Abstract
The MTG8 (ETO) locus is involved in a reciprocal exchange with runx1 in the t(8;21) of acute myeloid leukemia. It is a member of a small gene family encoding transcriptional regulators that interact with corepressors and histone deacetylase. However, the physiologic cellular processes controlled by MTG8 are not known. In order to gain an insight into the latter, we have generated mutant mice with an insertional inactivation at the locus, which disrupts transcription of exon 2. The postnatal viability of homozygous mutants was greatly reduced. In approximately 25% the midgut was missing, whereas practically all pups surviving past the first 2 days showed severe growth impairment, which was likely due to a gross disruption of the gut architecture. The latter phenotype could be traced back to late embryonic development. No difference in gut cell differentiation or proliferation was found compared to wild-type littermates. Levels of factors known to be involved in gut morphogenesis were also unchanged. MTG8 is expressed in the outermost layers of the developing gut from at least E9.5. Thus, MTG8 plays a novel, essential role in the gastrointestinal system.
The study of tumor-associated chromosomal translocations has led to the identification of genes that play a key role in controlling cell growth and differentiation (26). In the t(8;21) of acute myeloid leukemia (AML), early work showed the breakpoints to fall within the coding sequence of two previously unknown genes, runx1 at 21q22 (previously named AML1/CBFA2) (21) and MTG8 at 8q22 (also named ETO/CDR) (20). The former is related to the runt gene of Drosophila melanogaster and, together with it, defines a novel family of DNA-binding transcription factors (14, 33). Mouse runx1 has been shown to be essential for definitive hemopoiesis (23, 37) and haploinsufficiency at runx1 in humans is associated with diserythropoiesis and an increased risk of AML (32). Another runx member (runx2) is essential for osteogenesis in mammals, as shown both by gene targeting (16, 24) and by its mutations in cases of human cleidocranial dysplasia (22).
Relatively little is known about MTG8/ETO. It belongs to a small, phylogenetically conserved family, consisting of three members in humans and mice (4, 7, 10, 15) and one member in D. melanogaster (9). The latter (nervy) was identified as a target of the homeotic gene Ubx, and its expression in embryogenesis is largely restricted to precursors of the central and peripheral nervous system. However, there is no known phenotype associated with its mutations. Sequence comparison has identified four regions conserved among all MTG8-like polypeptides (4, 15). The COOH-terminal of these (NHR4, for nervy homology region 4) has the potential to fold as a double zinc finger, although it does not bind DNA (F. Calabi, unpublished results). The most NH2-proximal region (NHR1) is related to TAFII, a class of molecules involved in transcription initiation by RNA polymerase II (1). The notion that MTG8 is implicated in gene transcription is strengthened by two additional observations: first, MTG8 products are primarily localized to the nucleus (7, 8, 18) and, second, they interact with corepressors such as N-CoR and mSin3, leading to the recruitment of histone deacetylase to the transcription complex (11, 19, 36). Based on Northern blotting analysis, MTG8 in the adult is expressed mainly in the central nervous system, lungs, heart, and testis (20, 40). However, the cellular functions in which MTG8 is involved are not known.
In order to gain an insight into the latter, we have introduced a targeted mutation at the mouse MTG8 locus. Its phenotype reveals a crucial role in the gastrointestinal system.
MATERIALS AND METHODS
Gene targeting.
A mouse genomic library from strain 129/Sv in phage λ2001 (38) was screened with an MTG8 probe spanning exons 2 to 4 (Table 1). Two overlapping clones (λESMM5 and λESMM9) were chosen for further manipulations. A targeting vector was assembled in pUC18, by subcloning an ∼1.5-kb partial BglII/SstI fragment from λESMM5 spanning most of exon 2, and an ∼7-kb SstI fragment from λESMM9 spanning exon 3. An Escherichia coli lacZ gene (from the KpnI site at nucleotide (nt) 624 to the XbaI site at nt 4158 in plasmid pSV–β-galactosidase [Promega]) was inserted, after blunting, at the internal SstI site, giving an in-frame fusion to exon 2. A 1.1-kb fragment encoding the neor gene (from a modified pMC1neo PolyA vector [34]) was inserted at the BamHI site at the 3′ end of the lacZ gene, and a 2-kb fragment encoding the herpes simplex virus tk gene (38) was inserted at the 3′ end of the mouse sequence (with respect to the MTG8 transcriptional orientation).
TABLE 1.
Probes used in the present study
The SalI-linearized vector was transfected into CCB ES cells by electroporation as previously described (38). G418 and gangciclovir double-resistant clones were screened by Southern blotting using a 5′ flanking probe (a 0.3-kb PstI fragment, mapping ∼1.5 kb upstream of the region used to assemble the vector; Fig. 1A). Clones giving the expected pattern were further analyzed using a 3′ probe (a 0.6-kb RsaI/XbaI fragment mapping ∼5-kb downstream of the region used to assemble the vector; Fig. 1A) and a neo probe (the whole 1.1-kb fragment used for vector construction) to confirm single integration. Two clones were injected into C57BL/6 blastocysts, one of which (MTG8-lacZ222) yielded a high degree of chimerism and germ line transmission. Chimeras were backcrossed to C57BL/6, and further generations were produced by interbreeding.
FIG. 1.
Generation and expression of the MTG8Ex2/lacZ allele. (A) Diagram of the wt locus, the targeting vector and the mutant locus. Thick black line and boxes, murine MTG8; white box, lacZ; dark gray box, neor gene; light gray box, tk gene; thick striped lines, promoters (arrows indicate transcriptional starts) and 3′ UTR [lollipop symbols indicate poly(A) signals]; thin line, pUC18. The 5′ and 3′ bars indicate the positions of the probes flanking the targeted region and used in the Southern analysis. Relevant restriction sites are represented by capital letters (B, BamHI; H, HindIII; S, SstI, Sl, SalI). (B) Southern blotting analysis of wt (+/+) and MTG8Ex2/lacZ-targeted (+/−) CCB ES cells. Restriction enzymes are as in panel A. The probes are described in Materials and Methods. The numbers on the left indicate the molecular size markers in kilobase pairs. (C) S1 protection analysis of brain RNA from wt (+/+) and MTG8 exon 2-null (−/−) pups with a probe spanning MTG8 exons 2 to 4 (Table 1). tRNA, negative control for probe hybridization. The top band corresponds to residual undigested full-length probe. The double filled arrows point to the band resulting from full protection of MTG8 sequences; the single filled arrow points to the band resulting from protection of exons 3 and 4 only. For reference purposes, protection by an actin probe added to the same hybridization mixture is shown at the bottom (single open arrow). The numbers on the left indicate the molecular size markers in nucleotides. (D) Western blotting analysis of brain from wt (+/+) and MTG8 exon 2-null (−/−) mice with an antiserum against the C-terminal domain of MTG8. No bands were visible with normal rabbit serum (data not shown). Asterisks mark species that are absent in the mutant. The numbers on the right indicate the molecular size markers in kilodaltons.
Histological analysis.
Organ samples were removed immediately following sacrifice and fixed either in buffered formalin or in Bouin's solution overnight at room temperature, prior to embedding in paraffin and sectioning at 4 to 6 μm.
Staining with hematoxylin and eosin, or Alcian blue was performed according to standard protocols. For immunohistochemistry, the following monoclonal antibodies were used as primary reagents: anti-PCNA (clone sc-56 [Santa Cruz Biotechnology], 1 μg/ml), anti-human sucrase-isomaltase (clone MGlu2 [12], culture supernatant diluted 1:64), anti-α smooth muscle actin (clone 1A4 [Sigma], ascitic fluid diluted 1:800), and anti-β tubulin III (clone SDL.3D10 [Sigma], ascitic fluid diluted 1:1,600). A biotinylated horse anti-mouse immunoglobulin G (Vector Laboratories, 7.5 μg/ml) was used as a secondary reagent. Biotinylated Ulex europaeus agglutinin I (UEAI) lectin was purchased from Vector Laboratories and used at 7.5 μg/ml. Sections were deparaffinized, rehydrated, treated with 3% H2O2 in methanol for 10 min, heated to 95°C in a microwave oven for 10 min, and blocked in 10% horse serum in phosphate-buffered saline (PBS) for 30 min. Antibodies were diluted in 10% horse serum in PBS and incubated for 30 to 60 min at room temperature. The results were visualized with the Vectastain Elite kit (Vector Laboratories), using diaminobenzidine as the substrate, following the manufacturer's instructions. Sections were lightly counterstained with Gill's hematoxylin.
Embryos were fixed in 4% paraformaldehyde in PBS for 3 to 12 h and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) according to a published protocol (29) prior to embedding and sectioning as described above. Sections were counterstained with eosin.
RNA analysis.
Total RNA was extracted from fresh tissues by the guanidine-acid phenol method (6). For gene expression studies, probes (Table 1) were prepared by PCR amplification from mouse genomic DNA, cloned in M13 phage, and sequenced to confirm their identity.
High-specific activity, single-stranded DNA probes were prepared according to standard procedures (28). Ca. 5 × 104 cpm were mixed with ∼20 μg of total RNA in 20 μl of 50% formamide–0.5 M NaCl–1 mM Na2EDTA–25 mM PIPES (pH 6.8), heated at 50°C for 30 min, and then left to hybridize at 45°C for ∼18 h. S1 digestion was with 25 U for 30 min at 37°C.
Western blotting analysis.
Tissue extracts were prepared by homogenizing fresh organs in 10 volumes of 10% sodium dodecyl sulfate (SDS)–10 mM EDTA–25 mM Tris-Cl (pH 6.8) using an Ultra-Turrax homogenizer (IKA). Approximately 150 μg of total protein was fractionated by SDS-polyacrylamide gel electrophoresis on 7.5% gels and electroblotted onto polyvinylidene difluoride membranes (Hybond-P; Amersham Pharmacia Biotech) in 192 mM glycine–25 mM Tris–20% methanol at 125 V for 2 h. Blots were probed with a rabbit polyclonal antiserum raised against the C-terminal 212 amino acids of MTG8 (PC283; Oncogene Research Products; final concentration, 2.5 μg/ml), followed by horseradish peroxidase-coupled anti-rabbit antiserum (Amersham Pharmacia Biotech; 1:40,000), and developed using the ECL-Plus system (Amersham Pharmacia Biotech).
RESULTS
Gene targeting of mouse MTG8.
The human MTG8 locus consists of at least 13 exons spanning >87 kb (40; F.C., unpublished). We chose to target exon 2, since it represents the common splice acceptor for a number of alternative upstream exons (20; F. Calabi, unpublished data), and it is the exon to which 5′ runx1 sequences are most frequently spliced in transcripts deriving from the t(8;21) (30, 35).
Mouse genomic clones containing MTG8 exons 2 and 3 were isolated from an 129/Sv library. An E. coli lacZ coding sequence was inserted in frame in place of the 3′ end of exon 2 and of the adjoining intron, in order simultaneously to disrupt MTG8 transcription and to enable tracking of exon 2 expression by assaying for β-galactosidase activity. neor and tk cassettes were further inserted in order to allow selection of homologous recombinants according to a standard strategy. The structure of the resulting targeted allele, MTG8Ex2/lacZ, is illustrated in Fig. 1A.
Following electroporation in CCB ES cells, screening of 230 G418 plus gangciclovir double resistant clones by Southern blotting yielded three homologous recombinants. Only one, however, gave chimeras when injected into C57BL/6 blastocysts. Detailed genomic analysis of this clone by Southern blotting with probes flanking either end of the targeting vector, as well as for the inserted neor gene (Fig. 1B), confirmed correct and unique integration of the mutation.
The effect of the introduced mutation on MTG8 expression was investigated both at the RNA and at the protein level in extracts from brain, where the highest levels of MTG8 mRNA are found (20, 40). In nuclease protection experiments with a cDNA probe, a protected fragment corresponding to spliced exons 2 to 4 and representing the major species in the wild type (wt) is completely absent in the mutant (Fig. 1C). However, a fragment corresponding to alternative splicing upstream of exon 3, which is barely visible in the wt, is substantially increased in the mutant. Consistent with this result, a 3′ untranslated region (UTR) probe gives a comparable signal in both the mutant and the wt (data not shown).
On Western blotting analysis with an antiserum directed against the C-terminal domain of MTG8, two species of ca. 75 and 90 kDa are absent in the mutant, whereas a smaller species of ca. 55 kDa is increased, and a much larger polypeptide of >200 kDa is unchanged (Fig. 1D). The size of the two former polypeptides is consistent with that reported in human cell lines (8), whereas the other species must either result from alternative splicing or correspond to cross-reacting MTG8 paralogues. Altogether, the data confirm that the MTG8Ex2/lacZ mutation prevents the synthesis of wt MTG8 products carrying exon 2-encoded sequences.
Reduced viability of MTG8 exon 2-targeted mice.
The viability and fertility of MTG8Ex2/lacZ heterozygous mice were essentially identical to those of wt controls. Upon mating, heterozygous mice gave birth to the three expected genotypes in Mendelian ratios. However, postnatal viability of homozygous mutant pups was greatly reduced (Fig. 2A). Moreover, nearly all of those that survived past the first 2 days showed significantly reduced size and usually died before reaching puberty. Of the few adults, females were fertile, despite the reduced size, while no progeny were ever obtained from the males, despite successful mating, as judged by the formation of a vaginal plug.
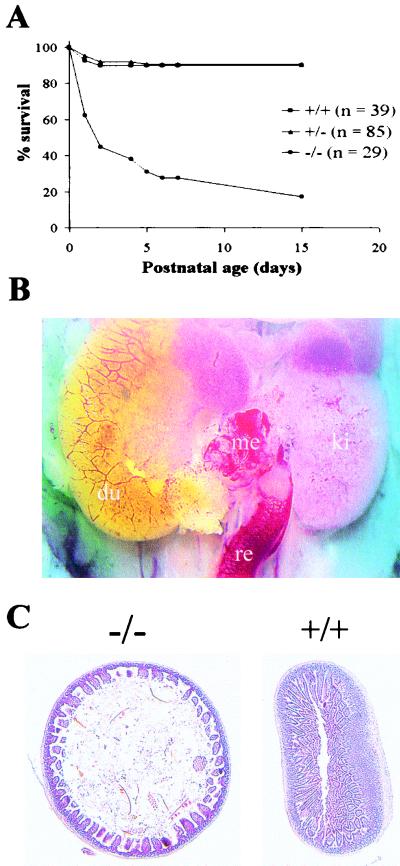
FIG. 2.
(A) Viability of the offspring from crosses between MTG8Ex2/lacZ heterozygous mice. The cumulative percent survival over the first 2 weeks after birth of 153 newborn pups representing all accounted offspring of 17 matings is shown. All five MTG8 exon 2-targeted homozygous mice surviving past day 15 were severely growth retarded. (B) ΔMidgut phenotype. An in situ view following sacrifice of a P0 homozygous mutant pup is shown. The defect extends from the distal portion of the duodenum to the rectum. The duodenal end is covered by an outgrowth of the mucosa, the rectal stump is surrounded by a prominent vascular network and the residual mesentery is hypervascularized. du, duodenum; ki, kidney; me, mesentery; re, rectum. (C) Abnormal gut structure in growth-impaired, MTG8 exon 2-targeted mice. A low-power view of hematoxylin and eosin-stained jejunal sections from mutant (−/−) and control (+/+) P29 mice is shown. Note in the mutant the reduction in gut wall thickness due primarily to disorganization of the villi; note also the dilation of the lumen.
Absence of the midgut in MTG8 exon 2-targeted mice.
While there was no significant difference in size and general appearance among P0/P1 pups born of heterozygous crosses, a fraction showed a distinctive pallor and no milk in their stomach. Upon sacrifice, these pups revealed a striking phenotype, consisting in the absence of most of the intestine, spanning from the distal duodenum to the greater part of the colon (Fig. 2B). The missing segments largely correspond to the districts supplied by the superior mesenteric artery, i.e., the midgut. On this basis, we operationally refer to this phenotype as Δmidgut (i.e., deletion of the midgut). It was never observed past P1, likely causing early postnatal death.
Of the residual intestinal segments, the proximal (duodenal) stump was pervious and showed an outgrowth of the mucosa, with villi projecting into the peritoneal cavity. The distal (rectal) stump was blind ended. On histopathological analysis, neither segment showed any significant anomaly, with the exception of a dilated vascular network, more prominent over the sigma-rectum, where microscopic examination occasionally revealed intraparietal and intraluminal hemorrhages (data not shown). While disjointed, the stumps were loosely held together by a short, fibrous, highly vascularized membrane in place of the mesentery. No gut structure was visible spanning the gap.
Genotyping showed the Δmidgut phenotype to occur nearly exclusively in homozygous mutant pups, at a frequency of ∼25%. It can thus account for most of the increased perinatal mortality of this class. Much rarer cases were observed in heterozygotes (∼1.3%), and none were seen in wt mice.
Growth impairment in MTG8 exon 2-targeted mice.
Of the offspring of heterozygous crosses surviving past the first 48 h, a number showed impaired growth, becoming progressively more marked during the subsequent 2 weeks. Typically, pups were 30 to 50% the size of normal littermates in weight, albeit well proportioned and normally active, except for the most extreme cases. Mortality was high. The few surviving mice gradually recovered with age after puberty, while remaining of below-average weight.
Nearly all affected mice were homozygous mutants. Conversely, all of the latter showed growth impairment. Thus, the phenotype was strongly associated with homozygosity for the MTG8 exon 2-null allele.
Upon sacrifice, growth-impaired mice did not show any obvious anomaly outside the gastrointestinal tract. The intestine, while of reduced length compared to control littermates, was proportionate to the lower body weight. However, the gut histology was grossly abnormal, particularly at the level of the jejunum (Fig. 2C). The intestinal wall was thinner, largely due to a reduction in the length of the villi, which also looked highly disorganized, thicker, and fewer in numbers. Moreover, the lumen was often dilated, probably reflecting a reduced tone of the muscle layers.
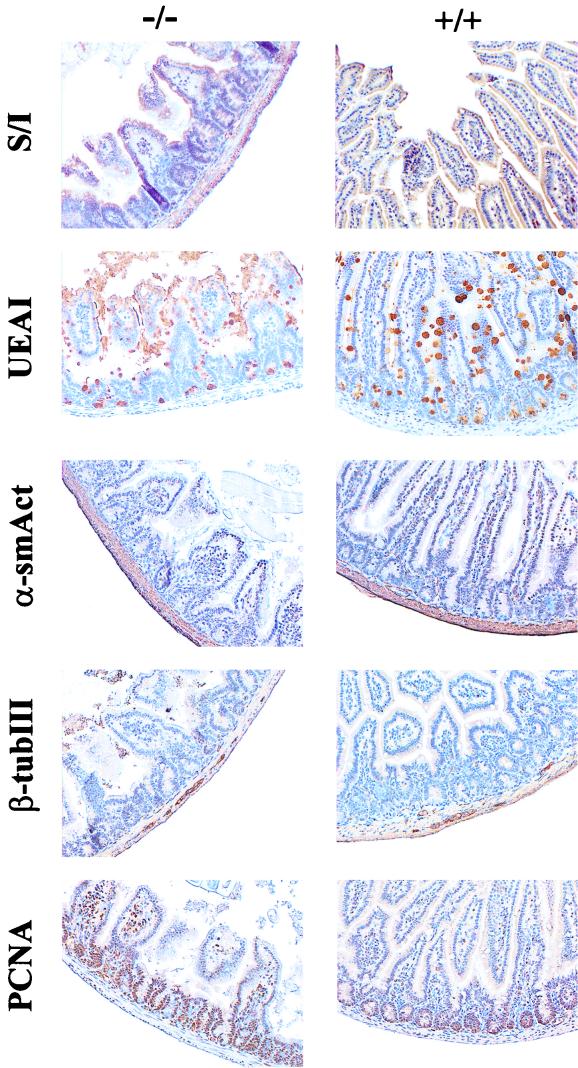
In order to investigate whether the abnormal architecture was associated with changes in cellular differentiation, sections from pathological and normal guts were stained for gut cell markers. Of the four main types of gut epithelial cells, enterocytes can be identified by the expression of sucrase-isomaltase and goblet and Paneth cells by a combination of Alcian blue and lectin UEAI staining. Contractile cells in the tunica muscularis, as well as in perivascular locations, express α-smooth muscle actin, and ENS cells express β-tubulin III. As shown in Fig. 3, all five cell types were present in homozygous mutant guts, in proportions and locations that were not significantly different from those of wt littermates.
FIG. 3.
Cell lineages and proliferation in the jejunum of growth-impaired MTG8 exon 2-targeted mice. Jejunal sections from the mice shown in Fig. 2 were stained for markers of differentiated and proliferating cells. Sucrase-isomaltase (S/I) is a brush border enzyme characteristic of enterocytes; UEAI lectin (UEAI) binds to α-linked fucose residues in polysaccharides secreted by goblet and Paneth cells; α-smooth muscle actin (α-smAct) is present in the muscle layers of the gastrointestinal tract, as well as in vascular smooth muscle cells and myofibroblasts; β-tubulin III (β-tubIII) identifies ENS cells, and PCNA is an antigen associated with actively cycling cells. A positive reaction (brown color) was developed by the immunoperoxidase-diaminobenzidine technique, followed by counterstaining with Gill's hematoxylin. Despite the disruption of the gut architecture, the rates of cell differentiation and cell proliferation in the mutant are not significantly different from those in the control.
The size of the intestinal villi results from the proliferative activity of cells in crypts. Quantitation of PCNA-positive, cycling cells in MTG8 exon 2-targeted and wt littermates (Fig. 3) does not show any significant difference, indicating that the shorter villi in the mutants are not the result of decreased epithelial stem cell activity.
Mouse MTG8 is expressed in the mesoderm of the developing gut.
While MTG8 has not been reported to be expressed in the adult gut, the phenotype of MTG8 exon 2-targeted mutants suggested it plays a key role in the gastrointestinal system. In order to test this hypothesis, MTG8 expression was studied during development, by staining heterozygous MTG8Ex2/lacZ embryos for β-galactosidase. Validation of the method was sought in preliminary experiments on adult tissues, in which the results obtained with the β-galactosidase stain were found to match faithfully those obtained by RNA analysis in wt mice (data not shown).
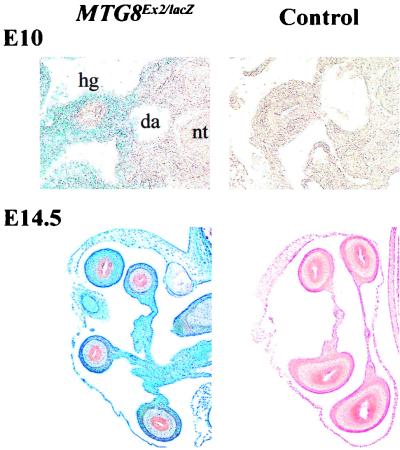
At the 26-somite stage, MTG8 is clearly expressed throughout the primitive gut, albeit at the highest levels in the hindgut (Fig. 4, top panels). Most of the LacZ signal is localized outside the epithelial layer lining the gut lumen, i.e., in the mesodermally derived component.
FIG. 4.
MTG8 expression in the embryonic gut. Transverse sections through MTG8Ex2/lacZ-heterozygous and wt embryos at E10 and E14.5, stained for β-galactosidase activity as a proxy for MTG8 expression, are shown. E14.5 sections were counterstained with eosin. MTG8 is expressed in the mesodermally, rather than in the endodermally, derived tissue and becomes restricted, at the later time point, to the outermost gut layers. fg, foregut; hg, hindgut; da, dorsal aorta.
This pattern becomes even more obvious at later stages. At E14.5 (Fig. 4, bottom panels), there is strong expression in the outermost gut tube layers and the contiguous mesentery, whereas almost no signal is detectable in the epithelium lining the lumen and in the subjacent lamina propria.
Developmental origin of the gut phenotype in MTG8 exon 2-targeted mice.
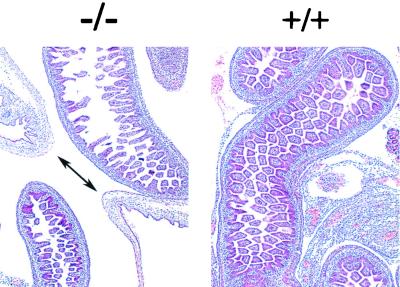
In order to define the developmental origin of the gut phenotype associated with the MTG8 exon 2-null allele, embryos from heterozygous crosses were collected between E9.5 and E17.5, corresponding to the stages at which most of the critical gut morphogenetic events occur. Compared to wt littermates, no significant difference was observed up to E15.5 although, at the latter time point, the size of the umbilical hernia appeared to be somewhat smaller than in controls and the complexity of the midgut loops was reduced (data not shown). However, a disruption of the villi similar to, albeit less extensive than, that seen in postnatal cases was clearly apparent at E17.5 (Fig. 5). This was associated with persistence of the umbilical hernia, normally disappearing entirely by E16.5. Although preliminary, the data indicate that the requirement for wt MTG8 in the gut starts in the late stages of prenatal development.
FIG. 5.
Developmental origin of the gut phenotype in MTG8 exon 2-targeted mice. Hematoxylin and eosin-stained transverse sections through the region of the umbilical hernia of MTG8 exon 2-targeted (−/−) and control (+/+) E17.5 embryos are shown. In the former, the double-headed arrow indicates the communication between the abdominal cavity and the umbilical hernia, which is still prominent, whereas it has normally disappeared at this stage in control littermates.
Expression of gut patterning factors in MTG8 exon 2-targeted mutants.
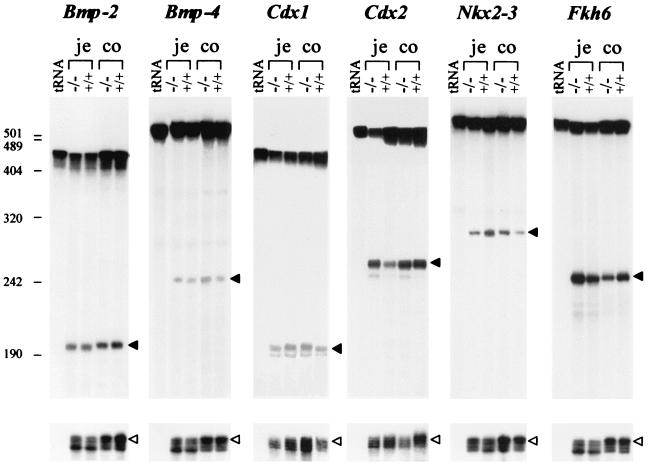
Gut development is known to be controlled by a number of factors which, as in other systems, can be distinguished into two classes: signaling factors, mediating cellular interactions, and transcription factors, directly controlling gene activity. The former includes mesodermally derived Bmp4 and endodermally derived shh/ihh (39). Among the latter class, in addition to Hox gene products (Hoxd13), knockout experiments have revealed a crucial role for the D. melanogaster caudal homologue Cdx2 (encoded in the ParaHox cluster) (5), as well as for Fkh6 (belonging to the forkhead family) (13) and Nkx2-3 (a homeobox gene product) (25). Since MTG8 is expressed in the mesoderm of the primitive gut and since its mutation has dramatic consequences on the gut structure, we sought to determine their relationship to other gut patterning factors by examining its expression in MTG8 exon 2-targeted mutants. RNA was extracted from proximal and distal gut segments of growth-impaired MTG8Ex2/lacZ-homozygous mice and wt littermates, and transcript levels were analyzed by nuclease protection. Representative results are shown in Fig. 6. Despite some occasional minor differences, there was no consistent change in RNA levels of Bmp2/4, Cdx1/2, Nkx2-3, or Fkh6 between the null mutant and the wt in either segment. Thus, the role of MTG8 in the gut is not mediated through one of the already-identified gut patterning factors.
FIG. 6.
Expression of gut morphogenetic factors in growth-impaired MTG8 exon 2-targeted mice. S1 protection analysis of RNA from proximal (je) and distal (co) gut segments from a P7 MTG8 exon 2-targeted homozygous pup (weight, 2.78 g) and a control (wt) littermate (weight, 5.98 g) was carried out. tRNA, negative control for probe hybridization. The probes are described in Table 1. The top band corresponds to residual undigested full-length probe. Filled arrows indicate the expected protected fragment for each probe. For reference purposes, protection by an actin probe added to the same hybridization mixture is shown at the bottom (white arrow). The numbers on the left indicate the molecular size markers in nucleotides. The apparent minor differences with some probes were inconsistent.
DISCUSSION
In order to investigate the function of the MTG8 locus, we generated a mutant allele, MTG8Ex2/lacZ, in which part of exon 2 and of the downstream intron have been replaced by sequences encoding β-galactosidase and neomycin phosphotransferase. The sequence encoded by exon 2, spanning 46 amino acids, is very close to the proposed alternative amino termini. While alternative splicing has been observed both 5′ and 3′, exon 2 has never been found missing either from wt MTG8 transcripts (20, 40; Calabi, unpublished) or from runx1/MTG8 fusion transcripts arising from the t(8;21) of AML (30, 35). This suggests that this exon plays a crucial function, although it does not encode any of the four regions (NHR1 to -4) that are conserved among all MTG8-like polypeptides, and database searches have yet to reveal any significant homology.
Lack of MTG8 polypeptides carrying exon 2-encoded sequences results in high mortality, either perinatally or associated with significant growth impairment during the first 2 weeks of life. Most of the early mortality is due to a massive defect in the gastrointestinal tract. An abnormal gut structure is also found in growth-impaired mice and is a plausible cause of the latter phenotype, given its likely effects on the absorption of nutrients. As in other cases of gene targeting, variable phenotypic penetrance and/or expression may be explained by genetic heterogeneity within the strain resulting from targeting and may indicate the existence of interacting genetic factors. Moreover, a gene dosage effect is suggested by the occurrence of a similar phenotype in heterozygous mice, albeit at a much reduced frequency.
Our results indicate that MTG8 has a crucial function in the gastrointestinal system. While no appreciable expression has been detected in the adult gut, our data show that the embryonic gut is, with the heart (data not shown), one of the main sites of expression at least from E9.5. The highest levels are found in the outer layers, in contrast to other factors so far found to be expressed in the developing gut, which are either endodermal (Cdx1 and -2, shh/ihh, Bmp2, and Tcf4) or primarily restricted to the subendodermal mesoderm (Nkx2-3, Fkh6, Gli1/Ptc, and Bmp4) (2, 13, 17, 25, 27). Intriguingly, the Drosophila homologue of MTG8 (nervy) was isolated as a downstream target of Ubx (9), which is known to play a role in gut patterning in the fruitfly (3). However, no phenotype is associated with nervy mutations, and the role of the latter in the fruitfly remains to be established, as is the potential existence of a Hox-MTG8 pathway in higher organisms.
The Δmidgut phenotype shows some analogies to intestinal atresias in humans, which are generally believed to result from vascular accidents, although a genetic origin has been implicated in some cases (31). The extent of the defect largely coincides with the districts supplied by the superior mesenteric artery. Moreover, while the latter seems to be properly formed, there is vascular congestion over the proximal and particularly the distal gastrointestinal stumps, albeit with no evidence of necrosis. Unlike human cases, however, there is no proximal atresia, while a peculiar mucosal outgrowth extends from the duodenal end. There are no remnants of the missing gut segments, and the mesentery, albeit greatly shortened, shows no gaps. The contribution of MTG8 mutations to gut defects in humans remains to be investigated.
The milder phenotype associated with the MTG8 exon 2 knockout has some superficial analogies with those recently described in other mice with targeted disruption of genes involved in gut development. Both Fkh6- and Nkx2-3-null embryos show delayed formation and slower growth of villi (13, 25). In both cases the changes are apparent from the time of the initial transition from pseudostratified to columnar gut epithelium, coincide with alterations in the proliferative compartment, and correlate with a reduction in the levels of Bmp2 and -4 mRNA, suggesting that they are mediated via a common signaling pathway. In Tcf4- and ihh-null mice (17, 27), which die at or shortly after birth, there is a substantial decrease in the size of the villi associated with a reduction or, respectively, a nearly complete absence of proliferating stem cells. Similarly to these other null mutants, the milder gut phenotype of MTG8 exon 2-targeted mice shows disorganization of the villi, which coexists with largely normal differentiation of gut cell lineages and is most pronounced in the proximal intestine (i.e., the jejunum). However, early midgut morphogenetic events (i.e., the formation of epithelial ridges) are not affected (data not shown), and cell proliferation is not reduced. Further proof that the MTG8 function in gut development and/or differentiation is independent of previously identified pathways is provided by the analysis of patterns of gene expression in the mutants: Bmp2/4, Cdx1/2, Nkx2-3, and Fkh6 mRNA levels are essentially unchanged in the MTG8 exon 2 knockout.
We hypothesize that the two distinct phenotypes of MTG8 exon 2 mutant mice represent different degrees of severity of the same condition, resulting from the lack of a single MTG8-controlled function. Such function is unlikely to be required for primary gut morphogenesis, since the gut was fully formed in a majority of mutants, and no gut anomaly was found in homozygous mutant embryos up to E15.5. Primary canalization of the gut tube in the Δmidgut phenotype is also indicated by the finding of meconium in the rectal stump (data not shown) and by the absence of concomitant abdominal wall defects indicative of a failure in the process of embryonic folding or ventral midline fusion.
We suggest that MTG8 is required for the maintenance of a normal gut structure from late embryonic development, since pathologic changes can be clearly detected in the mutants by E17.5. This function may be related to the blood supply of the midgut, leading in the most extreme cases to a complete regression (Δmidgut) and in less severe cases to dysplasia (causing malabsorption). Rescue of the latter phenotype may occur due to the postnatal triggering of compensatory mechanisms, similar to what has been reported in other knockouts. This hypothesis would be consistent with the localization of MTG8 expression to the outermost layers of the gut, containing the main submucosal vascular plexuses.
In addition to the gastrointestinal defects, sterility was consistently observed in the few male null mutants surviving into adulthood. While the basis of this phenotype remains to be clarified, X-Gal staining in MTG8Ex2/lacZ heterozygotes shows MTG8 to be mostly expressed by Leydig cells in the adult testis. This suggests that male sterility in homozygotes, despite apparently normal testis size and morphology, is due to hormonal insufficiency. Hind limb paresis and/or paralysis was rarely observed in adult mutant mice. By X-Gal staining in heterozygotes, we have been unable to detect MTG8 expression in the spinal cord, peripheral nerves, or skeletal muscles, and the cause of this phenotype remains to be investigated. In contrast, insertional inactivation of MTG8 exon 2 is phenotypically silent in the brain, lung, or heart, all major sites of expression. Histological examination has also so far failed to reveal any abnormality (data not shown). Thus, the function of MTG8 in these organs is likely to be at least potentially redundant, and its absence may be compensated for by an increase in alternative isoforms and/or by MTG8 paralogues.
Finally, our data do not support a role for MTG8 in haemopoiesis. Upon X-Gal staining, no significant expression of the MTG8Ex2/lacZ allele was found either in embryos or in the main hemopoietic lineages of adult mice (data not shown). The bone marrow Ly-6A/E+ subpopulation, containing hemopoietic stem cells, also scored negative. Moreover, no hemopoietic defect was observed in homozygous mutant mice. These data contrast with the report of MTG8 expression in human CD34+ cells (8). Apart from possible species-specific differences, the latter results may have rather been due to cross-reacting products encoded by MTG8 paralogues, which are known to be expressed in hemopoietic cells (4, 7, 10). We conclude that the role (if any) of MTG8 in leukemia may be at least partly related to its abnormal expression in hemopoietic precursors.
ACKNOWLEDGMENTS
We are particularly grateful to Terence Rabbitts for constant encouragement and strategic advice. We also thank Vania Cilli for help in the isolation of mouse MTG8 genomic clones, Dallas Swallow for the gift of the anti-human sucrase-isomaltase monoclonal antibody, Andy Copp and Patrizia Ferretti for comments, and the staff of the Royal Veterinary College, London, England, for expert mouse husbandry.
This work was supported by MRC PG9311737.
REFERENCES
- 1.Albright S R, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 2.Beck F, Tata F, Chawengsaksophak K. Homeobox genes and gut development. Bioessays. 2000;22:431–441. doi: 10.1002/(SICI)1521-1878(200005)22:5<431::AID-BIES5>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 3.Bienz M. Homeotic genes and positional signalling in the Drosophila viscera. Trends Genet. 1994;10:22–26. doi: 10.1016/0168-9525(94)90015-9. [DOI] [PubMed] [Google Scholar]
- 4.Calabi F, Cilli V. CBFA2T1, a gene rearranged in human leukemia, is a member of a multigene family. Genomics. 1998;52:332–341. doi: 10.1006/geno.1998.5429. [DOI] [PubMed] [Google Scholar]
- 5.Chawengsaksophak K, James R, Hammond V E, Kontgen F, Beck F. Homeosis and intestinal tumours in Cdx2 mutant mice. Nature. 1997;386:84–87. doi: 10.1038/386084a0. [DOI] [PubMed] [Google Scholar]
- 6.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 7.Davis J N, Williams B J, Herron J T, Galiano F J, Meyers S. ETO-2, a new member of the ETO-family of nuclear proteins. Oncogene. 1999;18:1375–1383. doi: 10.1038/sj.onc.1202412. [DOI] [PubMed] [Google Scholar]
- 8.Erickson P F, Dessev G, Lasher R S, Philips G, Robinson M, Drabkin H A. ETO and AML1 phosphoproteins are expressed in CD34+ hematopoietic progenitors—implications for t(8;21) leukemogenesis and monitoring residual disease. Blood. 1996;88:1813–1823. [PubMed] [Google Scholar]
- 9.Feinstein P G, Kornfeld K, Hogness D S, Mann R S. Identification of homeotic target genes in Drosophila melanogaster including nervy, a protooncogene homolog. Genetics. 1995;140:573–586. doi: 10.1093/genetics/140.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fracchiolla N S, Colombo G, Finelli P, Maiolo A T, Neri A. EHT, a new member of the MTG8/ETO gene family, maps on 20q11 region and is deleted in acute myeloid leukemias. Blood. 1998;92:3481–3484. [PubMed] [Google Scholar]
- 11.Gelmetti V, Zhang J S, Fanelli M, Minucci S, Pelicci P G, Lazar M A. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol Cell Biol. 1998;18:7185–7191. doi: 10.1128/mcb.18.12.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green F R, Greenwell P, Dickson L, Griffiths B, Noades J, Swallow D M. Expression of the ABH, Lewis and related antigens on the glycoproteins of the jejunal brush border. In: Harris J R, editor. Subcellular biochemistry. Vol. 12. New York, N.Y: Plenum Publishing; 1988. pp. 119–153. [DOI] [PubMed] [Google Scholar]
- 13.Kaestner K H, Silberg D G, Traber P G, Schutz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- 14.Kagoshima H, Shigesada K, Satake M, Ito Y, Miyoshi H, Ohki M, Pepling M, Gergen P. The runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993;9:338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- 15.Kitabayashi I, Ida K, Morohoshi F, Yokoyama A, Mitsuhashi N, Shimizu K, Nomura N, Hayashi Y, Ohki M. The AML1-MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol Cell Biol. 1998;18:846–858. doi: 10.1128/mcb.18.2.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of CBFA1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 17.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters P J, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- 18.Le X F, Claxton D, Kornblau S, Fan Y H, Mu Z M, Chang K S. Characterization of the ETO and AML1-ETO proteins involved in the 8;21 translocation in acute myelogenous leukemia. Eur J Haematol. 1998;60:217–225. doi: 10.1111/j.1600-0609.1998.tb01027.x. [DOI] [PubMed] [Google Scholar]
- 19.Lutterbach B, Westendorf J J, Linggi B, Patten A, Moniwa M, Davie J R, Huynh K D, Bardwell V J, Lavinsky R M, Rosenfeld M G, Glass C, Seto E, Hiebert S W. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol Cell Biol. 1998;18:7176–7184. doi: 10.1128/mcb.18.12.7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi H, Kozu T, Shimizu K, Enomoto K, Maseki N, Kaneko Y, Kamada N, Ohki M. The t(8;21) translocation in acute myeloid-leukemia results in production of an AML1-MTG8 fusion transcript. EMBO J. 1993;12:2715–2721. doi: 10.1002/j.1460-2075.1993.tb05933.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miyoshi H, Shimizu K, Kozu T, Maseki N, Kaneko Y, Ohki M. t(8;21) breakpoints on chromosome-21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci USA. 1991;88:10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mundlos S, Otto F, Mundlos C, Mulliken J B, Aylsworth A S, Albright S, Lindhout D, Cole W G, Henn W, Knoll J, Owen M J, Mertelsmann R, Zabel B U, Olsen B R. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 1997;89:773–779. doi: 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- 23.Okuda T, Vandeursen J, Hiebert S W, Grosveld G, Downing J R. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- 24.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G, Beddington R, Mundlos S, Olsen B R, Selby P B, Owen M J. CBFA1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 25.Pabst O, Zweigerdt R, Arnold H H. Targeted disruption of the homeobox transcription factor Nkx2–3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- 26.Rabbitts T H. Perspective: chromosomal translocations can affect genes controlling gene expression and differentiation—why are these functions targeted? J Pathol. 1999;187:39–42. doi: 10.1002/(SICI)1096-9896(199901)187:1<39::AID-PATH235>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Ramalho-Santos M, Melton D A, McMahon A P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. New York, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Sanes J R, Rubenstein J L, Nicolas J F. Use of a recombinant retrovirus to study post-implantation cell lineage in mouse embryos. EMBO J. 1986;5:3133–3142. doi: 10.1002/j.1460-2075.1986.tb04620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saunders M J, Tobal K, Keeney S, Yin J. Expression of diverse AML1/MTG8 transcripts is a consistent feature in acute myeloid-leukemia with t(8;21) irrespective of disease phase. Leukemia. 1996;10:1139–1142. [PubMed] [Google Scholar]
- 31.Skandalakis J E, Gray S W, Ricketts R, Richardson D D. The small intestine. In: Skandalakis J E, Gray S W, editors. Embryology for surgeons. 2nd ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. pp. 200–212. [Google Scholar]
- 32.Song W J, Sullivan M G, Legare R D, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende I C, Haworth C, Hock R, Loh M, Felix C, Roy D C, Busque L, Kurnit D, Willman C, Gewirtz A M, Speck N A, Bushweller J H, Li F P, Gardiner K, Poncz M, Maris J M, Gilliland D G. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- 33.Speck N A, Stacy T, Wang Q, North T, Gu T L, Miller J, Binder M, MarinPadilla M. Core-binding factor: a central player in hematopoiesis and leukemia. Cancer Res. 1999;59:S1789–S1793. [PubMed] [Google Scholar]
- 34.Thomas K R, Capecchi M R. Site-directed mutagenesis by gene targeting in mouse embryo-derived stem cells. Cell. 1987;51:503–512. doi: 10.1016/0092-8674(87)90646-5. [DOI] [PubMed] [Google Scholar]
- 35.Tighe J E, Calabi F. Alternative, out-of-frame runt/MTG8 transcripts are encoded by the derivative(8) chromosome in the t(8;21) of acute myeloid-leukemia M2. Blood. 1994;84:2115–2121. [PubMed] [Google Scholar]
- 36.Wang J X, Hoshino T, Redner R L, Kajigaya S, Liu J M. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc Natl Acad Sci USA. 1998;95:10860–10865. doi: 10.1073/pnas.95.18.10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Q, Stacy T, Binder M, Marinpadilla M, Sharpe A H, Speck N A. Disruption of the CBFA2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc Natl Acad Sci USA. 1996;93:3444–3449. doi: 10.1073/pnas.93.8.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warren A J, Colledge W H, Carlton M B, Evans M J, Smith A J, Rabbitts T H. The oncogenic cysteine-rich LIM domain protein rbtn2 is essential for erythroid development. Cell. 1994;78:45–57. doi: 10.1016/0092-8674(94)90571-1. [DOI] [PubMed] [Google Scholar]
- 39.Wells J M, Melton D A. Vertebrate endoderm development. Annu Rev Cell Dev Biol. 1999;15:393–410. doi: 10.1146/annurev.cellbio.15.1.393. [DOI] [PubMed] [Google Scholar]
- 40.Wolford J K, Prochazka M. Structure and expression of the human MTG8/ETO gene. Gene. 1998;212:103–109. doi: 10.1016/s0378-1119(98)00141-3. [DOI] [PubMed] [Google Scholar]