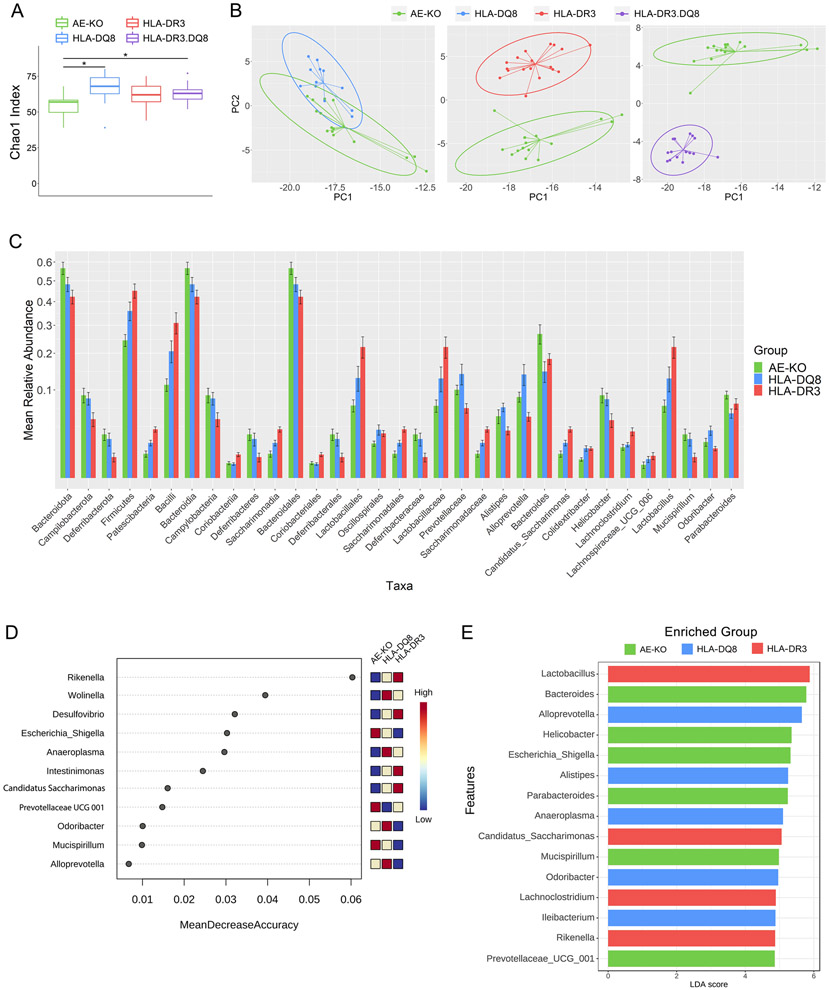
Alpha-diversity analysis for genus richness (Chao1 index) showed significantly decreased richness in AE-KO mice versus the other three groups (p-values < 0.03) (Fig 2A). There was no significant difference in alpha diversity among HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8. Euclidean distance-based beta-diversity analysis at the genus level between AE-KO and each of the other groups demonstrated a clear separation between AE-KO and both HLA-DR3 and HLA-DR3.DQ8 (Fig 2B) while AE-KO and HLA-DQ8 exhibited some compositional similarity.
Figure 2. Distinct gut microbiota among AE-KO and HLA transgenic mice.
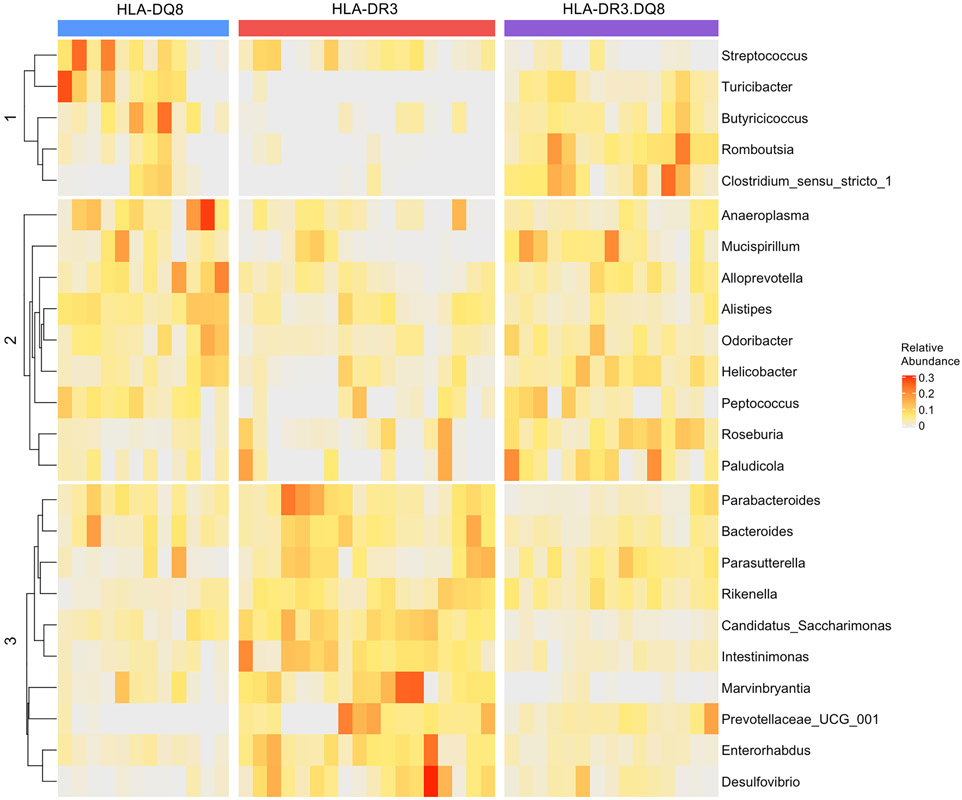
A) Boxplot showing unfiltered genus richness (Chao1 index) of AE-KO, HLA-DQ8, HLA-DR3, and HLA-DR3·DQ8 groups. Overall p-value: 0.0049. B) Pairwise principal coordinate analysis of beta-diversity using Euclidean distance at the genus level. Distinct separation between AE-KO and both HLA-DR3 and HLA-DR3·DQ8 while there is overlap between AE-KO and HLA-DQ8. PC1 and PC2 are the first and second principal components, respectively, and represent the most and second-most amount of variation in the bacterial abundance data between all the samples. C) Square root scaled bar plots of relative abundances of significant taxa at each taxonomic level with relative abundance >0.002. D) Top 15 important features selected by random forest. Removing Rikenella from the feature-set available to the model led to approximately a 16% decrease in classification accuracy. Removing Desulfovibrio led to around a 10% decrease in accuracy. E) Top 15 significant genera selected by LEfSe analysis, LDA score reflecting their effect sizes, and the heatmap on the right depicting whether they were high, medium, or low in AE-KO, HLA-DQ8, and HLA-DR3 groups from left to right.
A Kruskal-Wallis test was performed at all taxonomic levels from phylum to genus, restricted to taxa with relative abundance >0.002. At a false-discovery rate of 5%, 33 differentially abundant taxa were identified (Fig 2C and Table 1). AE-KO mice had a higher overall abundance of Bacteroidota (aka Bacteroidetes; the Silva database uses the Genome Taxonomy Database naming convention) when compared with HLA-DQ8 and HLA-DR3 mice. The AE-KO group also exhibited higher abundance of the phyla Campilobacterota and Deferribacterota when compared with HLA-DR3 but there was no difference when compared with HLA-DQ8. The Patescibacteria phylum was found in lower abundance in AE-KO mice when compared with HLA-DR3 mice, but not with HLA-DQ8 mice (Table 1).
Table 1:
Significant taxa at between AE-KO, HLA-DQ8, and HLA-DR3 at the phylum through genus levels
| Taxa | Mean (Standard Error) Relative Abundance | P-value | Q-value | ||
|---|---|---|---|---|---|
| AE-KO | HLA-DQ8 | HLA-DR3 | |||
| Phylum | |||||
| Bacteroidota (Bacteroides) | 5.66E-1 (3.32E-2) | 4.82E-1 (3.53E-2) | 4.22E-1 (3.21E-2) | 1.21E-02 | 1.93E-02 |
| Campilobacterota | 8.82E-2 (1.55E-2) | 8.15E-2 (1.29E-2) | 4.45E-2 (1.05E-2) | 3.45E-02 | 4.59E-02 |
| Deferribacterota | 2.45E-2 (6.33E-3) | 1.96E-2 (6.40E-3) | 5.65E-3 (2.33E-3) | 1.31E-03 | 2.62E-03 |
| Desulfobacterota | 1.83E-3 (4.62E-4) | 1.32E-3 (4.10E-4) | 9.07E-3 (2.39E-3) | 1.94E-04 | 6.17E-04 |
| Firmicutes | 2.44E-1 (2.12E-2) | 3.59E-1 (3.99E-2) | 4.50E-1 (3.43E-2) | 2.31E-04 | 6.17E-04 |
| Patescibacteria | 7.58E-3 (1.93E-3) | 1.59E-2 (2.55E-3) | 3.06E-2 (3.38E-3) | 2.07E-05 | 1.66E-04 |
| Proteobacteria | 6.11E-2 (1.15E-2) | 3.03E-2 (8.53E-3) | 2.46E-2 (4.55E-3) | 4.76E-02 | 5.44E-02 |
| Class | |||||
| Bacilli | 1.12E-1 (1.50E-2) | 2.07E-1 (3.55E-2) | 3.09E-1 (4.19E-2) | 1.07E-03 | 3.26E-03 |
| Bacteroidia | 5.66E-1 (3.32E-2) | 4.82E-1 (3.53E-2) | 4.22E-1 (3.21E-2) | 1.21E-02 | 2.01E-02 |
| Campylobacteria | 8.82E-2 (1.55E-2) | 8.15E-2 (1.29E-2) | 4.45E-2 (1.05E-2) | 3.45E-02 | 4.92E-02 |
| Coriobacteriia | 2.90E-3 (6.10E-4) | 2.57E-3 (4.55E-4) | 7.15E-3 (1.23E-3) | 1.63E-03 | 3.26E-03 |
| Deferribacteres | 2.45E-2 (6.33E-3) | 1.96E-2 (6.40E-3) | 5.65E-3 (2.33E-3) | 1.31E-03 | 3.26E-03 |
| Desulfovibrionia | 1.83E-3 (4.62E-4) | 1.32E-3 (4.10E-4) | 9.07E-3 (2.39E-3) | 1.94E-04 | 9.71E-04 |
| Gammaproteobacteria | 6.11E-2 (1.15E-2) | 3.03E-2 (8.53E-3) | 2.46E-2 (4.55E-3) | 4.76E-02 | 5.95E-02 |
| Saccharimonadia | 7.58E-3 (1.93E-3) | 1.59E-2 (2.55E-3) | 3.06E-2 (3.38E-3) | 2.07E-05 | 2.07E-04 |
| Order | |||||
| Acholeplasmatales | 4.47E-4 (4.47E-4) | 2.54E-2 (5.61E-3) | 8.05E-3 (2.20E-3) | 2.22E-07 | 4.00E-06 |
| Bacteroidales | 5.66E-1 (3.32E-2) | 4.82E-1 (3.53E-2) | 4.22E-1 (3.21E-2) | 1.21E-02 | 2.17E-02 |
| Clostridiales | 2.86E-3 (6.17E-4) | 1.05E-3 (4.84E-4) | 2.09E-3 (7.96E-4) | 2.54E-02 | 4.15E-02 |
| Coriobacterales | 2.90E-3 (6.10E-4) | 2.57E-3 (4.55E-4) | 7.15E-3 (1.23E-3) | 1.63E-03 | 4.19E-03 |
| Deferribacterales | 2.45E-2 (6.33E-3) | 1.96E-2 (6.40E-3) | 5.65E-3 (2.33E-3) | 1.31E-03 | 3.92E-03 |
| Desulfovibrionales | 1.83E-3 (4.62E-4) | 1.32E-3 (4.10E-4) | 9.07E-3 (2.39E-3) | 1.94E-04 | 8.74E-04 |
| Enterobacterales | 4.17E-2 (9.63E-3) | 1.40E-2 (7.07E-3) | 1.13E-4 (4.85E-5) | 6.41E-07 | 5.77E-06 |
| Lactobacillales | 6.74E-2 (1.13E-2) | 1.29E-1 (3.17E-2) | 2.21E-1 (3.60E-2) | 1.24E-03 | 3.92E-03 |
| Oscillospirales | 1.53E-2 (2.30E-3) | 3.00E-2 (6.35E-3) | 2.56E-2 (3.53E-3) | 1.02E-02 | 2.17E-02 |
| Peptococcales | 3.12E-4 (9.98E-5) | 3.25E-4 (6.94E-5) | 1.12E-4 (4.46E-5) | 1.09E-02 | 2.17E-02 |
| Saccharimonadales | 7.58E-3 (1.93E-3) | 1.59E-2 (2.55E-3) | 3.06E-2 (3.38E-3) | 2.07E-05 | 1.24E-04 |
| Family | |||||
| Acholeplasmataceae | 4.47E-4 (4.47E-4) | 2.54E-2 (5.61E-3) | 8.05E-3 (2.20E-3) | 2.22E-07 | 6.22E-06 |
| Deferribacteraceae | 2.45E-2 (6.33E-3) | 1.96E-2 (6.40E-3) | 5.65E-3 (2.33E-3) | 1.31E-03 | 5.23E-03 |
| Desulfovibrionaceae | 1.83E-3 (4.62E-4) | 1.32E-3 (4.10E-4) | 9.07E-3 (2.39E-3) | 1.94E-04 | 1.09E-03 |
| Eggerthellaceae | 1.86E-3 (3.42E-4) | 2.28E-3 (3.94E-4) | 6.94E-3 (1.17E-3) | 1.16E-04 | 8.09E-04 |
| Enterobacteriaceae | 4.17E-2 (9.63E-3) | 1.40E-2 (7.07E-3) | 1.13E-4 (4.85E-5) | 6.41E-07 | 8.97E-06 |
| Lactobacillaceae | 6.70E-2 (1.13E-2) | 1.27E-1 (3.17E-2) | 2.20E-1 (3.59E-2) | 1.24E-03 | 5.23E-03 |
| Peptococcaceae | 3.12E-4 (9.98E-5) | 3.25E-4 (6.94E-5) | 1.12E-4 (4.46E-5) | 1.09E-02 | 3.38E-02 |
| Prevotellaceae | 1.00E-1 (1.08E-2) | 1.39E-1 (2.76E-2) | 6.28E-2 (8.58E-3) | 9.70E-03 | 3.38E-02 |
| Saccharimonadaceae | 7.58E-3 (1.93E-3) | 1.59E-2 (2.55E-3) | 3.06E-2 (3.38E-3) | 2.07E-05 | 1.94E-04 |
| Genus | |||||
| Lachnospiraceae:Acetatifactor | 5.71E-5 (4.48E-5) | 1.73E-3 (5.41E-4) | 1.41E-3 (6.47E-4) | 1.65E-04 | 8.27E-04 |
| Rikenellaceae;Alistipes | 4.89E-2 (1.09E-2) | 6.44E-2 (7.74E-3) | 2.89E-2 (4.92E-3) | 1.50E-02 | 3.29E-02 |
| Prevotellaceae;Alloprevotella | 8.45E-2 (1.04E-2) | 1.38E-1 (2.77E-2) | 4.85E-2 (7.01E-3) | 2.13E-03 | 6.88E-03 |
| Bacteroidetes;Bacteroides | 2.67E-1 (3.56E-2) | 1.46E-1 (2.82E-2) | 1.83E-1 (1.67E-2) | 2.84E-02 | 4.88E-02 |
| Proteobacteria;Bilophila | 1.77E-3 (4.45E-4) | 2.96E-4 (1.00E-4) | 3.28E-4 (1.38E-4) | 2.83E-03 | 8.19E-03 |
| Clostridiaceae;Candidatus Arthromitus | 2.86E-3 (6.17E-4) | 1.05E-3 (4.84E-4) | 2.09E-3 (7.96E-4) | 2.54E-02 | 4.88E-02 |
| Eubacteriales;Colidextribacter | 4.47E-3 (8.08E-4) | 1.14E-2 (2.07E-3) | 1.12E-2 (1.42E-3) | 2.40E-04 | 1.10E-03 |
| Proteobacteria;Desulfovibrio | 6.95E-5 (3.34E-5) | 1.03E-3 (3.63E-4) | 8.75E-3 (2.40E-3) | 6.33E-08 | 1.74E-06 |
| Proteobacteria;Escherichia | 4.17E-2 (9.63E-3) | 1.40E-2 (7.07E-3) | 1.13E-4 (4.85E-5) | 6.41E-07 | 8.81E-06 |
| Helicobacteraceae;Helicobacter | 8.82E-2 (1.55E-2) | 8.00E-2 (1.27E-2) | 4.31E-2 (1.02E-2) | 2.70E-02 | 4.88E-02 |
| Firmicutes;Ileibacterium | 7.95E-4 (4.61E-4) | 1.61E-2 (5.81E-3) | 1.17E-2 (4.10E-3) | 1.64E-03 | 5.63E-03 |
| Lachnospiraceae;Lachnoclostridium | 1.22E-2 (2.33E-3) | 1.43E-2 (1.86E-3) | 2.78E-2 (4.73E-3) | 2.77E-02 | 4.88E-02 |
| Lachnospiraceae;Lachnospiraceae UCG 006 | 2.26E-3 (1.06E-3) | 4.56E-3 (1.44E-3) | 6.38E-3 (2.01E-3) | 6.59E-03 | 1.65E-02 |
| Lactobacillaceae;Lactobacillus | 6.70E-2 (1.13E-2) | 1.27E-1 (3.17E-2) | 2.20E-1 (3.59E-2) | 1.24E-03 | 4.80E-03 |
| Deferribacteraceae;Mucispirillum | 2.45E-2 (6.33E-3) | 1.96E-2 (6.40E-3) | 5.65E-3 (2.33E-3) | 1.31E-03 | 4.80E-03 |
| Porphyromonadaceae;Odoribacter | 1.67E-2 (3.68E-3) | 2.93E-2 (5.67E-3) | 1.12E-2 (1.62E-3) | 2.67E-02 | 4.88E-02 |
| Bacteroidetes;Parabacteroides | 8.89E-2 (8.27E-3) | 5.38E-2 (7.64E-3) | 7.11E-2 (1.00E-2) | 2.57E-02 | 4.88E-02 |
| Bacteroidetes;Rikenella | 2.20E-4 (1.83E-4) | 6.49E-3 (9.47E-4) | 1.49E-2 (1.98E-3) | 1.82E-08 | 1.00E-06 |
| Proteobacteria;Wolinella | 1.29E-5 (8.96E-6) | 1.50E-3 (3.16E-4) | 1.41E-3 (3.54E-4) | 2.76E-06 | 2.53E-05 |
Kruskall-Wallis test with Benjamini-Hochberg adjustment was performed on sample-scaled counts at a significance level of 0.05 to identify differentially abundant taxa. Q-values are the adjusted P-values.
HLA Class II Polymorphism Influences Gut Microbiota Profile
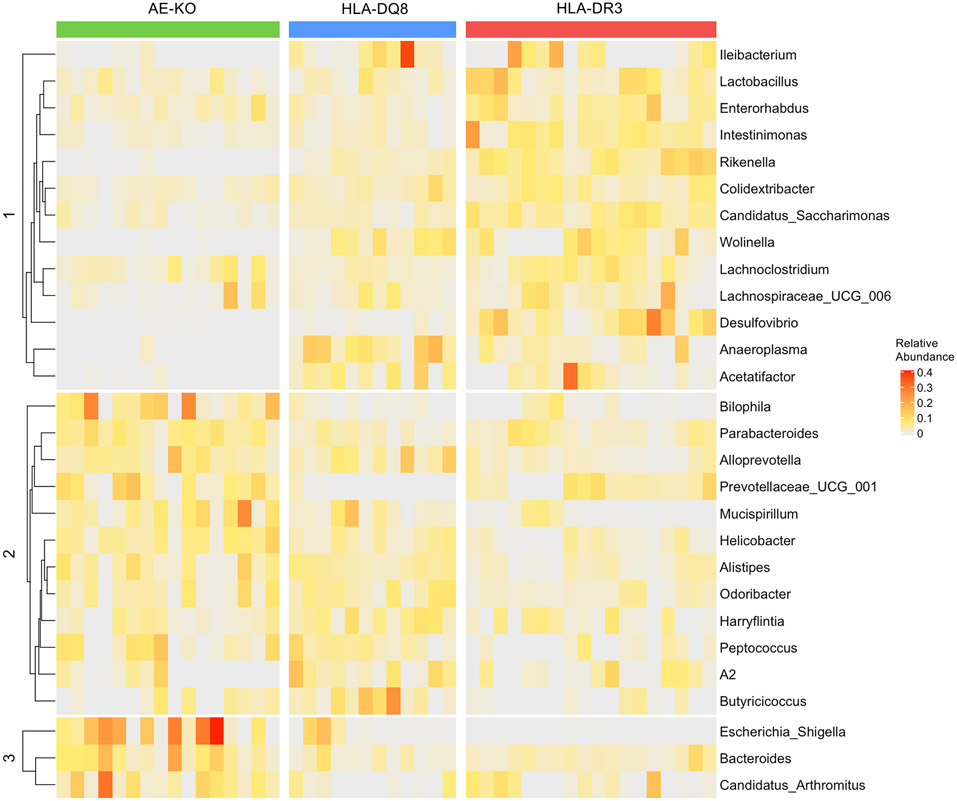
Numerous genera were depleted in AE-KO mice compared to both HLA-DQ8 and HLA-DR3, most of which belong to the Firmicutes phylum: Acetatifactor, Anaeroplasma, Ileibacterium, Colidextribacter, Intestinimonas, and Lachnospiraceae UCG-006. Additionally, Wolinella of the Campilobacterota phylum and Rikenella of the Bacteroidota (more often referred to as Bacteroidetes) phylum were depleted in AE-KO. In the Desulfobacterota phylum, while Desulfovibrio was depleted in AE-KO, Bilophila was significantly overabundant compared to the other two groups (Table 1 and Fig 3).
Figure 3. Differentially abundant bacteria in AE-KO, HLA-DQ8, and HLA-DR3 mice.
Colors indicate relative abundance increasing in value from gray to red. Bacteria were grouped into 3 groups based on k-means clustering.
While numerous bacteria were differentially abundant in AE-KO we wanted to identify specific bacteria that could best discriminate between AE-KO, HLA-DQ8, and HLA-DR3. To do this, we performed a random forest algorithm to classify the samples as either AE-KO, HLA-DQ8, or HLA-DR3 using the genus abundance data. Important bacteria were then identified based on how much the random forest classification accuracy decreased when that bacterium was removed from the feature-set (Fig 2D). Using this approach, Rikenella and Wolinella were identified as the top two important features in identifying the AE-KO genotype, with mean decrease accuracy of 6% and 4%, respectively.
In an effort to highlight bacteria most likely to have biological relevance in the differences between AE-KO, HLA-DQ8, and HLA-DR3 mice, linear discriminant analysis (LDA) effect size (LEfSe) was performed. The effect sizes, or how well the abundance of each bacteria separated the groups, of the 15 most significant genera are shown in Figure 2E. This analysis highlights Bacteroides, Parabacteroides, Helicobacter, and Escherichia as important features for distinguishing AE-KO from the other two groups. Each of these genera are overabundant in AE-KO, though Parabacteroides was only significantly overabundant in AE-KO compared to HLA-DQ8, while Helicobacter and Escherichia were only significantly overabundant compared to HLA-DR3.
Distinct Gut Microbiota in single vs double transgenic mice
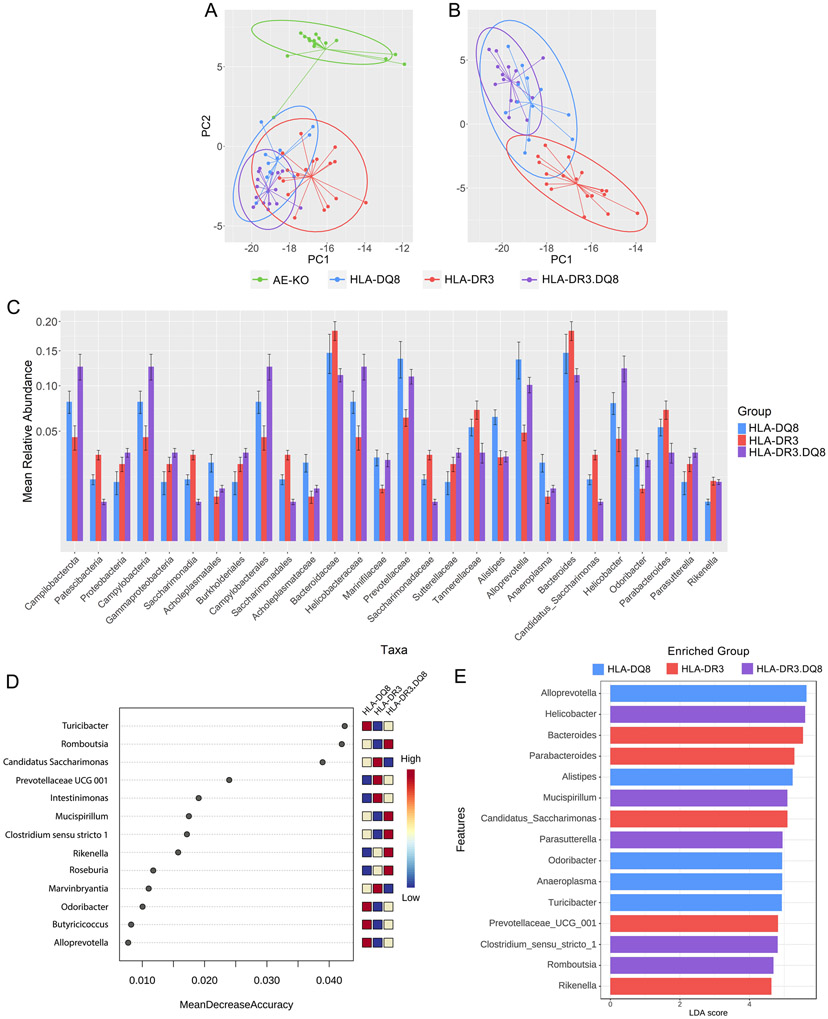
Next, microbial composition in HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 were characterized to highlight differences and similarities between single and double transgenic mice. First, beta-diversity analysis using Euclidean-based distance at the genus level was performed on all four groups, showing HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 clustering together, separate from AE-KO even when stratified by sex (Fig 4A and Supplemental Fig 1). Removing AE-KO from this analysis shows that HLA-DR3.DQ8 is compositionally distinct from HLA-DR3 but very similar to HLA-DQ8 (Fig 4B).
Figure 4. Distinct microbiota in single and double transgenic mice.
A) PCA plot comparing beta-diversity of all four groups at the genus level demonstrating separation of AE-KO from the other three groups. B) PCA plot comparing Euclidean distance-based beta-diversity of DQ8, DR3, and DR3.DQ8 at the genus level. C) Square root scaled bar plots of relative abundances of significant taxa at each taxonomic level with relative abundance >0.006. D) Top 15 important features selected by random forest. Removing Turicibacter from the feature-set available to the model leads to approximately an 11% decrease in accuracy. E) Top 15 significant genera selected by LEfSe analysis. LDA score reflects their effect sizes, and the heatmap on the right depicts whether they were high, medium, or low in HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 groups from left to right.
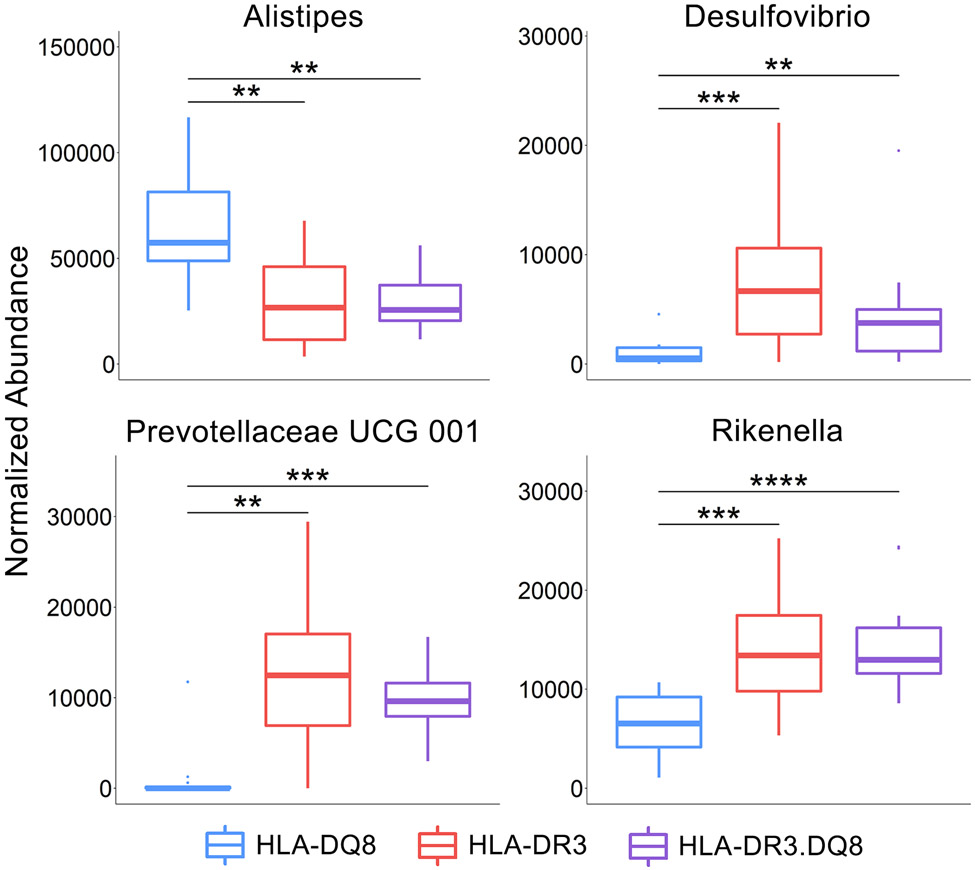
The Kruskal-Wallis test was performed to identify taxa that were differentially abundant between HLA-DQ8, HLA-DR3 and HLA-DR3.DQ8 groups at all taxonomic levels from phylum to genus. The p-values were adjusted using an FDR of 5%, and the 28 resulting taxa with relative abundance >0.006 are shown in Figure 4C. Within the Bacteroides phylum, the HLA-DQ8 group exhibited an overabundance of Alistipes but decreased abundance of Prevotellaceae UCG-001 at the genus level. The HLA-DQ8 group also had a higher abundance of Desulfovibrio (Desulfobacterota) and Rikenella (Bacteroidota) (Figs 4C, 5, & 6 and Table 2).
Figure 5. Differentially abundant bacteria among HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 mice.
Colors indicate relative abundance increasing in value from gray to red. Bacteria were grouped into 3 groups based on k-means clustering.
Figure 6. Differentially abundant bacteria in HLA-DQ8 mice.
Abundance values are Sum-scaled to one million. The three lines represent the lower quartile, median, and upper quartile from lowest to highest, respectively. Significance labels: **** indicates p-value < 0.001, *** indicates 0.01>p>0.001, ** indicates 0.05>p>0.01, and * indicates p<0.05.
Table 2:
Significant taxa at between HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 at the phylum through genus ranks
| Taxa | Mean (Standard Error) Relative Abundance | P-value | Q-value | ||
|---|---|---|---|---|---|
| HLA-DQ8 | HLA-DR3 | HLA-DR3.DQ8 | |||
| Phylum | |||||
| Campilobacterota | 8.04E-2 (1.28E-2) | 4.47E-2 (1.04E-2) | 1.26E-1 (1.87E-2) | 9.95E-04 | 1.99E-03 |
| Deferribacterota | 1.93E-2 (6.35E-3) | 5.65E-3 (2.32E-3) | 3.00E-2 (6.99E-3) | 4.32E-04 | 1.15E-03 |
| Desulfobacterota | 1.31E-3 (4.10E-4) | 9.15E-3 (2.39E-3) | 4.35E-3 (1.21E-3) | 3.91E-04 | 1.15E-03 |
| Patescibacteria | 1.57E-2 (2.54E-3) | 3.08E-2 (3.41E-3) | 6.35E-3 (8.80E-4) | 1.29E-06 | 1.03E-05 |
| Proteobacteria | 1.45E-2 (5.56E-3) | 2.46E-2 (4.57E-3) | 3.24E-2 (3.20E-3) | 5.95E-03 | 9.52E-03 |
| Class | |||||
| Campylobacteria | 8.04E-2 (1.28E-2) | 4.47E-2 (1.04E-2) | 1.26E-1 (1.87E-2) | 9.95E-04 | 1.99E-03 |
| Coriobacteriia | 2.22E-3 (3.69E-4) | 7.01E-3 (1.18E-3) | 2.25E-3 (3.47E-4) | 3.67E-04 | 1.08E-03 |
| Deferribacteres | 1.93E-2 (6.35E-3) | 5.65E-3 (2.32E-3) | 3.00E-2 (6.99E-3) | 4.32E-04 | 1.08E-03 |
| Desulfovibrionia | 1.31E-3 (4.10E-4) | 9.15E-3 (2.39E-3) | 4.35E-3 (1.21E-3) | 3.91E-04 | 1.08E-03 |
| Gammaproteobacteria | 1.45E-2 (5.56E-3) | 2.46E-2 (4.57E-3) | 3.24E-2 (3.20E-3) | 5.95E-03 | 9.92E-03 |
| Saccharimonadia | 1.57E-2 (2.54E-3) | 3.08E-2 (3.41E-3) | 6.35E-3 (8.80E-4) | 1.29E-06 | 1.29E-05 |
| Order | |||||
| Acholeplasmatales | 2.55E-2 (5.69E-3) | 8.12E-3 (2.21E-3) | 1.14E-2 (1.46E-3) | 5.24E-03 | 1.11E-02 |
| Burkholderiales | 1.45E-2 (5.56E-3) | 2.46E-2 (4.57E-3) | 3.24E-2 (3.20E-3) | 5.95E-03 | 1.12E-02 |
| Campylobacterales | 8.04E-2 (1.28E-2) | 4.47E-2 (1.04E-2) | 1.26E-1 (1.87E-2) | 9.95E-04 | 2.42E-03 |
| Clostridiales | 4.99E-3 (1.94E-3) | 2.50E-3 (8.39E-4) | 1.81E-2 (3.53E-3) | 5.47E-05 | 3.10E-04 |
| Coriobacteriales | 2.22E-3 (3.69E-4) | 7.01E-3 (1.18E-3) | 2.25E-3 (3.47E-4) | 3.67E-04 | 1.22E-03 |
| Deferribacterales | 1.93E-2 (6.35E-3) | 5.65E-3 (2.32E-3) | 3.00E-2 (6.99E-3) | 4.32E-04 | 1.22E-03 |
| Desulfovibrionales | 1.31E-3 (4.10E-4) | 9.15E-3 (2.39E-3) | 4.35E-3 (1.21E-3) | 3.91E-04 | 1.22E-03 |
| Peptococcales | 3.19E-4 (6.51E-5) | 1.13E-4 (4.46E-5) | 2.95E-4 (8.24E-5) | 8.71E-03 | 1.48E-02 |
| Peptostreptococcales_Tissierellales | 3.40E-3 (1.22E-3) | 7.74E-4 (1.68E-4) | 1.02E-2 (2.45E-3) | 3.47E-06 | 2.95E-05 |
| Saccharimonadales | 1.57E-2 (2.54E-3) | 3.08E-2 (3.41E-3) | 6.35E-3 (8.80E-4) | 1.29E-06 | 2.19E-05 |
| Family | |||||
| Acholeplasmataceae | 2.55E-2 (5.69E-3) | 8.12E-3 (2.21E-3) | 1.14E-2 (1.46E-3) | 5.24E-03 | 1.28E-02 |
| Bacteroidaceae | 1.47E-1 (3.04E-2) | 1.83E-1 (1.65E-2) | 1.14E-1 (9.09E-3) | 1.85E-02 | 3.24E-02 |
| Butyricicoccaceae | 2.33E-3 (6.39E-4) | 1.37E-3 (3.02E-4) | 3.24E-3 (4.03E-4) | 5.97E-03 | 1.28E-02 |
| Clostridiaceae | 4.99E-3 (1.94E-3) | 2.50E-3 (8.39E-4) | 1.81E-2 (3.53E-3) | 5.47E-05 | 5.10E-04 |
| Deferribacteraceae | 1.93E-2 (6.35E-3) | 5.65E-3 (2.32E-3) | 3.00E-2 (6.99E-3) | 4.32E-04 | 2.01E-03 |
| Desulfovibrionaceae | 1.31E-3 (4.10E-4) | 9.15E-3 (2.39E-3) | 4.35E-3 (1.21E-3) | 3.91E-04 | 2.01E-03 |
| Eggerthellaceae | 2.22E-3 (3.69E-4) | 7.01E-3 (1.18E-3) | 2.25E-3 (3.47E-4) | 3.67E-04 | 2.01E-03 |
| Helicobacteraceae | 8.04E-2 (1.28E-2) | 4.47E-2 (1.04E-2) | 1.26E-1 (1.87E-2) | 9.95E-04 | 3.48E-03 |
| Marinifilaceae | 2.89E-2 (5.44E-3) | 1.13E-2 (1.66E-3) | 2.73E-2 (4.44E-3) | 8.73E-04 | 3.48E-03 |
| Peptococcaceae | 3.19E-4 (6.51E-5) | 1.13E-4 (4.46E-5) | 2.95E-4 (8.24E-5) | 8.71E-03 | 1.63E-02 |
| Peptostreptococcaceae | 3.20E-3 (1.21E-3) | 3.28E-4 (1.46E-4) | 1.00E-2 (2.44E-3) | 6.94E-07 | 1.80E-05 |
| Prevotellaceae | 1.38E-1 (2.75E-2) | 6.32E-2 (8.57E-3) | 1.12E-1 (9.97E-3) | 2.58E-03 | 8.02E-03 |
| Saccharimonadaceae | 1.57E-2 (2.54E-3) | 3.08E-2 (3.41E-3) | 6.35E-3 (8.80E-4) | 1.29E-06 | 1.80E-05 |
| Streptococcaceae | 1.25E-3 (4.37E-4) | 4.93E-4 (9.65E-5) | 1.72E-4 (5.96E-5) | 6.39E-03 | 1.28E-02 |
| Sutterellaceae | 1.45E-2 (5.56E-3) | 2.46E-2 (4.57E-3) | 3.24E-2 (3.20E-3) | 5.95E-03 | 1.28E-02 |
| Tannerellaceae | 5.37E-2 (7.87E-3) | 7.14E-2 (9.97E-3) | 3.24E-2 (7.21E-3) | 4.83E-03 | 1.28E-02 |
| Genus | |||||
| Rikenellaceae;Alistipes | 6.39E-2 (7.60E-3) | 2.91E-2 (4.90E-3) | 2.96E-2 (3.61E-3) | 1.04E-03 | 3.29E-03 |
| Prevotellaceae;Alloprevotella | 1.37E-1 (2.76E-2) | 4.88E-2 (7.02E-3) | 1.01E-1 (1.01E-2) | 2.88E-04 | 1.83E-03 |
| Anaeroplasmataceae;Anaeroplasma | 2.55E-2 (5.69E-3) | 8.12E-3 (2.21E-3) | 1.14E-2 (1.46E-3) | 5.24E-03 | 1.30E-02 |
| Bacteroidetes;Bacteroides | 1.47E-1 (3.04E-2) | 1.83E-1 (1.65E-2) | 1.14E-1 (9.09E-3) | 1.85E-02 | 3.77E-02 |
| Clostridiaceae;Butyricicoccus | 1.63E-3 (6.27E-4) | 2.05E-4 (8.03E-5) | 1.21E-3 (2.59E-4) | 6.34E-05 | 5.17E-04 |
| Candidatus Saccharimonas | 1.57E-2 (2.54E-3) | 3.08E-2 (3.41E-3) | 6.35E-3 (8.80E-4) | 1.29E-06 | 3.67E-05 |
| Clostridiaceae;Clostridium sensu stricto 1 | 3.96E-3 (1.96E-3) | 3.87E-4 (3.21E-4) | 1.33E-2 (3.48E-3) | 1.05E-05 | 1.50E-04 |
| Proteobacteria;Desulfovibrio | 1.02E-3 (3.64E-4) | 8.82E-3 (2.41E-3) | 4.20E-3 (1.23E-3) | 3.22E-04 | 1.84E-03 |
| Eggerthellaceae;Enterorhabdus | 1.44E-3 (2.58E-4) | 3.71E-3 (7.55E-4) | 1.57E-3 (2.20E-4) | 3.79E-03 | 1.03E-02 |
| Helicobacteraceae;Helicobacter | 7.90E-2 (1.25E-2) | 4.33E-2 (1.02E-2) | 1.24E-1 (1.84E-2) | 1.04E-03 | 3.29E-03 |
| Eubacteriales;Intestinimonas | 8.09E-4 (1.34E-4) | 2.27E-3 (3.48E-4) | 8.77E-4 (9.55E-5) | 1.80E-03 | 5.39E-03 |
| Lachnospiraceae;Marvinbryantia | 2.69E-3 (8.47E-4) | 6.32E-3 (1.55E-3) | 6.30E-4 (2.91E-4) | 1.75E-05 | 2.00E-04 |
| Deferribacteraceae;Mucispirillum | 1.93E-2 (6.35E-3) | 5.65E-3 (2.32E-3) | 3.00E-2 (6.99E-3) | 4.32E-04 | 2.06E-03 |
| Porphyromonadaceae;Odoribacter | 2.89E-2 (5.44E-3) | 1.13E-2 (1.66E-3) | 2.73E-2 (4.44E-3) | 8.73E-04 | 3.11E-03 |
| Oscillospiraceae;Paludicola | 1.04E-4 (2.56E-5) | 1.21E-4 (5.75E-5) | 3.00E-4 (7.30E-5) | 1.40E-02 | 2.96E-02 |
| Bacteroidetes;Parabacteroides | 5.37E-2 (7.87E-3) | 7.14E-2 (9.97E-3) | 3.24E-2 (7.21E-3) | 4.83E-03 | 1.25E-02 |
| Sutterellaceae;Parasutterella | 1.45E-2 (5.56E-3) | 2.46E-2 (4.57E-3) | 3.24E-2 (3.20E-3) | 5.95E-03 | 1.41E-02 |
| Peptococcaceae;Peptococcus | 3.19E-4 (6.51E-5) | 1.13E-4 (4.46E-5) | 2.95E-4 (8.24E-5) | 8.71E-03 | 1.91E-02 |
| Prevotellaceae UCG 1 | 1.14E-3 (9.72E-4) | 1.44E-2 (3.02E-3) | 1.11E-2 (2.07E-3) | 4.46E-04 | 2.06E-03 |
| Bacteroidetes;Rikenella | 6.41E-3 (9.18E-4) | 1.50E-2 (1.97E-3) | 1.45E-2 (1.21E-3) | 1.18E-04 | 8.39E-04 |
| Peptostreptococcaceae;Romboutsia | 3.20E-3 (1.21E-3) | 3.28E-4 (1.46E-4) | 1.00E-2 (2.44E-3) | 6.94E-07 | 3.67E-05 |
| Lachnospiraceae;Roseburia | 2.55E-3 (5.37E-4) | 4.21E-3 (1.60E-3) | 9.73E-3 (1.56E-3) | 4.69E-04 | 2.06E-03 |
| Streptococcaceae;Streptococcus | 1.25E-3 (4.37E-4) | 4.93E-4 (9.65E-5) | 1.72E-4 (5.96E-5) | 6.39E-03 | 1.46E-02 |
| Erysipelotrichaceae;Turicibacter | 1.73E-2 (6.46E-3) | 2.00E-4 (2.00E-4) | 9.91E-3 (2.25E-3) | 2.13E-06 | 4.05E-05 |
Kruskall-Wallis test with Benjamini-Hochberg adjustment was performed on sample-scaled counts at a significance level of 0.05 to identify differentially abundant taxa. Q-values are the adjusted P-values.
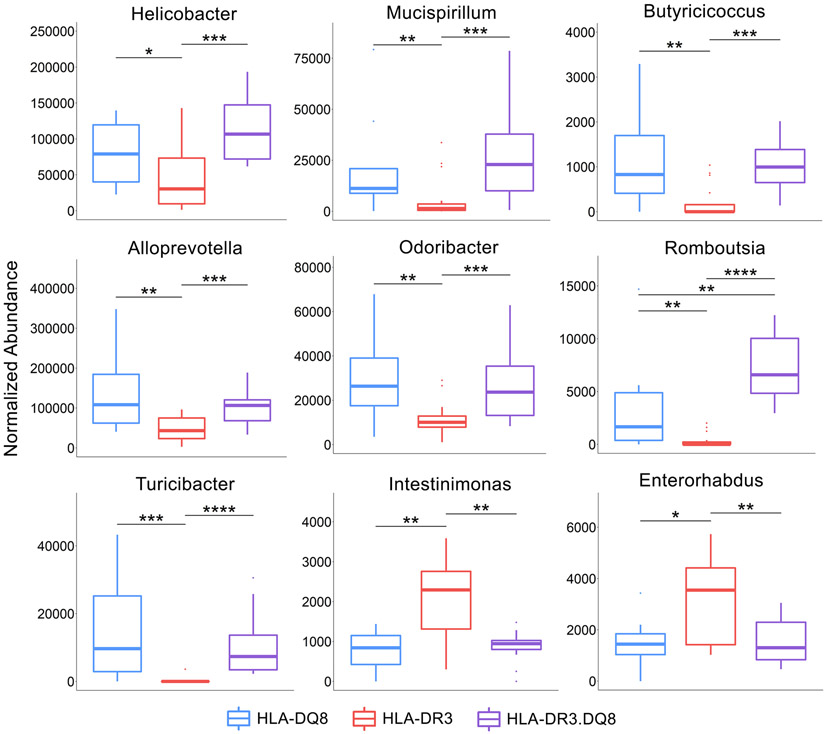
The phyla Campilobacterota and Proteobacteria were significantly underrepresented in HLA-DR3 while Patescibacteria were overabundant. Within the Proteobacteria phylum, Helicobacter was depleted in HLA-DR3 compared to the other two groups. Several genera within the Firmicutes class were depleted in HLA-DR3 – Butyricicoccus, Romboutsia, and Turicibacter – while Intestinimonas were overabundant. Within the Bacteroidota phylum, both Alloprevotella and Odoribacter were significantly depleted. Additionally, Mucispirillum of the Deferribacterota phylum was significantly depleted. Other than Intestinimonas, the other genus significantly overabundant in HLA-DR3 was Enterorhabdus of the Actinobacteriota phylum (Figs 4C, 5, & 7 and Table 2).
Figure 7. Differentially abundant bacteria in HLA-DR3.
Abundance values are sum-scaled to one million. The three lines represent the lower quartile, median, and upper quartile from lowest to highest, respectively. Significance labels: **** indicates p-value < 0.001, *** indicates 0.01>p>0.001, ** indicates 0.05>p>0.01, and * indicates p<0.05.
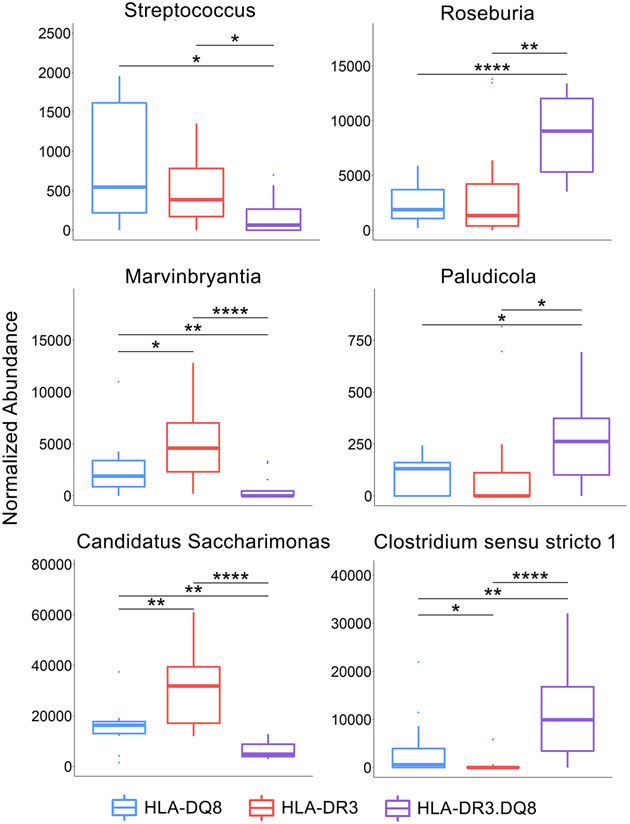
Interestingly, at the phylum level the HLA-DR3.DQ8 group demonstrated significant overabundance of Campilobacterota, Campylobacteria, and Proteobacteria, and depletion of the phylum Patescibacteria, mirroring the changes opposite to those observed in the HLA-DR3 group. At the genus level however, most of the significant genera belong to the Firmicutes phylum. Within this phylum, Streptococcus and Marvinbryantia are depleted in HLA-DR3.DQ8 compared to the other two groups, while Roseburia, Paludicola, and Clostridium sensu stricto 1 were overabundant (Figs 4C, 5, & 8 and Table 2).
Figure 8. Differentially abundant bacteria in HLA-DR3.DQ8.
Abundance values are sum-scaled to one million. The three lines represent the lower quartile, median, and upper quartile from lowest to highest, respectively. Significance labels: **** indicates p-value < 0.001, *** indicates 0.01>p>0.001, ** indicates 0.05>p>0.01, and * indicates p<0.05.
We again used a random forest model to identify the most discriminatory bacteria between HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8. Turicibacter, Roseburia, and Candidatus Saccharimonas were the most important features in separating these groups, all with around a 4% mean decrease accuracy, respectively (Figure 4D). Further analysis with LEfSe highlights four HLA-DR3-depleted genera with the greatest potential biological relevance – Alloprevotella, Helicobacter, Turicibacter, and Mucispirillum – with effect sizes greater than 4 (Fig 4E). Bacteroides and Parabacteroides were also identified as having a high effect size, though they are significantly overabundant in HLA-DR3.DQ8 only when compared to HLA-DR3 (Fig 4E).
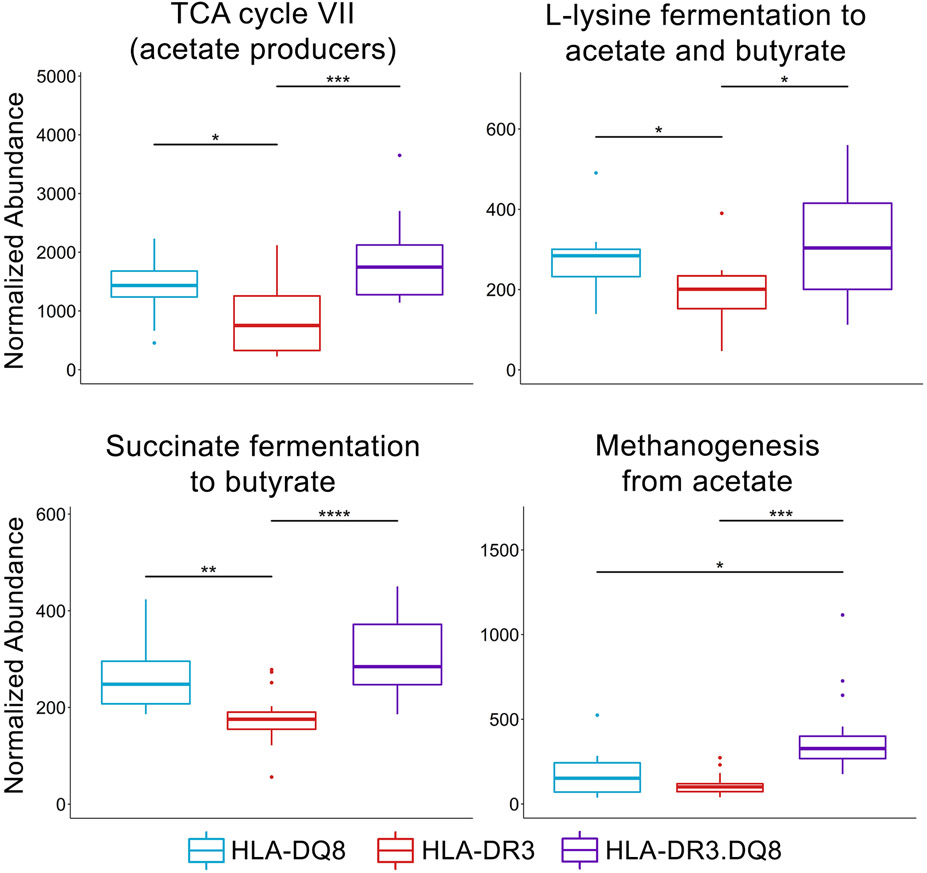
Altered Short-Chain Fatty Acid Metabolism and Methanogenesis in HLA-DR3.DQ8 Mice
In order to examine potential mechanisms behind different disease phenotypes, functional analysis of these microbiomes was conducted from pathway profiles generated using PICRUSt2. Numerous pathways were significantly differentially abundant in the three groups (Supplemental Table 1), including four pathways related to short-chain fatty acids (SCFAs) (Fig 9). Three of these pathways involved SCFA production and were increased in both HLA-DQ8 and HLA-DR3.DQ8 compared to HLA-DR3: TCA cycle VII (acetate producers) (padj = 4.3e-3), L-lysine fermentation to acetate and butyrate (padj = 0.024), and succinate fermentation to butyrate (padj = 2.3e-4) (Fig 9). The fourth SCFA-related pathway, methanogenesis from acetate (padj = 2.0e-4), was significantly more abundant in HLA-DR3.DQ8 compared to both HLA-DR3 and HLA-DQ8 (Fig 9). There were also several pathways predicted by PICRUSt2 to be relatively more abundant in the HLA-DQ8 group compared to both HLA-DR3 and HLA-DR3.DQ8 (Supplementary Table 1), and these are involved in the synthesis of widely prevalent proteins and biological compounds such as bacteriochlorophyll (pwy-5531 and pwy-7159) and polyamines (polyamsyn-pwy), and the degradation of nucleotides (pwy-6608 and pwy-6353). Overall, these results demonstrate an association of HLA class-II polymorphisms with gut bacterial functions including several SCFA metabolic pathways as well as methane production.
Figure 9. Short-chain fatty acid metabolism and methane production differ between HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 mice.
Abundance values are sum-scaled to one million. The three lines represent lower quartile, median, and upper quartile from lowest to highest, respectively. Significance labels: **** indicates p-value < 0.001, *** indicates 0.01>p>0.001, ** indicates 0.05>p>0.01, and * indicates p<0.05.
Discussion
Both genetic as well as environmental factors have been linked with risk of autoimmune diseases. In MS, genetic factors account for only 30% of disease risk while the remaining 70% is attributed to environmental factors. Development of disease only in a subset of individuals with susceptible HLA-class II genes points towards an important role of gene-environment interaction (GxE) in disease susceptibility versus protection. Given that the gut microbiome has recently emerged as a potential environmental factor linked with MS risk, it is imperative to analyze its interplay with genetic factors, especially HLA genotypes. Our study shows that HLA-class has a direct influence on the composition of gut microbiota as mice lacking MHC class-II showed lower bacterial diversity. Additionally, transgenic mice expressing either a single (HLA-DR3 or DQ8) or double HLA class-II genes (HLA-DR3.DQ8) showed distinct microbiota than AE-KO mice. Furthermore, clustering of the HLA-DQ8 microbiome with HLA-DR3.DQ8 rather than HLA-DR3 and severe disease in HLA-DR3.DQ8 mice points towards an important role of gut microbiota in disease modulation in EAE susceptible HLA-DR3 mice. Gene-environment interactions have been proposed to play significant role in disease susceptibility versus resistance (5) and our data suggest that such an interaction between HLA gene polymorphisms and gut microbiota contributes to disease outcome in MS.
The association of MHC class II genes with the risk of developing autoimmune diseases has been well-described. MHC Class II molecules play a central role in the maturation of T-cells, and thus can affect the autoreactivity of an individual’s T-cell population (44). Indeed, multiple HLA genes in humans such as HLA-DR2, HLA-DR3, HLA-DR4, HLA-DQ6, and HLA-DQ8 have been associated with MS susceptibility (11, 45). We have previously utilized transgenic mice expressing MS-linked HLA class II molecules to authenticate importance of HLA polymorphisms in the pathobiology of MS (14, 15, 46-48).
Examining the relationship between HLA genotype and the microbiome, the present study showed that HLA class II molecules influence the composition of the gut microbiome. Specifically, we showed that the microbiome composition of AE-KO mice is distinct from transgenic mice expressing human HLA class II molecules (HLA-DQ8, HLA-DR3, and HLA-DR3·DQ8 mice). Additionally, the microbiome of AE-KO mice exhibits lower genus richness than HLA-DQ8, HLA-DR3, and HLA-DR3.DQ8 mice, suggesting an important role of MHC class-II molecule in selection of gut microbiota. In the absence of MHC class-II molecule, mice are unable to maintain a diverse microbiome, allowing select bacteria to flourish at the cost of bacterial richness.
Our data also suggests that MHC class II polymorphisms might influence selection of specific bacteria in the gut. Previously, utilizing HLA class-II transgenic mice, we have validated the importance of HLA-class II genes in susceptibility vs resistance. Specifically, we found that HLA-DR3 mice are susceptible to disease and HLA-DQ8 mice are resistance to disease. Interestingly, double transgenic mice expressing both HLA-DR3 and HLA-DQ8 developed more severe disease suggesting that the disease resistant HLA-DQ8 gene also has a modulatory effect on disease severity (14, 46, 47). Thus, we hypothesized that the gut microbiome might play an important role in disease susceptibility, resistance, and/or severity in the context of the HLA-DR3 or -DQ8 molecule. EAE phenotypes can be broadly described by three characteristics, disease susceptibility or resistance and disease severity. As HLA-DR3 and HLA-DR3.DQ8 mice were susceptible for disease, they might be selecting for bacteria with a potential role in disease susceptibility. While HLA-DQ8 mice were resistant to disease development, the presence of HLA-DQ8 in a disease susceptible background can worsen the disease. Thus, we argued that HLA-DQ8 may be selecting for bacteria that protect from the development of disease, but also selecting for bacteria that can exacerbate existing disease. The distinct microbiome profile between HLA-DQ8 and HLA-DR3 bolsters the evidence that interaction between host genetics and microbiome composition may play a protective role in MS.
Desulfovibrio, Rikenella, and an uncultured genus of the Prevotellaceae family showed relatively higher abundances in disease susceptible HLA-DR3 and HLA-DR3.DQ8 transgenic mice but reduced in disease resistant HLA-DQ8 mice. Desulfovibrio has previously been shown to be associated with MS disease state (49), and other studies have shown that the abundance of Desulfovibrio is positively associated with symptoms and intestinal damage in DSS-induced colitis mice (50, 51). In one study comparing the microbiome of diabetic db/db mice with nondiabetic db/m mice Rikenella was decreased in the gut microbiota of diabetic mice (52). Furthermore, this depletion was reversed upon oral administration of resveratrol, a compound that is shown to ameliorate symptoms of diabetic nephropathy. Another study found a decrease in Rikenella in inflammatory bowel disease (IBD) patients compared to controls and other gastrointestinal diseases (53). The depletion of an uncultured Prevotellaceae family member is interesting since Prevotella, another member of that family, has been characterized numerous times as being a member of a healthy microbiome (19, 54). It is possible that these two Prevotellaceae members may compete over shared resources, and a high abundance of Prevotella confers protection via suppression of this uncultured genus. The depletion of these three genera suggest a potential role in the conferring susceptibility to EAE, where HLA-DQ8 may protect against EAE through suppression of these genera or the HLA-DR3 molecule may promote their growth in the other two groups.
In the disease resistant HLA-DQ8 group there was a higher relative abundance of Alistipes compared to the HLA-DR3 and HLA-DR3.DQ8 groups, and this genus has previously been found in decreased abundance in EAE mice (55) as well as diabetic mice (52). This suggests a potential protective role of Alistipes in certain inflammatory conditions including EAE.Meanwhile the HLA-DR3 mice had relatively lower abundance of Helicobacter and Mucispirillum. Due to the clinical importance of the Helicobacter pylori in gastritis and peptic ulcer disease, many studies have focused on this species’ role in autoimmune diseases as well, including MS and EAE. Most of these studies have measured the presence of H. pylori using levels of serum anti-H. pylori antibodies, however. Findings thus far suggest that H. pylori infection serves a protective role against the development of MS and the level of seropositivity was also negatively associated with disease severity (56, 57). However, studies focused on the abundance of the Helicobacter genus in the gut microbiome of patients with inflammatory diseases have demonstrated an increase in Helicobacter associated with EAE and IBD, and a decrease in butyrate-producing bacteria (55, 58). In fact, H. hepaticus and H. bilis have been comparable to the colitis-inducing chemical dextran sulfate sodium (DSS) in inducing IBD in mouse models (59, 60), and H. bilis abundance was associated with progression from IBD to colorectal cancer, suggesting a possible correlation with severity of inflammation or duration of disease (61). Similarly, Mucispirillum has been consistently associated with incidence and severity of DSS-induced colitis (50, 62). The relative increase of these bacteria in both HLA-DR3.DQ8 and HLA-DQ8 suggests that the HLA-DQ8 molecule may select for bacteria with context-dependent roles disease exacerbation. Similarly, while Butyricicoccus has been inversely correlated with MS symptoms and generally depleted in various inflammatory states including IBD (63, 64). Butyricicoccus is a known butyrate-producer in the human gut and is thus thought to be a generally beneficial bacteria, but one study suggests that these beneficial effects vary between individuals as a result of overall microbiome composition (65).
Alloprevotella, a close relative of Prevotella, was also significantly depleted in HLA-DR3 compared to the other two groups. Multiple studies have shown Prevotella to be depleted in MS patients and increased after treatment with disease modifying drugs (18, 19). Similarly, in EAE administration of Prevotella histicola has shown significant improvements in EAE (42, 66). Similar findings have been shown in patients with MS, and Prevotella has also been found to be negatively correlated with disease severity (18, 19). The relationship between Alloprevotella and MS is much less studied, and thus far studies have shown varying associations of this genus with inflammatory diseases (67, 68). Romboutsia, Odoribacter, and Turicibacter were all also relatively decreased in HLA-DR3. While they have not been studied with relation to EAE or MS, however they have been associated with various inflammatory diseases (69, 70).
Given the current findings in literature and our findings in this study, while bacteria such as Helicobacter and Mucispirillum are depleted in HLA-DR3 mice that experience moderate disease (Fig 1), their overabundance in mice with severe disease (HLA-DR3.DQ8) suggest they may be correlated with disease severity. The similar abundance in HLA-DR3.DQ8 and HLA-DQ8 mice further suggests that these bacteria may be positively selected for by the HLA-DQ8 phenotype and thus may play a specific role in modulating disease severity but not incidence of the disease itself.
There have been limited studies connecting Enterorhabdus and Intestinimonas – the two genera overabundant in HLA-DR3 mice – with autoimmune or inflammatory diseases. Two studies have isolated species of Enterorabdus from gut of colitis mouse models (71, 72), and a few other studies have found an association between Enterorhabdus and oxidative stress in mice (73, 74). Similarly, there are very limited studies examining the relationship between Streptococcus, Roseburia, Candidatus saccharimonas, Paludicola, or Clostridium sensu stricto 1 with autoimmune diseases. A few studies have shown Streptococcus to be overabundant in patients with MS and other inflammatory diseases (75, 76). Roseburia has been shown to be depleted in DSS-induced colitis and RA, and one study found that administration of Roseburia intestinalis flagellin protein decreased colitis-associated DAI (77). The epistatic interaction between HLA-DR3 and HLA-DQ8 could be selecting for bacteria that may worsen existing disease, such as Roseburia, and suppressing bacteria that may be protecting or ameliorating disease, such as Streptococcus.
Since we have previously observed that HLA-DQ8 caused an increase in disease severity through production of IL-17 and GM-CSF (14), we reasoned that gut microbiota can play an important role in disease modulating effect of HLA-DQ8 as the latter has been shown to regulate levels of Th17 cells in mice. A rapidly growing body of literature is elucidating the mechanistic connections between the gut microbiome and autoimmune diseases such as MS (78, 79). Another recent study demonstrated that a strain of the Erysipelotrichaceae family introduced into the gut of germ-free mice enhances Th17 response, and a strain of Lactobacillus reuteri may present peptides that mimic MOG (80). Additionally, H. hepaticus (increased in HLA-DR3.DQ8 mice at genus level) promotes the development of IL-17 and IFNγ producing CD4 T-cells as a mechanism of causing colitis in mice (55, 81). Other species of Helicobacter specifically H. hepaticus can induce severe IBD disease through IL-12 and IL-23 production (82). Similarly, a few studies have shown Mucispirillum associated with increased Th1 responses and the pro-inflammatory cytokine MCP-1 in the pathogenesis of DSS-induced colitis in mice (62, 83). Additionally, Desulfovibrio is a significant contributor to hydrogen sulfide production in the gut, enabling mucosal inflammation (84, 85), and has also been shown to induce a Th17 response in germ-free mice (86). Thus, HLA-DQ8 selected gut bacteria can modulate disease through induction of pro-inflammatory Th1 and Th17 pathways. Understanding how HLA polymorphisms affect the abundance of immunomodulatory bacteria in the gut could shed light on potential targets for affecting disease pathogenesis and progression.
Functional profiling with PICRUSt2 revealed differential abundance of four pathways related to short-chain fatty acid metabolism. Interestingly, the pathways involved in generating acetate and butyrate had higher relative abundance not only in HLA-DQ8, but HLA-DR3.DQ8 as well. Butyrate is generally regarded as a protective metabolite in inflammatory diseases (87-90). While serum acetate has been associated with higher Expanded Disability Status Scale (EDSS) scores in MS (91), it can be further metabolized to butyrate by various gut bacteria (92-95). However, the HLA-DR3.DQ8 group in this study possessed a uniquely high relative abundance of a pathway that uses acetate to generate methane, suggesting a potential shift towards higher gut methane-to-butyrate ratios compared to HLA-DQ8 and HLA-DR3. Previous studies have demonstrated increased methane production in a subset of MS patients using breath tests (18). Elevated gut methane has also been shown to increase colonic transit time (96, 97), and thus posited to increase nutrient absorption and heighten risk for obesity (98). Obesity is a known driver of systemic inflammation and has also been linked with higher EDSS scores in MS patients (99). These findings together raise the possibility that epistatic interaction between HLA class II alleles such as HLA-DR3 and HLA-DQ8 could contribute to MS susceptibility and severity via modulation of gut bacterial metabolic pathways. Potential relationships between inflammation and the pathways uniquely increased in HLA-DQ8 have yet to be elucidated.
There are some limitations to our findings on differences in microbiome among different strains. As AE-KO, HLA-DR3 and HLA-DQ8 mice were maintained through inbreeding for a prolonged period, there is a possibility that the observed differences in microbiota among these genetically modified strains were due to so-called strain founder effect (100, 101). Previously, Goodrich et al showed that although different TLR deficient mice strains and their respective wild-type littermate controls, harbored distinct microbiota, post-antibiotic treatment, all strains had similar microbial community (101). Additionally, microbiota of mice within same litter were more similar than mice from different litter. Based on these results, the study concluded that maternal transmission of microbiome is dominant over TLR signaling. However, all HLA-DR3.DQ8 transgenic mice used in the study were from F1 cross between HLA-DR3 to HLA-DQ8 mice. Thus, future studies exploring the role of antibiotics on gut microbiota among different HLA transgenic lines or rederivation of these transgenic lines in Germ-free environment will be able to help in determining the precise role of strain founder effect on the composition of gut microbiota.
Previously we have shown that HLA class II allele such as HLA-DR3 and HLA-DQ8 can modulate disease susceptibility, severity, and resistance through modulation of pro and anti-inflammatory cytokines. Future studies on dissecting the interaction between gut microbiota, immune system, cytokines0, and disease phenotypes will help in a better understanding the role of gut microbiome in the pathobiology of MS.
Supplementary Material
Funding
The authors acknowledge funding from the National Multiple Sclerosis Society (RG 5138A1/1T), NIAID/NIH (1R01AI137075-01), a Carver Trust Medical Research Initiative Grant, and the University of Iowa Environmental Health Sciences Research Center, NIEHS/NIH (P30 ES005605). SA was supported by the Emory Warner Fellowship, which provides medical students at the Carver College of Medicine the opportunity to take a full year out of their medical school curriculum to work in a laboratory in the University of Iowa Department of Pathology. Camille Jaime was supported by the Iowa Biosciences Academy (IBA) and an NIH - IMSD (Initiative for Maximizing Student Development) R25 training grant 5R25GM058939.
Footnotes
Disclosure of potential conflicts of interest:
AKM is an inventor of the use of Prevotella histicola for treatment of autoimmune disease and the patent is owned by Mayo Clinic Rochester, USA. The technology has been licensed by Mayo Clinic to Evelo Biosciences. AKM received royalties from Mayo Clinic (paid by Evelo Biosciences).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1.Sospedra M, and Martin R. 2005. Immunology of multiple sclerosis. Annu Rev Immunol 23: 683–747. [DOI] [PubMed] [Google Scholar]
- 2.Steinman L 1995. Multiple sclerosis. Presenting an odd autoantigen. Nature 375: 739–740. [DOI] [PubMed] [Google Scholar]
- 3.Waubant E, Lucas R, Mowry E, Graves J, Olsson T, Alfredsson L, and Langer-Gould A. 2019. Environmental and genetic risk factors for MS: an integrated review. Ann Clin Transl Neurol 6: 1905–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Multiple Sclerosis Genetics Consortium. Electronic address, c. c. y. e., and C. International Multiple Sclerosis Genetics. 2018. Low-Frequency and Rare-Coding Variation Contributes to Multiple Sclerosis Risk. Cell 175: 1679–1687 e1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montgomery TL, Kunstner A, Kennedy JJ, Fang Q, Asarian L, Culp-Hill R, D'Alessandro A, Teuscher C, Busch H, and Krementsov DN. 2020. Interactions between host genetics and gut microbiota determine susceptibility to CNS autoimmunity. Proc Natl Acad Sci U S A 117: 27516–27527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsson T, Barcellos LF, and Alfredsson L. 2017. Interactions between genetic, lifestyle and environmental risk factors for multiple sclerosis. Nat Rev Neurol 13: 25–36. [DOI] [PubMed] [Google Scholar]
- 7.Willer CJ, Dyment DA, Risch NJ, Sadovnick AD, Ebers GC, and G. Canadian Collaborative Study. 2003. Twin concordance and sibling recurrence rates in multiple sclerosis. Proc Natl Acad Sci U S A 100: 12877–12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramagopalan SV, Handel AE, Giovannoni G, Rutherford Siegel S, Ebers GC, and Chaplin G. 2011. Relationship of UV exposure to prevalence of multiple sclerosis in England. Neurology 76: 1410–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maccari G, Robinson J, Ballingall K, Guethlein LA, Grimholt U, Kaufman J, Ho CS, de Groot NG, Flicek P, Bontrop RE, Hammond JA, and Marsh SG. 2017. IPD-MHC 2.0: an improved inter-species database for the study of the major histocompatibility complex. Nucleic Acids Res 45: D860–D864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zivadinov R, Uxa L, Bratina A, Bosco A, Srinivasaraghavan B, Minagar A, Ukmar M, Benedetto S, and Zorzon M. 2007. HLA-DRB1*1501, -DQB1*0301, -DQB1*0302, -DQB1*0602, and -DQB1*0603 alleles are associated with more severe disease outcome on MRI in patients with multiple sclerosis. Int Rev Neurobiol 79: 521–535. [DOI] [PubMed] [Google Scholar]
- 11.Dyment DA, Herrera BM, Cader MZ, Willer CJ, Lincoln MR, Sadovnick AD, Risch N, and Ebers GC. 2005. Complex interactions among MHC haplotypes in multiple sclerosis: susceptibility and resistance. Hum Mol Genet 14: 2019–2026. [DOI] [PubMed] [Google Scholar]
- 12.Amirzargar A, Mytilineos J, Yousefipour A, Farjadian S, Scherer S, Opelz G, and Ghaderi A. 1998. HLA class II (DRB1, DQA1 and DQB1) associated genetic susceptibility in Iranian multiple sclerosis (MS) patients. Eur J Immunogenet 25: 297–301. [DOI] [PubMed] [Google Scholar]
- 13.Marrosu MG, Muntoni F, Murru MR, Costa G, Pischedda MP, Pirastu M, Sotgiu S, Rosati G, and Cianchetti C. 1992. HLA-DQB1 genotype in Sardinian multiple sclerosis: evidence for a key role of DQB1 *0201 and *0302 alleles. Neurology 42: 883–886. [DOI] [PubMed] [Google Scholar]
- 14.Mangalam A, Luckey D, Basal E, Jackson M, Smart M, Rodriguez M, and David C. 2009. HLA-DQ8 (DQB1*0302)-restricted Th17 cells exacerbate experimental autoimmune encephalomyelitis in HLA-DR3-transgenic mice. J Immunol 182: 5131–5139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Das P, Drescher KM, Geluk A, Bradley DS, Rodriguez M, and David CS. 2000. Complementation between specific HLA-DR and HLA-DQ genes in transgenic mice determines susceptibility to experimental autoimmune encephalomyelitis. Hum Immunol 61: 279–289. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, and Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139: 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colpitts SL, Kasper EJ, Keever A, Liljenberg C, Kirby T, Magori K, Kasper LH, and Ochoa-Reparaz J. 2017. A bidirectional association between the gut microbiota and CNS disease in a biphasic murine model of multiple sclerosis. Gut Microbes 8: 561–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, Patel B, Mazzola MA, Liu S, Glanz BL, Cook S, Tankou S, Stuart F, Melo K, Nejad P, Smith K, Topcuolu BD, Holden J, Kivisakk P, Chitnis T, De Jager PL, Quintana FJ, Gerber GK, Bry L, and Weiner HL. 2016. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun 7: 12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Chia N, Kalari KR, Yao JZ, Novotna M, Paz Soldan MM, Luckey DH, Marietta EV, Jeraldo PR, Chen X, Weinshenker BG, Rodriguez M, Kantarci OH, Nelson H, Murray JA, and Mangalam AK. 2016. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep 6: 28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tremlett H, Fadrosh DW, Faruqi AA, Hart J, Roalstad S, Graves J, Lynch S, Waubant E, and U. S. N. o. P. M. Centers. 2016. Gut microbiota composition and relapse risk in pediatric MS: A pilot study. J Neurol Sci 363: 153–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyake S, Kim S, Suda W, Oshima K, Nakamura M, Matsuoka T, Chihara N, Tomita A, Sato W, Kim SW, Morita H, Hattori M, and Yamamura T. 2015. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One 10: e0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ochoa-Reparaz J, and Kasper LH. 2014. Gut microbiome and the risk factors in central nervous system autoimmunity. FEBS Lett 588: 4214–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang K, Hornef MW, and Dupont A. 2015. The intestinal epithelium as guardian of gut barrier integrity. Cell Microbiol 17: 1561–1569. [DOI] [PubMed] [Google Scholar]
- 24.Rescigno M 2011. The intestinal epithelial barrier in the control of homeostasis and immunity. Trends Immunol 32: 256–264. [DOI] [PubMed] [Google Scholar]
- 25.Jarchum I, and Pamer EG. 2011. Regulation of innate and adaptive immunity by the commensal microbiota. Curr Opin Immunol 23: 353–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Blacher E, Levy M, Tatirovsky E, and Elinav E. 2017. Microbiome-Modulated Metabolites at the Interface of Host Immunity. J Immunol 198: 572–580. [DOI] [PubMed] [Google Scholar]
- 27.Forbes JD, Van Domselaar G, and Bernstein CN. 2016. The Gut Microbiota in Immune-Mediated Inflammatory Diseases. Front Microbiol 7: 1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kahrstrom CT, Pariente N, and Weiss U. 2016. Intestinal microbiota in health and disease. Nature 535: 47. [DOI] [PubMed] [Google Scholar]
- 29.Kinross JM, Darzi AW, and Nicholson JK. 2011. Gut microbiome-host interactions in health and disease. Genome Med 3: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khan MAW, Stephens WZ, Mohammed AD, Round JL, and Kubinak JL. 2019. Does MHC heterozygosity influence microbiota form and function? PLoS One 14: e0215946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L, Bell R, Ajami NJ, Petrosino JF, Morrison L, Potts WK, Jensen PE, O'Connell RM, and Round JL. 2015. MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun 6: 8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin P, Bach M, Asquith M, Lee AY, Akileswaran L, Stauffer P, Davin S, Pan Y, Cambronne ED, Dorris M, Debelius JW, Lauber CL, Ackermann G, Baeza YV, Gill T, Knight R, Colbert RA, Taurog JD, Van Gelder RN, and Rosenbaum JT. 2014. HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One 9: e105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madsen L, Labrecque N, Engberg J, Dierich A, Svejgaard A, Benoist C, Mathis D, and Fugger L. 1999. Mice lacking all conventional MHC class II genes. Proc Natl Acad Sci U S A 96: 10338–10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cardell S, Merkenschlager M, Bodmer H, Chan S, Cosgrove D, Benoist C, and Mathis D. 1994. The immune system of mice lacking conventional MHC class II molecules. Adv Immunol 55: 423–440. [DOI] [PubMed] [Google Scholar]
- 35.Nabozny GH, Baisch JM, Cheng S, Cosgrove D, Griffiths MM, Luthra HS, and David CS. 1996. HLA-DQ8 transgenic mice are highly susceptible to collagen-induced arthritis: a novel model for human polyarthritis. J Exp Med 183: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abraham RS, Kudva YC, Wilson SB, Strominger JL, and David CS. 2000. Co-expression of HLA DR3 and DQ8 results in the development of spontaneous insulitis and loss of tolerance to GAD65 in transgenic mice. Diabetes 49: 548–554. [DOI] [PubMed] [Google Scholar]
- 37.Strauss G, Vignali DA, Schonrich G, and Hammerling GJ. 1994. Negative and positive selection by HLA-DR3(DRw17) molecules in transgenic mice. Immunogenetics 40: 104–108. [PubMed] [Google Scholar]
- 38.Shahi SK, Zarei K, Guseva NV, and Mangalam AK. 2019. Microbiota Analysis Using Two-step PCR and Next-generation 16S rRNA Gene Sequencing. J Vis Exp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douglas GM, Maffei VJ, Zaneveld JR, Yurgel SN, Brown JR, Taylor CM, Huttenhower C, and Langille MGI. 2020. PICRUSt2 for prediction of metagenome functions. Nat Biotechnol 38: 685–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arndt D, Xia J, Liu Y, Zhou Y, Guo AC, Cruz JA, Sinelnikov I, Budwill K, Nesbo CL, and Wishart DS. 2012. METAGENassist: a comprehensive web server for comparative metagenomics. Nucleic Acids Res 40: W88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dhariwal A, Chong J, Habib S, King IL, Agellon LB, and Xia J. 2017. MicrobiomeAnalyst: a web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res 45: W180–W188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mangalam A, Shahi SK, Luckey D, Karau M, Marietta E, Luo N, Choung RS, Ju J, Sompallae R, Gibson-Corley K, Patel R, Rodriguez M, David C, Taneja V, and Murray J. 2017. Human Gut-Derived Commensal Bacteria Suppress CNS Inflammatory and Demyelinating Disease. Cell Rep 20: 1269–1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mangalam AK, Luckey D, Giri S, Smart M, Pease LR, Rodriguez M, and David CS. 2012. Two discreet subsets of CD8 T cells modulate PLP(91-110) induced experimental autoimmune encephalomyelitis in HLA-DR3 transgenic mice. J Autoimmun 38: 344–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unanue ER, Turk V, and Neefjes J. 2016. Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu Rev Immunol 34: 265–297. [DOI] [PubMed] [Google Scholar]
- 45.Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, Swerdlin A, Pericak-Vance MA, Haines JL, Hauser SL, and Multiple Sclerosis Genetics G. 2003. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am J Hum Genet 72: 710–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mangalam A, Luckey D, Basal E, Behrens M, Rodriguez M, and David C. 2008. HLA-DQ6 (DQB1*0601)-restricted T cells protect against experimental autoimmune encephalomyelitis in HLA-DR3.DQ6 double-transgenic mice by generating anti-inflammatory IFN-gamma. J Immunol 180: 7747–7756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luckey D, Bastakoty D, and Mangalam AK. 2011. Role of HLA class II genes in susceptibility and resistance to multiple sclerosis: studies using HLA transgenic mice. J Autoimmun 37: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mangalam AK, Taneja V, and David CS. 2013. HLA class II molecules influence susceptibility versus protection in inflammatory diseases by determining the cytokine profile. J Immunol 190: 513–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y, and Zhao L. 2010. Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice. ISME J 4: 232–241. [DOI] [PubMed] [Google Scholar]
- 50.Song H, Wang W, Shen B, Jia H, Hou Z, Chen P, and Sun Y. 2018. Pretreatment with probiotic Bifico ameliorates colitis-associated cancer in mice: Transcriptome and gut flora profiling. Cancer Sci 109: 666–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, Chen H, Kan J, Gou Y, Liu J, Zhang X, Wu X, Tang S, Sun R, Qian C, Zhang N, Niu F, and Jin C. 2020. Anti-inflammatory properties and gut microbiota modulation of an alkali-soluble polysaccharide from purple sweet potato in DSS-induced colitis mice. Int J Biol Macromol 153: 708–722. [DOI] [PubMed] [Google Scholar]
- 52.Cai TT, Ye XL, Li RR, Chen H, Wang YY, Yong HJ, Pan ML, Lu W, Tang Y, Miao H, Snijders AM, Mao JH, Liu XY, Lu YB, and Ding DF. 2020. Resveratrol Modulates the Gut Microbiota and Inflammation to Protect Against Diabetic Nephropathy in Mice. Front Pharmacol 11: 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopetuso LR, Petito V, Graziani C, Schiavoni E, Paroni Sterbini F, Poscia A, Gaetani E, Franceschi F, Cammarota G, Sanguinetti M, Masucci L, Scaldaferri F, and Gasbarrini A. 2018. Gut Microbiota in Health, Diverticular Disease, Irritable Bowel Syndrome, and Inflammatory Bowel Diseases: Time for Microbial Marker of Gastrointestinal Disorders. Dig Dis 36: 56–65. [DOI] [PubMed] [Google Scholar]
- 54.Shahi SK, Freedman SN, Murra AC, Zarei K, Sompallae R, Gibson-Corley KN, Karandikar NJ, Murray JA, and Mangalam AK. 2019. Prevotella histicola, A Human Gut Commensal, Is as Potent as COPAXONE(R) in an Animal Model of Multiple Sclerosis. Front Immunol 10: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li XL, Zhang B, Sun MJ, Bao CC, Yuan BY, Xie QF, Wang LJ, and Wang MX. 2019. Mechanism of gut microbiota and Axl/SOCS3 in experimental autoimmune encephalomyelitis. Biosci Rep 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yao G, Wang P, Luo XD, Yu TM, Harris RA, and Zhang XM. 2016. Meta-analysis of association between Helicobacter pylori infection and multiple sclerosis. Neurosci Lett 620: 1–7. [DOI] [PubMed] [Google Scholar]
- 57.Ranjbar R, Karampoor S, and Jalilian FA. 2019. The protective effect of Helicobacter Pylori infection on the susceptibility of multiple sclerosis. J Neuroimmunol 337: 577069. [DOI] [PubMed] [Google Scholar]
- 58.Yu W, Su X, Chen W, Tian X, Zhang K, Guo G, Zhou L, Zeng T, and Han B. 2019. Three types of gut bacteria collaborating to improve Kui Jie'an enema treat DSS-induced colitis in mice. Biomed Pharmacother 113: 108751. [DOI] [PubMed] [Google Scholar]
- 59.Willis CR, Seamons A, Maxwell J, Treuting PM, Nelson L, Chen G, Phelps S, Smith CL, Brabb T, Iritani BM, and Maggio-Price L. 2012. Interleukin-7 receptor blockade suppresses adaptive and innate inflammatory responses in experimental colitis. J Inflamm (Lond) 9: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Han Q, Kono TJY, Knutson CG, Parry NM, Seiler CL, Fox JG, Tannenbaum SR, and Tretyakova NY. 2020. Multi-Omics Characterization of Inflammatory Bowel Disease-Induced Hyperplasia/Dysplasia in the Rag2(−/−)/Il10(−/−) Mouse Model. Int J Mol Sci 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Peng W, Li H, Xu Y, Yan L, Tang Z, Hossein Mohseni A, Taghinezhad SS, Tang X, and Fu X. 2020. Association of Helicobacter bilis Infection with the Development of Colorectal Cancer. Nutr Cancer: 1–11. [DOI] [PubMed] [Google Scholar]
- 62.Caruso R, Mathes T, Martens EC, Kamada N, Nusrat A, Inohara N, and Nunez G. 2019. A specific gene-microbe interaction drives the development of Crohn's disease-like colitis in mice. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reynders T, Devolder L, Valles-Colomer M, Van Remoortel A, Joossens M, De Keyser J, Nagels G, D'Hooghe M, and Raes J. 2020. Gut microbiome variation is associated to Multiple Sclerosis phenotypic subtypes. Ann Clin Transl Neurol 7: 406–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Facchin S, Vitulo N, Calgaro M, Buda A, Romualdi C, Pohl D, Perini B, Lorenzon G, Marinelli C, D'Inca R, Sturniolo GC, and Savarino EV. 2020. Microbiota changes induced by microencapsulated sodium butyrate in patients with inflammatory bowel disease. Neurogastroenterol Motil 32: e13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Geirnaert A, Wang J, Tinck M, Steyaert A, Van den Abbeele P, Eeckhaut V, Vilchez-Vargas R, Falony G, Laukens D, De Vos M, Van Immerseel F, Raes J, Boon N, and Van de Wiele T. 2015. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol Ecol 91. [DOI] [PubMed] [Google Scholar]
- 66.Shahi SK, Jensen SN, Murra AC, Tang N, Guo H, Gibson-Corley KN, Zhang J, Karandikar NJ, Murray JA, and Mangalam AK. 2020. Human Commensal Prevotella histicola Ameliorates Disease as Effectively as Interferon-Beta in the Experimental Autoimmune Encephalomyelitis. Front Immunol 11: 578648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ren W, Chen S, Zhang L, Liu G, Hussain T, Hao X, Yin J, Duan J, Tan B, Wu G, Bazer FW, and Yin Y. 2016. Interferon Tau Affects Mouse Intestinal Microbiota and Expression of IL-17. Mediators Inflamm 2016: 2839232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sun Y, Chen Q, Lin P, Xu R, He D, Ji W, Bian Y, Shen Y, Li Q, Liu C, Dong K, Tang YW, Pei Z, Yang L, Lu H, Guo X, and Xiao L. 2019. Characteristics of Gut Microbiota in Patients With Rheumatoid Arthritis in Shanghai, China. Front Cell Infect Microbiol 9: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He H, Xu H, Xu J, Zhao H, Lin Q, Zhou Y, and Nie Y. 2020. Sodium Butyrate Ameliorates Gut Microbiota Dysbiosis in Lupus-Like Mice. Front Nutr 7: 604283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.El Mouzan MI, Winter HS, Assiri AA, Korolev KS, Al Sarkhy AA, Dowd SE, Al Mofarreh MA, and Menon R. 2018. Microbiota profile in new-onset pediatric Crohn's disease: data from a non-Western population. Gut Pathog 10: 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Clavel T, Charrier C, Braune A, Wenning M, Blaut M, and Haller D. 2009. Isolation of bacteria from the ileal mucosa of TNFdeltaARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int J Syst Evol Microbiol 59: 1805–1812. [DOI] [PubMed] [Google Scholar]
- 72.Clavel T, Duck W, Charrier C, Wenning M, Elson C, and Haller D. 2010. Enterorhabdus caecimuris sp. nov., a member of the family Coriobacteriaceae isolated from a mouse model of spontaneous colitis, and emended description of the genus Enterorhabdus Clavel et al. 2009. Int J Syst Evol Microbiol 60: 1527–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song R, Yao J, Shi Q, and Wei R. 2018. Nanocomposite of Half-Fin Anchovy Hydrolysates/Zinc Oxide Nanoparticles Exhibits Actual Non-Toxicity and Regulates Intestinal Microbiota, Short-Chain Fatty Acids Production and Oxidative Status in Mice. Mar Drugs 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feng YL, Cao G, Chen DQ, Vaziri ND, Chen L, Zhang J, Wang M, Guo Y, and Zhao YY. 2019. Microbiome-metabolomics reveals gut microbiota associated with glycine-conjugated metabolites and polyamine metabolism in chronic kidney disease. Cell Mol Life Sci 76: 4961–4978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zeng Q, Junli G, Liu X, Chen C, Sun X, Li H, Zhou Y, Cui C, Wang Y, Yang Y, Wu A, Shu Y, Hu X, Lu Z, Zheng SG, Qiu W, and Lu Y. 2019. Gut dysbiosis and lack of short chain fatty acids in a Chinese cohort of patients with multiple sclerosis. Neurochem Int 129: 104468. [DOI] [PubMed] [Google Scholar]
- 76.Schepici G, Silvestro S, Bramanti P, and Mazzon E. 2019. The Gut Microbiota in Multiple Sclerosis: An Overview of Clinical Trials. Cell Transplant 28: 1507–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu X, Pan S, Luo W, Shen Z, Meng X, Xiao M, Tan B, Nie K, Tong T, and Wang X. 2020. Roseburia intestinalisderived flagellin ameliorates colitis by targeting miR2233pmediated activation of NLRP3 inflammasome and pyroptosis. Mol Med Rep 22: 2695–2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shahi SK, Freedman SN, and Mangalam AK. 2017. Gut microbiome in multiple sclerosis: The players involved and the roles they play. Gut Microbes 8: 607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marietta E, Mangalam AK, Taneja V, and Murray JA. 2020. Intestinal Dysbiosis in, and Enteral Bacterial Therapies for, Systemic Autoimmune Diseases. Front Immunol 11: 573079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miyauchi E, Kim SW, Suda W, Kawasumi M, Onawa S, Taguchi-Atarashi N, Morita H, Taylor TD, Hattori M, and Ohno H. 2020. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 585: 102–106. [DOI] [PubMed] [Google Scholar]
- 81.Morrison PJ, Bending D, Fouser LA, Wright JF, Stockinger B, Cooke A, and Kullberg MC. 2013. Th17-cell plasticity in Helicobacter hepaticus-induced intestinal inflammation. Mucosal Immunol 6: 1143–1156. [DOI] [PubMed] [Google Scholar]
- 82.Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, Cua DJ, Powrie F, Cheever AW, Maloy KJ, and Sher A. 2006. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med 203: 2485–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shintouo CM, Mets T, Beckwee D, Bautmans I, Ghogomu SM, Souopgui J, Leemans L, Meriki HD, and Njemini R. 2020. Is inflammageing influenced by the microbiota in the aged gut? A systematic review. Exp Gerontol 141: 111079. [DOI] [PubMed] [Google Scholar]
- 84.Guo J, Han X, Zhan J, You Y, and Huang W. 2018. Vanillin Alleviates High Fat Diet-Induced Obesity and Improves the Gut Microbiota Composition. Front Microbiol 9: 2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kushkevych I, Dordevic D, and Vitezova M. 2021. Possible synergy effect of hydrogen sulfide and acetate produced by sulfate-reducing bacteria on inflammatory bowel disease development. J Adv Res 27: 71–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Figliuolo VR, Dos Santos LM, Abalo A, Nanini H, Santos A, Brittes NM, Bernardazzi C, de Souza HSP, Vieira LQ, Coutinho-Silva R, and Coutinho C. 2017. Sulfate-reducing bacteria stimulate gut immune responses and contribute to inflammation in experimental colitis. Life Sci 189: 29–38. [DOI] [PubMed] [Google Scholar]
- 87.Archer S, Meng S, Wu J, Johnson J, Tang R, and Hodin R. 1998. Butyrate inhibits colon carcinoma cell growth through two distinct pathways. Surgery 124: 248–253. [PubMed] [Google Scholar]
- 88.Christl SU, Eisner HD, Dusel G, Kasper H, and Scheppach W. 1996. Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa: a potential role for these agents in the pathogenesis of ulcerative colitis. Dig Dis Sci 41: 2477–2481. [DOI] [PubMed] [Google Scholar]
- 89.Csordas A 1996. Butyrate, aspirin and colorectal cancer. Eur J Cancer Prev 5: 221–231. [DOI] [PubMed] [Google Scholar]
- 90.Inan MS, Rasoulpour RJ, Yin L, Hubbard AK, Rosenberg DW, and Giardina C. 2000. The luminal short-chain fatty acid butyrate modulates NF-kappaB activity in a human colonic epithelial cell line. Gastroenterology 118: 724–734. [DOI] [PubMed] [Google Scholar]
- 91.Perez-Perez S, Dominguez-Mozo MI, Alonso-Gomez A, Medina S, Villarrubia N, Fernandez-Velasco JI, Garcia-Martinez MA, Garcia-Calvo E, Estevez H, Costa-Frossard L, Alvarez-Cermeno JC, Luque-Garcia JL, Arroyo R, Villar LM, and Alvarez-Lafuente R. 2020. Acetate correlates with disability and immune response in multiple sclerosis. PeerJ 8: e10220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Duncan SH, Barcenilla A, Stewart CS, Pryde SE, and Flint HJ. 2002. Acetate utilization and butyryl coenzyme A (CoA):acetate-CoA transferase in butyrate-producing bacteria from the human large intestine. Appl Environ Microbiol 68: 5186–5190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Diez-Gonzalez F, Bond DR, Jennings E, and Russell JB. 1999. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production, and phylogeny. Arch Microbiol 171: 324–330. [DOI] [PubMed] [Google Scholar]
- 94.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, and Bakker BM. 2013. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Duncan SH, Holtrop G, Lobley GE, Calder AG, Stewart CS, and Flint HJ. 2004. Contribution of acetate to butyrate formation by human faecal bacteria. Br J Nutr 91: 915–923. [DOI] [PubMed] [Google Scholar]
- 96.Jahng J, Jung IS, Choi EJ, Conklin JL, and Park H. 2012. The effects of methane and hydrogen gases produced by enteric bacteria on ileal motility and colonic transit time. Neurogastroenterol Motil 24: 185–190, e192. [DOI] [PubMed] [Google Scholar]
- 97.Pimentel M, Lin HC, Enayati P, van den Burg B, Lee HR, Chen JH, Park S, Kong Y, and Conklin J. 2006. Methane, a gas produced by enteric bacteria, slows intestinal transit and augments small intestinal contractile activity. Am J Physiol Gastrointest Liver Physiol 290: G1089–1095. [DOI] [PubMed] [Google Scholar]
- 98.Basseri RJ, Basseri B, Pimentel M, Chong K, Youdim A, Low K, Hwang L, Soffer E, Chang C, and Mathur R. 2012. Intestinal methane production in obese individuals is associated with a higher body mass index. Gastroenterol Hepatol (N Y) 8: 22–28. [PMC free article] [PubMed] [Google Scholar]
- 99.Stampanoni Bassi M, Iezzi E, Buttari F, Gilio L, Simonelli I, Carbone F, Micillo T, De Rosa V, Sica F, Furlan R, Finardi A, Fantozzi R, Storto M, Bellantonio P, Pirollo P, Di Lemme S, Musella A, Mandolesi G, Centonze D, and Matarese G. 2020. Obesity worsens central inflammation and disability in multiple sclerosis. Mult Scler 26: 1237–1246. [DOI] [PubMed] [Google Scholar]
- 100.Ubeda C, Lipuma L, Gobourne A, Viale A, Leiner I, Equinda M, Khanin R, and Pamer EG. 2012. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of TLR-deficient mice. J Exp Med 209: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, and Ley RE. 2014. Conducting a microbiome study. Cell 158: 250–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.