Abstract
Parkinson’s disease (PD) is one of the most common neurodegenerative diseases in the elderly population and is caused by the loss of dopaminergic neurons. PD has been predominantly attributed to mitochondrial dysfunction. The structural alteration of α-synuclein triggers toxic oligomer formation in the neurons, which greatly contributes to PD. In this article, we discuss the role of several familial PD-related proteins, such as α-synuclein, DJ-1, LRRK2, PINK1, and parkin in mitophagy, which entails a selective degradation of mitochondria via autophagy. Defective changes in mitochondrial dynamics and their biochemical and functional interaction induce the formation of toxic α-synuclein-containing protein aggregates in PD. In addition, these gene products play an essential role in ubiquitin proteasome system (UPS)-mediated proteolysis as well as mitophagy. Interestingly, a few deubiquitinating enzymes (DUBs) additionally modulate these two pathways negatively or positively. Based on these findings, we summarize the close relationship between several DUBs and the precise modulation of mitophagy. For example, the USP8, USP10, and USP15, among many DUBs are reported to specifically regulate the K48- or K63-linked de-ubiquitination reactions of several target proteins associated with the mitophagic process, in turn upregulating the mitophagy and protecting neuronal cells from α-synuclein-derived toxicity. In contrast, USP30 inhibits mitophagy by opposing parkin-mediated ubiquitination of target proteins. Furthermore, the association between these changes and PD pathogenesis will be discussed. Taken together, although the functional roles of several PD-related genes have yet to be fully understood, they are substantially associated with mitochondrial quality control as well as UPS. Therefore, a better understanding of their relationship provides valuable therapeutic clues for appropriate management strategies.
Keywords: Autophagy, Mitophagy, Parkinson’s disease, Ubiquitin, Ubi-quitination, Ubiquitin protease, Ubiquitin proteasome system
INTRODUCTION
Parkinson’s disease (PD) is the second most common neurodegenerative disorder (NDD) in elderly individuals aged above 60. It affects predominately dopaminergic neurons in a specific area of the brain called substantia nigra. PD is characterized by movement disorders, such as resting tremor, rigidity, slow movement, impaired posture and balance, and loss of automatic movements (1). The cause of PD is unknown, but several factors appear to play a role, including genetic and environmental factors. About 10-15% of patients diagnosed with PD are known to carry mutations in one of several specific genes, including α-synuclein, FBXO7, LRRK2, parkin, PINK1, and UCHL1. Similar to other NDDs, PD patients also display clumps of specific substances within the brain cells called Lewy bodies (LBs), which are the pathological hallmarks of PD and mainly composed of α-synuclein (α-Syn) (2). Mitochondrial dysfunction has also been implicated as the main causative factor in PD. Interestingly, familial PD-linked gene products and their binding partners, such as LRRK2, BAG5, Miro1, DJ-1, PINK1, and parkin, play a role in the process of mitochondrial quality control. Dysregulation of mitophagy, involving removal of damaged and surplus mitochondria, is also implicated in the pathogenesis of NDDs, including Alzheimer’s disease (AD) and PD (3).
Mitophagy is a subtype of macroautophagy, which selectively targets defective mitochondria to the lysosome for degradation. Mitophagy plays a pivotal role in cells, in preserving mitochondrial homeostasis, biogenesis, fission, and fusion for mitochondrial quality control (4). Two PD genes, the mitochondrial kinase PINK1 and ubiquitin ligase parkin, degrade damaged mitochondria via mitophagy. Under resting state, PINK1 is normally transported into the inner mitochondria membrane (IMM) of healthy mitochondria and cleaved by presenilin-associated rhomboid-like protein (PARL). The cleavage of PINK1 results in the formation of a 52-kDa fragment and release into cytosol. PINK1 fragment is then rapidly ubiquitinated and degraded by the ubiquitin proteasome system (UPS). However, various stimuli causing mitochondrial damage interfere with the processing and cytosolic translocation of PINK1, resulting in the accumulation of 63-kDa full-length PINK1 on the outer mitochondrial membrane (OMM). PINK1 is then activated by auto-phosphorylation during mitophagy, triggering the recruitment of parkin to the OMM and facilitating its E3 ligase activity (2, 5). Parkin and ubiquitin are then phosphorylated, resulting in the assembly of K6-, K11-, K48-, and K63-linked ubiquitin chains on the OMM target proteins for degradation (6). Finally, parkin builds ubiquitin chains on the damaged mitochondria for lysosomal degradation.
Deubiquitinating enzymes (DUBs) are members of cysteine and metalloprotease family that cleave ubiquitin from the protein substrates. DUB-mediated cleavage of ubiquitin chains from substrate plays a crucial role in various cellular processes by changing its biochemical properties, such as protein stability, function, and localization. DUBs can be classified into five families based on their sequence and structural homology: ubiquitin specific protease (USP), ubiquitin C-terminal hydrolase (UCH), otubain protease (OTU), Machado-Joseph Disease protease (MJD) and JAB1/MPN/Mov34 metalloenzyme (JAMM) (7). The majority of them belong to the ubiquitin-specific proteinase (USP) family. As expected, DUBs play a crucial role in the ubiquitin pathways and are responsible for the recycling of mono- or poly-ubiquitin. DUBs also reverse the ubiquitin-like modification of target proteins (7, 8). As autophagy entails the removal of protein aggregates, the turnover of organelles, as well as the elimination of intracellular pathogens, the potential targets should be selectively marked by the attachment of ubiquitin in order to be recognized by autophagy receptors. Thus, ubiquitination and ubiquitin-dependent autophagy (including mitophagy) are balanced by deubiquitination. In this article, we highlight the functional link between the most well-known degradation pathway of damaged mitochondria and PD. In addition, the role of a single subclass of deubiquitinating enzymes, USP, in regulating mitophagy and the pathologic features of PD will be discussed.
MITOCHONDRIAL DYSREGULATION, DEFECTIVE MITOPHAGY, AND PARKINSON’S DISEASE
Mitochondrial dysfunction in neurodegenerative diseases including PD
Mitophagy plays an important role in mitochondrial quality control, and the accurate clearance of the damaged mitochondria is critical for the maintenance of mitochondrial and cel-lular homeostasis. Therefore, mitochondria dysfunction results in several neurodegenerative diseases, including AD and PD. For example, oxidative stress and mitochondrial dysfunction are connected to genetic mutations in the mitochondrial DNAs, which are involved in AD pathogenesis (9). A defective mitochondrial respiratory chain, especially the reduced activity of complex I, was found in post-mortem brains obtained from sporadic PD patients. In addition, abnormal mitophagy was observed in several PD models, including environmental or genetic factors (10). While α-Syn is not only localized to mitochondria, it also directly regulates the mitochondrial morphology (11, 12) as well as Ca2+ signaling (13). Further, two PD-associated proteins, parkin and PINK1, primarily regulate the mitochondrial quality control. These findings suggest the need for prompt and precise mitophagy in mitochondrial homeostasis, and its alteration contributes to PD pathogenesis. When the mitochondria are damaged, PINK1 recruits parkin to the OMM (14). Once parkin is localized to OMM, PINK1 phosphorylates the UBL domain of parkin at S65 residue (14), which then initiates the clearance of damaged mitochondria via autophagy (15). Likewise, the PINK1-mediated mitophagic progression is critical for the regulation of parkin E3 ligase activity via a feed-forward mechanism. Thus, a better understanding of the mitochondrial quality control events is required to develop novel therapeutic interventions for PD.
The role of protein ubiquitination in mitophagy
In eukaryotes, UPS and autophagy are the two major intracellular degradation pathways responsible for eliminating unfolded/misfolded proteins. Ubiquitin (Ub) is a small regulatory protein consists of 76 amino acids, and can be attached to the target substrates (16). This protein is highly conserved among eukaryotes and the process of ubiquitin tagging to substrates is known as ubiquitination. Protein ubiquitination is a type of post-translational modifications (PTMs) in which three enzymes, E1 (ubiquitin activating enzyme), E2 (ubiquitin conjugating enzymes), and E3 (ubiquitin ligase), catalyze the sequential reaction of covalent ubiquitin conjugation (8). With regard to mitophagy, there are two different subtypes: receptor- and ubiquitin-mediated mitophagies. In this article, we focus on the ubiquitin-mediated mitophagy and review its role in controlling the PINK1/parkin-dependent mitophagy to maintain mitochondrial quality control. Besides UPS, parkin also uses ubiquitin as a key substrate to regulate the degradation of defective organelles. Upon mitochondria depolarization, both parkin and ubiquitin are activated via phosphorylation by PINK1 (17). Subsequently, the activated parkin is attached to target proteins in OMM via an active site of cysteine as an ubiquitin-thioester intermediate (18). The ubiquitinated OMM proteins, such as VDAC1, are promoted for degradation via K27- and K63-linked ubiquitin chains (19). Eventually, the conjugation of ubiquitin to parkin facilitates and completes the degradation of dysfunctional mitochondria in PINK1/parkin-dependent mitophagy.
SEVERAL PD-LINKED PROTEINS AFFECT MITOPHAGY
Given their widespread localization and biochemical properties, multiple PD-related genes have been associated with various cellular functions and signaling pathways, including mitophagy (Table 1). First, α-Syn aggregates compromise the autophagic mechanism by impairing the phagocytosis required for protein degradation in neuronal cell lines and α-Syn transgenic mice, which suggests a close association between α-Syn and autophagy (or/and mitophagy) (20, 21). Thus, the defective autophagy induced by α-Syn disrupts the mitochondrial clearance. Other studies have also proposed α-Syn-derived impairment in autophagy. For example, the overexpression of α-Syn in neuroblastoma cell line was shown to enhance the level of autophagic substrate p62, leading to a significant decrease in the levels of autophagy regulator LC3 as well as in the number of LC3-II-positive vesicles (21). In addition, autophagic activity was impaired by the aggregation of α-Syn (22). Interestingly, the effect of A53T mutation of α-Syn on autophagy may differ. An increase in lysosome-mediated mitophagy has been reported in dopaminergic neurons of α-Syn-A53T transgenic mice, which indicate a compensatory response to depletion of defective mitochondria (23). The PD-linked α-Syn mutants, but not wild-type α-Syn, bind to the LAMP2 transporter in the lysosomal membrane and block protein uptake in the chaperone-mediated autophagy pathway, thereby inhibiting self-degradation and that of other substrates (24). Thus, α-Syn overexpression or the physiological effect of α-Syn mutations contributes to impaired autophagy via diverse mechanisms, and in turn compromise the clearance of abnormal mitochondria by mitophagy.
Table 1.
Diverse functions of many PD-related gene products in a number of cellular pathways, including mitophagy
| Gene | Genetic functions | Regulatory role in the mitophagy and PD | References |
|---|---|---|---|
| α-Synuclein | Accumulated in Lewy bodies and its pathogenic aggregation negatively affects mitophagy | Impairing membrane engulfing process and ultimately leading to the dysfunction in autophagy and mitochondrial clearance | Guhathakurta S et al. |
| Sugiura A et al. | |||
| Parkin | Acts as an E3 ubiquitin ligase which interacts with PINK1 and recruits Ub chains to target substrate to activate mitophagy | Amplifies a damage detection signal from PINK1 by facilitating ubiquitin chain formation | Narendra DP et al. |
| McLelland GL et al. | |||
| DJ-1 | Acts as a redox sensor to regulate autophagy as well as mitochondrial dynamics | Knockdown of DJ-1 recruits the parkin in PINK1/parkin-dependent mitophagy to maintain mitochondrial function | Thomas KJ et al. |
| Joselin AP et al. | |||
| LRRK2 | Large multifunctional protein containing the kinase and GTPase domain and regulates mitophagy | G2019S mutant upregulates intracellular α-synuclein level for altering the lysosome morphology and reduces the mitophagy | Walter J et al. |
| Obergasteiger J et al. | |||
| BAG5 | Regulates both cell death and survival pathway; BAG5 enhances dopaminergic neurodegeneration and physically interacts with parkin | BAG5 suppresses the parkin recruitment to damaged mitochondria, consequently reducing mitophagy; it also interacts with PINK1 | Kalia SK et al. |
| De Snoo ML et al. | |||
| Wang X et al. | |||
| Miro1 | Regulates mitochondrial homeostasis, apoptosis, and mediates mitochondrial motility | Knockdown of Miro1 reduces the translocation of parkin and finally down-regulates the mitophagy | Berenguer-Escuder C et al. |
| Safiulina D et al. |
Second, parkin is an E3 ubiquitin ligase, which recruits Ub chains to the target substrate. PINK1 is a Ser/Thr kinase that activates parkin in mitophagy to maintain mitochondrial homeostasis, apoptosis, and oxidative stress (25). Many stressors, such as membrane depolarization, mitochondrial complex dysfunction, mutagenic stress, and proteotoxicity, lead to accumulation of PINK1 on the OMM. Subsequent homodimerization of PINK1 on the OMM leads to auto-phosphorylation, which promotes the kinase activity of PINK1 and facilitates its binding to substrates, parkin and ubiquitin. PINK1 then activates parkin via phosphorylation of ubiquitin on Ser65 as well as direct phosphorylation of parkin on Ser65. These results suggest that parkin amplifies a damage detection signal from PINK1 by facilitating the formation of ubiquitin chains, which recruit additional parkin to the mitochondria (26). Once recruited to the mitochondria, parkin mediates the ubiquitination of multiple targets in OMM, IMM, and mitochondrial matrix. Another report revealed that PINK1 and parkin are associated with the mitochondria-derived vesicles (MDVs), ultimately targeting into the lysosomes for degradation (21). Parkin is ultimately released into the cytosol after carrying the target protein to the lysosome (22).
DJ-1 protein plays an essential role in various cellular pathways, including transcription regulation, mitochondrial homeostasis, and cellular apoptosis. Especially, DJ-1 acts as a redox sensor and regulates autophagy and mitochondrial dynamics during oxidative stress (27). Knockdown of DJ-1 induces loss of mitochondrial integrity, mitochondrial fragmentation, and polarization; however, antioxidant treatment reverses these effects (28). DJ-1 also regulates mitophagy via association with ER-mitochondria and physically interacts with the IP3R3-Grp75-VDAC1, the mitochondria-associated membrane (MAM)-essential complex. However, the familial PD-associated L166P mutant of DJ-1 displayed a remarkable reduction of the MAM complex. As a result, the knockdown of DJ-1 induced mitophagy (29), enhanc-ing the mitochondrial mass and activities including the complex I activity (30). In addition, the loss of DJ-1 increased the parkin recruitment to damaged mitochondria and mitophagy, whereas DJ-1 levels accumulated on the mitochondria under oxidative stress conditions were also dependent on parkin and PINK1. These results suggest a close link between DJ-1 and the PINK1/parkin-mediated pathway (31). This hypothesis was further supported by DJ-1 functions in parallel with the PINK1/parkin pathway to maintain mitochondrial function under an oxidative environment (32). Further, Hao et al. have shown the mitochondrial defect in DJ-1 knockout flies, which is similar to that of PINK1- and parkin-mutants (33).
Mutations in leucine-rich repeat kinase 2 (LRRK2) gene are the most frequent cause of autosomal PD. LRRK2 is a large and multi-domain protein containing two catalytic domains: a Roc GTPase-domain and a kinase domain. Many studies revealed that LRRK2 also plays a key role in regulating mitophagy. For example, patients with familial PD expressing G2019S-LRRK2 mutation, which results in enhanced kinase activity, displayed abnormal mitochondrial function and morphology, and a reduction of mitophagy (34). This LRRK2-2019S mutation also upregulates the intracellular α-Syn level, consequently altering the morphology of lysosomes (35). In addition, LRRK2 activates MAPK/ERK pathway and upregulates transcription of α-Syn in HEK293 cells (36). Further, G2019S-LRRK2 mutant affects the PINK1/parkin-dependent mitophagy pathway that disturbs mitochondrial function, which may lead to accumulation of damaged mitochondria and ultimately increase cellular vulnerability to external stressors (37). In parallel, conflicting findings involving idiopathic and genetic PD suggest that the G2019S mutation of LRRK2 activates class III HDACs to increase mitophagy in familial PD patients, whereas idiopathic PD cases exhibit considerable downregulation in the clearance of those defective mitochondria (38).
Molecular chaperones regulate intracellular proteostasis by promoting efficient folding and expediting the refolding or degradation of misfolded proteins under environmental and physiological stress, including heat, oxidative stress, and inflammation. The Bcl-2 associated athanogene (BAG) family of proteins acts co-chaperones in cell survival and cell death pathways. BAG5 enhances dopaminergic neurodegeneration in rodent models of PD (39). While physically interacting with parkin, BAG5 impairs mitophagy by suppressing parkin recruitment to damaged mitochondria and reducing the movement of damaged mitochondria into the lysosomes (40). BAG5 also enhanced parkin-mediated Mcl-1 degradation and cell death following severe mitochondrial insult. Recently, BAG5 was found to interact with PINK1 (41), suggesting a possible role for this co-chaperone in the regulation of the PINK1/parkin-dependent mitophagy. Two other BAG family members, BAG2 and BAG4, have been shown to differentially modulate parkin recruitment to depolarized mitochondria. In addition, BAG3 has been identified as a risk locus for PD (42).
Lastly, mitochondrial Rho GTPase 1 (Miro1) protein is encoded by RHOT1 gene and regulates mitochondrial homeostasis and apoptosis. Miro1 is localized on the mitochondrial surface and mediates mitochondrial motility. The link between Miro1 dysfunction and PD was established by studies demonstrating Miro1 as a target of mitochondrial quality control via PINK1/parkin-mediated mitophagy and mitochondrial transport. Miro1 is removed from depolarized mitochondria to facilitate their clear-ance via mitophagy. Miro1 was also identified as an important regulator of mitochondria-ER contact sites (MERCs), where it acts as a sensor of cytosolic calcium levels (43). Recently, there was a report that a group of PD patients expressed RHOT1 mutations. In addition, the overexpression of Miro1-T351A or -T610A mutants in fibroblasts resulted in calcium dysfunction and reduction in MERCs, both of which play an important role in mitochondrial function. Further, Miro1 interferes with the function of MERCs, indicating that Miro1 mutants may trigger PD pathology (44). Knockdown of Miro1 reduces the translocation of parkin to the mitochondria and downregulates mitophagy in a calcium-dependent manner (45). While Miro1 is a component of parkin receptor complex, the interaction of Miro1 with parkin and the following K27-linked ubiquitination of downstream targets are dependent on Ser65 residue of parkin (46). Overall, these results suggest that Miro1 interacts with parkin to regulate mitochondrial dynamics.
DYNAMIC REGULATION OF MITOPHAGY BY PROTEIN DEUBIQUITINATION ENZYMES AND ITS DEFECT IN PD
The role of DUBs in mitophagy
Protein ubiquitination is a reversible pathway, and ubiquitin is removed by DUBs upon the completion of signaling events leading to the covalent conjugation of ubiquitin. In this sense, DUBs act as proteases to cleave ubiquitin or ubiquitin-like pr-oteins from the target proteins (47). As mitophagy is triggered by ubiquitin modification of proteins residing on the surface of mitochondria, it is also subject to the modulation or suppression via deubiquitination and DUBs. Several studies demonstrated that specific DUBs are associated with multiple types of auto-phagic pathways and NDDs (Fig. 1) (48). For example, USP15, USP30, and USP35 are known to eliminate the ubiquitin- and parkin-mediated signals, consequently delaying or disrupting mitophagy (5). In addition, several E3 ligases, including parkin, undergo autoubiquitination, and DUBs can counteract this activity (18). For instance, USP8 deubiquitinates the K6-linked ubiquitin conjugates from parkin, contributing to the release of parkin autoinhibition to promote CCCP-induced mitochondrial translocation of parkin and parkin-dependent mitophagy (49).
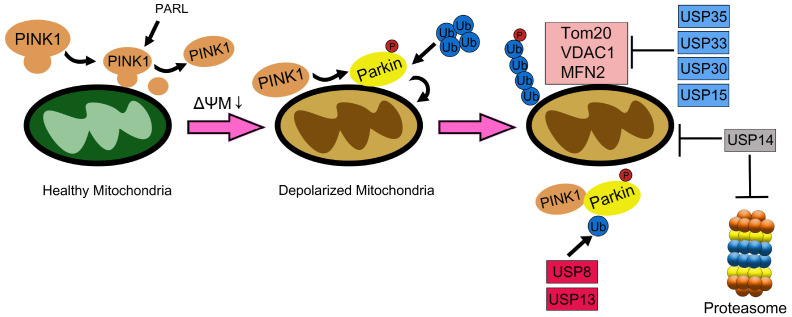
Fig. 1.
Many DUBs regulate mitophagy in a positive or negative manner. In healthy mitochondria, PINK1 is constitutively imported into the mitochondria for processing and release into the cytosolic area, followed by rapid degradation. However, when the mitochondrial membrane potential (ΔΨm) dissipates, PINK1 is stabilized on the OMM and forms a large complex on the OMM surface where it recruits parkin to the damaged mitochondria. The accumulated PINK1 phosphorylates parkin and attaches ubiquitin chains to several mitochondrial substrates, such as translocase of the outer membrane 20 (Tom20), voltage-dependent anion-selective channel 1 (VDAC1), and mitofusin-2 (MFN2). Such ubiquitinated proteins may act as adaptors for sequestosome-1 (SQSTM1/p62) and promote the translocation of defective mitochondria to the autophagosome followed by sequential steps of mitophagy. Multiple DUBs regulate mitophagy in a positive/negative manner. For example, USP8 and USP13 promote mitophagy by directly detaching ubiquitin from parkin, whereas USP15, USP30, USP33, and USP35 inhibit parkin-mediated ubiquitination of OMM proteins. Accordingly, these DUBs promote or suppress mitophagy during the removal of ubiquitin from the parkin. USP14 negatively regulates proteasome activity, and also acts as a negative regulator of mitophagy.
The relationship between DUBs and PD-related genes
α-Synuclein and DUBs: Patients with sporadic PD generally show extensive mitochondrial dysfunction with toxic accumulation of α-Syn aggregates (12). α-Syn is the main component of LB and its mutation, duplication, or triplication results in autosomal-dominant PD (50). Since ubiquitination plays an essen-tial role in regulating both α-Syn levels and mitochondrial quality control, the degradation of defective mitochondria should be accurately regulated to prevent accumulation of misfolded α-Syn. A few studies reported DUBs targeting to the α-Syn (Table 2). For example, USP8 deubiquitinated K63-linked ubiquitin chains on α-Syn, and knockdown of endogenous USP8 prevented α-Syn-induced toxicity in a Drosophila model (51). These data demonstrated that altered deubiquitination of α-Syn by USP8 contributes to PD pathogenesis. In addition, USP9X was thought to be a key regulator in altering the mono-ubiquitination level of α-Syn, which also reduces the α-Syn toxicity (52). Based on these findings, the regulation of USP9X deubiquitinase activity might play a role in decreasing the levels of toxic α-Syn ag-gregates, and their cellular toxicity. Moreover, parkin mediates the clearance of α-Syn (53) and knockdown of USP13 increases parkin ubiquitination, whereas the clearance of α-Syn is promoted in a parkin-independent manner (54). Although parkin ubiquitination is not directly related to α-Syn degradation, USP13 has a direct effect on the cellular activity of α-Syn and its neuronal death.
Table 2.
The functional link between DUBs and three PD-related gene products
| PD genes | DUBs | The role of DUBs in the regulation of PD-related genes | References |
|---|---|---|---|
| α-Synuclein | USP8 | Deubiquitinates K63-linked ubiquitin chains of α-synuclein and ameliorates α-synuclein induced toxicity | Alexopoulou Z et al. |
| USP9X | Regulates mono-ubiquitination of α-synuclein to reduce its aggregation and cellular toxicity | Rott R et al. | |
| USP13 | Knockdown shows clearance of α-synuclein in a parkin-independent manner but directly regulates α-synuclein-mediated neuronal death | Liu X et al. | |
| PINK1/Parkin | USP15 | Attenuates the clearance of dysfunctional mitochondria but doesn’t affect the ubiquitination status of parkin | Bingol B et al. |
| Cornelissen T et al. | |||
| USP30 | Eliminates the parkin-mediated signals and reduces clearance of damaged mitochondria | Wang Y et al. | |
| USP35 | |||
| USP33 | Removes several kinds of lysine-linked ubiquitin chains from parkin, whereas its knockdown increases the protein stability of parkin | Niu K et al. | |
| Chakraborty J et al. | |||
| USP14 | Negatively regulates proteasome activity, leading to the inhibition of mitochondrial clearance | Chakraborty J et al. | |
| Wang L et al. |
Many studies have reported the mitochondrial translocation of α-Syn via its N-terminus, impairing the mitochondrial function (55, 56). Further, α-Syn was also found to impair autophagy, particularly mitophagy. α-Syn impairs mitophagy in numerous ways. In the neurons of PD patients, α-Syn interacts with Miro via its N-terminus and upregulates Miro protein levels, leading to excessive and abnormal accumulation of Miro on the mitochondrial surface and delayed mitophagy. These results suggest that Miro is a target of α-Syn-associated mitochondrial injury (57). In addition, the overexpression of A53T α-Syn mutant results in p38 MAPK activation, and directly induces the phosphorylation of parkin at serine 131, disturbing the function of parkin and mitophagy (58). In A53T α-Syn-overexpressing mice, α-Syn accumulates on mitochondria, resulting in increased mitophagy and neuronal death, and these mitochondrial deficits can be rescued by silencing parkin and overexpressing MFN2 or a dominant-negative variant of Drp1 (53, 59). Yeast overexpressing both the human wild-type α-Syn gene and A53T mutant resulted in enhanced mitophagy (60). These studies indicate the role of abnormal mitophagy in α-syn-mediated toxicity.
Another DUB, UCH-L1, which is also associated with familial PD, affects mitophagy. UCH-L1 alters the polyubiquitin chain and increases the availability of free monomeric ubiquitin to the UPS, thus increasing proteasome-dependent proteolysis (61). Interestingly, the I94M mutation in UCH-L1 has been found in patients carrying autosomal dominant PD (62). Reduced mRNA and protein levels of this DUB were found in samples obtained from frontal cortex and medulla oblongata in patients dying from PD. This UCH-L1 directly interacts with chaperone-mediated autophagy (CMA), by physically binding to LAMP-2A, Hsp70, and Hsp90 (63). It should be noted that a protective S18Y-UCH-L1 variant is negatively correlated with disease onset (64).
PINK1/Parkin and DUBs: Duncan et al. demonstrated that USP8 directly deubiquitinates parkin and abrogates its autoubiquitination effect (49). RNAi-mediated knockdown of USP8 in various cell lines resulted in delayed parkin recruitment to depolarized mitochondria and in the accumulation of ubiqui-tinated parkin, which persisted longer in the damaged mitochondria and induced a delay in their clearance by mitophagy. In addition, USP8 was shown to selectively remove K6-linked ubiquitin chains from parkin, and was critical for efficient mitophagy (65). Three additional DUBs were found to be linked with mitophagy (Table 2). USP15 and two mitochondrial DUBs, USP30 and USP35, were shown to oppose parkin-mediated ubiquitination of OMM proteins and attenuate the subsequent clearance of depolarized mitochondria (66, 67). In contrast to USP8, neither USP15 nor USP30 affected the ubiquitination status of parkin or its recruitment to damaged mitochondria. While USP30 delays parkin-mediated mitophagy by interfering with its recruitment to the mitochondria, USP35 does not delay parkin recruitment, suggesting differential regulation of mitophagy via distinct mechanisms (68). Interestingly, USP35 only associates with polarized mitochondria, and rapidly translocates to the cytosol during CCCP-induced mitophagy (Fig. 1).
Recently, another mitochondrial DUB, USP33, present at the OMM was found to deubiquitinate parkin by removing the K6-, K11-, K48- and K63-linked ubiquitin conjugates from parkin (69; Table 2). USP33 knockdown increased both parkin protein stability and its translocation to depolarized mitochondria, resulting in enhanced mitophagy and eventual protection of human neuroblastoma cells from the neurotoxin MPTP-induced apoptotic cell death. Interestingly, pharmacological or genetic inhibition of USP14 leads to increased mitochondrial clearance in the absence of PINK1 and parkin (70). Mitochondrial fragmentation and membrane rupture exposes the autophagy receptor prohibitin 2 and the formation of mitophagic vesicles, which are key elements in USP14-induced mitophagy (71). In addition, genetic or pharmacological inhibition of USP14 in vivo corrected mitochondrial dysfunction and rectified the impaired locomotion in the established PINK1- and parkin-mutant Drosophila model of PD. Thus, USP14 is closely related to proteasome activity and negatively affects proteasome-mediated proteolysis, thereby inhibiting mitophagy via deubiquitinating activity of USP14 (71).
CONCLUSION
Investigation of cellular mechanisms underpinning mitochondrial quality control in CNS has led to the development of potential therapeutics for neurological disorders. Elucidation of the mitophagy pathway and underlying regulatory mechanisms has revealed the close relationship between the coordinated mitochondrial dynamics and several PD-associated genes. Several genes, such as LRRK2, PINK1, parkin, and a-synuclein, are causally linked to idiopathic and familiar PD. These gene products directly or indirectly modulate the mitochondrial quality control system, e.g., mitochondrial fission and fusion, biogenesis, and maintenance of ER/mitochondrial Ca2+ balance. Many cases have reported the mitochondrial dysfunction in PD patients. These results consistently support the hypothesis that the balance between mitochondria function and clearance is important for homeostasis in normal cells. Defective mitochondrial function and clearance lead to many NDDs, including PD. Although the development of therapeutic strategies targeting PD are based on diverse molecular mechanisms, selective regulation of autophagy and mitophagy system represents a promising strategy to delay the progression of PD. Moreover, multiple ubiquitin proteases have shown to regulate PINK1/parkin-dependent mitophagy positively or negatively. Several DUBs counteract the auto-ubiquitination pathway of parkin and its target in the sequential reactions of mitophagy. For example, USP8 eliminated K6-linked ubiquitin conjugates from parkin (18), and the overexpression of USP30 and USP35 delayed PINK/parkin-mediated mitophagy (68). In contrast, knockdown of some DUBs facilitates mitophagy in a PINK1/parkin-dependent manner, resulting in a neuroprotective effect. Based on the diverse and important roles of DUBs in mitophagy, elimi-nating abnormal mitochondria via UPS- and DUBs-controlled mitophagy is a key strategy to delay the onset of PD. Collectively, understanding of DUBs functions and their regulation in cellular mitophagic pathway provides a novel but effective therapeutic approach against PD.
ACKNOWLEDGEMENTS
This work was supported by a grant from the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT, No. 2021R1A2C1005469 to KCC). We apologize to several researchers whose work could not be cited in this review due to space limitations.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Henderson MX, Trojanowski JQ, Lee VMY. α-Synuclein pathology in Parkinson's disease and related α-synucleinopathies. Neurosci Lett. 2019;709:134316. doi: 10.1016/j.neulet.2019.134316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan BJ, Hoek S, Fon EA, Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson's disease: from mechanism to therapy. Trends Biochem Sci. 2020;40:200–210. doi: 10.1016/j.tibs.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Wang W, Wang X, Fujioka H, et al. Parkinson's disease-associated mutant VPS35 causes mitochondrial dysfunction by recycling DLP1 complexes. Nat Med. 2016;22:54–63. doi: 10.1038/nm.3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Um JH, Yun JH. Emerging role of mitophagy in human diseases and physiology. BMB Rep. 2017;50:299. doi: 10.5483/BMBRep.2017.50.6.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palikaras K, Lionaki E, Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol. 2018;20:1013–1022. doi: 10.1038/s41556-018-0176-2. [DOI] [PubMed] [Google Scholar]
- 6.Heo JM, Ordureau A, Paulo JA, Rinehart J, Harper JW. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol Cell. 2015;60:7–20. doi: 10.1016/j.molcel.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nijman SM, Luna-Vargas MP, Velds A, et al. Genomic and functional inventory of deubiquitinating enzymes. Cell. 2015;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiqui-tinating enzymes. Annu Rev Biochem. 2019;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006;443:787–795. doi: 10.1038/nature05292. [DOI] [PubMed] [Google Scholar]
- 10.Marella M, Seo BB, Yagi T, Matsuno-Yagi A. Parkinson's disease and mitochondrial complex I: a perspective on the Ndi1 therapy. J Bioenerg Biomembr. 2009;41:493–497. doi: 10.1007/s10863-009-9249-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrucci M, Pasquali L, Ruggieri S, Paparelli A, Fornai F. Alpha-synuclein and autophagy as common steps in neurodegeneration. Parkinsonism Relat Disord. 2008;14:S180–S184. doi: 10.1016/j.parkreldis.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Devoto VT, Falzone TL. Mitochondrial dynamics in Parkinson's disease: a role for α-synuclein. Dis Model Mech. 2017;10:1075–1087. doi: 10.1242/dmm.026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vicario M, Cieri D, Brini M, Calì T. The close encounter between alpha-synuclein and mitochondria. Front Neurosci. 2018;12:1–13. doi: 10.3389/fnins.2018.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kane LA, Lazarou M, Fogel AI, et al. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J Cell Biol. 2014;205:143–153. doi: 10.1083/jcb.201402104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Durcan TM, Fon EA. The three 'P's of mitophagy: PARKIN, PINK1, and post-translational modifications. Genes Dev. 2015;29:989–999. doi: 10.1101/gad.262758.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eiyama A, Okamoto K. PINK1/Parkin-mediated mitophagy in mammalian cells. Curr Opin Cell Biol. 2015;33:95–101. doi: 10.1016/j.ceb.2015.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Swatek KN, Komander D. Ubiquitin modifications. Cell Res. 2016;26:399–422. doi: 10.1038/cr.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan T, Zimmermann M, Reichert AS. Controlling quality and amount of mitochondria by mitophagy: insights into the role of ubiquitination and deubiquitination. Biol Chem. 2016;397:637–647. doi: 10.1515/hsz-2016-0125. [DOI] [PubMed] [Google Scholar]
- 19.Shaid S, Brandts CH, Serve H, Dikic I. Ubiquitination and selective autophagy. Cell Death Differ. 2013;20:121–130. doi: 10.1038/cdd.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guhathakurta S, Kim J, Adams L, et al. Targeted attenuation of elevated histone marks at SNCA alleviates α‐synuclein in Parkinson's disease. EMBO Mol Med. 2021;13:e12188. doi: 10.15252/emmm.202012188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sugiura A, McLelland GL, Fon EA, McBride HM. A new pathway for mitochondrial quality control: mitochondrial-derived vesicles. EMBO J. 2014;33:2142–2156. doi: 10.15252/embj.201488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanik SA, Schultheiss CE, Volpicelli-Daley LA, Brunden KR, Lee VM. Lewy body-like α-synuclein aggregates resist degradation and impair macroautophagy. J Biol Chem. 2013;288:15194–15210. doi: 10.1074/jbc.M113.457408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chinta SJ, Mallajosyula JK, Rane A, Andersen JK. Mitochondrial alpha-synuclein accumulation impairs complex I function in dopaminergic neurons and results in increased mitophagy in vivo. Neurosci Lett. 2010;486:235–239. doi: 10.1016/j.neulet.2010.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. Impaired degradation of mutant α-synuclein by chaperone-mediated autophagy. Science. 2004;305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 25.Narendra DP, Jin SM, Tanaka A, et al. PINK1 is selectively stabilized on impaired mitochondria to activate Parkin. PLoS Biol. 2010;8:e1000298. doi: 10.1371/journal.pbio.1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McLelland GL, Soubannier V, Chen CX, McBride HM, Fon EA. Parkin and PINK 1 function in a vesicular trafficking pathway regulating mitochondrial quality control. EMBO J. 2014;33:282–295. doi: 10.1002/embj.201385902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thomas KJ, McCoy MK, Blackinton J, et al. DJ-1 acts in parallel to the PINK1/Parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Y, Ma X, Fujioka H, Liu J, Chen S, Zhu X. DJ-1 regulates the integrity and function of ER-mitochondria association through interaction with IP3R3-Grp75-VDAC1. Proc Natl Acad Sci U S A. 2019;116:25322–25328. doi: 10.1073/pnas.1906565116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Gong XG, Wang ZZ, et al. Overexpression of DJ‐1/PARK7, the Parkinson's disease‐related protein, improves mitochondrial function via Akt phosphorylation on threonine 308 in dopaminergic neuron‐like cells. Eur J Neurosci. 2016;43:1379–1388. doi: 10.1111/ejn.13216. [DOI] [PubMed] [Google Scholar]
- 30.Thomas HE, Zhang Y, Stefely JA, et al. Mitochon-drial complex I activity is required for maximal autophagy. Cell Rep. 2018;24:2404–2417. doi: 10.1016/j.celrep.2018.07.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joselin AP, Hewitt SJ, Callaghan SM, et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21:4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- 32.Thomas KJ, McCoy MK, Blackinton J, et al. DJ-1 acts in parallel to the PINK1/Parkin pathway to control mitochondrial function and autophagy. Hum Mol Genet. 2011;20:40–50. doi: 10.1093/hmg/ddq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao LY, Giasson BI, Bonini NM. DJ-1 is critical for mitochondrial function and rescues PINK1 loss of function. Proc Natl Acad Sci U S A. 2010;107:9747–9752. doi: 10.1073/pnas.0911175107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walter J, Bolognin S, Antony PM, et al. Neural stem cells of Parkinson's disease patients exhibit aberrant mitochondrial morphology and functionality. Stem Cell Reports. 2019;12:878–889. doi: 10.1016/j.stemcr.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Obergasteiger J, Frapporti G, Lamonaca G, et al. Kinase inhibition of G2019S-LRRK2 enhances autolysosome formation and function to reduce endogenous alpha-synuclein intracellular inclusions. Cell Death Discov. 2020;6:1–13. doi: 10.1038/s41420-020-0279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carballo-Carbajal I, Weber-Endress S, Rovelli G, et al. Leucine-rich repeat kinase 2 induces α-synuclein expression via the extracellular signal-regulated kinase pathway. Cell Signal. 2010;22:821–827. doi: 10.1016/j.cellsig.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonello F, Hassoun SM, Mouton-Liger F, et al. LRRK2 impairs PINK1/Parkin-dependent mitophagy via its kinase activity: pathologic insights into Parkinson's disease. Hum Mol Genet. 2019;28:1645–1660. doi: 10.1093/hmg/ddz004. [DOI] [PubMed] [Google Scholar]
- 38.Yakhine-Diop SM, Niso-Santano M, Rodríguez-Arribas M. Impaired mitophagy and protein acetylation levels in fibroblasts from Parkinson's disease patients. Mol Neurobiol. 2019;56:2466–2481. doi: 10.1007/s12035-018-1206-6. [DOI] [PubMed] [Google Scholar]
- 39.Kalia SK, Lee S, Smith PD, et al. BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron. 2004;44:931–945. doi: 10.1016/j.neuron.2004.11.026. [DOI] [PubMed] [Google Scholar]
- 40.De Snoo ML, Friesen EL, Zhang YT, et al. Bcl-2-associated athanogene 5 (BAG5) regulates Parkin-dependent mitophagy and cell death. Cell Death Dis. 2019;10:907. doi: 10.1038/s41419-019-2132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Guo J, Fei E, et al. BAG5 protects against mitochondrial oxidative damage through regulating PINK1 degradation. PLoS One. 2014;9:e86276. doi: 10.1371/journal.pone.0086276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qu D, Hage A, Don-Carolis K, et al. BAG2 gene-mediated regulation of PINK1 protein is critical for mitochondrial translocation of PARKIN and neuronal survival. J Biol Chem. 2015;290:30441–30452. doi: 10.1074/jbc.M115.677815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Grossmann D, Berenguer-Escuder C, Chemla A, Arena G, Krüger R. The emerging role of RHOT1/Miro1 in the pathogenesis of Parkinson's disease. Front Neurol. 2020;11:587. doi: 10.3389/fneur.2020.00587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berenguer-Escuder C, Grossmann D, Massart F, et al. Variants in Miro1 cause alterations of ER-mitochondria contact sites in fibroblasts from Parkinson's disease patients. J Clin Med. 2019;8:2226. doi: 10.3390/jcm8122226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Safiulina D, Kuum M, Choubey V, et al. Miro proteins prime mitochondria for Parkin translocation and mitophagy. EMBO J. 2019;38:e99384. doi: 10.15252/embj.201899384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birsa N, Norkett R, Wauer T, et al. Lysine 27 ubiquitination of the mitochondrial transport protein Miro is dependent on serine 65 of the Parkin ubiquitin ligase. J Biol Chem. 2014;289:14569–14582. doi: 10.1074/jbc.M114.563031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Todi SV, Paulson HL. Balancing act: deubiquitinating enzymes in the nervous system. Trends Neurosci. 2011;34:370–382. doi: 10.1016/j.tins.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Magraoui FE, Reidick C, Meyer HE, Platta HW. Autophagy-related deubiquitinating enzymes involved in health and disease. Cells. 2015;4:596–621. doi: 10.3390/cells4040596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Durcan TM, Tang MY, Pérusse JR, et al. USP8 regulates mitophagy by removing K6-linked ubiquitin conju-gates from parkin. EMBO J. 2014;33:2473–2491. doi: 10.15252/embj.201489729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Polymeropoulos MH. Autosomal dominant Parkinson's disease and alpha-synuclein. Ann Neurol. 1998;44:S63–64. doi: 10.1002/ana.410440710. [DOI] [PubMed] [Google Scholar]
- 51.Alexopoulou Z, Lang J, Perrett RM, et al. Deubiquitinase USP8 regulates α-synuclein clearance and modifies its toxicity in Lewy body disease. Proc Natl Acad Sci U S A. 2016;113:E4688–E4697. doi: 10.1073/pnas.1523597113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rott R, Szargel R, Haskin J, et al. α-Synuclein fate is determined by USP9X-regulated monoubiquitination. Proc Proc Natl Acad Sci U S A. 2011;108:18666–18671. doi: 10.1073/pnas.1105725108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lonskaya I, Desforges NM, Hebron ML, Moussa CE. Ubiquitination increases parkin activity to promote autophagic α-synuclein clearance. PLoS One. 2013;8:e83914. doi: 10.1371/journal.pone.0083914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Liu X, Hebron M, Shi W, Lonskaya I, Moussa CE. Ubiquitin specific protease-13 independently regulates parkin ubiquitination and alpha-synuclein clearance in alpha-synucleinopathies. Hum Mol Genet. 2019;28:548–560. doi: 10.1093/hmg/ddy365. [DOI] [PubMed] [Google Scholar]
- 55.Devi L, Raghavendran V, Prabhu BM, Avadhani NG, Anandatheerthavarada HK. Mitochondrial import and accumulation of alpha-synuclein impair complex I in human dopaminergic neuronal cultures and Parkinson disease brain. J Biol Chem. 2008;283:9089–9100. doi: 10.1074/jbc.M710012200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen J, Du T, Wang X, et al. α-Synuclein amino terminus regulates mitochondrial membrane permeability. Brain Res. 2014;1591:14–26. doi: 10.1016/j.brainres.2014.09.046. [DOI] [PubMed] [Google Scholar]
- 57.Liu J, Liu W, Li R, Yang H. Mitophagy in Parkinson's disease: From pathogenesis to treatment. Cells. 2019;8:712. doi: 10.3390/cells8070712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hirota Y, Yamashita S, Kurihara Y, et al. Mitophagy is primarily due to alternative autophagy and requires the MAPK1 and MAPK14 signaling pathways. Autophagy. 2015;11:332–343. doi: 10.1080/15548627.2015.1023047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rojas-Charr L, Cookson MR, Niño A, Arboleda H, Arboleda G. Downregulation of PINK1 influences mitochondrial fusion-fission machinery and sensitizes to neurotoxins in dopaminergic cells. Neurotoxicology. 2014;44:140–148. doi: 10.1016/j.neuro.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dixon C, Mathias N, Zweig RM, Davis DA, Gross DS. Alpha-synuclein targets the plasma membrane via the secretory pathway and induces toxicity in yeast. Genetics. 2005;170:47–59. doi: 10.1534/genetics.104.035493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bishop P, Rocca D, Henley JM. Ubiquitin C-terminal hydrolase L1 (UCH-L1): structure, distribution and roles in brain function and dysfunction. Biochem J. 2016;473:2453–2462. doi: 10.1042/BCJ20160082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakraborty J, Ziviani E. Deubiquitinating enzymes in Parkinson's disease. Front Physiol. 2020;11:535. doi: 10.3389/fphys.2020.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kabuta T, Furuta A, Aoki S, Furuta K, Wada K. Aberrant interaction between Parkinson disease-associated mutant UCH-L1 and the lysosomal receptor for chaperone-mediated autophagy. J Biol Chem. 2008;283:23731–23738. doi: 10.1074/jbc.M801918200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carmine Belin A, Westerlund M, Bergman O, et al. S18Y in ubiquitin carboxy-terminal hydrolase L1 (UCH-L1) associated with decreased risk of Parkinson's disease in Sweden. Parkinsonism Relat Disord. 2007;13:295–298. doi: 10.1016/j.parkreldis.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 65.Faesen AC, Luna-Vargas MP, Geurink PP. The differential modulation of USP activity by internal regulatory domains, interactors and eight ubiquitin chain types. Chem Biol. 2011;18:1550–1561. doi: 10.1016/j.chembiol.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 66.Bingol B, Tea JS, Phu L, et al. The mitochondrial deubiquitinase USP30 opposes Parkin-mediated mitophagy. Nature. 2014;510:370–375. doi: 10.1038/nature13418. [DOI] [PubMed] [Google Scholar]
- 67.Cornelissen T, Haddad D, Wauters F, et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Y, Serricchio M, Jauregui M, et al. Deubiquitinating enzymes regulate PARK2-mediated mitophagy. Autophagy. 2015;11:595–606. doi: 10.1080/15548627.2015.1034408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Niu K, Fang H, Chen Z, et al. USP33 deubiquitinates PRKN/Parkin and antagonizes its role in mitophagy. Autophagy. 2020;16:724–734. doi: 10.1080/15548627.2019.1656957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chakraborty J, von Stockum S, Marchesan E, et al. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol Med. 2018;10:e9014. doi: 10.15252/emmm.201809014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Qi H, Tang Y, Shen HM. Post-translational modifications of key machinery in the control of mitophagy. Trends Biochem Sci. 2020;45:58–75. doi: 10.1016/j.tibs.2019.08.002. [DOI] [PubMed] [Google Scholar]