Abstract
Aptamers are oligonucleotides selected from large pools of random sequences based on their affinity for bioactive molecules and are used in similar ways to antibodies. Aptamers provide several advantages over antibodies, including their small size, facile, large-scale chemical synthesis, high stability, and low immunogenicity. Amyloidogenic proteins, whose aggregation is relevant to neurodegenerative diseases, such as Alzheimer’s, Parkinson’s, and prion diseases, are among the most challenging targets for aptamer development due to their conformational instability and heterogeneity, the same characteristics that make drug development against amyloidogenic proteins difficult. Recently, chemical tethering of aptagens (equivalent to antigens) and advances in high-throughput sequencing-based analysis have been used to overcome some of these challenges. In addition, internalization technologies using fusion to cellular receptors and extracellular vesicles have facilitated central nervous system (CNS) aptamer delivery. In view of the development of these techniques and resources, here we review antiamyloid aptamers, highlighting preclinical application to CNS therapy.
Keywords: oligonucleotide, RNA, DNA, amyloid, Alzheimer's disease, Parkinson's disease, prion, blood-brain barrier, oligomer, exosome
Abbreviations: αSyn, α-synuclein; Aβ, amyloid β-protein; AD, Alzheimer's disease; ADDLs, Aβ-derived diffusible ligands; AI, artificial intelligence; CD, circular dichroism; HT-SELEX, high throughput-systematic evolution of ligands by exponential enrichment; PD, Parkinson's disease; PICUP, photo-induced cross-linking of unmodified protein; RVG, rabies viral glycoprotein; SDS-PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; SELEX, systematic evolution of ligands by exponential enrichment
Neurodegenerative diseases are characterized by a progressive loss of neuronal function. Most of these diseases are age-related, and self-assembly of amyloidogenic proteins is thought to be a cause or a major deleterious mechanism in many of them. Examples include Alzheimer’s disease (AD) and various other tauopathies, Parkinson’s diseases (PD) and other synucleinopathies, prion diseases, and many other sporadic or genetic proteinopathies. The amyloidogenic proteins involved in these diseases are prone to self-association into neurotoxic oligomers and amyloid fibrils (Fig. 1). In many cases, the oligomers, which have metastable structures, have been shown to play pivotal roles in the pathogenesis of the associated diseases and to be more toxic than the structurally stable fibrils (1). The abnormal protein assemblies cause neurotoxicity by a variety of mechanisms, including apoptosis, oxidative stress, inflammation, and disruption of proteostasis through blockage of proteasomal and lysosomal protein degradation. Given the significance of oligomers in the pathogenesis of many neurodegenerative diseases (2, 3, 4), they have been the focus of attention as molecular targets of both diagnostic and therapeutic research and development. Therefore, generation of specific oligomer-binding reagents is a promising approach for development of early detection tools and redirecting the self-assembly process into dissociation back to nontoxic monomers, formation of nontoxic and nonamyloidogenic assemblies amenable to degradation, or in some cases, accelerating the aggregation process to reduce steady-state levels of oligomers in favor of less-toxic fibrils.
Figure 1.
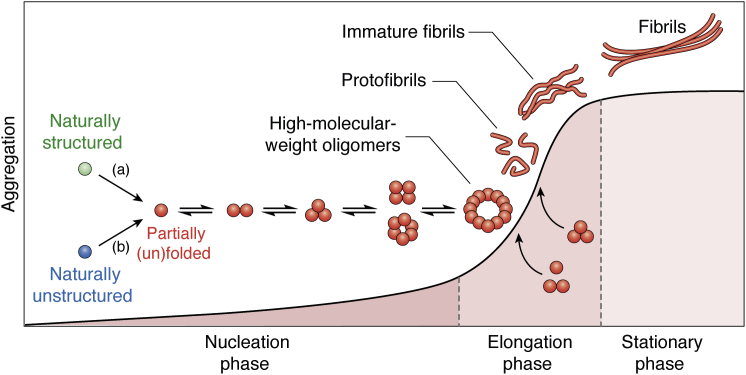
A Schematic diagram of amyloidogenic proteins aggregation. Amyloidogenic proteins can be naturally structured or unstructured. Naturally structured proteins (a), e.g., PrP undergo partial unfolding, whereas naturally unstructured proteins (b), such as Aβ, αSyn, or tau, undergo partial folding under pathological conditions, initiating the self-assembly process. Both cases lead to formation of partially (un)folded monomers, which self-associate into increasing-size oligomers until a quasi-stable nucleus forms leading to the elongation phase. Elongation typically proceeds at a fast rate compared with the nucleation and may involve formation of quasi-stable high-molecular-weight oligomers, protofibrils, and eventually fibrils. Finally, the monomers are consumed and the system reaches a stationary phase in which no more growth is observed.
Aptamers are molecular-recognition agents comprising single-stranded DNA or RNA oligonucleotides that similar to antibodies, bind specifically to diverse targets, including small molecules, peptides, proteins, and nucleic acids (5). Amyloidogenic proteins inherently tend to bind nucleic acids (6, 7, 8) making it difficult to select aptamers specific for just one protein and even more so for distinct assembly states of these proteins. Nonetheless, aptamers offer several advantages compared with antibodies including their small size, facile chemical synthesis, including in large scale, high stability, and low immunogenicity, making them attractive for researchers aiming at developing molecular recognition tools for amyloidogenic proteins. Aptamers typically are obtained by selection from a random-sequence oligonucleotide library based on their affinity for the target of interest using a method called systematic evolution of ligands by exponential enrichment (SELEX, Fig. 2). The oligonucleotides usually span 30 to 100 nucleotides in length and their dissociation constants in complexes with their targets range from pM to mM. The sequence also contains constant regions required for enzymatic manipulation, such as PCR-primer binding and in vitro transcription.
Figure 2.
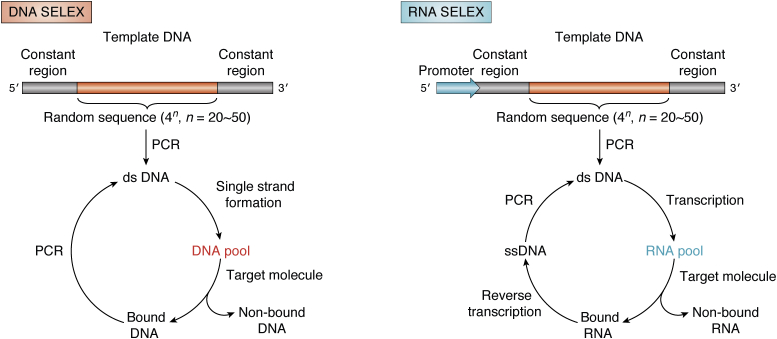
Generation of aptamers by SELEX. The process of SELEX can be used for selection of DNA or RNA aptamers. After the initial PCR amplification of the template DNA, single-strand nucleic acid sequences need to be prepared from the double-strand DNA for binding to the target and selection of high-affinity sequences. This is done using NaOH denaturation or enzyme digestion for DNA aptamers and by in vitro transcription for RNA aptamers. Unbound sequences are discarded and the bound oligonucleotide pool is released from the target. DNA sequences are subjected to PCR amplification to produce double-strand DNA for the next round of selection, whereas RNA sequences are reverse-transcribed first and then amplified by PCR for the next cycle.
A systematic survey by Dumontier, DeRosa, and their colleagues (9) based on 492 published aptamer-related papers has found that the use of DNA aptamers is increasing compared with RNA aptamers. Each nucleic acid has its own advantages and limitations. DNA oligonucleotides are more stable than their RNA counterparts to enzymatic and chemical degradation and are therefore easier to work with. On the other hand, the presence of the 2′-OH in ribose, as opposed to deoxyribose, and the absence of the 5′-methyl group in uracil compared with thymine allow higher conformational stability of RNA, potentially increasing their affinity for the target (10). Deciding whether to use DNA or RNA aptamers is thus a crucial step in the beginning of every aptamer-based research project.
The application of aptamers in the neurodegenerative-disease field has been reviewed in the past (11, 12, 13), yet to our knowledge, there are no systematic reviews on aptamers targeting amyloidogenic proteins. The metastable nature of intrinsically disordered or misfolded proteins, which are prone to form toxic oligomers and eventually amyloid, makes these proteins one of the most challenging targets for aptamer generation. In the case of oligomers, the aptagens (equivalent to antigens) presented to the oligonucleotide library constantly change, whereas in the amyloid fibrils, the conformation of the aptagens can be both variable for the same protein due to formation of different strains and similar for different proteins in the core cross-β structure shared by most amyloid fibrils.
Aptamers selected against amyloidogenic proteins can have various applications, including sensitive detection of biomarkers, as selective inhibitors of the self-assembly process, and as tools for probing molecular mechanisms. The first application—using aptamers as probes for biomarkers—has been developing rapidly and is too large to include in this review. Therefore, we focus here on the challenges and potential solutions in this field and on preclinical therapeutic applications of aptamers specific for amyloidogenic proteins.
To overcome the difficulty in selecting aptamers against metastable protein assemblies, approaches using stable mimics of these assemblies could be useful if the stabilized molecules represent accurately the metastable target. Here, we provide an update on the development of specific aptamers against amyloidogenic proteins, including amyloid β-protein (Aβ), tau, α-synuclein (αSyn), and prion protein (PrP). These proteins represent a range of sizes spanning an order of magnitude—from 40- to 441-amino acid residues, and the two main mechanisms of initial misfolding and aggregation—partial folding of an unstructured protein, such as Aβ, αSyn, or tau, and partial unfolding of a structured protein—PrP (Fig. 1) (14). We discuss how bioinformatics-assisted approaches or artificial-intelligence-based technologies are used to assist the aptamer selection and optimization processes. We also examine approaches for analysis of the secondary and tertiary structures of aptamers and aptamer–target interaction and strategies for CNS-targeting delivery of aptamers for future development of therapeutics in neurodegenerative diseases.
Development of aptamers against amyloidogenic proteins
Several dozen amyloidogenic proteins play major deleterious roles in over 50 proteinopathies. However, studies of aptamers against these proteins have concentrated primarily on four proteins, Aβ, tau, αSyn, and PrP, and therefore, these proteins are also the focus of this review.
Aptamers against Aβ
AD is the most common neurodegenerative disease. Amyloid fibrils in senile plaques in the AD brain consist mainly of the 42-amino acid residue form of Aβ, Aβ42, whereas vascular deposits comprise predominantly the 40-residue form, Aβ40. Both forms are generated from the Aβ-protein precursor by the somewhat promiscuous protease, γ-secretase (15, 16, 17). Aβ42 aggregates faster (18), forms higher-molecular-weight oligomers (19), and is more neurotoxic than Aβ40 (20), and the oligomers of both isoforms are more neurotoxic than the corresponding fibrils (21, 22), making Aβ42 oligomers a primary target for therapy development. In this context, it is important to consider, however, that fibrils might sequester the more toxic Aβ oligomers and possibly are a way cells attempt to reduce the damage caused by the oligomers (23). Thus, if strategies targeting fibril dissociation are considered, inadvertent increase in the concentration of the toxic oligomers, leading to exacerbation, rather than amelioration, of the disease might occur, and one must ensure that this is not the case.
A complicating factor is that “oligomer” is a loosely defined term used for anything from a dimer to large assemblies consisting of hundreds of monomers (22, 24, 25, 26) as long as these assemblies are soluble in aqueous solutions, as opposed to fibrils and other insoluble aggregates. Many different types of Aβ oligomers have been reported, including paranuclei (5,6-mers) (19), Aβ∗56 (56 kDa, 12-mer) (27), protofibrils (PFs, 24–700-mer) (28, 29, 30), globulomers (38/48 kDa, ∼12-mer) (31), AβO (∼90 kDa, 15–20-mer) (32), Aβ-derived diffusible ligands (ADDLs, ∼90 kDa, ∼24-mer) (33), annuli (150–250 kDa, ∼50-mer) (34), and amylospheroids (ASPD; 158–669 kDa, ∼100-mer) (35). Thus, when one contemplates selection of aptamers against Aβ oligomers, an important step is definition of the oligomers used as a target for the selection process.
Protofibrils and ADDLs were the first types of Aβ oligomers described, in 1997 (28, 30) and 1998 (33), respectively. Follow-up studies have generated antibodies against these assemblies, including mAb158 (36) and NU-1 (37), which bind protofibrils and ADDLs, respectively. Many other oligomer-selective antibodies have been reported over the last 2 decades (36, 37, 38, 39, 40, 41) as reviewed elsewhere (25), though due to the metastable nature of the targets, in all cases the specificity of the antibody was lower than what is typically expected for antibodies against stable antigens. Thus, stabilization of the antigens, for example, by attachment of Aβ to gold particles (38), has been an important strategy for generation of antioligomer antibodies. However, an important consideration is that the relevance of the stabilized antigens to the pathological species in the AD brain must be established in each case. Of the many clinical trials using anti-Aβ antibodies, more recent ones have examined antibodies selective against Aβ oligomers, including Biogen’s aducanumab, which recently was approved by the Food and Drug Administration (FDA), and BAN2401, a humanized version of mAb158.
The first aptamers targeting Aβ were reported in 2002 by Ylera et al. The aptamers were RNA oligonucleotides screened against Aβ40 monomers tethered to a Sepharose support. All the selected aptamers recognized Aβ fibrils but not monomers (42). In a later study by Hyman and coworkers (43), one of the aptamers reported by Ylera et al., β55, was used to visualize senile plaques using multiphoton microscopy in brain tissue from patients with AD and the APP/PS1 mouse model (44). The characteristics of these anti-Aβ aptamers and all the subsequent aptamers against amyloidogenic proteins included in this review are summarized in Table 1.
Table 1.
Characteristics of aptamers against Aβ, tau, αSyn, and PrP
| Name | Target | Nucleic acid | KD (testing method) | Selectivity | Year | Ref. |
|---|---|---|---|---|---|---|
| Anti-Aβ aptamers | ||||||
| β55 | Aβ40 monomers | RNA | 29 nM (affinity chromatography) | fibrils | 2002 | (42) |
| KM33 | Aβ40 trimers | RNA | N.T.a | fibrils | 2009 | (48) |
| T-SO508 | αSyn oligomers | DNA | 25 nM for Aβ40 oligomers (ELONAb) | oligomersc of αSyn and Aβ40 | 2012 | (51) |
| E2 | Aβ40 | RNA | 10.9 μM (fluorescence anisotropy) | fibrils | 2009 | (53) |
| RNV95 | Aβ40 | DNA | N.T. | oligomers (∼75 kDa and ∼150 kDa) | 2018 | (55) |
| E22P-AbD43 | Aβ42 protofibrils | RNA | 20 nM (BLId) | dimers | 2020 | (58) |
| Aβ7-92-1H1 | Aβ42 | DNA | 53.3 nM (SPRe) | oligomers | 2020 | (64) |
| Antitau aptamers | ||||||
| ssDNA1 | 1N3R-tau | DNA | 190 nM (capillary electrophoresis) | monomers | 2005 | (66) |
| 3146 | 2N4R-tau | DNA | 13 nM (SPR) | monomers | 2018 | (69) |
| tau-1 | 2N4R-tau | RNA | N.T. | dimers and trimers | 2016 | (70) |
| Anti-αSyn aptamers | ||||||
| M5-15 | αSyn | DNA | N.T. | monomer and oligomers | 2010 | (80) |
| T-SO530 | αSyn oligomers | DNA | 63 nM for αSyn oligomers (ELONA) | oligomers of αSyn and Aβ40 | 2012 | (51) |
| F5R1 | αSyn | DNA | 2.4 nM (SPR) | N.T. | 2018 | (82) |
| Antiprion aptamers | ||||||
| Ap1 | hamster PrP23–231 | RNA | N.T. | hamster, mouse, cattle PrPC | 1997 | (93) |
| 60–3 | mouse PrP23–230 | RNA | 5.6 nM for PrPC (competitive assay) | mouse and bovine PrPC | 2006 | (94) |
| RM312 | sheep PrP23–234 | RNA | 15 nM for PrPC (SPR) | N.T. | 2006 | (95) |
| 4–9 | mouse PrP23–230 | DNA | 113 nM for PrPC, 100 nM for PrPSc (SPR) | PrPC ≅ β-isoform of PrPSc | 2007 | (96) |
| 4C26 | mouse PrP90–231 | DNA | 18 nM for PrPC (affinity assay) | PrPC (N.T. for PrPSc) | 2008 | (97) |
| SAF-93 | PrP fibris from infected hamster brain | RNA | 23.4 nM for PrPSc (affinity assay) | PrPSc > PrPC | 2003 | (98) |
| DP7 | human PrP90–129 | RNA | 1.7 μM for PrPC (affinity assay) | human, hamster, mouse PrPC | 2002 | (100) |
| SSAP3-10 | human PrP23–231 | DNA | N.T. | human, sheep, calf, piglet, deer PrPC, but not PrPSc | 2006 | (101) |
| R14 (from apt#1) | bovine PrP25–241 | RNA | 8.5 nM for PrPC, 280 nM for PrPSc (affinity assay) | PrPC > PrPSc | 2008 | (102) |
Not tested.
Enzyme-linked oligonucleotide assay.
Size is unspecified, unless stated otherwise.
BioLayer interferometry.
Surface plasmon resonance.
To select aptamers against Aβ oligomers, several chemical tethering approaches have been used in later studies. Rahimi et al. prepared covalently cross-linked Aβ40 oligomers using Photo-Induced Cross-linking of Unmodified Proteins (PICUP) (45, 46), which creates covalent bonds at unspecified positions, primarily at Tyr10 in Aβ (Fig. 3A) (47). Of several Aβ40 oligomers stabilized by this technique, they then isolated the most abundant type, trimers, using SDS-PAGE and used the cross-linked trimers to isolate RNA aptamers out of a 77-nucleotide library including 49 randomized nucleotides (A:U:G:C at equal ratios). This size library contains 3.2 × 1029 unique sequences theoretically, yet in reality, if every sequence indeed was included in the library, the mass of such a library would be too high, and the actual number of unique sequences is lower.
Figure 3.
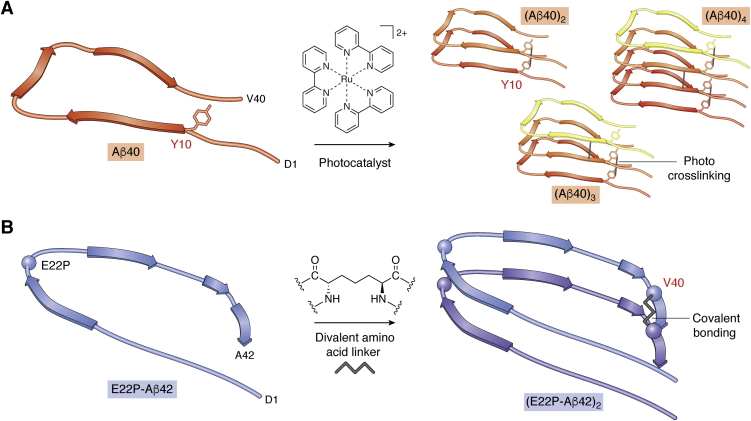
Strategies for stabilization of Aβ oligomers for generation of aptamers.A, photo-cross-linking of Aβ40 by PICUP. Cross-linking is induced by visible-light irradiation in the presence of the photocatalyst tris(bipyridyl)ruthenium(II) ([Ru(bpy)3]2+) and the electron acceptor ammonium persulfate. This method leads to “zero-length” cross-linking directly between amino acid residues, primarily tryptophane and tyrosine. In Aβ, the main cross-link is at Y10, though other bonds also can form. B, a turn structure in Aβ42 is stabilized by an E22P substitution. Dimer stabilization is achieved through substituting V40 by a divalent amino acid (e.g., 2,6-diaminopimelic acid).
Although the selection of the RNA aptamers was against Aβ40 trimers, the final aptamers, KM33 and KM41, were not selective for trimers or other oligomers, but bound to Aβ40 fibrils and did not display a higher affinity for the fibrils than the naive oligonucleotide library used for the selection before enrichment (48, 49). Part of the explanation of these results was that the PICUP-immobilized Aβ40 trimers themselves might have aggregated during the selection process to form fibrillar structures. Further analysis suggested that oligonucleotides have a high, nonspecific affinity for amyloid fibrils, as KM33 and KM41 also recognized fibrils of other amyloidogenic proteins, including calcitonin, islet amyloid polypeptide, insulin, lysozyme, and PrP106–126 (48), which share a cross-β structure and fibrillar morphology with Aβ fibrils (50). Thus, Rahimi et al. (48) demonstrated that the aptamers could be used to monitor fibril formation in a similar manner to thioflavin T (ThT) fluorescence.
Interestingly, a DNA aptamer called T-SO517, originally developed by Ikebukuro and colleagues against oligomeric αSyn, a key protein involved in PD and other synucleinopathies, bound not only to their intended target, αSyn oligomers, but also to Aβ40 oligomers (51), demonstrating the difficulty in obtaining highly specific aptamers against assemblies of amyloidogenic proteins. Considering the high affinity of the KM aptamers mentioned above for fibrils, these findings likely reflect the presence of common structures in such assemblies, i.e., not only the common cross-β structure of the fibrils, but also in the oligomers, as has been suggested by binding of antioligomer antibodies, such as A11, to oligomers of multiple amyloidogenic proteins (38). In an attempt to develop a detection system for Aβ40 oligomers, T-SO517 was applied to a fluorescence detection system using abasic site-containing DNA oligonucleotides (52). When monitoring the Aβ40 aggregation process, this system showed preference for detecting oligomers over monomers or fibrils in vitro, which was confirmed by transmission electron microscopy.
Using a different selection approach, Takahashi et al. (53) tethered Aβ40 to colloidal gold nanoparticles as a target for selection of RNA aptamers using the same method previously developed by Kayed et al. (38) for preparation of oligomer-specific antibodies. The RNA aptamers, N2 and E2, bound to Aβ40 monomers and inhibited Aβ40 fibril formation assessed using the ThT fluorescence assay and transmission electron micrography. However, as was found by Rahimi et al. (48), Babu et al. (54) demonstrated later by using atomic force microscopy that these aptamers also bound to Aβ40 fibrils. Binding to Aβ- or other protein oligomers was not described in either study. A DNA aptamer named RNV95, selected using column-immobilized Aβ40 by Chakravarthy et al. with the goal of detecting low-molecular-weight Aβ40, recognized human brain Aβ oligomers at both ∼75 kDa and ∼150 kDa. However, it was not tested for binding to Aβ fibrils or fibrils of other amyloidogenic proteins so its selectivity for oligomers was not established (55).
To our knowledge, to date, only two studies have explored development of aptamers against Aβ42, likely because it is an even more difficult target than Aβ40. Aβ42 has been shown to aggregate faster (18), form distinct oligomers (19, 56), and cause stronger neurotoxicity than Aβ40 (21, 57). The first characteristic, substantially faster aggregation, makes working with Aβ42 particularly difficult because the preparation of the protein changes during the experiment making obtaining reproducible data highly challenging.
The first example of a successful generation of aptamers against Aβ oligomers we are aware of is by Murakami et al., who developed RNA aptamers termed E22P-AbD4, -AbD31, and -AbD43 against Aβ42 protofibrils (58, 59). To select the aptamers, a dimer of E22P-Aβ42 was used in which the monomers were tethered covalently by a bivalent amino acid linker in place of Val40, within the C-terminal hydrophobic region of Aβ42 (Fig. 3B) (60). Upon incubation in phosphate buffer at 37 °C without agitation for 48 h, this dimer construct, in which a turn near Glu22 was stabilized by an E22P substitution (61), formed protofibrils, the morphology of which was confirmed by transmission electron microscopy. The protofibrils then were used in SELEX to obtain the aptamers from a 77-nucleotide RNA library including a 49-nucleutide random sequence. When used in histological experiments, all three aptamers stained diffuse oligomeric aggregates in two mouse models of AD, Tg2576/PS2 (62) and AppNL-G-F/NL-G-F (63), suggesting that the Aβ assemblies formed in the brain of these two mouse models contained similar protofibril-derived aptatopes. Incubation of the E22P-Aβ42 dimer in the presence of aptamer E22P-AbD43 showed that the aptamer inhibited the nucleation phase of the protofibril formation. The aptamer also inhibited dose-dependently the neurotoxicity of both the E22P-Aβ42 dimer and Aβ42 in the neuroblastoma cell line SH-SY5Y cells. Computational and two-dimensional structure analysis of E22P-AbD43 suggested that preferential binding of the aptamer to Aβ42 protofibrils compared with fibrils might be related to formation of a G-quadruplex structure, implying the presence of a common structure in protofibrils made of either the synthetic dimer or native Aβ42 protofibrils (59).
For the purpose of developing inhibitors of Aβ42 aggregation, Zheng et al. selected an anti-Aβ42 DNA aptamer (Aβ7-92-1H1) by incubating a library of DNA-oligonucleotide-coated beads with nonaggregated Aβ42. This library consisted of 100-nucleotide-long sequences including two separate 18-nucleotide random sequences. Aβ7-92-1H1 bound Aβ42 and neither Aβ40 nor other amyloids. The affinity of the aptamer for Aβ42 oligomers was slightly higher than for Aβ42 monomers, measured by surface plasmon resonance (SPR). The aptamer inhibited Aβ42 fibril formation as evidenced by atomic force microscopy and β-sheet formation measured using CD spectra (64).
Aptamers against tau protein
Tauopathies are neurodegenerative diseases caused by formation of toxic oligomers and aggregates of the microtubule-associated protein tau. Tau is produced as six different isoforms in humans, due to alternative splicing of exons 2, 3, and 10 leading to polypeptides ranging from 352 to 441 amino acid residues. Translation of neither, one, or both exons 2 and 3 is marked as 0N, 1N, or 2N isoforms of tau, whereas the alternative splicing of exon 10, which encodes part of the microtubule binding, repeat domain of tau leads to isoforms containing three (3R) or four (4R) repeats.
The most prevalent tauopathy is AD (65). Other examples of tauopathies include frontotemporal lobar degeneration, progressive supranuclear palsy, and chronic traumatic encephalopathy. Aggregation and deposition of tau, e.g., as neurofibrillary tangles in AD, are associated with hyperphosphorylation and other aberrant posttranslational modifications of the protein.
In the course of testing whether tau binds single-strand DNA, Krylova et al. (66) first found DNA sequences binding recombinant 1N3R-tau or 2N3R-tau using nonequilibrium capillary electrophoresis of equilibrium mixtures, in which the gel shift caused by binding of the DNA oligonucleotides to tau was evaluated. In follow-up studies, these DNA sequences were used as aptamers for detection of 1N3R-tau in human plasma using SPR and were shown to reach femtomolar level sensitivity (67). Selection of aptamers without amplification between selection rounds shortens the time and saves costs compared with conventional SELEX (68). However, this is not a standard method in the amyloid field. Lisi et al. applied this method coupled with capillary electrophoresis for partitioning of bound DNA from unbound DNA to isolate DNA aptamers targeting several tau isoforms. The aptamer 3146 was obtained from a 5 × 1012 DNA oligonucleotide library in only three rounds within 1 day. 3146 bound tau isoforms in the order 2N4R (KD = 13 nM) > 0N4R (49 nM) > 0N3R (84 nM) > 1N3R (116 nM) (69).
An RNA aptamer called tau-1 was obtained against 2N4R-tau, by applying SELEX to a 90-nucleotide RNA library containing a 40-nucleotide random region. This aptamer prevented the formation of 2N4R-tau dimers and trimers assessed by SDS-PAGE/western blotting (70). It is important to note, however, that SDS-PAGE is not a reliable method for assessing oligomer formation by amyloidogenic proteins, as SDS perturbs protein conformation and can both dissociate existing assemblies and induce formation of different assemblies (26). Nonetheless, in an experiment using a HEK293-derived cell line expressing human 2N4R-tau under control of doxycycline induction (71), reduced cell viability resulting from tau expression was recovered by treating with tau-1 compared to a random RNA library (70).
Aptamers against α-synuclein
αSyn, a causative agent of synucleinopathies, such as PD, dementia with Lewy bodies, and multiple system atrophy, oligomerizes and aggregates in the brain of patients leading to neurotoxicity and neurodegeneration (72). αSyn is a 140-amino-acid long protein mainly located at presynaptic terminals. The amphipathic N-terminal region (residues 1–60), which includes four 11-residue imperfect repeats, and the hydrophobic middle region (residues 61–95) are more important for aggregation than the acidic and proline-rich C-terminal region (residues 96–140) of αSyn (73). The C-terminal region of αSyn is susceptible to cleavage upon aggregation (74), yet most antibodies bind to this region and therefore may miss αSyn aggregates in pathological analysis of patient brains or animal models. This problem is particularly important because the most common form of αSyn used to detect pathological aggregates is phosphorylated at Ser129 (pS129-αSyn), yet some cleavage sites are N-terminal to position 129 and eliminate the epitope for antibodies against pS129-αSyn.
Similar to Aβ and tau, αSyn oligomers, rather than fibrils, are thought to be the primary neurotoxic form of the protein. αSyn oligomers cause neurotoxicity, synaptic impairment, mitochondrial dysfunction, endoplasmic reticulum stress, neuroinflammation, proteostasis dysregulation, and apoptosis, culminating in neuronal death (75). Recent cryo-electron microscopy analyses have deciphered the atomic structure of αSyn filaments (76) and structure–activity analysis suggested that the N-terminus controls αSyn aggregation (77). The details of αSyn oligomer structures are not known, yet β-sheet structure has been reported in rigid regions of toxic αSyn oligomers, whereas in nontoxic oligomers, these regions are unstructured (78). The mechanism underlying oligomer toxicity may involve insertion into lipid bilayers, disrupting membrane integrity (78). Bioinformatic and NMR studies have supported an important role for the N-terminus in modulating the aggregation of αSyn (79).
Ikebukuro and colleagues reported a first DNA aptamer against αSyn, called M5-15, which showed affinity for both monomers and oligomers (80). In follow-up studies, the same group applied counterselection of monomers and fibrils, leading to a more selective aptamer, T-SO530, against αSyn oligomers, whose selectivity was confirmed by dot blots (51). CD analysis suggested that T-SO530 formed a G-quadruplex structure, which might have contributed to its affinity. Indeed, G-quadruplex stabilizers, such as L1H1-7OTD and TmPyP4 (Fig. 4), enhanced the binding of T-SO530 to αSyn oligomers (81), suggesting that such stabilizers can be promising synthetic modulators/cofactors for applications of aptamers against amyloidogenic proteins in neurodegenerative diseases.
Figure 4.
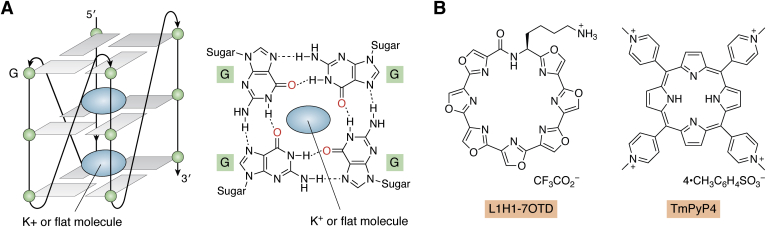
G-quadruplex structures in aptamers and their stabilization.A, a schematic structure of a G-quadruplex, which can be stabilized by a metal ion, e.g., K+, or flat molecules, for example those shown in panel B. B, structures of L1H1-7OTD and TmPyP4, which have been used as stabilizers of a G-quadruplex in the anti-αSyn aptamer T-SO530.
Zheng et al. (82) developed 58-nucleotide DNA aptamers they termed F5R1 and F5R2, starting with a 40-nucleotide random single-strand DNA library, which bound αSyn with a high affinity and inhibited αSyn aggregation. To enhance cell membrane permeability of the aptamers, they were modified by attachment of a peptide carrier, CADY, which had been reported previously to form stable complexes with nucleic acids, leading to improvement of their delivery into cultured cells (83). The modified aptamers reduced intracellular aggregation of αSyn, synaptic protein loss, and neuronal death caused by αSyn overexpression.
Aptamers against prion protein
The accumulation of misfolded PrP characterizes prionoses, such as Creutzfeldt-Jakob disease, Kuru, Gerstmann–Sträussler–Scheinker Syndrome, and fatal familial insomnia in humans, bovine spongiform encephalopathy in cattle, scrapie in sheep, and chronic wasting disease in deer and elk (84, 85, 86). PrP exists normally as PrPC (cellular form), which is involved in neuroprotection and trophic signaling (87, 88, 89), yet this form can misfold due to genetic, environmental, or yet unknown causes into a toxic and infectious form called PrPSc (scrapie form). The term PrPSc does not describe one particular structure but refers to many conformational strains that are self-propagating, transmissible from cell to cell, and are infectious within the same species and sometimes across species. PrPSc infectivity to other organisms depends on the specific strain and species barrier. PrPC is rich in α-helix, whereas PrPSc contains the typical cross-β structure of amyloid fibrils. The aggregation process, including oligomerization of PrPSc, plays a central role in the prion’s propagation and neurotoxicity (90). Biochemically, PrPSc is a highly stable form of the protein, which is resistant to proteinases and denaturing agents.
Interestingly, one of the biological roles of PrPC is a receptor of Aβ oligomers. Binding of Aβ oligomers to PrPC causes synaptotoxicity and neuritic dystrophy, possibly also leading to tauopathy (91, 92). Thus, both PrPC and PrPSc are potential therapeutic targets for development of various drug modalities, including aptamers.
Weiss et al. reported the first RNA aptamer, Ap1, against a recombinant Syrian golden hamster prion protein PrP23–231, a common model of full-length PrPC. The aptamer recognized PrPC in brain extracts of scrapie-infected mice, hamsters, and cattle, though its binding affinity was not measured (93). An RNA aptamer called “60-3”, selected against mouse PrP23–230, was reported by Sekiya et al. (94) and also bound bovine PrP25–241. Mercey et al. (95) developed an RNA aptamer, RM312, against sheep PrP23–234, which bound with 15 nM affinity to PrPC. In another study, Ikebukuro and colleagues developed a DNA aptamer, termed “4-9”, against mouse PrP23–230 that bound to both PrPC and PrPSc equally (96). A DNA aptamer called 4C26 was prepared by Garson and colleagues against mouse PrP90–231 and exhibited a higher binding affinity for mouse prion than “4-9” (97) suggesting that shortening the target length in this case improved the affinity.
To pursue aptamers selective for PrPSc versus PrPC, an RNA aptamer, SAF-93, was developed against scrapie-associated fibrils from the brains of infected hamsters. In a competitive binding assay using bovine PrPSc fibrils, SAF-93 showed >10-fold higher affinity for PrPSc than for PrPC (98). Uniquely, SAF-93 was obtained by SELEX using 2′-fluoro-modified pyrimidine triphosphate nucleotides together with unmodified purine nucleotides. Enzymatic probing and gel footprinting, in addition to computer-assisted secondary structure analysis, identified multiple binding sites of SAF-93 on PrPSc (99).
Several aptamers also have been selected against human prion proteins. An RNA aptamer termed DP7 was prepared by Proske et al. against human PrP90–129, an important domain for the conversion of PrPC into PrPSc. The aptamer also bound PrP derived from hamster and mouse. The utility of this aptamer for reducing the ratio of PrPSc to PrPC was demonstrated in prion-infected mouse neuroblastoma N2a cells (100). Takemura et al. (101) reported a DNA aptamer called SSAP3-10 against human PrP23–231, which bound other mammalian PrPC (sheep, calf, piglet, and deer) in addition to human PrPC, but not to PrPSc.
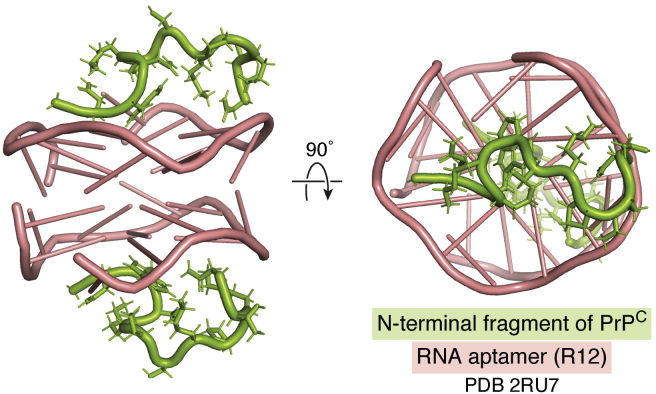
Structural studies of prion aptamers have been advanced compared with other amyloidogenic proteins. Murakami et al. developed an RNA aptamer, apt#1, selected against bovine PrP25–241. The binding was deduced to be due to a G-quadruplex structure using circular dichroism (CD) spectroscopy. An extracted sequence from apt#1 (R14 aptamer, GGAGGUUUUGGAGG) was identified using mutagenesis and showed improved binding affinity and ∼30-fold selectivity for PrPC compared with PrPSc (102). Katahira and colleagues further optimized the sequence to deduce a tandem repeat sequence (R12 aptamer, (GGA)4), and found using NMR measurements that it formed an intramolecular parallel G-quadruplex (103). R12 bound to two sites in the N-terminal half of PrPC by forming a dimer (104, 105) (Fig. 5). Treatment with R12 reduced the level of PrPC (104) and blocked the pathological conformational conversion of PrPC into PrPSc in scrapie-infected mouse neuronal GT1-7 cells (106).
Figure 5.
Stabilization of a complex between the antiprion RNA aptamer R12 and the N-terminal region of PrPC. A dimer of R12 [pink, r(GGAGGAGGAGGA)] associates with two N-terminal PrPC peptides (green, Gly-Gln-Trp-Asn-Lys-Pro-Ser-Lys-Pro-Lys-Thr-Asn) providing a 1:1 stoichiometric ratio. The illustration was created using PyMOL from PDB ID: 2RU7.
Computer-assisted development and structure prediction of aptamers
Selection and clustering of aptamers by high-throughput sequencing
Despite the large potential of aptamers as therapeutic agents, biosensors, and research tools, technical issues can require major time and money investments, limiting development and progress. One such technical issue is the use of classic Sanger sequencing of the selected oligonucleotides. Although the selection process removes the vast majority of the sequences, the final pool still may contain thousands of distinct oligonucleotide sequences, making sequencing of this pool and identifying the sequences possessing the best affinity and/or specificity labor-intensive and time-consuming.
In silico approaches, previously reviewed by Hamada (107), have been useful for aptamer design. Artificial intelligence (AI) coupled with machine learning algorithms assists in identifying potential aptamer candidates from the selected sequences expeditiously, leading to improvement compared with classical prediction tools (108). As only the oligonucleotide pool of the last round of selection typically is cloned and sequenced in conventional SELEX, superior aptamers may be overlooked. In this section, we discuss the role of bioinformatics in aptamer development, including in sequencing, secondary-structure prediction, and simulation of aptamer–target interaction, and highlight the use of these advances in selection of aptamers against amyloidogenic proteins.
With the advent of new sequencing technologies, such as Next-Generation sequencing, exhaustive parallel sequencing of all the selection rounds has facilitated high-throughput SELEX (HT-SELEX) (109, 110, 111). In the first report of this technique, a KD in the picomolar range was achieved after only three rounds of selection for aptamers against the BB subunits of platelet-derived growth factor (109). The abundance of sequences read in all the SELEX pools can be ranked by various parameters in HT-SELEX, e.g., sequence enrichment (112), and meta-Z-score (113). The meta-Z-score is a statistical scoring method for high prediction accuracy of binding potential. This method allows gleaning insight into whether sequence enrichment occurs, rather than whether the affinity for the target is enhanced at substantially earlier stages of the aptamer-selection process, resulting in considerable time saving. Additionally, a decreased number of selection rounds helps reduce PCR bias (nonspecific amplification) caused by over selection or excessive cycle numbers in PCR (114). Comprehensive analysis of very large sequence datasets by such bioinformatics methods has enabled not only accurate characterization of aptamers, data alignment, and clustering, but also improved prediction of aptamer structure and aptamer–target interaction mode.
After collecting and ranking datasets of sequences, data clustering typically is a next step, which can be achieved using free software tools, such as AptaCluster (115) (https://www.ncbi.nlm.nih.gov/CBBresearch/Przytycka/index.cgi#aptatools) and FASTAptamer (116) (https://burkelab.missouri.edu/fastaptamer.html), both of which use RNA sequence, but not structural information. Ikebukuro and colleagues proposed a genetic methodology named evolution-mimicking algorithm, to identify optimized aptamer sequences in rugged sequence spaces. Using this methodology, by combining several in vitro assays, e.g., binding affinity, inhibitory activity, specificity, and 3D structure, in silico maturation that included selection and duplication, recombination, and point mutation has facilitated identifying several aptamers against PrP and αSyn (117, 118, 119).
Analysis of secondary and tertiary structure of aptamers by motif finding and structure optimization
When conducting an aptamer discovery campaign, the three-dimensional structures of the selected aptamers and aptamer–target complexes are important considerations. The relatively simpler conformational space of nucleic acids compared with proteins makes computer-aided calculation and modeling of secondary and tertiary structures particularly useful for aptamers. The secondary structure of oligonucleic acids is determined by canonical Watson–Crick base-paring interactions or noncanonical Hoogsteen base pairs in single or double strands (Fig. 6). Due to the additional hydroxyl group in ribose compared with deoxyribose, RNA single strands are believed to form more complex and diverse structures than DNA, such as stem loops and pseudoknots.
Figure 6.
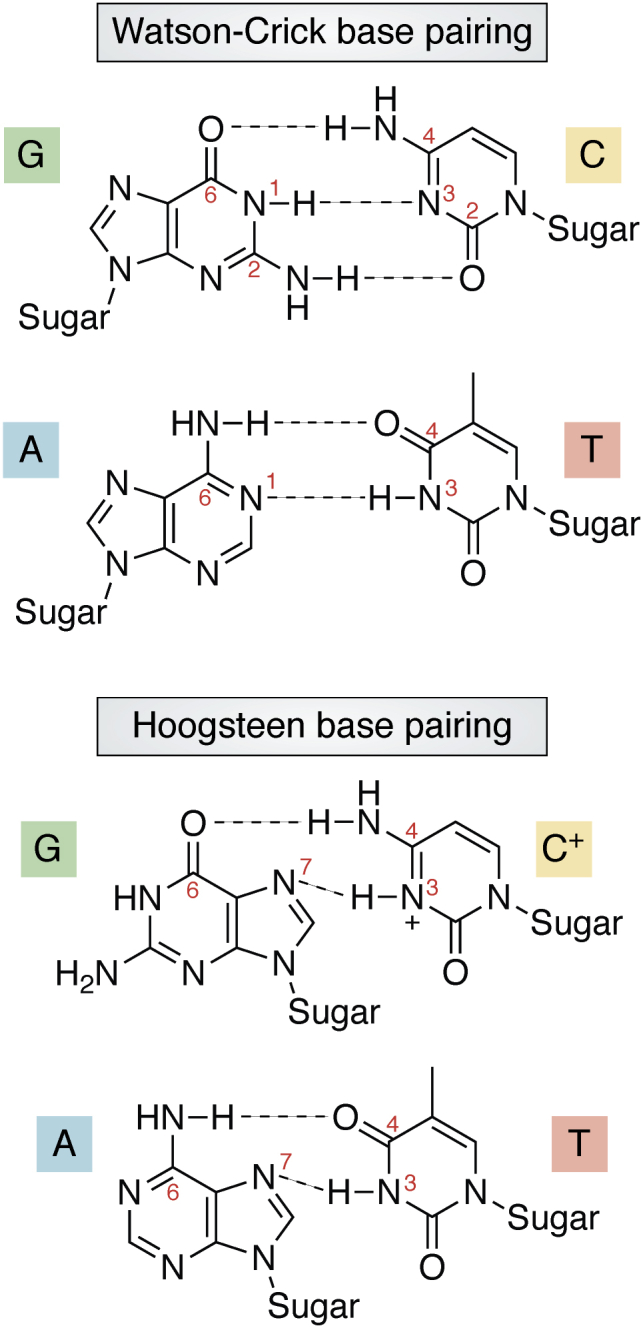
Canonical and noncanonical stabilization of natural base-pairs. Canonical, Watson–Crick interactions have three hydrogen bonds between guanine and cytosine (O6–N4, N1–N3, and N2–O2), and two hydrogen bonds between adenine and thymine (N6–O4 and N1–N3). In noncanonical Hoogsteen interactions, the adenine or guanine is rotated 180° around the glycosidic bond, resulting in alternative hydrogen base pairs—two hydrogen bonds between guanine and cytosine (O6–N4 and N7–N3), and two hydrogen bonds between adenine and thymine (N6–O4 and N7–N3). Hoogsteen interactions are minor compared with the Watson–Crick structures but may contribute to formation of unique DNA or RNA structures.
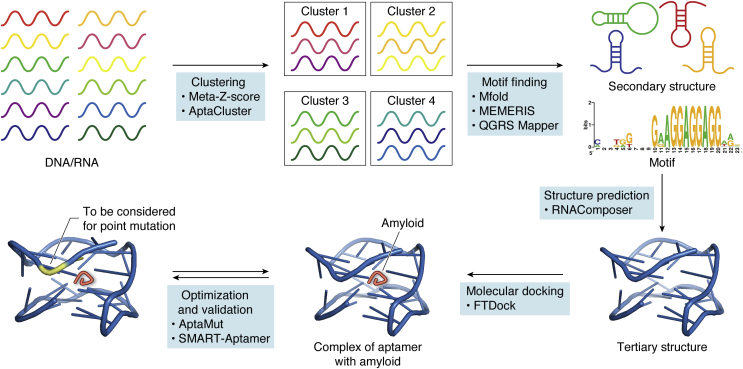
The program Mfold, originally developed by Zuker (120, 121) and later modified by incorporating into the University of Wisconsin Genetics Computer Group software suites (122), uses minimal free energy optimization and is one of the most popular tools for determination of nucleic acids’ secondary structure. Typically used after Mfold, MEMERIS is a general motif-finding algorithm for integration of secondary structures (123). AptaMotif is a program designed for identifying binding motifs in aptamers identified through SELEX and is based on structural processing, including suboptimal secondary structures, for prediction of sequence motifs in the loop regions (124). The tertiary structures of aptamers can be predicted based on the calculated information of secondary structures. Online programs and servers are available for this purpose, including RNAComposer (125) and SimRNAweb (126). A putative flow from an identified sequence to computer-aided structural optimization of aptamers is shown in Figure 7. To allow convenient selection of the appropriate tool for each task, Rtools (http://rtools.cbrc.jp/) was developed as an integrated web-server hosting existing prediction algorithms for RNA secondary structure analysis (127).
Figure 7.
A recommended workflow for in silico aptamer design, analysis, and optimization. Candidates of DNA or RNA aptamers are subjected to clustering by high-throughput sequence analysis programs, such as meta-Z-score or AptaCluster. To predict secondary structures and motifs, the resulting clusters are processed by secondary-structure and motif-finding software tools such as Mfold, MEMERIS, and/or QGRS Mapper. Next, prediction of the tertiary structure of the aptamers is performed by RNAComposer. Then, the binding site(s) may be deciphered by molecular docking of the aptamers with their target, e.g., an amyloid protein, using machine learning tools, such as FTDock. Finally, the resulting complex structure is further tested by SMART-Aptamer, followed by AptaMut analysis for considering point mutations that may improve the aptamer’s affinity and/or specificity.
Structural prediction programs have begun being applied in the amyloid field. Mfold likely is most used program in the field due to its free availability. Mfold analysis of the anti-Aβ40 DNA aptamer RNV95 suggested a stem-loop structure, which was validated by CD spectroscopy measurements (55). Bunka et al. (128) utilized Mfold for the prediction of RNA aptamer structure against β2-microglobulin, a protein whose deposition as amyloid in joints is associated with symptoms of dialysis-related amyloidosis, to reveal an enzyme cleavage site in a stem-loop structure, facilitating the identification of the aptamer’s aptatope.
G-quadruplex is a noncanonical structure in both RNA and DNA aptamers, which, as mentioned in previous sections, has been found in several cases to play key roles in aptamer binding to amyloid–protein targets. The G-quadruplex structure includes a stable planar core comprising four guanine bases in the same plane forming G-tetrads (Fig. 4A) stabilized by π–π interactions (129, 130). The structure can be stabilized further by metal ions (131) or by flat molecule-induced chelation (Fig. 4B) (81). However, prediction of G-quadruplex motifs is particularly difficult because of the need to bring together four noncontiguous guanines that may be far apart from each other in the sequence (132).
Several software tools have been developed to address this challenge, such as QGRS (Quadruplex forming G-Rich Sequences) Mapper (133), GRSdb (134), and QuadBase2 (135), which are specific to prediction of G-quadruplexes in RNA and DNA sequences. For example, the formation of G-quadruplex in an RNA aptamer against Aβ42 protofibrils predicted by QGRS Mapper was validated by detection of a negative peak at ∼240 nm and a positive peak at ∼265 nm in the CD spectrum (136), and by observation of a 1650 cm−1 absorbance peak in the Attenuated Total Reflection-FTIR spectrum (137), both of which are unique to guanine carbonyl groups in a G-quadruplex (58, 59). Similarly, a DNA aptamer (T-SO508) against αSyn oligomers was predicted to contain a G-quadruplex by QGRS Mapper, and the prediction was confirmed by CD spectroscopy (81).
In the course of sequence optimization after structure prediction, to increase the affinity for the targets, the program AptaMut (112) can provide clues for the potential effects of point mutations on affinity and structure. To enhance the resistance of RNA aptamers to chemical and enzymatic degradation, inclusion of modified nucleotides, such as 2′-O-methyl, 2′-fluoro, or other unnatural nucleotides (138, 139) in place of the natural 2′-OH ribose are common approaches. A step forward was introduced independently by Hirao and colleagues (140, 141), and Tan, Benner and coworkers (142), who incorporated modified nucleotides originally designed to form unnatural base pairs for synthetic xenobiology, into aptamers, increasing the resistance of the oligonucleotides to degradation (Fig. 8).
Figure 8.
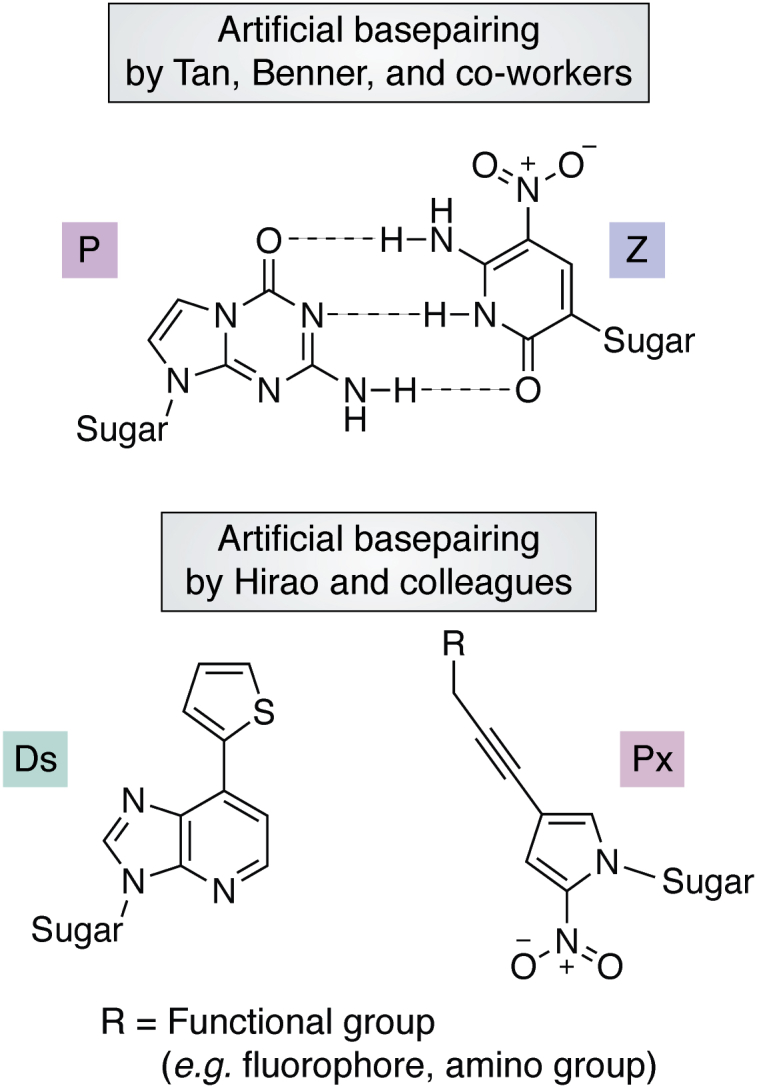
Noncanonical artificial base pairs used in aptamers. Artificial base pairs amenable to PCR amplification and aptamer development together with the natural ones: P, 2-amino-8-(1′-β-D-2-deoxyribofuranosyl)-imidazo[1,2-a]-1,3,5-triazin-4(8H)one; Z, 6-amino-5-nitro-3-(1′-β-D-2′-deoxyribofuranosyl)-2(1H)-pyridone; Ds, 7-(2-thienyl)-imidazo[4,5-b]pyridine; and Px, 2-nitro-4-propynylpyrrole. Hydrogen-bond stabilization for the Ds-Px pair has not been described.
Among the challenges in translating aptamers against amyloidogenic proteins into therapeutic applications is also their potential toxicity, similar to other nucleic-acid-based drugs. For example, in the development of antisense therapy, acute hepatotoxicity of 2′-fluoro-modified antisense oligonucleotides was found in 6- to 8-week-old wild-type mice, in which the oligonucleotides were administered subcutaneously at 20 mg/kg three times a week (143). In contrast, in other studies, a 2′-fluoro nucleotide-stabilized siRNA was administered subcutaneously as a single, 30-mg/kg bolus for 2 years in a rat or at daily 7.5-mg/kg injections for 5 days in healthy volunteers without apparent toxicity (144). Similarly, antisense oligonucleotides containing 2′-O-methyl nucleotides were administered to cynomolgus macaques (Macaca fascicularis) at 20 mg/kg by subcutaneous injections for 9 months and to patients with hereditary transthyretin amyloidosis in a phase 2/3 clinical study at 300 mg for 15 months without cytotoxic effects (145). Another concern is that as is the case with antibodies (see, e.g., an interesting report by Hatami et al. (146)), aptamers may cross-react with off-targets. In particular, aptamers against amyloid proteins might cross-react with functional human amyloids (147, 148) and disrupt their function, leading to cytotoxicity. Thus, in addition to characterization of their binding and efficacy, aptamers intended for in vivo use must be evaluated for their safety and be applied only if they have a sufficiently large therapeutic index. In this context, an advantage of aptamers as chemotypes for drug development is the ease with which they can be modified and subjected to structure–activity relationship studies compared with small molecules or protein biologics.
Prediction of aptamer–protein interaction by machine learning and molecular docking
After determination of the secondary and tertiary structures of new aptamers, deciphering the aptamer–protein complex structure often is the next goal. Molecular docking of aptamers and target proteins on each other is a forecasting method that typically consists of searching all the potential binding modes between the pair of molecules in silico and scoring each complex based on its thermodynamic stability. As an example, a docking simulation of multiple aptamer–protein pairs was carried out to optimize the aptamer sequence using different proteins in combination either with RNA aptamers or with ribosomal RNA by Zhang et al. (149) using FTDock. The report was primarily methodological, but we expect that this methodology will be useful for future drug development.
Recently, a new Sequential Multidimensional Analysis algoRiThm for aptamer discovery (SMART-Aptamer) using HT-SELEX based on machine learning has been described (150). This system of molecular docking allowed obtaining high-affinity aptamers with low false-positive and false-negative rates. This new system coupled with modeling of the tertiary structure has high promise for optimization of the interaction mode between aptamers and amyloid–protein targets. However, although these approaches have helped optimizing aptamers in different cases, such as for human embryonic stem cells, epithelial cell adhesion molecule, and cell-surface vimentin (150), they have not yet been used in the amyloid field.
Brain-targeting delivery of aptamers by exosomes and nanoliposomes
Oligonucleic acids are too large and negatively charged to pass through the blood–brain barrier (BBB). To exert the functions of amyloid-targeting aptamers in the CNS, the development of effective delivery systems is an important challenge. Since bare nucleic acid delivered systemically also would be subject to enzymatic degradation and may not even reach the BBB in sufficient amounts (151), innovative engineering of CNS-targeting delivery methods of aptamers using nanocarriers, such as exosomes or nanoliposome, has been a major focus in recent years.
Exosomes are nanovesicles, 30 to 200 nm in diameter, secreted by virtually all cell types and are thought to mediate intercellular and interorgan communication, as well as serve a mechanism for removal of cellular stressors (152, 153, 154). They can be used as a delivery mechanism when the cargo is expressed in cultured cells and the exosomes containing the cargo are isolated from the cell-culture medium.
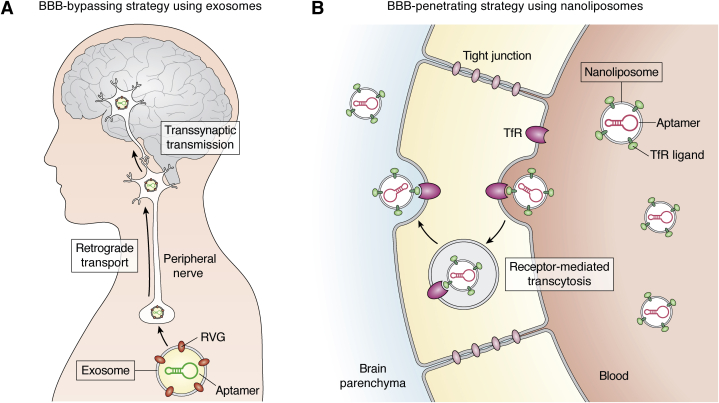
A follow-up study on the DNA aptamer F5R1 (82) mentioned above revealed that it bound preferably to fibrillar, rather than monomeric αSyn (155). To investigate a therapeutic potential of F5R1, it was encapsuled, following polyethylenimine-assisted transfection, in exosomes isolated from the culture medium of HEK293 cells. It was then delivered into the brain of wild-type mice, which received intrastriatal injection of preformed recombinant αSyn fibrils prepared from recombinant αSyn, by taking advantage of a virus-transmission system (Fig. 9A) (155). The cells also expressed rabies virus glycoprotein (RVG) fused with lamp2B, which positions the RVG on the outer membrane of exosomes secreted by these cells. Intraperitoneal administration of the RVG-decorated exosomes containing F5R1 led to retrograde transport and transsynaptic transmission into the CNS through the axons and synapses of peripheral neurons, circumventing the BBB. The treatment decreased αSyn aggregation in the substantia nigra and ameliorated motor dysfunction in the treated mice (155). This pioneering work provided proof of concept for application of DNA–aptamer therapy delivered into the CNS using an innovative delivery system that bypasses the BBB, resulting in reduction of the aggregation, transmission, and toxicity of αSyn in a preclinical model of synucleinopathy.
Figure 9.
Strategies for delivering aptamers into the CNS by encapsulation in nanovesicles.A, aptamers encapsulated in exosome expressing rabies virus glycoprotein (RVG) on their surface are retrogradely and transsynaptically transported from the peripheral nervous system into the brain, circumventing the BBB. B, aptamers included in nanoliposome carrying ligands of transferrin receptor (TfR) are transported from the blood to the brain parenchyma by receptor-mediated transcytosis.
A different aptamer-delivery method into the CNS using nanoliposomes was reported by McConnell et al. (Fig. 9B) (156). Like exosomes, nanoliposomes are nanovesicles, yet they are prepared synthetically, as opposed to the natural origin of exosomes, allowing a greater degree of control over their composition and cargo. Similar to exosomes, the nanoliposomes are enclosed by a lipid bilayer surrounding trapped water in which hydrophilic compounds can be transported. McConnell et al. encapsuled a dopamine-specific DNA aptamer, Da20m, into nanoliposomes conjugated on their surface by a different aptamer, specific for the transferrin receptor (TfR). TfR is expressed on endothelial cells in the luminal surface of the BBB. Binding of the anti-TfR aptamer to TfR followed by clathrin-mediated transcytosis of TfR allowed delivery of the nanoliposomes across the BBB and release of the antidopamine aptamer cargo on the CNS-parenchymal side. Compared with unencapsulated Da20m, which had little or no effect, systemic administration of Da20m via the functionalized nanoliposomes potently attenuated abnormal behavior of wild-type mice triggered by aberrantly increased extracellular dopamine following exposure to cocaine (156). This system appears to be highly promising, yet to our knowledge it has not been used yet for delivery of aptamers against amyloidogenic proteins.
To date, over 20 liposomal products have been approved by the FDA, including anticancer drugs and vaccine formulations against SARS-CoV-2 containing RNA molecules (157, 158). Encapsulation of RNA (and DNA) in liposomes promotes the biosafety and stability of the exogenous nucleic acids inside the body. Both exosomes and nanoliposomes are attractive delivery systems for aptamers. Exosome cargo and surface markers are easier to manipulate genetically, compared with those of nanoliposomes, yet nanoliposomes can be produced more easily on large scales (159). In addition, because exosomes are produced in cells and may contain unintended biological material, it is crucial to establish ways of controlling and monitoring their clinical safety if they are going to be approved for therapeutic applications (160). Although exosomes are less immunogenic than cell-based therapies, a concern in using them as drug carriers is that they may be recognized as nonself by the immune system and induce an immune response, potentially causing damaging inflammation. Should this become a problem, it could be overcome by extracting the patient’s own cells and preparing the exosomes using the patient-derived cells. Though more time-consuming and potentially expensive, such a procedure could be justified for life-saving treatments.
Conclusions and future perspectives
In 2004, the first aptamer drug, the RNA aptamer Macugen (pegaptanib sodium), which acts as an antagonist of vascular endothelial growth factor, was approved by FDA for treatment of age-related macular degeneration. Several additional aptamers have been tested in clinical trials for cancer, cardiovascular disease, and eye diseases, yet to date, despite the growing number of reports on antiamyloid aptamers and the advances in their design and delivery discussed above, aptamers have not yet advanced to clinical trials in the neurodegenerative disease field.
To advance the development of aptamers targeting amyloidogenic protein for biomedical applications, several issues should be addressed. First, the conformational metastability and heterogeneity of the targets, particularly oligomers of amyloidogenic proteins, are major impediments for aptamer selection. This challenge can be addressed by covalent tethering of the protein monomers to create a more stable aptagen that mimics the metastable oligomers, as was demonstrated in the case of Aβ42 (58), by identification of conformations or sequences responsible for pathology, as was done for prion (98) or tau (70), and by competitive selection of target assemblies in combination with counterselection of other species, as was successfully carried out for αSyn (51).
Second, isolating aptamers with high affinity and specificity for the desired target is a labor-intensive and time-consuming process that is always accompanied by a level of uncertainty. Next-Generation sequencing and structural prediction programs coupled with AI and machine learning algorithms are promising approaches for increasing both the speed of the process and the likelihood of success. Application of these modern techniques has been limited in the amyloid field but is beginning to catch up. Additional advances are expected when detailed structures of aptamers and their complexes with their respective targets become available and support structure prediction algorithms. To our knowledge, to date, only two X-ray crystallography studies reporting such structures have been published (161, 162). However, the increasing popularity of high-resolution structural determination by cryo-electron microscopy suggests that additional data will be forthcoming. High-resolution structures and improved structural prediction technology not only will facilitate obtaining improved aptamers, but also will help clarify the molecular basis of target recognition by aptamers, including in the amyloid field.
Finally, the inability of aptamers to cross the BBB on their own can now be addressed by at least two delivery systems, including exosomes and nanoliposomes, as discussed above. Each approach has its advantages and limitations, yet we expect that future research will address the current shortcomings and lead to successful application of these techniques in delivering aptamers targeting amyloidogenic proteins into the CNS, as has been demonstrated recently for aptamer F5R1 in a mouse model of synucleinopathy (155). Recent advances in both computational and experimental approaches suggest that using aptamers as antiamyloid therapeutics is achievable in the near future.
Conflicts of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
Author contributions
K. M. and G. B. conceptualization; K. M. supervision; K. M. and N. I. writing—original draft; G. B. writing—review and editing.
Funding and additional information
This study was supported in part by the JSPS KAKENHI, grant number 20KK0126 to K. M. and N. I, and by NIH/NIA grant RF1AG054000, The Alzheimer's Association, The Michael J. Fox Foundation, Weston Brain Institute, and Alzheimer’s Research UK Biomarkers Across Neurodegenerative Diseases (BAND 3) grant 17990, and CurePSP grant 665-2019-07 to G. B.
Edited by Paul Fraser
Contributor Information
Kazuma Murakami, Email: murakami.kazuma.4v@kyoto-u.ac.jp.
Gal Bitan, Email: gbitan@mednet.ucla.edu.
References
- 1.Iadanza M.G., Jackson M.P., Hewitt E.W., Ranson N.A., Radford S.E. A new era for understanding amyloid structures and disease. Nat. Rev. Mol. Cell Biol. 2018;19:755–773. doi: 10.1038/s41580-018-0060-8. [DOI] [PubMed] [Google Scholar]
- 2.Hartl F.U. Protein misfolding diseases. Annu. Rev. Biochem. 2017;86:21–26. doi: 10.1146/annurev-biochem-061516-044518. [DOI] [PubMed] [Google Scholar]
- 3.Chuang E., Hori A.M., Hesketh C.D., Shorter J. Amyloid assembly and disassembly. J. Cell Sci. 2018;131 doi: 10.1242/jcs.189928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otzen D., Riek R. Functional amyloids. Cold Spring Harb. Perspect. Biol. 2019;11 doi: 10.1101/cshperspect.a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou J., Rossi J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ginsberg S.D., Galvin J.E., Chiu T.S., Lee V.M., Masliah E., Trojanowski J.Q. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 1998;96:487–494. doi: 10.1007/s004010050923. [DOI] [PubMed] [Google Scholar]
- 7.Ginsberg S.D., Crino P.B., Hemby S.E., Weingarten J.A., Lee V.M., Eberwine J.H., Trojanowski J.Q. Predominance of neuronal mRNAs in individual Alzheimer's disease senile plaques. Ann. Neurol. 1999;45:174–181. [PubMed] [Google Scholar]
- 8.Hua Q., He R.Q., Haque N., Qu M.H., del Carmen Alonso A., Grundke-Iqbal I., Iqbal K. Microtubule associated protein tau binds to double-stranded but not single-stranded DNA. Cell. Mol. Life Sci. 2003;60:413–421. doi: 10.1007/s000180300034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKeague M., McConnell E.M., Cruz-Toledo J., Bernard E.D., Pach A., Mastronardi E., Zhang X., Beking M., Francis T., Giamberardino A., Cabecinha A., Ruscito A., Aranda-Rodriguez R., Dumontier M., DeRosa M.C. Analysis of in vitro aptamer selection parameters. J. Mol. Evol. 2015;81:150–161. doi: 10.1007/s00239-015-9708-6. [DOI] [PubMed] [Google Scholar]
- 10.Shu Y., Pi F., Sharma A., Rajabi M., Haque F., Shu D., Leggas M., Evers B.M., Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev. 2014;66:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qu J., Yu S., Zheng Y., Zheng Y., Yang H., Zhang J. Aptamer and its applications in neurodegenerative diseases. Cell. Mol. Life Sci. 2017;74:683–695. doi: 10.1007/s00018-016-2345-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouvier-Muller A., Duconge F. Nucleic acid aptamers for neurodegenerative diseases. Biochimie. 2018;145:73–83. doi: 10.1016/j.biochi.2017.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Rahimi F. Aptamers selected for recognizing amyloid β-protein-a case for cautious optimism. Int. J. Mol. Sci. 2018;19:668. doi: 10.3390/ijms19030668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fezoui Y., Teplow D.B. Kinetic studies of amyloid β-protein fibril assembly. Differential effects of α-helix stabilization. J. Biol. Chem. 2002;277:36948–36954. doi: 10.1074/jbc.M204168200. [DOI] [PubMed] [Google Scholar]
- 15.Glenner G.G., Wong C.W. Alzheimer's disease: Initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem. Biophys. Res. Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 16.Masters C.L., Simms G., Weinman N.A., Multhaup G., McDonald B.L., Beyreuther K. Amyloid plaque core protein in Alzheimer disease and Down syndrome. Proc. Natl. Acad. Sci. U. S. A. 1985;82:4245–4249. doi: 10.1073/pnas.82.12.4245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karran E., Mercken M., De Strooper B. The amyloid cascade hypothesis for Alzheimer's disease: An appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 2011;10:698–712. doi: 10.1038/nrd3505. [DOI] [PubMed] [Google Scholar]
- 18.Jarrett J.T., Berger E.P., Lansbury P.T., Jr. The carboxy terminus of the β amyloid protein is critical for the seeding of amyloid formation: Implications for the pathogenesis of Alzheimer's disease. Biochemistry. 1993;32:4693–4697. doi: 10.1021/bi00069a001. [DOI] [PubMed] [Google Scholar]
- 19.Bitan G., Kirkitadze M.D., Lomakin A., Vollers S.S., Benedek G.B., Teplow D.B. Amyloid β-protein (Aβ) assembly: Aβ40 and Aβ42 oligomerize through distinct pathways. Proc. Natl. Acad. Sci. U. S. A. 2003;100:330–335. doi: 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haass C., Selkoe D.J. Soluble protein oligomers in neurodegeneration: Lessons from the Alzheimer's amyloid β-peptide. Nat. Rev. Mol. Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- 21.Dahlgren K.N., Manelli A.M., Stine W.B., Jr., Baker L.K., Krafft G.A., LaDu M.J. Oligomeric and fibrillar species of amyloid-β peptides differentially affect neuronal viability. J. Biol. Chem. 2002;277:32046–32053. doi: 10.1074/jbc.M201750200. [DOI] [PubMed] [Google Scholar]
- 22.Roychaudhuri R., Yang M., Hoshi M.M., Teplow D.B. Amyloid β-protein assembly and Alzheimer disease. J. Biol. Chem. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meyer-Luehmann M., Spires-Jones T.L., Prada C., Garcia-Alloza M., de Calignon A., Rozkalne A., Koenigsknecht-Talboo J., Holtzman D.M., Bacskai B.J., Hyman B.T. Rapid appearance and local toxicity of amyloid-β plaques in a mouse model of Alzheimer's disease. Nature. 2008;451:720–724. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benilova I., Karran E., De Strooper B. The toxic Aβ oligomer and Alzheimer's disease: An emperor in need of clothes. Nat. Neurosci. 2012;15:349–357. doi: 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- 25.Murakami K. Conformation-specific antibodies to target amyloid β oligomers and their application to immunotherapy for Alzheimer's disease. Biosci. Biotechnol. Biochem. 2014;78:1293–1305. doi: 10.1080/09168451.2014.940275. [DOI] [PubMed] [Google Scholar]
- 26.Bitan G., Fradinger E.A., Spring S.M., Teplow D.B. Neurotoxic protein oligomers--what you see is not always what you get. Amyloid. 2005;12:88–95. doi: 10.1080/13506120500106958. [DOI] [PubMed] [Google Scholar]
- 27.Lesnè S., Koh M.T., Kotilinek L., Kayed R., Glabe C.G., Yang A., Gallagher M., Ashe K.H. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 28.Walsh D.M., Lomakin A., Benedek G.B., Condron M.M., Teplow D.B. Amyloid β-protein fibrillogenesis. Detection of a protofibrillar intermediate. J. Biol. Chem. 1997;272:22364–22372. doi: 10.1074/jbc.272.35.22364. [DOI] [PubMed] [Google Scholar]
- 29.Walsh D.M., Hartley D.M., Kusumoto Y., Fezoui Y., Condron M.M., Lomakin A., Benedek G.B., Selkoe D.J., Teplow D.B. Amyloid β-protein fibrillogenesis. Structure and biological activity of protofibrillar intermediates. J. Biol. Chem. 1999;274:25945–25952. doi: 10.1074/jbc.274.36.25945. [DOI] [PubMed] [Google Scholar]
- 30.Harper J.D., Wong S.S., Lieber C.M., Lansbury P.T. Observation of metastable Aβ amyloid protofibrils by atomic force microscopy. Chem. Biol. 1997;4:119–125. doi: 10.1016/s1074-5521(97)90255-6. [DOI] [PubMed] [Google Scholar]
- 31.Barghorn S., Nimmrich V., Striebinger A., Krantz C., Keller P., Janson B., Bahr M., Schmidt M., Bitner R.S., Harlan J., Barlow E., Ebert U., Hillen H. Globular amyloid β-peptide oligomer - a homogenous and stable neuropathological protein in Alzheimer's disease. J. Neurochem. 2005;95:834–847. doi: 10.1111/j.1471-4159.2005.03407.x. [DOI] [PubMed] [Google Scholar]
- 32.Deshpande A., Mina E., Glabe C., Busciglio J. Different conformations of amyloid β induce neurotoxicity by distinct mechanisms in human cortical neurons. J. Neurosci. 2006;26:6011–6018. doi: 10.1523/JNEUROSCI.1189-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lambert M.P., Barlow A.K., Chromy B.A., Edwards C., Freed R., Liosatos M., Morgan T.E., Rozovsky I., Trommer B., Viola K.L., Wals P., Zhang C., Finch C.E., Krafft G.A., Klein W.L. Diffusible, nonfibrillar ligands derived from Aβ1-42 are potent central nervous system neurotoxins. Proc. Natl. Acad. Sci. U. S. A. 1998;95:6448–6453. doi: 10.1073/pnas.95.11.6448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caughey B., Lansbury P.T. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 35.Hoshi M., Sato M., Matsumoto S., Noguchi A., Yasutake K., Yoshida N., Sato K. Spherical aggregates of β-amyloid (amylospheroid) show high neurotoxicity and activate tau protein kinase I/glycogen synthase kinase-3β. Proc. Natl. Acad. Sci. U. S. A. 2003;100:6370–6375. doi: 10.1073/pnas.1237107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tucker S., Moller C., Tegerstedt K., Lord A., Laudon H., Sjodahl J., Soderberg L., Spens E., Sahlin C., Waara E.R., Satlin A., Gellerfors P., Osswald G., Lannfelt L. The murine version of BAN2401 (mAb158) selectively reduces amyloid-β protofibrils in brain and cerebrospinal fluid of tg-ArcSwe mice. J. Alzheimers Dis. 2015;43:575–588. doi: 10.3233/JAD-140741. [DOI] [PubMed] [Google Scholar]
- 37.Lambert M.P., Velasco P.T., Chang L., Viola K.L., Fernandez S., Lacor P.N., Khuon D., Gong Y., Bigio E.H., Shaw P., De Felice F.G., Krafft G.A., Klein W.L. Monoclonal antibodies that target pathological assemblies of Aβ. J. Neurochem. 2007;100:23–35. doi: 10.1111/j.1471-4159.2006.04157.x. [DOI] [PubMed] [Google Scholar]
- 38.Kayed R., Head E., Thompson J.L., McIntire T.M., Milton S.C., Cotman C.W., Glabe C.G. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 39.Hillen H., Barghorn S., Striebinger A., Labkovsky B., Muller R., Nimmrich V., Nolte M.W., Perez-Cruz C., van der Auwera I., van Leuven F., van Gaalen M., Bespalov A.Y., Schoemaker H., Sullivan J.P., Ebert U. Generation and therapeutic efficacy of highly oligomer-specific β-amyloid antibodies. J. Neurosci. 2010;30:10369–10379. doi: 10.1523/JNEUROSCI.5721-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goñi F., Marta-Ariza M., Peyser D., Herline K., Wisniewski T. Production of monoclonal antibodies to pathologic β-sheet oligomeric conformers in neurodegenerative diseases. Sci. Rep. 2017;7:9881. doi: 10.1038/s41598-017-10393-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kayed R., Canto I., Breydo L., Rasool S., Lukacsovich T., Wu J., Albay R., 3rd, Pensalfini A., Yeung S., Head E., Marsh J.L., Glabe C. Conformation dependent monoclonal antibodies distinguish different replicating strains or conformers of prefibrillar Aβ oligomers. Mol. Neurodegener. 2010;5:57. doi: 10.1186/1750-1326-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ylera F., Lurz R., Erdmann V.A., Furste J.P. Selection of RNA aptamers to the Alzheimer's disease amyloid peptide. Biochem. Biophys. Res. Commun. 2002;290:1583–1588. doi: 10.1006/bbrc.2002.6354. [DOI] [PubMed] [Google Scholar]
- 43.Farrar C.T., William C.M., Hudry E., Hashimoto T., Hyman B.T. RNA aptamer probes as optical imaging agents for the detection of amyloid plaques. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankowsky J.L., Fadale D.J., Anderson J., Xu G.M., Gonzales V., Jenkins N.A., Copeland N.G., Lee M.K., Younkin L.H., Wagner S.L., Younkin S.G., Borchelt D.R. Mutant presenilins specifically elevate the levels of the 42 residue β-amyloid peptide in vivo: Evidence for augmentation of a 42-specific γ secretase. Hum. Mol. Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- 45.Bitan G. Structural study of metastable amyloidogenic protein oligomers by photo-induced cross-linking of unmodified proteins. Methods Enzymol. 2006;413:217–236. doi: 10.1016/S0076-6879(06)13012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bitan G., Teplow D.B. Rapid photochemical cross-linking--a new tool for studies of metastable, amyloidogenic protein assemblies. Acc. Chem. Res. 2004;37:357–364. doi: 10.1021/ar000214l. [DOI] [PubMed] [Google Scholar]
- 47.Maji S.K., Ogorzalek Loo R.R., Inayathullah M., Spring S.M., Vollers S.S., Condron M.M., Bitan G., Loo J.A., Teplow D.B. Amino acid position-specific contributions to amyloid β-protein oligomerization. J. Biol. Chem. 2009;284:23580–23591. doi: 10.1074/jbc.M109.038133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rahimi F., Murakami K., Summers J.L., Chen C.H., Bitan G. RNA aptamers generated against oligomeric Aβ40 recognize common amyloid aptatopes with low specificity but high sensitivity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahimi F., Bitan G. Selection of aptamers for amyloid β-protein, the causative agent of Alzheimer's disease. J. Vis. Exp. 2010;39:1955. doi: 10.3791/1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sipe J.D., Cohen A.S. Review: History of the amyloid fibril. J. Struct. Biol. 2000;130:88–98. doi: 10.1006/jsbi.2000.4221. [DOI] [PubMed] [Google Scholar]
- 51.Tsukakoshi K., Abe K., Sode K., Ikebukuro K. Selection of DNA aptamers that recognize α-synuclein oligomers using a competitive screening method. Anal. Chem. 2012;84:5542–5547. doi: 10.1021/ac300330g. [DOI] [PubMed] [Google Scholar]
- 52.Zhu L., Zhang J., Wang F., Wang Y., Lu L., Feng C., Xu Z., Zhang W. Selective amyloid β oligomer assay based on abasic site-containing molecular beacon and enzyme-free amplification. Biosens. Bioelectron. 2016;78:206–212. doi: 10.1016/j.bios.2015.11.048. [DOI] [PubMed] [Google Scholar]
- 53.Takahashi T., Tada K., Mihara H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol. Biosyst. 2009;5:986–991. doi: 10.1039/b903391b. [DOI] [PubMed] [Google Scholar]
- 54.Babu E., Muthu Mareeswaran P., Sathish V., Singaravadivel S., Rajagopal S. Sensing and inhibition of amyloid-β based on the simple luminescent aptamer-ruthenium complex system. Talanta. 2015;134:348–353. doi: 10.1016/j.talanta.2014.11.020. [DOI] [PubMed] [Google Scholar]
- 55.Chakravarthy M., AlShamaileh H., Huang H., Tannenberg R.K., Chen S., Worrall S., Dodd P.R., Veedu R.N. Development of DNA aptamers targeting low-molecular-weight amyloid-β peptide aggregates in vitro. Chem. Commun. 2018;54:4593–4596. doi: 10.1039/c8cc02256a. [DOI] [PubMed] [Google Scholar]
- 56.Bernstein S.L., Dupuis N.F., Lazo N.D., Wyttenbach T., Condron M.M., Bitan G., Teplow D.B., Shea J.E., Ruotolo B.T., Robinson C.V., Bowers M.T. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer's disease. Nat. Chem. 2009;1:326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murakami K., Irie K., Morimoto A., Ohigashi H., Shindo M., Nagao M., Shimizu T., Shirasawa T. Neurotoxicity and physicochemical properties of Aβ mutant peptides from cerebral amyloid angiopathy: Implication for the pathogenesis of cerebral amyloid angiopathy and Alzheimer's disease. J. Biol. Chem. 2003;278:46179–46187. doi: 10.1074/jbc.M301874200. [DOI] [PubMed] [Google Scholar]
- 58.Murakami K., Obata Y., Sekikawa A., Ueda H., Izuo N., Awano T., Takabe K., Shimizu T., Irie K. An RNA aptamer with potent affinity for a toxic dimer of amyloid β42 has potential utility for histochemical studies of Alzheimer's disease. J. Biol. Chem. 2020;295:4870–4880. doi: 10.1074/jbc.RA119.010955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Obata Y., Murakami K., Kawase T., Hirose K., Izuo N., Shimizu T., Irie K. Detection of amyloid β oligomers with RNA aptamers in AppNL-G-F/NL-G-F mice: A model of Arctic Alzheimer's disease. ACS Omega. 2020;5:21531–21537. doi: 10.1021/acsomega.0c02134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Murakami K., Tokuda M., Suzuki T., Irie Y., Hanaki M., Izuo N., Monobe Y., Akagi K., Ishii R., Tatebe H., Tokuda T., Maeda M., Kume T., Shimizu T., Irie K. Monoclonal antibody with conformational specificity for a toxic conformer of amyloid β42 and its application toward the Alzheimer's disease diagnosis. Sci. Rep. 2016;6:29038. doi: 10.1038/srep29038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Irie K. New diagnostic method for Alzheimer's disease based on the toxic conformation theory of amyloid β. Biosci. Biotechnol. Biochem. 2020;84:1–16. doi: 10.1080/09168451.2019.1667222. [DOI] [PubMed] [Google Scholar]
- 62.Toda T., Noda Y., Ito G., Maeda M., Shimizu T. Presenilin-2 mutation causes early amyloid accumulation and memory impairment in a transgenic mouse model of Alzheimer's disease. J. Biomed. Biotechnol. 2011;2011:617974. doi: 10.1155/2011/617974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saito T., Matsuba Y., Mihira N., Takano J., Nilsson P., Itohara S., Iwata N., Saido T.C. Single app knock-in mouse models of Alzheimer's disease. Nat. Neurosci. 2014;17:661–663. doi: 10.1038/nn.3697. [DOI] [PubMed] [Google Scholar]
- 64.Zheng Y., Wang P., Li S., Geng X., Zou L., Jin M., Zou Q., Wang Q., Yang X., Wang K. Development of DNA aptamer as a β-amyloid aggregation inhibitor. ACS Appl. Bio Mater. 2020;3:8611–8618. doi: 10.1021/acsabm.0c00996. [DOI] [PubMed] [Google Scholar]
- 65.Arendt T., Stieler J.T., Holzer M. Tau and tauopathies. Brain Res. Bull. 2016;126:238–292. doi: 10.1016/j.brainresbull.2016.08.018. [DOI] [PubMed] [Google Scholar]
- 66.Krylova S.M., Musheev M., Nutiu R., Li Y., Lee G., Krylov S.N. Tau protein binds single-stranded DNA sequence specifically--the proof obtained in vitro with non-equilibrium capillary electrophoresis of equilibrium mixtures. FEBS Lett. 2005;579:1371–1375. doi: 10.1016/j.febslet.2005.01.032. [DOI] [PubMed] [Google Scholar]
- 67.Kim S., Wark A.W., Lee H.J. Femtomolar detection of tau proteins in undiluted plasma using surface plasmon resonance. Anal. Chem. 2016;88:7793–7799. doi: 10.1021/acs.analchem.6b01825. [DOI] [PubMed] [Google Scholar]
- 68.Berezovski M.V., Musheev M.U., Drabovich A.P., Jitkova J.V., Krylov S.N. Non-SELEX: Selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006;1:1359–1369. doi: 10.1038/nprot.2006.200. [DOI] [PubMed] [Google Scholar]
- 69.Lisi S., Fiore E., Scarano S., Pascale E., Boehman Y., Duconge F., Chierici S., Minunni M., Peyrin E., Ravelet C. Non-SELEX isolation of DNA aptamers for the homogeneous-phase fluorescence anisotropy sensing of tau proteins. Anal. Chim. Acta. 2018;1038:173–181. doi: 10.1016/j.aca.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 70.Kim J.H., Kim E., Choi W.H., Lee J., Lee J.H., Lee H., Kim D.E., Suh Y.H., Lee M.J. Inhibitory RNA aptamers of tau oligomerization and their neuroprotective roles against proteotoxic stress. Mol. Pharm. 2016;13:2039–2048. doi: 10.1021/acs.molpharmaceut.6b00165. [DOI] [PubMed] [Google Scholar]
- 71.Bandyopadhyay B., Li G., Yin H., Kuret J. Tau aggregation and toxicity in a cell culture model of tauopathy. J. Biol. Chem. 2007;282:16454–16464. doi: 10.1074/jbc.M700192200. [DOI] [PubMed] [Google Scholar]
- 72.Breydo L., Wu J.W., Uversky V.N. α-Synuclein misfolding and Parkinson's disease. Biochim. Biophys. Acta. 2012;1822:261–285. doi: 10.1016/j.bbadis.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 73.Tripathi T. A master regulator of α-synuclein aggregation. ACS Chem. Neurosci. 2020;11:1376–1378. doi: 10.1021/acschemneuro.0c00216. [DOI] [PubMed] [Google Scholar]
- 74.Sorrentino Z.A., Giasson B.I. The emerging role of α-synuclein truncation in aggregation and disease. J. Biol. Chem. 2020;295:10224–10244. doi: 10.1074/jbc.REV120.011743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Du X.Y., Xie X.X., Liu R.T. The role of α-synuclein oligomers in Parkinson's disease. Int. J. Mol. Sci. 2020;21:8645. doi: 10.3390/ijms21228645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schweighauser M., Shi Y., Tarutani A., Kametani F., Murzin A.G., Ghetti B., Matsubara T., Tomita T., Ando T., Hasegawa K., Murayama S., Yoshida M., Hasegawa M., Scheres S.H.W., Goedert M. Structures of α-synuclein filaments from multiple system atrophy. Nature. 2020;585:464–469. doi: 10.1038/s41586-020-2317-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.McGlinchey R.P., Ni X., Shadish J.A., Jiang J., Lee J.C. The N terminus of α-synuclein dictates fibril formation. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2023487118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fusco G., Chen S.W., Williamson P.T.F., Cascella R., Perni M., Jarvis J.A., Cecchi C., Vendruscolo M., Chiti F., Cremades N., Ying L., Dobson C.M., De Simone A. Structural basis of membrane disruption and cellular toxicity by α-synuclein oligomers. Science. 2017;358:1440–1443. doi: 10.1126/science.aan6160. [DOI] [PubMed] [Google Scholar]
- 79.Doherty C.P.A., Ulamec S.M., Maya-Martinez R., Good S.C., Makepeace J., Khan G.N., van Oosten-Hawle P., Radford S.E., Brockwell D.J. A short motif in the N-terminal region of α-synuclein is critical for both aggregation and function. Nat. Struct. Mol. Biol. 2020;27:249–259. doi: 10.1038/s41594-020-0384-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tsukakoshi K., Harada R., Sode K., Ikebukuro K. Screening of DNA aptamer which binds to α-synuclein. Biotechnol. Lett. 2010;32:643–648. doi: 10.1007/s10529-010-0200-5. [DOI] [PubMed] [Google Scholar]
- 81.Tsukakoshi K., Ikuta Y., Abe K., Yoshida W., Iida K., Ma Y., Nagasawa K., Sode K., Ikebukuro K. Structural regulation by a G-quadruplex ligand increases binding abilities of G-quadruplex-forming aptamers. Chem. Commun. 2016;52:12646–12649. doi: 10.1039/c6cc07552e. [DOI] [PubMed] [Google Scholar]
- 82.Zheng Y., Qu J., Xue F., Zheng Y., Yang B., Chang Y., Yang H., Zhang J. Novel DNA aptamers for Parkinson's disease treatment inhibit α-synuclein aggregation and facilitate its degradation. Mol. Ther. Nucleic Acids. 2018;11:228–242. doi: 10.1016/j.omtn.2018.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Crombez L., Aldrian-Herrada G., Konate K., Nguyen Q.N., McMaster G.K., Brasseur R., Heitz F., Divita G. A new potent secondary amphipathic cell-penetrating peptide for siRNA delivery into mammalian cells. Mol. Ther. 2009;17:95–103. doi: 10.1038/mt.2008.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Prusiner S.B., Scott M.R., DeArmond S.J., Cohen F.E. Prion protein biology. Cell. 1998;93:337–348. doi: 10.1016/s0092-8674(00)81163-0. [DOI] [PubMed] [Google Scholar]
- 85.Collinge J. Prion diseases of humans and animals: Their causes and molecular basis. Annu. Rev. Neurosci. 2001;24:519–550. doi: 10.1146/annurev.neuro.24.1.519. [DOI] [PubMed] [Google Scholar]
- 86.Aguzzi A., Polymenidou M. Mammalian prion biology: One century of evolving concepts. Cell. 2004;116:313–327. doi: 10.1016/s0092-8674(03)01031-6. [DOI] [PubMed] [Google Scholar]
- 87.McLennan N.F., Brennan P.M., McNeill A., Davies I., Fotheringham A., Rennison K.A., Ritchie D., Brannan F., Head M.W., Ironside J.W., Williams A., Bell J.E. Prion protein accumulation and neuroprotection in hypoxic brain damage. Am. J. Pathol. 2004;165:227–235. doi: 10.1016/S0002-9440(10)63291-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Spudich A., Frigg R., Kilic E., Kilic U., Oesch B., Raeber A., Bassetti C.L., Hermann D.M. Aggravation of ischemic brain injury by prion protein deficiency: Role of ERK-1/-2 and STAT-1. Neurobiol. Dis. 2005;20:442–449. doi: 10.1016/j.nbd.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 89.Mitteregger G., Vosko M., Krebs B., Xiang W., Kohlmannsperger V., Nolting S., Hamann G.F., Kretzschmar H.A. The role of the octarepeat region in neuroprotective function of the cellular prion protein. Brain Pathol. 2007;17:174–183. doi: 10.1111/j.1750-3639.2007.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sengupta I., Udgaonkar J.B. Structural mechanisms of oligomer and amyloid fibril formation by the prion protein. Chem. Commun. 2018;54:6230–6242. doi: 10.1039/c8cc03053g. [DOI] [PubMed] [Google Scholar]
- 91.Lauren J. Cellular prion protein as a therapeutic target in Alzheimer's disease. J. Alzheimers Dis. 2014;38:227–244. doi: 10.3233/JAD-130950. [DOI] [PubMed] [Google Scholar]
- 92.Lauren J., Gimbel D.A., Nygaard H.B., Gilbert J.W., Strittmatter S.M. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weiss S., Proske D., Neumann M., Groschup M.H., Kretzschmar H.A., Famulok M., Winnacker E.L. RNA aptamers specifically interact with the prion protein PrP. J. Virol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sekiya S., Noda K., Nishikawa F., Yokoyama T., Kumar P.K., Nishikawa S. Characterization and application of a novel RNA aptamer against the mouse prion protein. J. Biochem. 2006;139:383–390. doi: 10.1093/jb/mvj046. [DOI] [PubMed] [Google Scholar]
- 95.Mercey R., Lantier I., Maurel M.C., Grosclaude J., Lantier F., Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Arch. Virol. 2006;151:2197–2214. doi: 10.1007/s00705-006-0790-3. [DOI] [PubMed] [Google Scholar]
- 96.Ogasawara D., Hasegawa H., Kaneko K., Sode K., Ikebukuro K. Screening of DNA aptamer against mouse prion protein by competitive selection. Prion. 2007;1:248–254. doi: 10.4161/pri.1.4.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bibby D.F., Gill A.C., Kirby L., Farquhar C.F., Bruce M.E., Garson J.A. Application of a novel in vitro selection technique to isolate and characterise high affinity DNA aptamers binding mammalian prion proteins. J. Virol. Methods. 2008;151:107–115. doi: 10.1016/j.jviromet.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 98.Rhie A., Kirby L., Sayer N., Wellesley R., Disterer P., Sylvester I., Gill A., Hope J., James W., Tahiri-Alaoui A. Characterization of 2'-fluoro-RNA aptamers that bind preferentially to disease-associated conformations of prion protein and inhibit conversion. J. Biol. Chem. 2003;278:39697–39705. doi: 10.1074/jbc.M305297200. [DOI] [PubMed] [Google Scholar]
- 99.Sayer N.M., Cubin M., Rhie A., Bullock M., Tahiri-Alaoui A., James W. Structural determinants of conformationally selective, prion-binding aptamers. J. Biol. Chem. 2004;279:13102–13109. doi: 10.1074/jbc.M310928200. [DOI] [PubMed] [Google Scholar]
- 100.Proske D., Gilch S., Wopfner F., Schatzl H.M., Winnacker E.L., Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. ChemBioChem. 2002;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 101.Takemura K., Wang P., Vorberg I., Surewicz W., Priola S.A., Kanthasamy A., Pottathil R., Chen S.G., Sreevatsan S. DNA aptamers that bind to PrP(C) and not PrP(Sc) show sequence and structure specificity. Exp. Biol. Med. (Maywood) 2006;231:204–214. doi: 10.1177/153537020623100211. [DOI] [PubMed] [Google Scholar]
- 102.Murakami K., Nishikawa F., Noda K., Yokoyama T., Nishikawa S. Anti-bovine prion protein RNA aptamer containing tandem GGA repeat interacts both with recombinant bovine prion protein and its β isoform with high affinity. Prion. 2008;2:73–80. doi: 10.4161/pri.2.2.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mashima T., Matsugami A., Nishikawa F., Nishikawa S., Katahira M. Unique quadruplex structure and interaction of an RNA aptamer against bovine prion protein. Nucleic Acids Res. 2009;37:6249–6258. doi: 10.1093/nar/gkp647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mashima T., Nishikawa F., Kamatari Y.O., Fujiwara H., Saimura M., Nagata T., Kodaki T., Nishikawa S., Kuwata K., Katahira M. Anti-prion activity of an RNA aptamer and its structural basis. Nucleic Acids Res. 2013;41:1355–1362. doi: 10.1093/nar/gks1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hayashi T., Oshima H., Mashima T., Nagata T., Katahira M., Kinoshita M. Binding of an RNA aptamer and a partial peptide of a prion protein: Crucial importance of water entropy in molecular recognition. Nucleic Acids Res. 2014;42:6861–6875. doi: 10.1093/nar/gku382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mashima T., Lee J.H., Kamatari Y.O., Hayashi T., Nagata T., Nishikawa F., Nishikawa S., Kinoshita M., Kuwata K., Katahira M. Development and structural determination of an anti-PrP(C) aptamer that blocks pathological conformational conversion of prion protein. Sci. Rep. 2020;10:4934. doi: 10.1038/s41598-020-61966-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Hamada M. In silico approaches to RNA aptamer design. Biochimie. 2018;145:8–14. doi: 10.1016/j.biochi.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 108.Chen Z., Hu L., Zhang B.T., Lu A., Wang Y., Yu Y., Zhang G. Artificial intelligence in aptamer-target binding prediction. Int. J. Mol. Sci. 2021;22:3605. doi: 10.3390/ijms22073605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cho M., Xiao Y., Nie J., Stewart R., Csordas A.T., Oh S.S., Thomson J.A., Soh H.T. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jolma A., Kivioja T., Toivonen J., Cheng L., Wei G., Enge M., Taipale M., Vaquerizas J.M., Yan J., Sillanpaa M.J., Bonke M., Palin K., Talukder S., Hughes T.R., Luscombe N.M., et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kupakuwana G.V., Crill J.E., 2nd, McPike M.P., Borer P.N. Acyclic identification of aptamers for human α-thrombin using over-represented libraries and deep sequencing. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hoinka J., Berezhnoy A., Dao P., Sauna Z.E., Gilboa E., Przytycka T.M. Large scale analysis of the mutational landscape in HT-SELEX improves aptamer discovery. Nucleic Acids Res. 2015;43:5699–5707. doi: 10.1093/nar/gkv308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jiang P., Meyer S., Hou Z., Propson N.E., Soh H.T., Thomson J.A., Stewart R. MPBind: A meta-motif-based statistical framework and pipeline to predict binding potential of SELEX-derived aptamers. Bioinformatics. 2014;30:2665–2667. doi: 10.1093/bioinformatics/btu348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nguyen Quang N., Perret G., Duconge F. Applications of high-throughput sequencing for in vitro selection and characterization of aptamers. Pharmaceuticals (Basel) 2016;9:76. doi: 10.3390/ph9040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hoinka J., Berezhnoy A., Sauna Z.E., Gilboa E., Przytycka T.M. AptaCluster - a method to cluster HT-SELEX aptamer pools and lessons from its application. Res. Comput. Mol. Biol. 2014;8394:115–128. doi: 10.1007/978-3-319-05269-4_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alam K.K., Chang J.L., Burke D.H. FASTAptamer: A bioinformatic toolkit for high-throughput sequence analysis of combinatorial selections. Mol. Ther. Nucleic Acids. 2015;4 doi: 10.1038/mtna.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ikebukuro K., Okumura Y., Sumikura K., Karube I. A novel method of screening thrombin-inhibiting DNA aptamers using an evolution-mimicking algorithm. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Noma T., Ikebukuro K. Aptamer selection based on inhibitory activity using an evolution-mimicking algorithm. Biochem. Biophys. Res. Commun. 2006;347:226–231. doi: 10.1016/j.bbrc.2006.06.083. [DOI] [PubMed] [Google Scholar]
- 119.Hasegawa H., Savory N., Abe K., Ikebukuro K. Methods for improving aptamer binding affinity. Molecules. 2016;21:421. doi: 10.3390/molecules21040421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]
- 121.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hiller M., Pudimat R., Busch A., Backofen R. Using RNA secondary structures to guide sequence motif finding towards single-stranded regions. Nucleic Acids Res. 2006;34 doi: 10.1093/nar/gkl544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoinka J., Zotenko E., Friedman A., Sauna Z.E., Przytycka T.M. Identification of sequence-structure RNA binding motifs for SELEX-derived aptamers. Bioinformatics. 2012;28:i215–i223. doi: 10.1093/bioinformatics/bts210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Biesiada M., Purzycka K.J., Szachniuk M., Blazewicz J., Adamiak R.W. Automated RNA 3D structure prediction with RNAComposer. Methods Mol. Biol. 2016;1490:199–215. doi: 10.1007/978-1-4939-6433-8_13. [DOI] [PubMed] [Google Scholar]
- 126.Magnus M., Boniecki M.J., Dawson W., Bujnicki J.M. SimRNAweb: A web server for RNA 3D structure modeling with optional restraints. Nucleic Acids Res. 2016;44:W315–W319. doi: 10.1093/nar/gkw279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hamada M., Ono Y., Kiryu H., Sato K., Kato Y., Fukunaga T., Mori R., Asai K. Rtools: A web server for various secondary structural analyses on single RNA sequences. Nucleic Acids Res. 2016;44:W302–W307. doi: 10.1093/nar/gkw337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bunka D.H., Mantle B.J., Morten I.J., Tennent G.A., Radford S.E., Stockley P.G. Production and characterization of RNA aptamers specific for amyloid fibril epitopes. J. Biol. Chem. 2007;282:34500–34509. doi: 10.1074/jbc.M703679200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Davis J.T. G-quartets 40 years later: From 5'-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. doi: 10.1002/anie.200300589. [DOI] [PubMed] [Google Scholar]
- 130.Gatto B., Palumbo M., Sissi C. Nucleic acid aptamers based on the G-quadruplex structure: Therapeutic and diagnostic potential. Curr. Med. Chem. 2009;16:1248–1265. doi: 10.2174/092986709787846640. [DOI] [PubMed] [Google Scholar]
- 131.Wu Y., Shi Y., Deng S., Wu C., Deng R., He G., Zhou M., Zhong K., Gao H. Metal-induced G-quadruplex polymorphism for ratiometric and label-free detection of lead pollution in tea. Food Chem. 2021;343:128425. doi: 10.1016/j.foodchem.2020.128425. [DOI] [PubMed] [Google Scholar]
- 132.Kwok C.K., Merrick C.J. G-quadruplexes: Prediction, characterization, and biological application. Trends Biotechnol. 2017;35:997–1013. doi: 10.1016/j.tibtech.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 133.Kikin O., D'Antonio L., Bagga P.S. QGRS mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kikin O., Zappala Z., D'Antonio L., Bagga P.S. GRSDB2 and GRS_UTRdb: Databases of quadruplex forming G-rich sequences in pre-mRNAs and mRNAs. Nucleic Acids Res. 2008;36:D141–D148. doi: 10.1093/nar/gkm982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dhapola P., Chowdhury S. QuadBase2: Web server for multiplexed guanine quadruplex mining and visualization. Nucleic Acids Res. 2016;44:W277–W283. doi: 10.1093/nar/gkw425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Andrushchenko V., Tsankov D., Krasteva M., Wieser H., Bour P. Spectroscopic detection of DNA quadruplexes by vibrational circular dichroism. J. Am. Chem. Soc. 2011;133:15055–15064. doi: 10.1021/ja204630k. [DOI] [PubMed] [Google Scholar]
- 138.Duffy K., Arangundy-Franklin S., Holliger P. Modified nucleic acids: Replication, evolution, and next-generation therapeutics. BMC Biol. 2020;18:112. doi: 10.1186/s12915-020-00803-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hirao I., Kimoto M., Yamashige R. Natural versus artificial creation of base pairs in DNA: Origin of nucleobases from the perspectives of unnatural base pair studies. Acc. Chem. Res. 2012;45:2055–2065. doi: 10.1021/ar200257x. [DOI] [PubMed] [Google Scholar]
- 140.Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic αbet. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 141.Kimoto M., Hirao I. Genetic alphabet expansion technology by creating unnatural base pairs. Chem. Soc. Rev. 2020;49:7602–7626. doi: 10.1039/d0cs00457j. [DOI] [PubMed] [Google Scholar]
- 142.Sefah K., Yang Z., Bradley K.M., Hoshika S., Jimenez E., Zhang L., Zhu G., Shanker S., Yu F., Turek D., Tan W., Benner S.A. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. U. S. A. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shen W., De Hoyos C.L., Sun H., Vickers T.A., Liang X.H., Crooke S.T. Acute hepatotoxicity of 2' fluoro-modified 5-10-5 gapmer phosphorothioate oligonucleotides in mice correlates with intracellular protein binding and the loss of DBHS proteins. Nucleic Acids Res. 2018;46:2204–2217. doi: 10.1093/nar/gky060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Janas M.M., Zlatev I., Liu J., Jiang Y., Barros S.A., Sutherland J.E., Davis W.P., Liu J., Brown C.R., Liu X., Schlegel M.K., Blair L., Zhang X., Das B., Tran C., et al. Safety evaluation of 2'-deoxy-2'-fluoro nucleotides in GalNAc-siRNA conjugates. Nucleic Acids Res. 2019;47:3306–3320. doi: 10.1093/nar/gkz140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yu R.Z., Wang Y., Norris D.A., Kim T.W., Narayanan P., Geary R.S., Monia B.P., Henry S.P. Immunogenicity assessment of inotersen, a 2'-O-(2-methoxyethyl) antisense oligonucleotide in animals and humans: Effect on pharmacokinetics, pharmacodynamics, and safety. Nucleic Acid Ther. 2020;30:265–275. doi: 10.1089/nat.2020.0867. [DOI] [PubMed] [Google Scholar]
- 146.Hatami A., Monjazeb S., Glabe C. The anti-amyloid-β monoclonal antibody 4G8 recognizes a generic sequence-independent epitope associated with α-synuclein and islet amyloid polypeptide amyloid fibrils. J. Alzheimers Dis. 2016;50:517–525. doi: 10.3233/JAD-150696. [DOI] [PubMed] [Google Scholar]
- 147.Fowler D.M., Koulov A.V., Balch W.E., Kelly J.W. Functional amyloid--from bacteria to humans. Trends Biochem. Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 148.Maji S.K., Perrin M.H., Sawaya M.R., Jessberger S., Vadodaria K., Rissman R.A., Singru P.S., Nilsson K.P., Simon R., Schubert D., Eisenberg D., Rivier J., Sawchenko P., Vale W., Riek R. Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science. 2009;325:328–332. doi: 10.1126/science.1173155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang Z., Lu L., Zhang Y., Hua Li C., Wang C.X., Zhang X.Y., Tan J.J. A combinatorial scoring function for protein-RNA docking. Proteins. 2017;85:741–752. doi: 10.1002/prot.25253. [DOI] [PubMed] [Google Scholar]
- 150.Song J., Zheng Y., Huang M., Wu L., Wang W., Zhu Z., Song Y., Yang C. A sequential multidimensional analysis algorithm for aptamer identification based on structure analysis and machine learning. Anal. Chem. 2020;92:3307–3314. doi: 10.1021/acs.analchem.9b05203. [DOI] [PubMed] [Google Scholar]
- 151.Reichmuth A.M., Oberli M.A., Jaklenec A., Langer R., Blankschtein D. mRNA vaccine delivery using lipid nanoparticles. Ther. Deliv. 2016;7:319–334. doi: 10.4155/tde-2016-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018;19:213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 153.Hill A.F. Extracellular vesicles and neurodegenerative diseases. J. Neurosci. 2019;39:9269–9273. doi: 10.1523/JNEUROSCI.0147-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Vassileff N., Cheng L., Hill A.F. Extracellular vesicles - propagators of neuropathology and sources of potential biomarkers and therapeutics for neurodegenerative diseases. J. Cell Sci. 2020;133 doi: 10.1242/jcs.243139. [DOI] [PubMed] [Google Scholar]
- 155.Ren X., Zhao Y., Xue F., Zheng Y., Huang H., Wang W., Chang Y., Yang H., Zhang J. Exosomal DNA aptamer targeting α-synuclein aggregates reduced neuropathological deficits in a mouse Parkinson's disease model. Mol. Ther. Nucleic Acids. 2019;17:726–740. doi: 10.1016/j.omtn.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McConnell E.M., Ventura K., Dwyer Z., Hunt V., Koudrina A., Holahan M.R., DeRosa M.C. In vivo use of a multi-DNA aptamer-based payload/targeting system to study dopamine dysregulation in the central nervous system. ACS Chem. Neurosci. 2019;10:371–383. doi: 10.1021/acschemneuro.8b00292. [DOI] [PubMed] [Google Scholar]
- 157.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., Brachtendorf S., Lorks V., Sikorski J., Hilker R., Becker D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 158.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., McGettigan J., Khetan S., Segall N., Solis J., Brosz A., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Raemdonck K., Braeckmans K., Demeester J., De Smedt S.C. Merging the best of both worlds: Hybrid lipid-enveloped matrix nanocomposites in drug delivery. Chem. Soc. Rev. 2014;43:444–472. doi: 10.1039/c3cs60299k. [DOI] [PubMed] [Google Scholar]
- 160.Zhu X., Badawi M., Pomeroy S., Sutaria D.S., Xie Z., Baek A., Jiang J., Elgamal O.A., Mo X., Perle K., Chalmers J., Schmittgen T.D., Phelps M.A. Comprehensive toxicity and immunogenicity studies reveal minimal effects in mice following sustained dosing of extracellular vesicles derived from HEK293T cells. J. Extracell. Vesicles. 2017;6:1324730. doi: 10.1080/20013078.2017.1324730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Nomura Y., Sugiyama S., Sakamoto T., Miyakawa S., Adachi H., Takano K., Murakami S., Inoue T., Mori Y., Nakamura Y., Matsumura H. Conformational plasticity of RNA for target recognition as revealed by the 2.15 angstrom (Å) crystal structure of a human IgG-aptamer complex. Nucleic Acids Res. 2010;38:7822–7829. doi: 10.1093/nar/gkq615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kato K., Ikeda H., Miyakawa S., Futakawa S., Nonaka Y., Fujiwara M., Okudaira S., Kano K., Aoki J., Morita J., Ishitani R., Nishimasu H., Nakamura Y., Nureki O. Structural basis for specific inhibition of Autotaxin by a DNA aptamer. Nat. Struct. Mol. Biol. 2016;23:395–401. doi: 10.1038/nsmb.3200. [DOI] [PubMed] [Google Scholar]