Abstract
African swine fever (ASF) is a viral hemorrhagic disease that affects domestic pigs and wild boar and is caused by the African swine fever virus (ASFV). The ASFV virion contains a long double-stranded DNA genome, which encodes more than 150 proteins. However, the immune escape mechanism and pathogenesis of ASFV remain poorly understood. Here, we report that the pyroptosis execution protein gasdermin D (GSDMD) is a new binding partner of ASFV-encoded protein S273R (pS273R), which belongs to the SUMO-1 cysteine protease family. Further experiments demonstrated that ASFV pS273R-cleaved swine GSDMD in a manner dependent on its protease activity. ASFV pS273R specifically cleaved GSDMD at G107-A108 to produce a shorter N-terminal fragment of GSDMD consisting of residues 1 to 107 (GSDMD-N1–107). Interestingly, unlike the effect of GSDMD-N1–279 fragment produced by caspase-1-mediated cleavage, the assay of LDH release, cell viability, and virus replication showed that GSDMD-N1–107 did not trigger pyroptosis or inhibit ASFV replication. Our findings reveal a previously unrecognized mechanism involved in the inhibition of ASFV infection-induced pyroptosis, which highlights an important function of pS273R in inflammatory responses and ASFV replication.
Keywords: African swine fever virus, pS273R, gasdermin D, pyroptosis, viral replication
Abbreviations: ASF, African swine fever; ASFV, African swine fever virus; Co-IP, coimmunoprecipitation; GSDMD, gasdermin D; hpt, hours posttransfection; MOI, multiplicity of infection; PAMs, porcine alveolar macrophages
African swine fever (ASF) is a viral hemorrhagic disease that affects domestic pigs and wild boar and is characterized by a series of different clinical symptoms, including anorexia, moribundity, bloody diarrhea, vomiting, high fever, and hemorrhagic symptoms (1, 2). Because ASF was first discovered in Kenya in the 1920s, it has rapidly spread to many areas in sub-Saharan Africa, the Caucasus, and Eastern Europe (2). The first ASF case was reported in August of 2018 in China and caused serious economic losses (3). To date, there are no effective drugs or commercial vaccines against American swine fever virus (ASFV) (4).
ASFV is the only member of the family Asfarviridae, genus Asfivirus. The ASFV virion is a large, icosahedral structure of approximately 200 nm. The ASFV genome contains a linear double-stranded DNA (170–190 kbp) that encodes approximately 150 proteins (5). Similar to other nucleocytoplasmic large DNA viruses (NCLDVs), ASFV encodes many proteins involved in not only the viral life cycle, including viral entry, DNA replication and repair, viral assembly, and egress (6) but also the evasion of host defenses, including the inhibition of host innate immune responses (such as type I-interferon production and inflammatory responses) and cell death pathways (7).
ASFV encodes two large polyprotein precursors, pp220 and pp62, which are proteolytically cleaved by ASFV pS273R to yield the structural proteins required for virus assembly (8, 9). The cysteine protease pS273R encoded by ASFV is synthesized at the late stages of viral infection and is localized within cytoplasmic viral factories (9). The overall structure of the ASFV pS273R is represented by two domains named the N-terminal “arm domain” and the “core domain” (10). The “arm domain” contains the residues from M1 to N83, which is unique to ASFV and plays an important role in maintaining the enzyme activity of ASFV pS273R (10). The “core domain” contains the residues from N84 to A273, which shares a high degree of structural similarity with chlamydial-deubiquitinating enzyme, sentrin-specific protease, and adenovirus protease.
Pyroptosis is a recently discovered form of programmed cell death that is activated in response to diverse microbial ligands, including bacterial flagellin, toxins, lipopolysaccharide, and DNA that gains access to the cell cytosol (11). Pyroptosis is characterized by the activation of inflammatory caspases (such as caspase-1 and caspase-4/5/11) and pore formation in the cellular plasma membrane, resulting in the release of a large number of proinflammatory cytokines (12). Recently, gasdermin family members, such as gasdermin D (GSDMD), were found to be cleaved by inflammatory-related caspases to execute pyroptosis. For example, GSDMD, a critical mediator of pyroptosis, can be cleaved within a linker between the N-terminal domain and the C-terminal domain by activated caspase-1 (13, 14), caspase-4/5/11 (15, 16), and caspase-8 (17, 18). Subsequently, the N-terminal domain of human GSDMD (GSDMD-N1–275) oligomerizes to be inserted into the plasma membrane, resulting in pore formation (19). It is widely accepted that the pore formation by human GSDMD-N1–275 results in the loss of osmotic homeostasis, leading to cell swelling and death, which releases inflammatory factors to inhibit and clear intracellular pathogens (20, 21). Recently, the caspase-3-mediated cleavage of GSDME (22) and the granzyme A-mediated cleavage of GSDMB were also reported (23).
Here, we report that swine GSDMD is a novel binding partner of the ASFV-encoded pS273R. ASFV infection decreased the expression of GSDMD by cleavage of GSDMD at the G107-A108 pair to yield a nonfunctional GSDMD-N fragment consisting of amino acids (aa) 1 to 107 (GSDMD-N1–107). Mechanistically, unlike the GSDMD-N1–279 produced by caspase-1, the GSDMD-N1–107 produced by pS273R did not induce pyroptosis and inhibit ASFV replication. Our findings reveal a previously unrecognized novel mechanism by which ASFV evades the host antiviral innate immune responses.
Results
GSDMD is a novel target of the ASFV pS273R protease
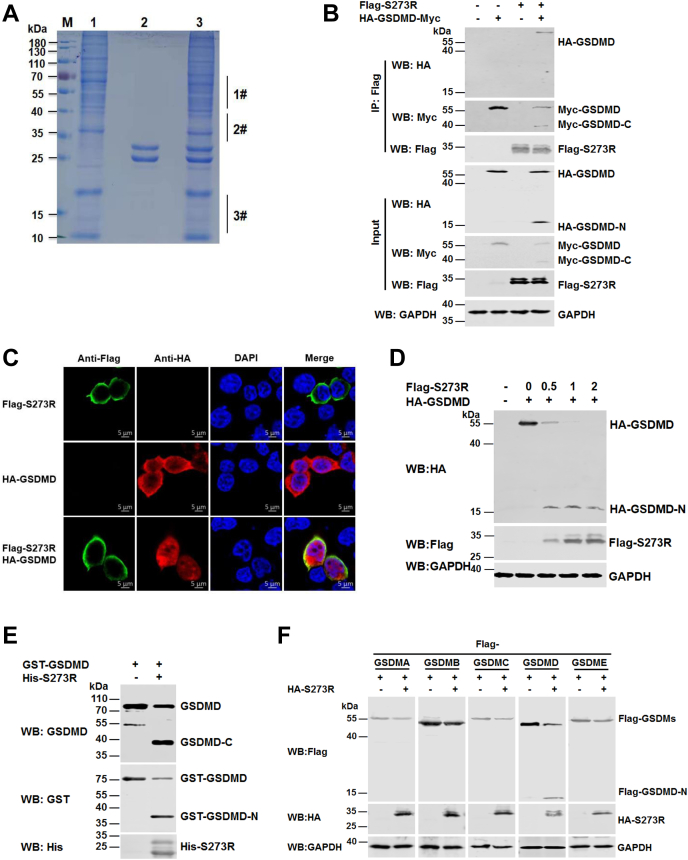
To investigate the novel function of ASFV-encoded pS273R (ASFV pS273R) on the host cellular immune responses, we first screened and identified the host proteins that interact with ASFV pS273R by pull down-mass spectrometry. The cell lysates of porcine alveolar macrophages (PAMs) were incubated with His-pS273R protein bound to Ni Sepharose. ASFV pS273R-binding proteins were eluted and stained with Coomassie brilliant blue. We found that several specific bands were observed in the eluted proteins in the His-pS273R group (lane 3) compared with the control group (lane 1 and lane 2) (Fig. 1A). Then, the gels containing these specific bands (lane 3) were cut into three pieces as indicated and analyzed by mass spectrometry. As shown in Table 1, 12 pS273R-interacting proteins were identified in the sample 1# (sample 2# and 3# data not shown). Among them, GSDMD, a pyroptosis executor, was selected for subsequent studies due to its score and role in pyroptosis.
Figure 1.
ASFV pS273R interacts with and cleaves GSDMD in vitro.A, the cell lysates of PAMs were incubated with Ni Sepharose bound with His-pS273R (lane 3), whereas Ni Sepharose incubated with the cell lysates of PAMs (lane 1) was used as a negative control. Then, His-pS273R and its binding partners were eluted with elution buffer, resolved by SDS-PAGE, and stained with Coomassie brilliant blue. Lane M, protein marker. Lane 1, cell lysates of PAMs. Lane 2, purified ASFV pS273R. The 1#, 2#, and 3# in lane 3 indicates that the specific bands containing regions were cut for mass spectrometry analysis. B, HEK293T cells were transfected with a plasmid encoding HA-GSDMD-Myc, in combination with a plasmid expressing Flag-tagged ASFV pS273R or empty vector as indicated. The cell lysates were coimmunoprecipitated with an anti-Flag antibody. The immunoprecipitants and aliquots of the whole-cell lysates were subjected to Western blot analysis with anti-HA, anti-Flag, anti-Myc, or anti-GAPDH antibodies. C, HEK293T cells were transfected with a plasmid expressing Flag-pS273R or HA-tagged GSDMD alone or both. The cells were probed with rabbit anti-HA monoclonal antibody (mAb) (red) and mouse anti-Flag mAb (green). The cell nuclei (blue) were stained with DAPI. D, HEK293T cells were transfected with a plasmid expressing HA-GSDMD alone or together with increasing doses (0, 0.5, 1, or 2 μg) of a plasmid encoding Flag-pS273R. The cell lysates were analyzed with the indicated antibodies. E, recombinant GST-tagged GSDMD was incubated with purified His-tagged pS273R (6 × His-pS273R). The action products were analyzed with anti-GST and anti-GSDMD antibodies. F, HEK293T cells were transfected with a plasmid expressing Flag-tagged GSDMA, GSDMB, GSDMC, GSDMD, or GSDME, along with a plasmid encoding HA-pS273R. At 36 hpt, the cell lysates were analyzed by Western blot with the indicated antibodies. ASFV, African swine fever virus; DAPI, 4,6-diamidino-2-phenylindole; GSDMD, gasdermin D; hpt, hours posttransfection; PAMs, porcine alveolar macrophages.
Table 1.
Results of mass spectrometry analysis of the host proteins that interact with pS273R
| Sample name | Accession | Mass (Da) | Score | Matches | Sequences | emPAI | Protein description |
|---|---|---|---|---|---|---|---|
| 1 | GSDMD | 51,941 | 613 | 30 (21) | 12 (10) | 2.22 | Gasdermin D |
| 2 | ARHGAP45 | 126,235 | 277 | 24 (11) | 20 (9) | 0.36 | Minor histocompatibility protein HA-1 isoform X3 |
| 3 | NOP9 | 70,389 | 160 | 8 (3) | 7 (2) | 0.15 | Uncharacterized protein |
| 4 | WDR7 | 165,837 | 116 | 13 (4) | 9 (4) | 0.08 | WD repeat domain 7 |
| 5 | RUFY3 | 68,422 | 60 | 4 (2) | 3 (1) | 0.1 | Protein RUFY3 isoform X4 |
| 6 | CHERP | 104,279 | 53 | 5 (1) | 5 (1) | 0.03 | Calcium homeostasis endoplasmic reticulum protein |
| 7 | ABCE1 | 68,254 | 39 | 7 (1) | 6 (1) | 0.05 | Uncharacterized protein |
| 8 | RPL4 | 48,287 | 39 | 3 (2) | 2 (2) | 0.14 | Ribosomal protein L4 |
| 9 | ECE2 | 172,699 | 35 | 3 (1) | 3 (1) | 0.04 | Uncharacterized protein |
| 10 | FLII | 150,898 | 31 | 2 (0) | 2 (0) | 0.02 | FLII actin remodeling protein |
| 11 | CRIP1 | 8926 | 21 | 1 (0) | 1 (0) | 0.39 | Cysteine-rich intestinal protein 1 |
| 12 | STAT3 | 81,896 | 20 | 7 (0) | 5 (0) | 0.04 | Signal transducer and activator of transcription |
To further characterize the interaction between ASFV pS273R and swine GSDMD, Flag-pS273R or HA-GSDMD alone or both were coexpressed in HEK293T cells, and the interaction and the subcellular colocalization of the two proteins were examined. As shown in Figure 1, B and C, ASFV pS273R interacted with and colocalized with GSDMD in the cytoplasm. In addition, we noticed that ASFV S273R interacted with the C-terminal of swine GSDMD and GSDMD in a low activity proteolysis. We also observed that the reduced abundance of full-length GSDMD (55 kDa) was accompanied by the appearance of an N-terminal fragment of GSDMD with apparent molecular masses of approximately 10 and 15 kDa (designated GSDMD-N), suggesting that ASFV pS273R may cleave swine GSDMD in vitro. To further confirm whether GSDMD is cleaved by ASFV pS273R, HEK293T cells were cotransfected with a plasmid encoding Flag-GSDMD and increasing amounts of a plasmid expressing ASFV pS273R. As shown in Figure 1D, the intensity of Flag-GSDMD diminished as the protein levels of ASFV pS273R were increased, and an N-terminal fragment of GSDMD began to appear, suggesting that GSDMD is cleaved by ASFV pS273R in a dose-dependent manner. To further confirm that ASFV pS273R directly cleaves GSDMD in vitro, purified ASFV pS273R (6 × His-pS273R) and GST-GSDMD were incubated, and the reaction products were detected with anti-His and anti-GSDMD antibodies. As shown in Figure 1E, we found that the purified ASFV pS273R cleaved GST-GSDMD and produced a GST-GSDMD-N fragment with a molecular mass of approximately 39 kDa. To further investigate whether GSDMD is specifically cleaved by ASFV pS273R, HEK293T cells were transfected with a plasmid expressing Flag-tagged GSDMA, GSDMB, GSDMC, GSDMD, or GSDME in combination with an empty vector or a plasmid expressing HA-pS273R. As shown in Figure 1F, GSDMD, but none of the other GSDMDs chosen in this study, was cleaved by ASFV pS273R.
ASFV pS273R interacts with and cleaves GSDMD in ASFV-infected PAMs
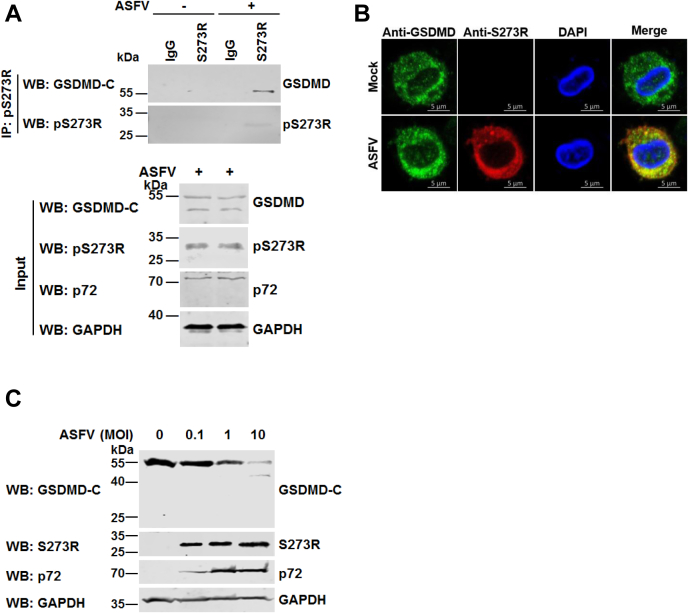
To test whether ASFV pS273R interacts with endogenous GSDMD in PAMs upon ASFV infection. PAMs were infected with ASFV HLJ/18 and a Co-IP assay was performed. We found that ASFV pS273R interacted with endogenous GSDMD in PAMs infected with ASFV (Fig. 2A). Meanwhile, we also noticed that the ASFV pS273R colocalized with endogenous GSDMD in the cytoplasm in PAMs following ASFV infection (Fig. 2B). These data suggested that ASFV pS273R specifically interacts with GSDMD. To investigate the impact of ASFV infection on the integrity of GSDMD, we analyzed the expression of GSDMD in PAMs either mock-infected or infected with ASFV at different multiplicities of infection (MOIs). As shown in Figure 2C, upon viral infection with increasing doses of ASFV, the levels of endogenous GSDMD decreased in a dose-dependent manner, and only an approximately 40 kDa cleavage product (designated GSDMD-C) was recognized by the anti-GSDMD antibody against the C-terminus of GSDMD.
Figure 2.
ASFV-encoded pS273R targets endogenous GSDMD in ASFV-infected PAMs.A, PAMs were infected with ASFV at an MOI of 0.1 for 36 h, and then the cell lysates were coimmunoprecipitated with mouse IgG or anti-pS273R polyclonal antibody (pAb). The cell lysates and the immunoprecipitants were analyzed by Western blot with anti-GSDMD and anti-pS273R pAbs. B, PAM cells were infected with ASFV at an MOI of 0.1 for 36 h. The cells were probed with rabbit anti-GSDMD monoclonal antibody (mAb) (green) and mouse anti-pS273R mAb (red). The cell nuclei (blue) were stained with DAPI. C, PAMs were mock-infected or infected with ASFV at MOIs of 0.1, 1, and 10. At the indicated time points, the cell lysates were analyzed by Western blot with antibodies against GSDMD, p72, pS273R, and GAPDH. ASFV, African swine fever virus; DAPI, 4,6-diamidino-2-phenylindole; GSDMD, gasdermin D; MOI, multiplicity of infection; PAMs, porcine alveolar macrophages.
ASFV pS273R protease activity is required for GSDMD cleavage
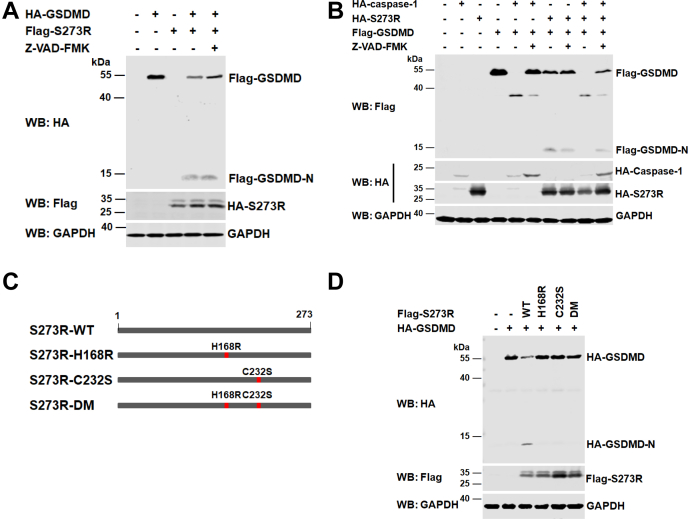
To explore whether the protease activity of ASFV pS273R is required for the cleavage of GSDMD, we evaluated the impact of Z-VAD-FMK, a pan-caspase inhibitor, on GSDMD cleavage by pS273R. As shown in Figure 3, A and B, Z-VAD-FMK did not affect ASFV pS273R-mediated GSDMD cleavage although it inhibited caspase-1-mediated GSDMD cleavage, suggesting that ASFV pS273R-mediated GSDMD cleavage was not dependent on the activities of host caspases. His168 and Cys232 are key amino acids that form the two catalytic sites of ASFV pS273R (9). To further verify this issue, we constructed three plasmids expressing different mutant forms of ASFV pS273R, such as pS273R-H168R, C232S, and H168R/C232S (Fig. 3C), and used them to analyze GSDMD cleavage. As shown in Figure 3D, ASFV pS273R with H168R, C232S, or H168R/C232S (DM) substitutions within the active site disrupted its protease activity, resulting in loss of the cleavage of GSDMD. Hence, these results demonstrate that the protease activity of ASFV pS273R is essential for GSDMD cleavage.
Figure 3.
The protease activity of ASFV pS273R is required for the cleavage of GSDMD.A, HEK293T cells were transfected with a plasmid encoding HA-tagged GSDMD, along with a Flag-vector or a plasmid expressing Flag-pS273R. At 4 hpt, the cells were incubated with the caspase inhibitor Z-VAD-FMK (50 μM) as indicated and cultured for another 30 h. The cell lysates were then processed for Western blot. B, HEK293T cells were transfected with a plasmid expressing Flag-tagged GSDMD alone or together with a plasmid expressing individual HA-pS273R protein or HA-caspase-1 protein. The cells were treated with the caspase inhibitor Z-VAD-FMK (50 μM) or not. At 24 hpt, the cell lysates were analyzed by Western blot with antibodies against HA, Flag, and GAPDH. C, a schematic of ASFV pS273R constructs showing the positions of ASFV pS273R protease activity sites (labeled in red) at H168R and C232S. D, HEK293T cells were transfected with a plasmid encoding HA-tagged GSDMD alone or together with a plasmid expressing Flag-pS273R or its mutants. The cell lysates were prepared and analyzed by Western blot. ASFV, African swine fever virus; GSDMD, gasdermin D; hpt, hours posttransfection.
GSDMD is cleaved at Gly-107 by ASFV pS273R
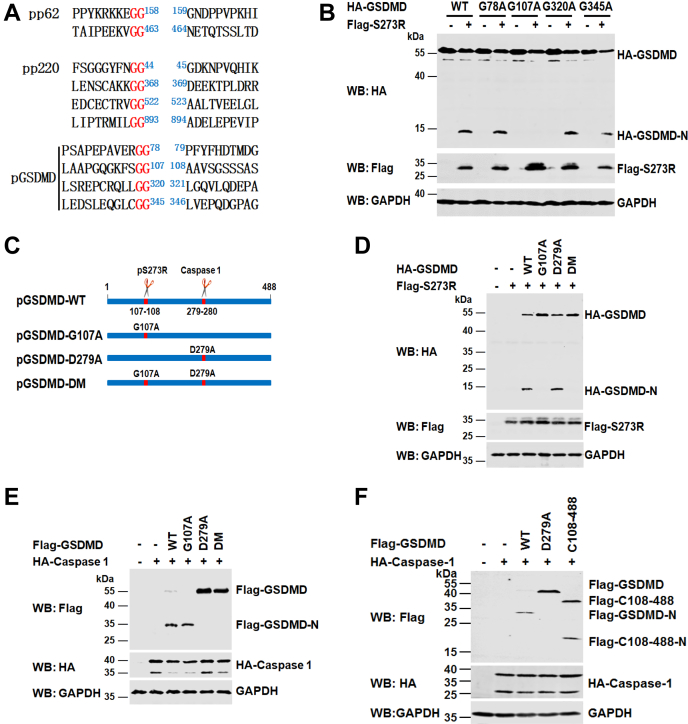
Previous reports demonstrated that ASFV pS273R preferentially cleaves Gly-Gly (G-G) amino acid pairs within pp62 and pp220 (9). According to the features of the pS273R cleavage sites with pp62 and pp220 (Fig. 4A, above), we examined the amino acid sequence of the swine GSDMD for potential ASFV pS273R cleavage sites and found that four regions bearing several glycines (G) resemble the signature G-G sequences of the proteolytic sites of ASFV pS273R (Fig. 4A, bottom). Therefore, we inferred that at least four potential ASFV pS273R cleavage sites may exist in swine GSDMD. To define the putative cleavage site, we constructed a series of GSDMD mutants, in which the glycine was replaced with alanine (such as G78A, G107A, G320A, and G345A). GSDMD or its mutants were coexpressed with ASFV pS273R in HEK293T cells to identify the actual cleavage sites. As shown in Figure 4B, GSDMD-G107A was absolutely resistant to ASFV pS273R. However, other GSDMD mutants did not prevent ASFV pS273R cleavage.
Figure 4.
ASFV pS273R cleaves GSDMD at Gly107.A, alignment of the ASFV S273R cleavage sites within pp62 and pp220 (above) and putative ASFV pS273R cleavage sites within swine GSDMD (bottom). The Glycine residues (G) at 78, 107, 320, or 345 within swine GSDMD were substituted with alanine residues (A). B, HEK293T cells were transfected with a plasmid expressing HA-tagged GSDMD or its mutants alone or in combination with a plasmid expressing Flag-pS273R. At 36 hpt, the cell lysates were analyzed by Western blot. C, a schematic of swine GSDMD showing the positions of the ASFV pS273R cleavage site at Gly107 (G107) and the caspase-1 cleavage site at Asp279 (D279). D, HEK293T cells were cotransfected with a plasmid expressing HA-tagged GSDMD or its mutants (G107A, D279A, and DM) together with a plasmid expressing Flag-pS273R or empty vector. At 36 hpt, the cell lysates were analyzed with anti-Flag, HA, and GAPDH antibodies as indicated. E, HEK293T cells were cotransfected with a plasmid expressing Flag-GSDMD or its mutants (G107A, D279A, and DM) in combination with a plasmid expressing HA-caspase-1 or empty vector. At 24 hpt, the cell lysates were analyzed with anti-Flag, HA, and GAPDH antibodies as indicated. F, HEK293T cells were transfected with a plasmid expressing Flag-tagged GSDMD, GSDMD-D279A, or GSDMD-C108–488, along with a plasmid encoding HA-caspase-1. At 24 hpt, the cell lysates were analyzed by Western blot with the indicated antibodies. ASFV, African swine fever virus; GSDMD, gasdermin D; hpt, hours posttransfection.
Compared with the human GSDMD sequence, we predicted that Asp279 (D279) is a site at which swine GSDMD is cleaved by caspase-1 (Fig. 4C). To further test whether G107 is indeed the cleavage site, we generated a GSDMD-G107A/D279A double mutant (GSDMD-DM). ASFV pS273R was coexpressed with GSDMD, GSDMD-G107A, GSDMD-D279A, and GSDMD-DM. As shown in Figure 4D, we found that GSDMD and GSDMD-D279A were still cleaved by ASFV pS273R. However, GSDMD-G107A and GSDMD-DM were completely resistant to ASFV pS273R cleavage. In contrast, GSDMD-D279A and GSDMD-DM were resistant to caspase-1 (Fig. 4E). Consistent with these results, we also noticed that GSDMD-C108–488 could be cleaved by caspase-1 (Fig. 4F). Taken together, these findings suggest that swine GSDMD is cleaved at G107 by ASFV pS273R and at D279 by caspase-1.
The cleavage fragments of GSDMD by pS273R are unable to induce pyroptosis
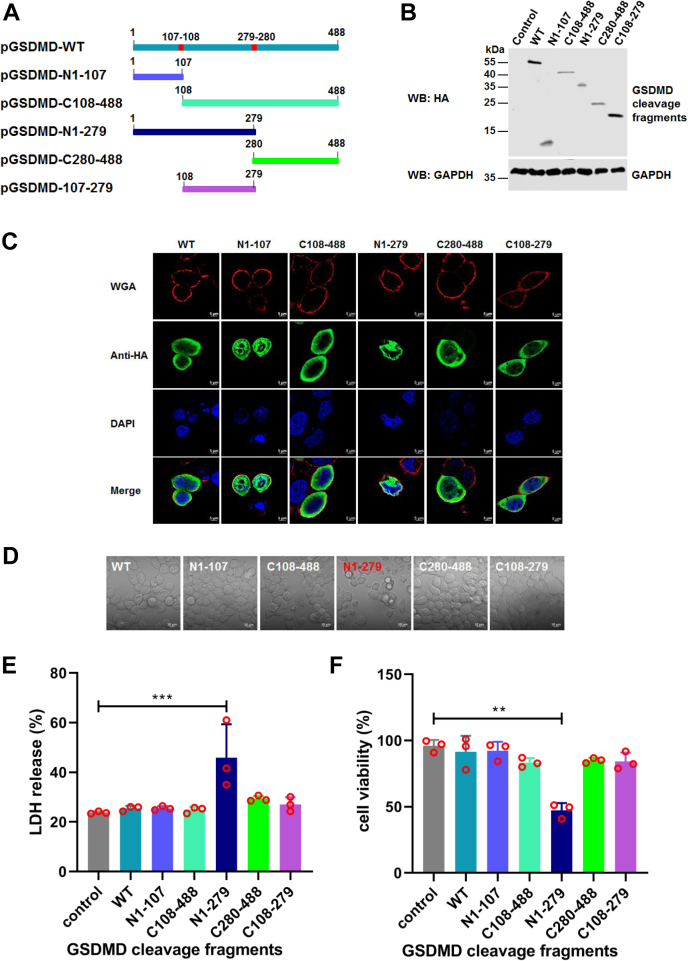
Previous reports showed that the cleavage product (GSDMD-N1–275) of human GSDMD by active caspase-1 induces pyroptosis by forming a large pore in the cell plasma membrane (9, 19). We constructed two plasmids expressing swine GSDMD-N1–107 and GSDMD-C108–488 that mimic cleavage products generated by ASFV pS273R (Fig. 5A). We also constructed two other plasmids expressing swine GSDMD-N1–279 and GSDMD-C280–488 that mimic the cleavage products generated by active caspase-1. In addition, we constructed a plasmid expressing GSDMD-C108–279, a cleavage product of GSDMD by ASFV S273R and caspase-1. The expression levels of these proteins were verified (Fig. 5B). To visualize the cellular distribution of transiently expressed WT GSDMD and the cleavage products of GSDMD, HEK293T cells were transfected with a plasmid expressing GSDMD-WT and its cleaved fragments, including GSDMD-N1–107, GSDMD-C108–488, GSDMD-C280–488, GSDMD-C108–279, and GSDMD-N1–279, respectively. Alexa Fluor 594-conjugated wheat germ agglutinin was used to stain the plasma membrane. We found that GSDMD-WT and GSDMD-N1–107 diffusely localized in the cytosol and nuclear, GSDMD-C108–488, GSDMD-C280–488, and GSDMD-C108–279 localized in the cytosol whereas only GSDMD-N1–279 localized in the cell membrane (Fig. 5C). Consistent with these results, GSDMD-N1–279, but not GSDMD and other cleavaged fragments, induces cell death with typical pyroptosis morphological features (Fig. 5D). Furthermore, we found that only GSDMD-N1–279 induced LDH release (Fig. 5E) and decreased cell viability (Fig. 5F). Taken together, these results indicate that pS273R may cleave GSDMD to produce GSDMD-N1–107 to lose its function.
Figure 5.
The cleavage fragments of GSDMD by ASFV pS273R are unable to induce pyroptosis.A, schematic diagrams of GSDMD and its cleavage fragments produced by caspase-1, ASFV pS273R protease alone, or both. B, detection of the expression of GSDMD and its deletion mutants in HEK293T cells. C, HeLa cells were transfected with a plasmid encoding HA-tagged GSDMD or its cleaved fragments, including GSDMD-N1–107, GSDMD-C108–488, GSDMD-N1–279, GSDMD-C280–488, and GSDMD-C108–279. Alexa Fluor 594-conjugated wheat germ agglutinin (WGA) was added at 37 °C for 10 min to stain the plasma membrane. The colocalization of HA-GSDMD (green) and its cleavage fragments with the cell plasma membrane (red) and cell nuclei (blue) stained with DAPI was observed by confocal microscopy, and GSDMD-N1–279 was used as the positive control. D, HEK293T cells were transfected with HA-tagged GSDMD or its cleaved fragments. The representative views of pyroptosis morphology are shown in (D). E and F, detection of LDH release and cell viability of HEK293T cells expressing HA-tagged GSDMD and its cleaved fragments in pyroptosis. LDH release and ATP-based cell viability are expressed as means ± SD from three technical replicates. All the data shown are representative of three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001. ASFV, African swine fever virus; DAPI, 4,6-diamidino-2-phenylindole; GSDMD, gasdermin D.
The effect of GSDMD mutations on ASFV replication
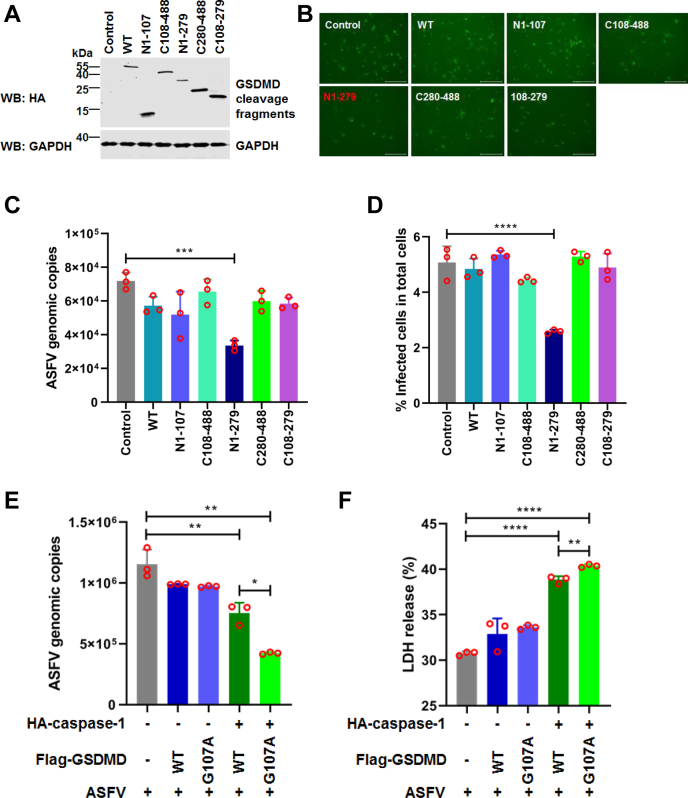
Previous studies reported that several viral proteases can cleave GSDMD to affect pyroptosis, resulting in the regulation of viral replication (24, 25). Therefore, we first tested whether the swine GSDMD cleavage products generated by ASFV pS273R and caspase-1 are involved in ASFV replication. The plasmids expressing GSDMD, GSDMD-N1–107, GSDMD-C108–488, GSDMD-N1–279, and GSDMD-C280–488, GSDMD-C108–279 were ectopically transfected into MA-104 cells (Fig. 6A), a suitable cell line for ASFV growth (26). Meanwhile, the cells were infected with ASFV for 24 h, and ASFV genomic DNA was extracted for quantitative PCR analysis. As shown in Figure 6C, ASFV replication significantly decreased in the cells expressing GSDMD-N1–279. However, other cleavage products of GSDMD had no inhibitory effect on ASFV replication. Based on the above results, we further tested whether the cleavage of GSDMD by endogenous pS273R disrupts the GSDMD-N1–279-mediated pyroptosis and further affects ASFV replication. MA-104 cells transfected with plasmids coexpressing GSDMD and caspase-1 were chosen as positive control, which induces pyroptosis. MA-104 cells were transfected with a plasmid coexpressing GSDMD or its mutants, together with an empty vector or a plasmid expressing caspase-1 as indicated, and then infected with ASFV. Compared with the cells coexpressing GSDMD and caspase-1, ASFV replication has a slight decrease (Fig. 6E). Meanwhile, the LDH release significantly increased in the cell supernatants of the cells coexpressing GSDMD-G107A and caspase-1 (Fig. 6F). Taken together, our findings reveal that pS273R cleaves GSDMD-N1–279 to generate GSDMD-N1–107, which inhibits GSDMD-N1–279-mediated pyroptosis, resulting in the inhibition of the host antiviral innate immune responses and the enhancement of ASFV replication.
Figure 6.
The effect of GSDMD and its cleaved fragments on ASFV replication.A, detection of the expression of GSDMD and its deletion mutants in MA-104 cells. B, PAMs were infected with cell samples from (C), and then fluorescence focus assays were performed. The scale bar represents 650 μm. C, MA-104 cells were transfected with plasmids encoding GSDMD and its cleaved fragments as indicated. An empty vector was used as a control. At 24 hpt, the cells were infected with ASFV (5 MOI). After 24 hpi, total DNA was extracted, and the viral genome copies of ASFV were evaluated by quantitative PCR (qPCR) using TaqMan. The data are expressed as the fold change of the ASFV genome copies. The data were expressed as means ± SD of the triplicate well from one independent experiment. D, MA-104 cells were transfected with plasmids encoding GSDMD and its cleaved fragments as indicated. An empty vector was used as a control. At 24 hpt, the cells were infected with ASFV at 1 MOI. After 36 hpi, the cells were fixed and then the percent of infected cells was analyzed. E, MA-104 cells were transfected with plasmids encoding GSDMD and empty vector, GSDMD-G107A and empty vector, GSDMD and caspase-1, and GSDMD-G107A and caspase-1 as indicated. An empty vector was used as a control. At 8 hpt, the cells were infected with ASFV (5 MOI) for. After 18 hpi, total DNA was extracted, and the viral genomic copies of ASFV were evaluated by quantitative PCR (qPCR) using TaqMan. The data were expressed as means ± SD of the triplicate well from one independent experiment. F, the supernatants were harvested from (E), and the level of cell death was measured by LDH assay. The data were expressed as means ± SD of the triplicate well from one independent experiment. Statistical significance was determined by one-way ANOVA and Student’s t test with a multiple comparison correction. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001. ASFV, African swine fever virus; GSDMD, gasdermin D; hpt, hours posttransfection; MOI, multiplicity of infection; PAM, porcine alveolar macrophages.
GSDMD is cleaved by several RNA viral proteases
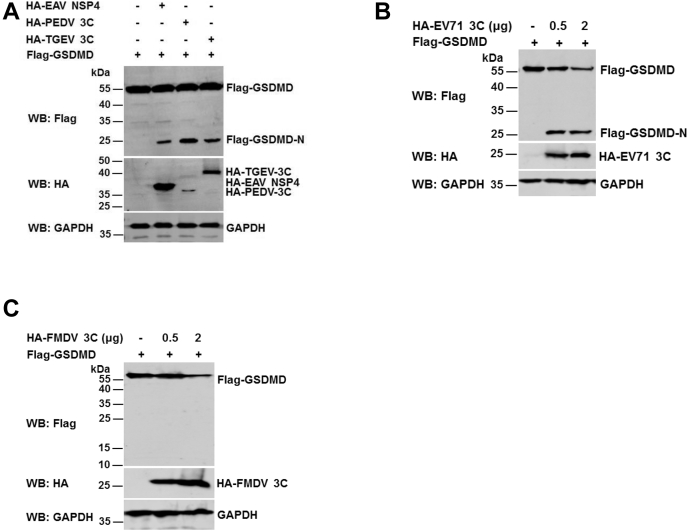
The RNA virus genome can be translated into a polyprotein, which can be processed into mature functional proteins by viral protease(s) (27, 28). It is well known that several viral proteases can cleave host proteins to regulate host innate immune responses (29). To test whether swine GSDMD is cleaved by the indicated viral proteases, GSDMD was coexpressed with some major proteases encoded by several viruses belonging to the Coronaviridae, Arteriviridae, and Picornaviridae. As shown in Figure 7, A–C, we found that GSDMD was cleaved to produce a 25-KD fragment by porcine epidemic diarrhea virus (PEDV) Nsp4, porcine transmissible gastroenteritis virus (TGEV) Nsp4, equine arteritis virus (EAV) Nsp4 (Fig. 7A), and by enterovirus type 71 (EV71) 3C protease (Fig. 7B), but not by foot-and-mouth disease virus (FMDV) 3C protease (Fig. 7C). These results suggest that GSDMD is also a new target of some RNA viral proteases, which may be required for these RNA viruses infection-mediated pyroptosis.
Figure 7.
GSDMD is cleaved by several RNA viral proteases.A–C, HEK293T cells were transfected with a plasmid encoding Flag-tagged GSDMD together with a plasmid expressing viral proteases encoded by Arteriviridae and Coronaviridae (A) and Picornaviridae (B and C). The cell lysates were subjected to Western blot analysis with anti-HA, Flag, and GAPDH antibodies. GSDMD, gasdermin D.
Discussion
The ASFV genome contains 150 to 167 ORFs encoding more than 150 viral proteins (30). ASFV-encoded proteins execute different functions, including virus entry, viral gene transcription and replication, genome integrity maintenance, viral assembly, and egress. Similar to other viruses, ASFV infection induces apoptosis (31, 32), the Endoplasmic reticulum (ER) stress response (33), and the inflammatory responses (34) to antagonize viral infection. In addition, previous studies have reported that ASFV has evolved series of mechanisms to evade host antiviral responses. For example, it has been experimentally shown that several immunoregulatory proteins suppress the host antiviral immune responses by reducing interferon production (35, 36, 37), activating NF-κB (38), and inhibiting apoptosis in ASFV-infected macrophages (39, 40). In this study, we found that GSDMD, an executor of pyroptosis, is a new target of ASFV pS273R (Fig. 2). ASFV pS273R interacted with and cleaved GSDMD at the G107 site to produce GSDMD-N1–107, which may disrupt the pyroptosis induced by GSDMD-N1–279 to benefit ASFV replication.
Pyroptosis has been defined as an inflammatory cell death process that is characterized by lytic forms of death resulting in the release of cytokines and other cellular factors to drive inflammation and alert immune cells to a pathogenic or sterile insult. As one of the six gasdermin family members (GSDMs), GSDMD has been identified as a substrate of several inflammatory caspases (41). For example, GSDMD can be cleaved by active caspases to produce the GSDMD-N1–275 and C-terminal repressor domain (GSDMD-N276–484) (13, 14). GSDMD-N1–275 oligomerizes to form large pores in the cell membrane, which drives cell swelling and membrane rupture to release inflammatory cytokines (19, 41). Recently, other GSDMD family members, such as GSDME, have also been shown to be cleaved and form pores, resulting in pyroptosis (22).
ASFV pS273R belongs to the SUMO-1-specific protease family (9). It has been reported that ASFV pS273R is involved in the cleavage and maturation of pp220 and pp62 polyprotein precursors into core-shell proteins (9). However, whether ASFV pS273R participates in the regulatory function of the host immune responses is still unknown. In this study, GSDMD was found to be an ASFV pS273R-binding partner. Subsequently, we found that pS273R cleaved GSDMD in a dose-dependent manner when the two proteins were expressed in mammalian cells (Fig. 1). We tested all the members of the gasdermin family in swine and found that only GSDMD could be specific cleaved by pS273R (Fig. 1F). Consistently, we found that the cleavage of GSDMD by pS273R was dependent on its protease activity but not host caspases activity (Fig. 3). Our findings indicated that pS273R may have a specific role in the regulation of ASFV infection-mediated pyroptosis.
Compared with the specific cleavage sites of pS273R in pp220 and pp62, we noticed that the cleavage sites in pp220 and pp62 have a highly conserved Gly-Gly signature: Gly-Gly-Xaa (9). Therefore, we speculated that the cleavage sites on GSDMD may have the same characteristics. Consistent with our speculation, mutation analysis showed that the cleavage of GSDMD by S273R occurred at the site G107-A108 (Fig. 4). The results suggest that once GSDMD is cleaved by caspase-1 to produce GSDMD-N1–279 and GSDMD-C280–488, GSDMD-N1–279 may be cleaved by pS273R to produce GSDMD-N1–107 and GSDMD-C108–279 (Fig. 4F). Subsequently, our findings revealed that overexpression of the three GSDMD truncated fragments (GSDMD-N1–107, GSDMD-C108–279, and GSDMD-C280–488) did not cause pyroptosis (Fig. 5), suggesting S273R may inhibit GSDMD-N1–279-induced pyroptosis by the cleavage of GSDMD-N1–279. Previous results revealed that several viral proteases can convert pyroptosis by the cleavage of GSDMD-N1–275. For example, enterovirus 71 (EV71) protease 3C directly cleaves human GSDMD to produce a shorter N-terminal fragment (1–193) of GSDMD, which results in disruption of the pore formation, thereby avoiding pyroptosis and ensuring viral persistence (25). Our results showed that the swine GSDMD-N1–279 significantly promoted cell death and affected the ASFV replication (Fig. 6), whereas the GSDMD-N1–107 and GSDMD-C108–279 fragments produced by ASFV S273R did not affect cell death.
It has been well documented that virus infection can activate pyroptosis-related cell death, which contributes to viral replication and transmission. For instance, SARS-CoV-2 infection activates the NLRP3 inflammasome to induce pyroptosis, which leads to GSDMD cleavage and inflammatory cytokines release to generate a cytokine storm (42, 43), suggesting that GSDMD is a promising target for the treatment of severe coronavirus-related diseases (44). Recently, several viral proteases have been found to cleave host GSDMD. For example, Zika virus (ZIKV) NS2B3 directly cleaves GSDMD to produce the N-terminus of GSDMD-N1–249, which promotes virus release through ZIKV-induced pyroptosis (24). In this study, we also noticed that the proteases from several other RNA viruses, including the EAV NSP4, PEDV 3C, and TGEV 3C proteases, but not the FMDV 3C protease, could also cleave GSDMD (Fig. 7). Taken together, different viruses may use their specific proteases to cleave GSDMD to regulate pyroptosis to affect viral replication, which may be likely to occur widely in the process of host cell innate immunity.
In this study, we found and verified the cleavage of swine GSDMD by ASFV pS273R, which is a novel mechanism to disrupt the pyroptosis mediated by GSDMD-N1–279 (Fig. 8). Interestingly, we also noticed that pS273R inhibited the type I interferon signaling pathway to inhibit interferon-stimulated genes production, suggesting that pS273R might interact with and cleave the core components in the interferon signaling pathway to subvert interferon responses (data not shown). Therefore, it is necessary to continue to screen and identify pS273R binding partners and then elucidate their roles in ASFV replication and pathogenesis.
Figure 8.
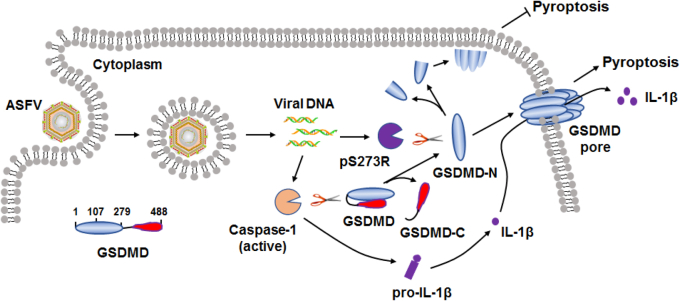
ASFV pS273R inhibits pyroptosis by noncanonically cleaving GSDMD. The ASFV-encoded pS273R is expressed in an late stage before viral assembly. ASFV pS273R cleaves swine GSDMD at G107-A108 in a manner that is dependent on its protease activity. After the cleavage of GSDMD by virus-activated caspase-1, the structure of GSDMD-N1–279, which can be polymerized to form membrane pores, is disrupted by pS273R cleavage. This is a direct and effective way for the virus to regulate the pyroptosis of the host cells, which may facilitate the subsequent viral replication. This may be a ubiquitous mechanism by which viruses regulate the pyroptosis of host cells. ASFV, African swine fever virus; GSDMD, gasdermin D.
Experimental procedures
Cell lines and viruses
PAMs were isolated from the lung lavage fluid from 4-week-old healthy specific pathogen-free piglets, as previously described (45) and maintained in RPMI-1640 medium containing 10% fetal bovine serum (Gibco), 100 U/ml penicillin, 50 mg/ml streptomycin, and nonessential amino acids (Gibco). HEK293T cells and HeLa cells obtained from ATCC were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum. All the cells were maintained at 37 °C with 5% CO2. The ASFV HLJ/18 strain (GenBank accession number: MK333180.1) was isolated from the pigs, as previously described (46). A HAD assay was performed, as described previously (47). Briefly, primary peripheral blood mononuclear cells were seeded in 96-well plates. The samples were then added to the plates and titrated in triplicate using 10× dilutions. The quantity of ASFV was determined by the identification of characteristic rosette formation, which represents the hemadsorption of erythrocytes around the infected cells. HAD was observed for 7 days, and 50% HAD doses (HAD50) were calculated by using the method of Reed and Muench (48).
Plasmids and reagents
To construct plasmids expressing HA-tagged and Flag-tagged pS273R, cDNA corresponding to the ASFV S273R gene was cloned into the pCAGGS-HA (pHA) and pCAGGS-Flag (pFlag) vectors. Three plasmids expressing ASFV S273R and its mutants (S273R-H168R, C232S, and H168R/C232S (DM)) were constructed based on a plasmid expressing ASFV S273R by site-directed mutagenesis. The cDNAs corresponding to the swine GSDMA, GSDMB, GSDMC, GSDMD, and GSDME proteins were synthesized by GenScript and then cloned into the pHA and pFlag vectors. The cDNAs corresponding to the point mutants of swine GSDMD (G78A, G107A, G320A, G345A, D279A, and G107A/D279A (DM)) and the deleted mutants of GSDMD, including 1 to 107 amino acid (aa), 108 to 488 aa, 1 to 279 aa, and 280 to 488 aa of GSDMD, were cloned into the pHA vector. The pET-22b-S273R plasmid was a gift from Prof. Yu Guo of Nankai University, and the cDNA of GSDMD was cloned into the pGEX-6P1 vector to express and purify the recombinant GST-GSDMD fusion protein. The sequences of the primers used in this study are available upon request. All the constructs were validated by DNA sequencing. Alexa Fluor 594-conjugated wheat germ agglutinin (W11262) was purchased from Thermo Fisher Scientific. The pan-caspase inhibitor Z-VAD-FMK was purchased from Sigma-Aldrich.
Antibodies
Anti-Flag (14793S) and anti-HA (3724S) were purchased from Cell Signaling Technology. Anti-GAPDH (10494-1-AP) was purchased from Proteintech. The rabbit anti-GSDMDC1 polyclonal antibody (NBP2-33422) was purchased from Novus. IRDye 800CW goat anti-mouse IgG (H + L) (926-32210) was purchased from Sera Care, and IRDye 800CW goat anti-rabbit IgG (H + L) (925-32211) was purchased from LI-COR. Polyclonal antibodies against ASFV pS273R, p72, and p30 were prepared by immunizing mice using the purified recombinant ASFV pS273R, p72, and p30 proteins as immunogens. All animal procedures were approved by Harbin Veterinary Research Institute Animal Ethics Committee of CAAS.
Pull-down assay
PAMs (1 × 106) were washed three times with PBS and lysed with cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 5 mM MgCl2, 1 mM EDTA, and 10% glycerol) containing 1 mM PMSF and 1× protease inhibitor mixture (Roche Diagnostics). A Ni Sepharose column was washed three times with PBS. Ni Sepharose bound with His-pS273R or Ni Sepharose were incubated with the cell lysates of PAMs at 4 °C overnight. The resin was washed five times with wash buffer (20 mM Tris, 300 mM NaCl, and 20 mM imidazole (pH 8.0)), and the candidate proteins were eluted with elution buffer (20 mM Tris, 300 mM NaCl, and 500 mM imidazole (pH 8.0)). Then, the eluted samples were resolved by SDS-PAGE and stained with Coomassie brilliant blue.
MS analysis
The gels containing His-pS273R-binding proteins were cut and named 1#, 2#, and 3# and then processed for LC-MS/MS to identify pS273R binding host proteins (45). Briefly, the samples were mixed with trypsin and digested at 37 °C overnight. LC-MS/MS was implemented by using a Dionex ultimate 3000 nano-LC system (Dionex) coupled with an Q Exactive mass spectrometer (Thermo Fisher Scientific) at Beijing Protein Innovation in China. The MS/MS signals were then processed against the uni_sus_9822 9822 database (122,086 sequences; 73,310,586 residues) using the Mascot software (Version 2.3.01, Matrix Science) with the following parameters: Fixed modification: Carbamidomethyl (C); Variable modification: Oxidation (M), Gln→Pyro-Glu (N-term Q); and maximum missed cleavages, 2; filter by score ≥31.
Coimmunoprecipitation and Western blot analysis
Co-IP and Western blot analysis were performed, as described previously (49). At 36 h posttransfection (hpt), the cells transfected with the different plasmids were lysed with the cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 5 mM MgCl2, 1 mM EDTA, and 10% glycerol) containing 1 mM PMSF and 1× protease inhibitor mixture (Roche Diagnostics). The cell lysates were incubated with anti-Flag agarose beads (A2220-5ML, Sigma) overnight at 4 °C on a roller. The immunoprecipitants were subjected to electrophoresis. To identify the interactions between endogenous proteins, PAMs were either mock-infected or infected with ASFV (1 MOI) for 36 h. The cell lysates then were incubated with anti-S273R monoclonal antibody or IgG for 8 h at 4 °C, and S273R complexes were captured using protein A+G-Sepharose (Pierce Protein A/G Plus Agarose, Thermo). The equal amounts of cell lysates and immunoprecipitants were resolved by 12% SDS-PAGE and then transferred to polyvinylidine difluoride membranes (ISEQ00010, Merck-Millipore). After incubation with primary and secondary antibodies as indicated, the membranes were visualized by an Odyssey two-color infrared fluorescence imaging system (LI-COR).
Fluorescence microscopy
To test the cellular localization of GSDMD and S273R, HEK293T cells were transfected with a plasmid expressing HA-GSDMD or Flag-S273R alone or both plasmids for 24 h. To test the cellular localization of endogenous GSDMD and S273R, PAM cells were infected with ASFV at an MOI of 0.1 for another 36 h. To test the cellular localization GSDMD or its deletion mutants, HeLa cells were transfected with a plasmid expressing HA-GSDMD or its deletion mutants. At 24 h posttransfection, the cells were fixed with 4% paraformaldehyde, permeabilized with 0.3% Triton X-100 and then probed with primary antibodies as indicated and 4,6-diamidino-2-phenylindole. The samples were visualized with a Zeiss LSM-800 laser scanning fluorescence microscope (Carl Zeiss AG).
Expression of recombinant proteins and cleavage assay in vitro
The purification of recombinant proteins and the cleavage assay were performed in vitro, as described previously (14, 50). In brief, His-pS273R protein was purified from clarified bacterial lysates, as described previously (10). The GST-GSDMD protein was purified on a glutathione-agarose column. The two proteins were dialyzed and stored at −80 °C. To examine GSDMD cleavage in vitro, the aliquots of recombinant ASFV pS273R (His-pS273R) and GST-GSDMD were incubated in Buffer A (50 mM Hepes (pH 7.5), 3 mM EDTA, 150 mM NaCl, 0.005% (vol/vol) Tween-20, and 10 mM DTT). The reaction was incubated for 2 h at 37 °C, and the reactions were terminated by adding 1× loading buffer and then subjected to Western blot analysis.
Cell viability and lactate dehydrogenase assay
HEK293T cells (1.5 × 104) grown in 96-well plates were transfected with the indicated plasmids using Lipofectamine 3000 (Thermo Fisher Scientific). After 36 hpt, cell viability was determined by the CellTiter-Glo 2.0 Assay (Promega, G9242). Cell death was measured by LDH assay using a CytoTox 96 NonRadioactive Cytotoxicity Assay kit (Promega, G1780).
MA-104 cells (1 × 105) grown in 24-well plates were transfected with the indicated plasmids for 8 h after ASFV infection with 5 MOI. At 18 hpi, the supernatants were harvested, and the cell death was measured by LDH assay.
Quantitative PCR
To test the GSDMD cleavage fragments for ASFV replication, MA-104 cells (1 × 105) were seeded on 24-well plates and cultured for 8 h after transfection with indicated plasmids (1 μg per well) for 24 h and then followed ASFV infection with 5 MOI. At 24 hpi, the cells were harvested.
To test the endogenous S273R to cleaving GSDMD and GSDMD mutant (GSDMD-G107A) for ASFV replication, MA-104 cells (1 × 105) were seeded on 24-well plates and cultured for 12 h after transfection with indicated plasmids (1 μg per well) for 8 h and then followed ASFV infection with 5 MOI. At 18 hpi, the cells were harvested. ASFV genomic DNA was extracted from MA-104 cells using a Qiagen DNA Mini Kit (Qiagen). Quantitative PCR was carried out on a QuantStudio5 system (Applied Biosystems) according to the OIE-recommended procedure.
Flow cytometry
MA-104 cells (2 × 105) were seeded on 24-well plates and cultured for 12 h after transfection with indicated plasmids (0.5 μg per well) for 24 h and then followed ASFV infection with 1 MOI. At 36 hpi, the cells were harvested and then fixed with 4% paraformaldehyde and were analyzed by a Cytomics FC 500 flow cytometer (Beckman).
Statistics
All the statistical analyses were performed using one-way ANOVA and Student’s t test using GraphPad Prism 8 software (GraphPad Software Inc). The data are presented as the mean ± SD. The data are presented as the mean ± SD, ∗ indicates a significant difference (p < 0.05), ∗∗ indicates a highly significant difference (p < 0.01), and ∗∗∗ indicates an extremely significant difference (p < 0.001).
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (51) with the dataset identifier PXD028692.
Supporting information
This article contains supporting information. All primers used in this study are listed in Table S1.
Conflict of interest
The authors declared that they have no conflicts of interest with the contents of this article.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (31941002 and 32172874), National Key Research and Development Program of China (Grant No. 2021YFD1800100), and State Key Laboratory of Veterinary Biotechnology Program (SKLVBP202101).
Author contributions
J. Z. and C. W. methodology; G. Z., T. L., X. L., T. Z., Z. Z., L. K., J. S., S. Z., X. C., X. W., J. L., L. H., and C. L. investigation; G. Z., T. L., X. L., T. Z., Z. Z., L. K., J. S., S. Z., X. C., X. W., J. L., L. H., and C. L. data analysis; J. Z. and C. W. writing–original draft.
Edited by Craig Cameron
Contributor Information
Jun Zheng, Email: zhengjun01@caas.cn.
Changjiang Weng, Email: wengchangjiang@caas.cn.
Supporting information
References
- 1.Dixon L.K., Sun H., Roberts H. African swine fever. Antiviral Res. 2019;165:34–41. doi: 10.1016/j.antiviral.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 2.Sánchez-Cordón P., Montoya M., Reis A., Dixon L. African swine fever: A re-emerging viral disease threatening the global pig industry. Vet. J. 2018;233:41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang T., Sun Y., Huang S., Qiu H.J. Multifaceted immune responses to African swine fever virus: Implications for vaccine development. Vet. Microbiol. 2020;249:108832. doi: 10.1016/j.vetmic.2020.108832. [DOI] [PubMed] [Google Scholar]
- 4.Teklue T., Sun Y., Abid M., Luo Y., Qiu H.J. Current status and evolving approaches to African swine fever vaccine development. Transbound. Emerg. Dis. 2020;67:529–542. doi: 10.1111/tbed.13364. [DOI] [PubMed] [Google Scholar]
- 5.Alejo A., Matamoros T., Guerra M., Andrés G. A proteomic atlas of the African swine fever virus particle. J. Virol. 2018;92 doi: 10.1128/JVI.01293-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alonso C., Galindo I., Cuesta-Geijo M.A., Cabezas M., Hernaez B., Muñoz-Moreno R. African swine fever virus-cell interactions: From virus entry to cell survival. Virus Res. 2013;173:42–57. doi: 10.1016/j.virusres.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reis A.L., Netherton C., Dixon L.K. Unraveling the armor of a killer: Evasion of host defenses by African swine fever virus. J. Virol. 2017;91 doi: 10.1128/JVI.02338-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andrés G., Alejo A., Salas J., Salas M.L. African swine fever virus polyproteins pp220 and pp62 assemble into the core shell. J. Virol. 2002;76:12473–12482. doi: 10.1128/JVI.76.24.12473-12482.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrés G., Alejo A., Simón-Mateo C., Salas M.L. African swine fever virus protease, a new viral member of the SUMO-1-specific protease family. J. Biol. Chem. 2001;276:780–787. doi: 10.1074/jbc.M006844200. [DOI] [PubMed] [Google Scholar]
- 10.Li G., Liu X., Yang M., Zhang G., Wang Z., Guo K., Gao Y., Jiao P., Sun J., Chen C., Wang H., Deng W., Xiao H., Li S., Wu H., et al. Crystal structure of African swine fever virus pS273R protease and implications for inhibitor design. J. Virol. 2020;94 doi: 10.1128/JVI.02125-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broz P., Dixit V.M. Inflammasomes: Mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 12.Jorgensen I., Miao E. Pyroptotic cell death defends against intracellular pathogens. Immunol. Rev. 2015;265:130–142. doi: 10.1111/imr.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.He W.T., Wan H., Hu L., Chen P., Wang X., Huang Z., Yang Z.H., Zhong C.Q., Han J. Gasdermin D is an executor of pyroptosis and required for interleukin-1β secretion. Cell Res. 2015;25:1285–1298. doi: 10.1038/cr.2015.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 15.Kayagaki N., Warming S., Lamkanfi M., Vande Walle L., Louie S., Dong J., Newton K., Qu Y., Liu J., Heldens S., Zhang J., Lee W., Roose-Girma M., Dixit V. Non-canonical inflammasome activation targets caspase-11. Nature. 2011;479:117–121. doi: 10.1038/nature10558. [DOI] [PubMed] [Google Scholar]
- 16.Shi J., Zhao Y., Wang Y., Gao W., Ding J., Li P., Hu L., Shao F. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 17.Orning P., Weng D., Starheim K., Ratner D., Best Z., Lee B., Brooks A., Xia S., Wu H., Kelliher M.A., Berger S.B., Gough P.J., Bertin J., Proulx M.M., Goguen J.D., et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362:1064–1069. doi: 10.1126/science.aau2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarhan J., Liu B.C., Muendlein H.I., Li P., Nilson R., Tang A.Y., Rongvaux A., Bunnell S.C., Shao F., Green D.R., Poltorak A. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. U. S. A. 2018;115:e10888–e10897. doi: 10.1073/pnas.1809548115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V.G., Wu H., Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia S., Hollingsworth L.R.T., Wu H. Mechanism and regulation of gasdermin-mediated cell death. Cold Spring Harb. Perspect. Biol. 2020;12 doi: 10.1101/cshperspect.a036400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S., Ruan J., Wu H. Monitoring gasdermin pore formation in vitro. Methods Enzymol. 2019;625:95–107. doi: 10.1016/bs.mie.2019.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y., Gao W., Shi X., Ding J., Liu W., He H., Wang K., Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 23.Zhou Z., He H., Wang K., Shi X., Wang Y., Su Y., Wang Y., Li D., Liu W., Zhang Y., Shen L., Han W., Shen L., Ding J., Shao F. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368 doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 24.Yamaoka Y., Matsunaga S., Jeremiah S.S., Nishi M., Miyakawa K., Morita T., Khatun H., Shimizu H., Okabe N., Kimura H., Hasegawa H., Ryo A. Zika virus protease induces caspase-independent pyroptotic cell death by directly cleaving gasdermin D. Biochem. Biophys. Res. Commun. 2021;534:666–671. doi: 10.1016/j.bbrc.2020.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Lei X., Zhang Z., Xiao X., Qi J., He B., Wang J. Enterovirus 71 inhibits pyroptosis through cleavage of gasdermin D. J. Virol. 2017;91 doi: 10.1128/JVI.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rai A., Pruitt S., Ramirez-Medina E., Vuono E., Silva E., Velazquez-Salinas L., Carrillo C., Borca M., Gladue D. Identification of a continuously stable and commercially available cell line for the identification of infectious African swine fever virus in clinical samples. Viruses. 2020;12:820. doi: 10.3390/v12080820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Y., Snijder E.J. The PRRSV replicase: Exploring the multifunctionality of an intriguing set of nonstructural proteins. Virus Res. 2010;154:61–76. doi: 10.1016/j.virusres.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snijder E.J., Kikkert M., Fang Y. Arterivirus molecular biology and pathogenesis. J. Gen. Virol. 2013;94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 29.Huang L., Liu Q., Zhang L., Zhang Q., Hu L., Li C., Wang S., Li J., Zhang Y., Yu H., Wang Y., Zhong Z., Xiong T., Xia X., Wang X., et al. Encephalomyocarditis virus 3C protease relieves TRAF family member-associated NF-κB activator (TANK) inhibitory effect on TRAF6-mediated NF-κB signaling through cleavage of TANK. J. Biol. Chem. 2015;290:27618–27632. doi: 10.1074/jbc.M115.660761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dixon L., Chapman D., Netherton C., Upton C. African swine fever virus replication and genomics. Virus Res. 2013;173:3–14. doi: 10.1016/j.virusres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Alonso C., Miskin J., Hernáez B., Fernandez-Zapatero P., Soto L., Cantó C., Rodríguez-Crespo I., Dixon L., Escribano J. African swine fever virus protein p54 interacts with the microtubular motor complex through direct binding to light-chain dynein. J. Virol. 2001;75:9819–9827. doi: 10.1128/JVI.75.20.9819-9827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon L.K., Islam M., Nash R., Reis A.L. African swine fever virus evasion of host defences. Virus Res. 2019;266:25–33. doi: 10.1016/j.virusres.2019.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia N., Wang H., Liu X., Shao Q., Ao D., Xu Y., Jiang S., Luo J., Zhang J., Chen N., Meurens F., Zheng W., Zhu J. African swine fever virus structural protein p17 inhibits cell proliferation through ER stress-ROS mediated cell cycle arrest. Viruses. 2020;13:21. doi: 10.3390/v13010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borca M., O'Donnell V., Holinka L., Ramírez-Medina E., Clark B., Vuono E., Berggren K., Alfano M., Carey L., Richt J., Risatti G., Gladue D. The L83L ORF of African swine fever virus strain Georgia encodes for a non-essential gene that interacts with the host protein IL-1β. Virus Res. 2018;249:116–123. doi: 10.1016/j.virusres.2018.03.017. [DOI] [PubMed] [Google Scholar]
- 35.Henriques E.S., Brito R.M., Soares H., Ventura S., de Oliveira V.L., Parkhouse R.M. Modeling of the toll-like receptor 3 and a putative toll-like receptor 3 antagonist encoded by the African swine fever virus. Protein Sci. 2011;20:247–255. doi: 10.1002/pro.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D., Yang W., Li L., Li P., Ma Z., Zhang J., Qi X., Ren J., Ru Y., Niu Q., Liu Z., Liu X., Zheng H. African swine fever virus MGF-505-7R negatively regulates cGAS-STING-mediated signaling pathway. J. Immunol. 2021;206:1844–1857. doi: 10.4049/jimmunol.2001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Wu J., Wu Y., Chen H., Zhang S., Li J., Xin T., Jia H., Hou S., Jiang Y., Zhu H., Guo X. Inhibition of cGAS-STING-TBK1 signaling pathway by DP96R of ASFV China 2018/1. Biochem. Biophys. Res. Commun. 2018;506:437–443. doi: 10.1016/j.bbrc.2018.10.103. [DOI] [PubMed] [Google Scholar]
- 38.Correia S., Ventura S., Parkhouse R.M. Identification and utility of innate immune system evasion mechanisms of ASFV. Virus Res. 2013;173:87–100. doi: 10.1016/j.virusres.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 39.Brun A., Rivas C., Esteban M., Escribano J.M., Alonso C. African swine fever virus gene A179L, a viral homologue of bcl-2, protects cells from programmed cell death. Virology. 1996;225:227–230. doi: 10.1006/viro.1996.0592. [DOI] [PubMed] [Google Scholar]
- 40.Dixon L.K., Sanchez-Cordon P.J., Galindo I., Alonso C. Investigations of pro- and anti-apoptotic factors affecting African swine fever virus replication and pathogenesis. Viruses. 2017;9:241. doi: 10.3390/v9090241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kovacs S.B., Miao E.A. Gasdermins: Effectors of pyroptosis. Trends Cell Biol. 2017;27:673–684. doi: 10.1016/j.tcb.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., Schreiner P., Neale G., Vogel P., Webby R., Jonsson C.B., et al. Synergism of TNF-α and IFN-γ triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ratajczak M.Z., Kucia M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells. Leukemia. 2020;34:1726–1729. doi: 10.1038/s41375-020-0887-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yap J.K.Y., Moriyama M., Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID-19. J. Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J., Hu L., Liu Y., Huang L., Mu Y., Cai X., Weng C. DDX19A senses viral RNA and mediates NLRP3-dependent inflammasome activation. J. Immunol. 2015;195:5732–5749. doi: 10.4049/jimmunol.1501606. [DOI] [PubMed] [Google Scholar]
- 46.Zhao D., Liu R., Zhang X., Li F., Wang J., Zhang J., Liu X., Wang L., Zhang J., Wu X., Guan Y., Chen W., Wang X., He X., Bu Z. Replication and virulence in pigs of the first African swine fever virus isolated in China. Emerg. Microbes Infect. 2019;8:438–447. doi: 10.1080/22221751.2019.1590128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malmquist W.A., Hay D. Hemadsorption and cytopathic effect produced by African swine fever virus in swine bone marrow and buffy coat cultures. Am. J. Vet. Res. 1960;21:104–108. [PubMed] [Google Scholar]
- 48.Reed L.J. A simple method of estimating fifty percent endpoints. Am. J. Epidemiol. 1938;27:493–497. [Google Scholar]
- 49.Huang L., Liu H., Zhang K., Meng Q., Hu L., Zhang Y., Xiang Z., Li J., Yang Y., Chen Y., Cui S., Tang H., Pei H., Bu Z., Weng C. Ubiquitin-conjugating enzyme 2S enhances viral replication by inhibiting type I IFN production through recruiting USP15 to deubiquitinate TBK1. Cell Rep. 2020;32:108044. doi: 10.1016/j.celrep.2020.108044. [DOI] [PubMed] [Google Scholar]
- 50.Huang L., Xiong T., Yu H., Zhang Q., Zhang K., Li C., Hu L., Zhang Y., Zhang L., Liu Q., Wang S., He X., Bu Z., Cai X., Cui S., et al. Encephalomyocarditis virus 3C protease attenuates type I interferon production through disrupting the TANK-TBK1-IKKε-IRF3 complex. Biochem. J. 2017;474:2051–2065. doi: 10.1042/BCJ20161037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ma J., Chen T., Wu S., Yang C., Bai M., Shu K., Li K., Zhang G., Jin Z., He F., Hermjakob H., Zhu Y. iProX: An integrated proteome resource. Nucleic Acids Res. 2019;47:D1211–D1217. doi: 10.1093/nar/gky869. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the iProX partner repository (51) with the dataset identifier PXD028692.