Abstract
Cyclic GMP‐AMP synthase (cGAS) functions as a key sensor for microbial invasion and cellular damage by detecting emerging cytosolic DNA. Here, we report that GTPase‐activating protein‐(SH3 domain)–binding protein 1 (G3BP1) primes cGAS for its prompt activation by engaging cGAS in a primary liquid‐phase condensation state. Using high‐resolution microscopy, we show that in resting cells, cGAS exhibits particle‐like morphological characteristics, which are markedly weakened when G3BP1 is deleted. Upon DNA challenge, the pre‐condensed cGAS undergoes liquid–liquid phase separation (LLPS) more efficiently. Importantly, G3BP1 deficiency or its inhibition dramatically diminishes DNA‐induced LLPS and the subsequent activation of cGAS. Interestingly, RNA, previously reported to form condensates with cGAS, does not activate cGAS. Accordingly, we find that DNA – but not RNA – treatment leads to the dissociation of G3BP1 from cGAS. Taken together, our study shows that the primary condensation state of cGAS is critical for its rapid response to DNA.
Keywords: cGAS, G3BP1, LLPS
Subject Categories: Immunology; Microbiology, Virology & Host Pathogen Interaction; Signal Transduction
Host cell encoded cGAS is a critical DNA sensor to detect invading pathogens. The stress‐granule protein G3BP1 engages cGAS in a primary condensation state to enable a rapid response to free DNA.
Introduction
A common event of pathogen infection is the introduction of foreign nucleic acids, such as DNA, into the cytoplasm of the host cell (Barbalat et al, 2011; Gurtler & Bowie, 2013). Cytosolic DNA detecting is one of the fundamental mechanisms for the host to sense invading pathogens (Stetson & Medzhitov, 2006; Takeuchi & Akira, 2010; Gurtler & Bowie, 2013; Wu & Chen, 2014). Among the identified intracellular DNA sensors, cGAS plays a pivotal role in innate immunity (Li et al, 2013b). When it binds to DNA, cGAS is activated and, in turn, catalyzes the synthesis of the second messenger molecule cyclic GMP‐AMP (cGAMP) (Ablasser et al, 2013; Gao et al, 2013a; Li et al, 2013b; Sun et al, 2013). As a ligand of the endoplasmic reticulum (ER)–associated adaptor protein STING (also known as ERIS; MITA; MPYS) (Ishikawa & Barber, 2008; Zhong et al, 2008; Sun et al, 2009), cGAMP binds to STING and elicits a series of downstream events that lead to the extensive production of type I interferons (IFNs) and other cytokines (Bowie, 2012; Gao et al, 2013a; Gao et al, 2013b). cGAS thus executes a crucial function in transducing signals by sensing invading pathogens to trigger the activation of anti‐infection immune responses. Besides microbial infections, reverse transcription of endogenous retroviruses or cellular damage can also result in the presence of free DNA in the cytoplasm (O'Neill, 2013; Ahn & Barber, 2014; Kassiotis & Stoye, 2016). The chronical activation of cGAS by such self‐DNA is a major cause for several autoimmune diseases, such as lupus and Aicardi–Goutières syndrome (AGS; Aicardi & Goutieres, 1984; Ahn & Barber, 2014; Lisnevskaia et al, 2014; Crow & Manel, 2015). Therefore, it is critical to understand how cGAS activation is precisely regulated.
It was recently reported that upon activation, DNA robustly induces the formation of liquid droplets containing cGAS (Du & Chen, 2018; Xie et al, 2019). These cGAS‐DNA condensates are formed via LLPS. LLPS is a physicochemical process that allows macromolecules, such as proteins and nucleic acids, to condensate into a dense phase (Jiang et al, 2015; Boeynaems et al, 2018; Fox et al, 2018; Martin et al, 2019). Accumulating evidence demonstrates that LLPS is a key mechanism underlying the formation of membrane‐less organelles (Feric et al, 2016; Lee et al, 2016; Fei et al, 2017; Mitrea et al, 2018; Fujioka et al, 2020; Yasuda et al, 2020). These temporarily assembled cellular compartments confer an important capacity for the cells to dynamically and efficiently organize functional units or reactions in response to different stresses (Kroschwald et al, 2015; Riback et al, 2017; Franzmann et al, 2018; Franzmann & Alberti, 2019). In the case of DNA detection by cGAS, the liquid droplets provide microreactors, in which cGAS, the enzyme, and the reactants, DNA, ATP and GTP, are highly concentrated (Du & Chen, 2018). This process allows the efficient activation of cGAS.
Another well‐studied membrane‐less organelle is the stress granule (SG) (Molliex et al, 2015; Protter & Parker, 2016; Boeynaems et al, 2017; Youn et al, 2019). SGs are cytoplasmic puncta, mainly containing RNAs and proteins, assembled through LLPS unpon different types of stresses, such as oxidative stress and RNA virus infection (Buchan & Parker, 2009; Onomoto et al, 2014; McCormick & Khaperskyy, 2017). Among the known proteins in SG, G3BP1 is believed to be a critical organizer of SG assembly and regulates properties and composition of SGs (Tourriere et al, 2003; Yang et al, 2020). As a core protein for SG regulation, G3BP1 contains a special amino acid sequence feature called the intrinsically disordered region (IDR) (Guillen‐Boixet et al, 2020; Sanders et al, 2020; Yang et al, 2020). For instance, the arginine–glycine–glycine (RGG) motif of G3BP1 is a IDR (Chong et al, 2018). The IDR in G3BP1 is critical for mediating weak multivalent protein–protein or protein–nucleic acid interactions and facilitating the LLPS process (Guillen‐Boixet et al, 2020; Sanders et al, 2020; Yang et al, 2020). We recently found that G3BP1 is critical for DNA binding and activation of cGAS (Liu et al, 2019). In the current study, we further show that G3BP1 primes cGAS activation by forming primary condensates with cGAS in the resting state, while DNA engagement leads to the dissociation of G3BP1 from cGAS. Multiple regions of G3BP1, including the nuclear transporter factor 2 (NTF2)–like domain, the RNA recognition module (RRM) and the RGG domain, were required for cGAS‐G3BP1 binding and the formation of cGAS‐G3BP1 condensates. In the presence of G3BP1, DNA was much more efficient at inducing LLPS and activation of cGAS. In addition, zinc ions that are known to enhance cGAS activity (Du & Chen, 2018), also induced the formation of cGAS condensates both in vitro and in vivo. Thus, our study shows that the G3BP1‐mediated formation of the primary condensation state of cGAS is a critical step for an expeditious response to cytosolic DNA.
Results
G3BP1 engages cGAS in a condensed state
We previously reported that G3BP1 promotes the DNA binding and activation of cGAS (Liu et al, 2019). Interestingly, when recombinant cGAS was incubated with G3BP1, but not when it was incubated with bovine serum albumin (BSA), the control protein, the solution became turbid (Fig 1A). We then observed the solution under a microscope and found that many droplets appeared when cGAS and G3BP1 were mixed, and the formation of these droplets was further increased in the presence of DNA (Fig 1A). cGAS activity can be examined using an in vitro cGAMP synthesis assay, in which recombinant cGAS protein is incubated with DNA, ATP, and GTP (Li et al, 2013a; Dai et al, 2019). Using this assay, we showed that with G3BP1, cGAS produced much higher amounts of cGAMP (Fig 1B). To further analyze the cGAS‐G3BP1 droplets, we incubated cGAS with G3BP1 at different concentrations and found that G3BP1 promoted cGAS condensation in a dosage‐dependent manner (Fig 1C). Because cGAS is known to undergo LLPS upon DNA engagement, we further purified recombinant cGAS and G3BP1 proteins with heparin affinity chromatography followed by Ni‐agarose purification to remove potential nucleic acid contaminations. With the purified proteins, we obtained similar results showing that G3BP1 alone can form condensates with cGAS (Fig EV1A).
Figure 1. G3BP1 engages cGAS in a condensed state.
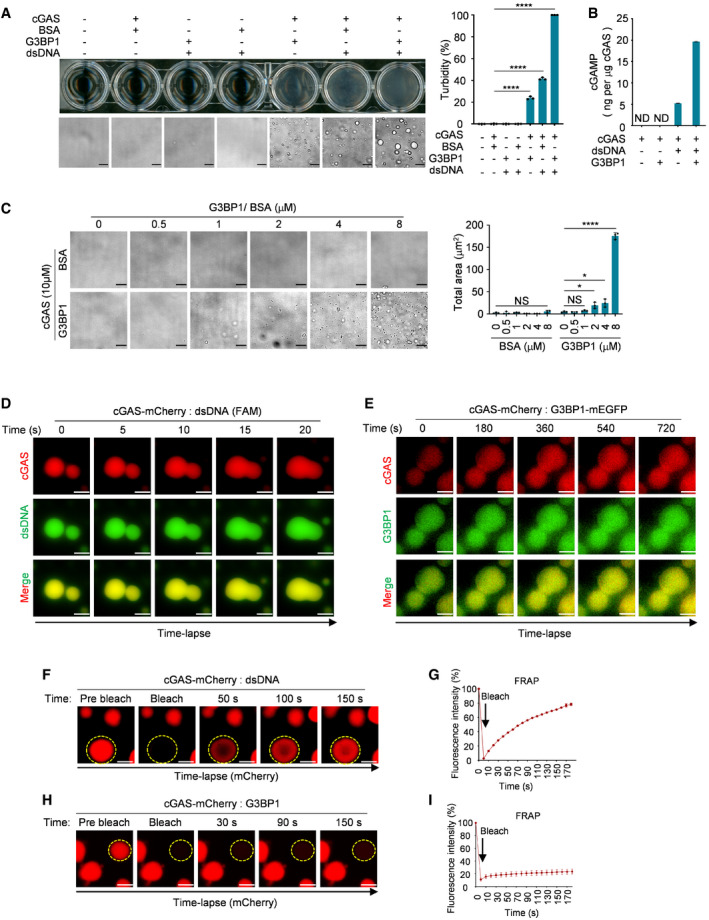
- Turbidity and bright‐field microscope images of indicated groups (left) and the quantitative analysis of turbidity of samples were measured by absorbance at 600 nm (right). 10 μM cGAS, 5 μM G3BP1, 5 μM BSA and 100 nM dsDNA (60 bp) were used in these assays. n = 3 biological replicates.
- cGAMP production assays. Recombinant cGAS (10 μM) and G3BP1 (10 μM) were incubated with 100 nM dsDNA. The produced cGAMP was quantitatively analyzed by liquid chromatography–mass spectrometry/multiple reaction monitoring (LC‐MS/MRM). ND, not detected. n = 3 technical replicates.
- Bright‐field microscope images of cGAS (10 μM) with indicated concentrations of BSA or G3BP1 (left) and a quantitative analysis of the total area of condensates (right). n = 3 biological replicates.
- Time‐lapse imaging of liquid droplets fusion after mixing 10 μM cGAS‐mCherry with 1 μM FAM‐labeled dsDNA.
- Time‐lapse imaging of liquid droplets fusion after mixing 40 μM cGAS‐mCherry with 10 μM G3BP1‐mEGFP.
- FRAP analysis of DNA‐induced droplets of cGAS (1 μM dsDNA, 10 μM cGAS‐mCherry), the yellow dotted circle indicated the region of photobleaching.
- FRAP curve of (F). n = 3 biological replicates.
- FRAP analysis of cGAS‐G3BP1 droplets. 10 μM cGAS‐mCherry and 5 μM G3BP1 were used. The yellow dotted circle indicated the region of photobleaching.
- FRAP curve of (H). n = 3 biological replicates.
Data information: Representative images are shown (A, C–F and H). Error bars, mean with s.d. (A–C, G and I), *P < 0.05, ****P < 0.0001, two‐tailed t‐test. NS, non‐significant; FRAP, fluorescence recovery after photobleaching. Scale bars, 10 μm (A and C), 5 μm (D), 3 μm (E), 2 μm (F and H). See also Fig EV1.
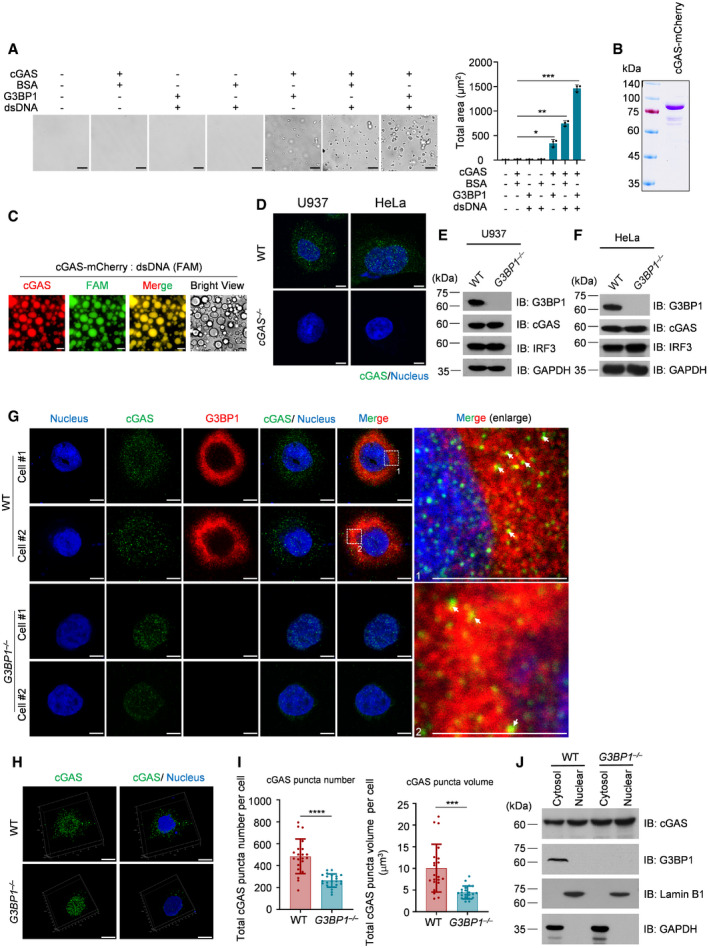
Figure EV1. G3BP1 engages cGAS in a condensed state.
-
ABright‐field microscope images of indicated groups (left) and a quantitative analysis of total area of the condensates. n = 3 biological replicates. 10 μM cGAS, 5 μM G3BP1, 5 μM BSA and 500 nM dsDNA were used in these assays.
-
BCoomassie blue staining of purified recombinant cGAS‐mCherry protein.
-
CFluorescent images of liquid droplets after mixing 10 μM cGAS‐mCherry with 1 μM FAM‐labeled dsDNA.
-
DImmunofluorescent staining of cGAS in WT and cGAS −/− cells. Images were acquired with Leica TCS SP8 Confocal Microscopy.
-
E, FImmunoblotting of WT and G3BP1 −/− cells with indicated antibodies.
-
GImmunofluorescent staining of cGAS and G3BP1 in both WT and G3BP1 −/− HeLa cells. Images were acquired with Leica TCS SP8 Confocal microscopy. The white arrows indicate the cGAS‐G3BP1 condensates.
-
HImmunofluorescent staining of cGAS and G3BP1 in both WT and G3BP1 −/− HeLa cells. 3D images were reconstituted by Leica LAS X software.
-
IQuantitative analysis of total cGAS puncta number (left) and volume (right) per cell of (H). n = 21 cells.
-
JImmunoblotting of fractionated lysates from WT and G3BP1 −/− HeLa cells. GAPDH and Lamin B1 blots were used as controls for cytoplasmic and nuclear fractions, respectively.
Data information: Representative images are shown (A, C–H, and J). Error bars, mean with s.d. (A and I). Scale bars, 10 μm (A), 14 μm (C), 5 μm (D, G and H). WT, wild type; IB, immunoblotting; NS, non‐significant. Hoechst (blue), nuclear staining. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two‐tailed t‐test.
Source data are available online for this figure.
We then examined if the G3BP1‐mediated formation of cGAS droplets was a similar process to DNA‐triggered cGAS LLPS. We first purified mCherry‐tagged recombinant cGAS (Fig EV1B) and induced cGAS LLPS with a 60 base‐pair (bp) FAM (6‐carboxy‐fluorescein)–labeled double‐stranded DNA (dsDNA). As shown in Fig EV1C, mCherry‐tagged cGAS robustly formed droplets with dsDNA. By recording the dynamic formation process of these droplets, we found that the smaller cGAS‐DNA droplets can fuse into bigger ones (Fig 1D, Movie EV1). In contrast, the fusion of cGAS‐G3BP1 droplets was barely detected (Fig 1E, Movie EV2). Next, using fluorescence recovery after photobleaching (FRAP), we showed that the fluorescence intensity of cGAS‐DNA droplets recovered soon after bleaching (Fig 1F and G, Movie EV3). These data indicate that the cGAS‐DNA droplets are dynamically exchanging molecules with the environment, which is a hallmark of LLPS. The cGAS‐G3BP1 droplets, however, exhibited much lower fluorescence recovery rate (Fig 1H and I, Movie EV4). Thus, the G3BP1‐mediated formation of cGAS droplets was not as dynamic as for the droplets induced by DNA. These observations suggested that cGAS may undergo gel‐like transitions without the engagement of DNA. We termed this G3BP1‐engaged gel‐like transition of the cGAS ‘primary condensation state’.
G3BP1 engages cGAS in a condensed state in vivo
To confirm the above findings in cells, we analyzed cGAS in both wild‐type (WT) and G3BP1‐deficient U937 cells, a monocytic cell line that is widely used to study cGAS activation (Watson et al, 2015). To do so, we first confirmed the specificity of our anti‐cGAS antibodies in immunofluorescence experiments using cGAS‐deleted U937 and HeLa cells (Fig EV1D). We then generated G3BP1‐null U937 and HeLa cells (Fig EV1E and F). Using high‐resolution microscopy, we found that cGAS showed particle‐like morphological characteristics in U937 cells (Fig 2A). Consistent with our previous findings (Liu et al, 2019), cGAS and G3BP1 were colocalized in these cells (Fig 2A). Importantly, the particle‐like characteristics of cGAS was markedly weakened when G3BP1 was deleted (Fig 2A–C). This phenomenon was further confirmed in G3BP1‐deficient HeLa cells (Fig EV1G–I). The above data showed that in G3BP1‐deficient cells, cGAS appeared to predominantly localize to the nucleus. To examine the cGAS subcellular distribution upon G3BP1 deletion, we detected cGAS expression in the cytosolic and nuclear fractions from both WT and G3BP1 −/− cells and found that G3BP1 deficiency did not obviously affect the subcellular distribution of cGAS (Figs 2D and Fig EV1J). Thus, the ablation of G3BP1 resulted in the disorganization of cGAS primary condensation in cytoplasm. The G3BP1‐engaged primary condensation state of cGAS seemed to be important, because the deficiency of G3BP1 resulted in dampened cGAS activation (Fig 2E–G).
Figure 2. G3BP1 engages cGAS in a condensed state in vivo .
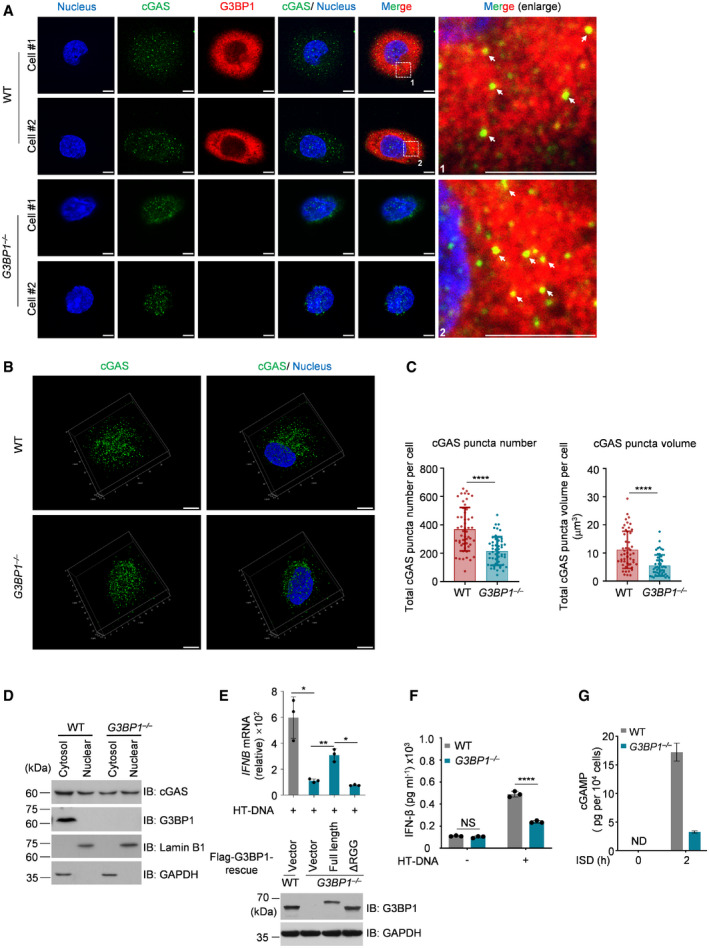
- Immunofluorescent staining of cGAS in both wild‐type (WT) and G3BP1 −/− U937 cells. Leica TCS SP8 Confocal Microscopy was used to acquire images. The white arrows indicate the cGAS‐G3BP1 condensates.
- Immunofluorescent staining of cGAS in both WT and G3BP1 −/− U937 cells. 3D images were reconstituted with Leica LAS X software.
- Quantitative analysis of total cGAS puncta number (left) and volume (right) per cell of (B). n = 51 cells.
- Immunoblotting of fractionated lysates from WT and G3BP1 −/− U937 cells. GAPDH and Lamin B1 blots were used as controls for cytoplasmic and nuclear fractions, respectively.
- qPCR analysis of IFNB mRNA levels in HT‐DNA–treated WT and the indicated rescued cells (U937) (top). Immunoblotting analysis of indicated proteins (bottom). n = 3 biological replicates.
- ELISA analysis of the secreted IFN‐β in HT‐DNA‐treated WT or G3BP1 −/− U937 cells. n = 3 biological replicates.
- cGAMP production in ISD‐treated WT or G3BP1 −/− U937 cells were analyzed by LC‐MS/MRM. ND, not detected. n = 3 technical replicates.
Data information: Representative images are shown (A and B). Scale bars, 5 μm. Hoechst (blue), nuclear staining. Error bars, mean with s.d (C, E–G), *P < 0.05, **P < 0.01, ****P < 0.0001, two‐tailed t‐test. NS, non‐significant; WT, wild type; IB, immunoblotting; HT‐DNA, Herring testes DNA; ISD, interferon stimulatory DNA. See also Figs EV1 and EV2.
Source data are available online for this figure.
It has been reported that cancer cells exhibit basal levels of cytoplasmic DNA (Shen et al, 2015). To rule out the possibility that the cGAS condensation we observed in U937 and HeLa cells may be attributed to existing cytoplasmic DNA in these cells, we measured the cytoplasmic dsDNA levels in HeLa cells and U937 cells using anti‐dsDNA antibodies. We also included a human fibroblast cell line, Hs27, in this experiment. Using immunofluorescence staining, we detected cytosolic dsDNA in both U937 and HeLa cells, but not in Hs27 cells (Fig EV2A). Using Hs27, we found that, consistently, the deficiency of G3BP1 significantly dampened cytosolic cGAS condensation (Fig EV2B–D).
Figure EV2. G3BP1 engages cGAS in a condensed state in human fibroblast cell line.
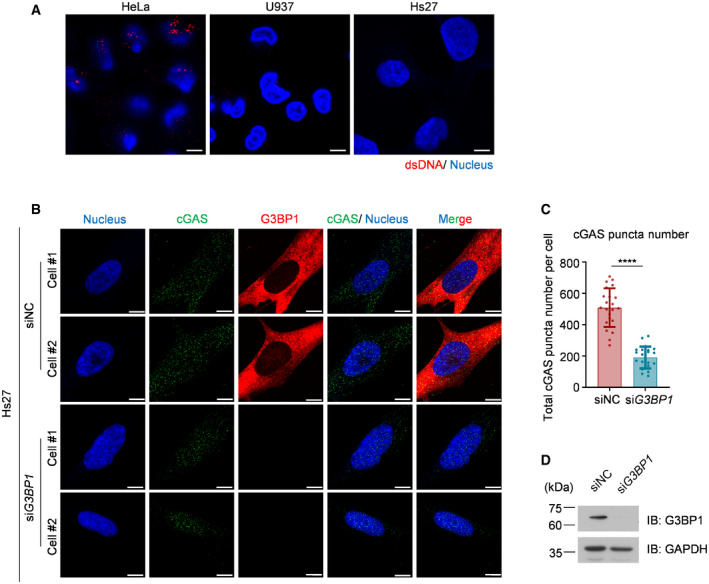
- Immunofluorescent staining of cytosolic dsDNA in indicated cells.
- Immunofluorescent staining of cGAS in both WT and G3BP1‐knockdown Hs27 cells.
- Quantitative analysis of total cGAS puncta number per cell in (B). n = 22 cells. Error bar, mean with s.d. ****P < 0.0001, two‐tailed t‐test.
- Knockdown effect of G3BP1 was analyzed by immunoblotting.
Data information: Representative images are shown (A, B and D). Scale bars, 5 µm (A and B). Hoechst (blue), nuclear staining. WT, wild type; IB, immunoblotting; NC, negative control.
Source data are available online for this figure.
Together, our data indicate that G3BP1 engages cGAS in a primary condensation state in cells.
The engagement with DNA, but not RNA, leads to the dissociation of G3BP1 from cGAS
Our previous study suggested that DNA treatment gradually disrupted the interaction of G3BP1 with cGAS (Liu et al, 2019). Here, we further investigated whether G3BP1 participated in DNA‐triggered LLPS of cGAS. We first incubated mEGFP‐tagged G3BP1 with mCherry‐tagged cGAS and analyzed cGAS‐G3BP1 condensation at early time points. Our data showed that G3BP1 formed condensates with cGAS immediately after the two proteins were mixed (Fig 3A, Movie EV5). By measuring the fluorescence intensity distribution in individual condensate (Fig 3B) and fluorescence intensity changes over time (Fig EV3A), we further confirmed this finding. Interestingly, when dsDNA was added, cGAS quickly condensed with DNA and dissociated from G3BP1 (Figs 3C and EV3B, Movie EV6). Further analysis on the fluorescence intensity distributions and fluorescence changes of cGAS‐DNA droplets yielded consistent data (Figs 3D and EV3C). Given that RNA was recently reported to induce cGAS LLPS without activating cGAS (Du & Chen, 2018), we then examined the effect of RNAs on cGAS‐G3BP1 primary condensates. We found that both dsRNA and single‐stranded RNA (ssRNA) condensed with cGAS and G3BP1, but the stimulation with RNAs did not trigger the dissociation of G3BP1 from cGAS (Figs 3E and F, and EV3D–H, Movie EV7). These data indicated that the engagement with DNA, but not with RNA, leads to the dissociation of G3BP1 from cGAS.
Figure 3. The engagement with DNA, but not with RNA, leads to the dissociation of G3BP1 from cGAS.
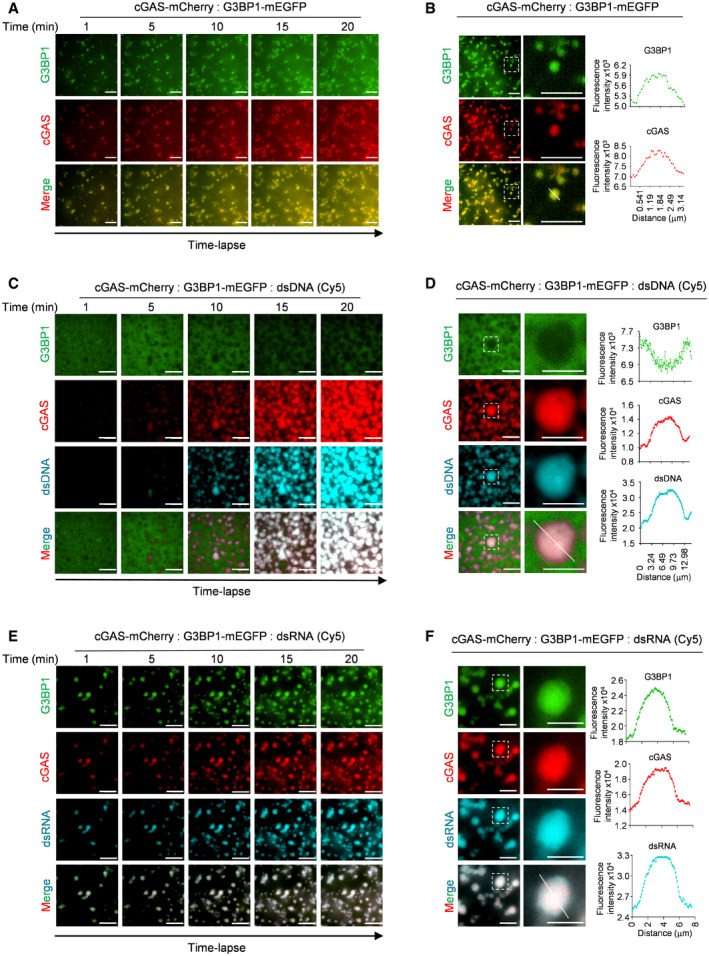
- Time‐lapse imaging of cGAS‐G3BP1 condensates.
- Fluorescence intensity distribution on cGAS‐G3BP1 droplets. A quantitative analysis of a representative cGAS‐G3BP1 droplet is shown. Along the white line on the merged image, the fluorescence intensity of both G3BP1 and cGAS channels were recorded.
- Time‐lapse imaging of cGAS‐DNA droplets in the presence of G3BP1‐mEGFP.
- cGAS, G3BP1, and Cy5‐dsDNA were incubated. A quantitative analysis of a representative cGAS‐DNA droplet is shown. Along the white line on the merged image, the fluorescence intensity of G3BP1, cGAS, and dsDNA channels were recorded.
- Time‐lapse imaging of condensates formed by cGAS, G3BP1, and dsRNA.
- cGAS, G3BP1, and Cy5‐dsRNA were incubated. Similar quantification analysis as in D was performed.
Data information: Representative images are shown (A–F). Scale bars, 10 μm (A–E), 5 μm (F). 45 μM cGAS‐mCherry, 10 μM G3BP1‐mEGFP, 2 μM dsDNA and 2 μM dsRNA were used in the assays. See also Fig EV3.
Figure EV3. The engagement with DNA, but not with RNA, leads to the dissociation of G3BP1 from cGAS.
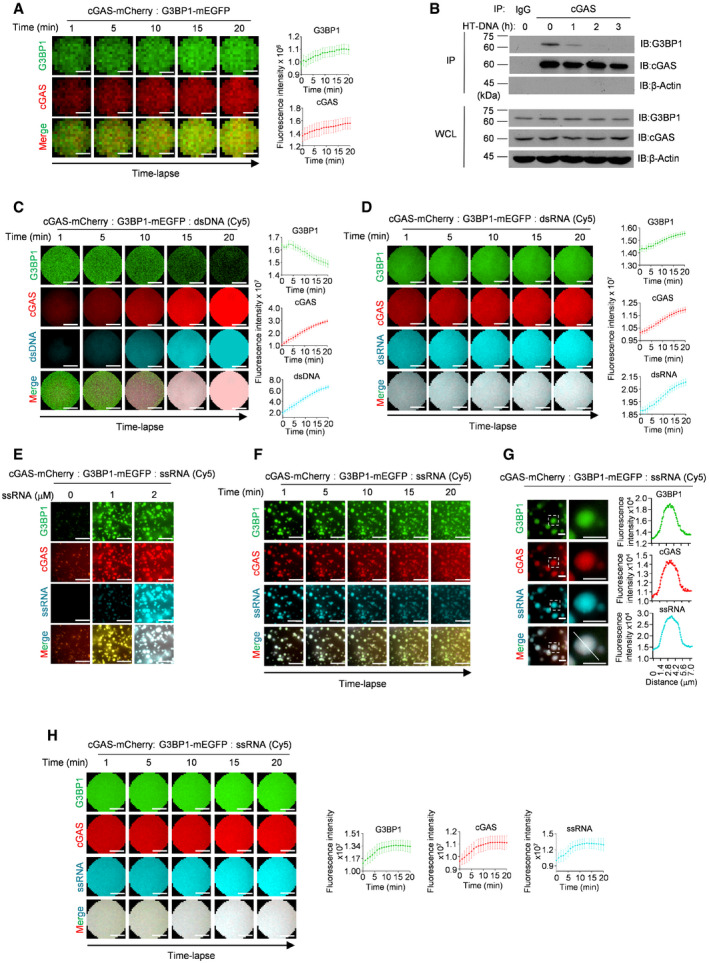
- Time‐lapse imaging of a single cGAS‐G3BP1 droplet (left). A quantitative analysis of the time course of the droplet fluorescence intensity is shown (right). n = 20 condensates.
- cGAS‐G3BP1 interaction analysis. cGAS was precipitated with anti‐cGAS antibodies in U937 cells treated with HT‐DNA as indicated. IgG served as control. The cGAS‐G3BP1 interaction was further analyzed with immunoblotting (IB).
- Time‐lapse imaging of a single cGAS‐DNA droplet in the presence of G3BP1‐mEGFP (left). A quantitative analysis of the time course of the droplet fluorescence intensity is shown (right). n = 20 condensates.
- Time‐lapse imaging of a single droplet formed by cGAS, G3BP1, and dsRNA (left). A quantitative analysis of the time course of the droplet fluorescence intensity is shown (right). n = 20 condensates.
- cGAS, G3BP1, and Cy5‐labled ssRNA were incubated. Indicated concentrations of Cy5‐labeled ssRNA were used.
- Time‐lapse imaging of condensates formed by cGAS, G3BP1, and ssRNA.
- cGAS, G3BP1, and Cy5‐ssRNA were incubated. A quantitative analysis of a representative cGAS‐DNA condensate is shown. Along the white line on the merged image, the fluorescence intensity of G3BP1, cGAS, and ssRNA channels were recorded.
- Time‐lapse imaging of a single droplet formed by cGAS, G3BP1, and ssRNA (left). A quantitative analysis of the time course of the droplet fluorescence intensity is shown (right). n = 20 condensates.
Data information: Representative images are shown (A, C–H). 45 μM cGAS‐mCherry, 10 μM G3BP1‐mEGFP, 2 μM dsDNA, dsRNA ssRNA were used in the assays. Error bars, mean with s.d. (A, C, H) and s.e.m. (D). Scale bars, 0.28 μm (A), 1 μm (C, D and H), 10 μm (E and F), 5 μm (G).
Source data are available online for this figure.
G3BP1 promotes DNA‐induced LLPS and activation of cGAS
To further study how G3BP1 regulates DNA‐induced LLPS of cGAS, we incubated different amounts of dsDNA with cGAS, in the presence or absence of G3BP1. Our data showed that dsDNA induced cGAS LLPS in a time‐ and dosage‐dependent manner (Fig EV4A–C). In the presence of G3BP1, this process was dramatically promoted, especially when limiting amounts of DNA were used (Fig 4A–C, Movies EV8). The effect of G3BP1 on cGAS LLPS was also dosage dependent (Fig EV4D and E). We further confirmed the above findings using heparin affinity chromatography–purified proteins (Fig EV4F and G). As a result, G3BP1 significantly enhanced the synthesis of cGAMP by cGAS (Fig 4D). Consistent with these data, G3BP1 deficiency suppressed DNA‐induced cGAS foci formation (Fig 4E and F) and IFN expression (Fig 4G). Collectively, G3BP1 facilitated DNA‐induced LLPS of cGAS and ensured efficient cGAS activation.
Figure EV4. G3BP1 promotes DNA‐induced LLPS and activation of cGAS.
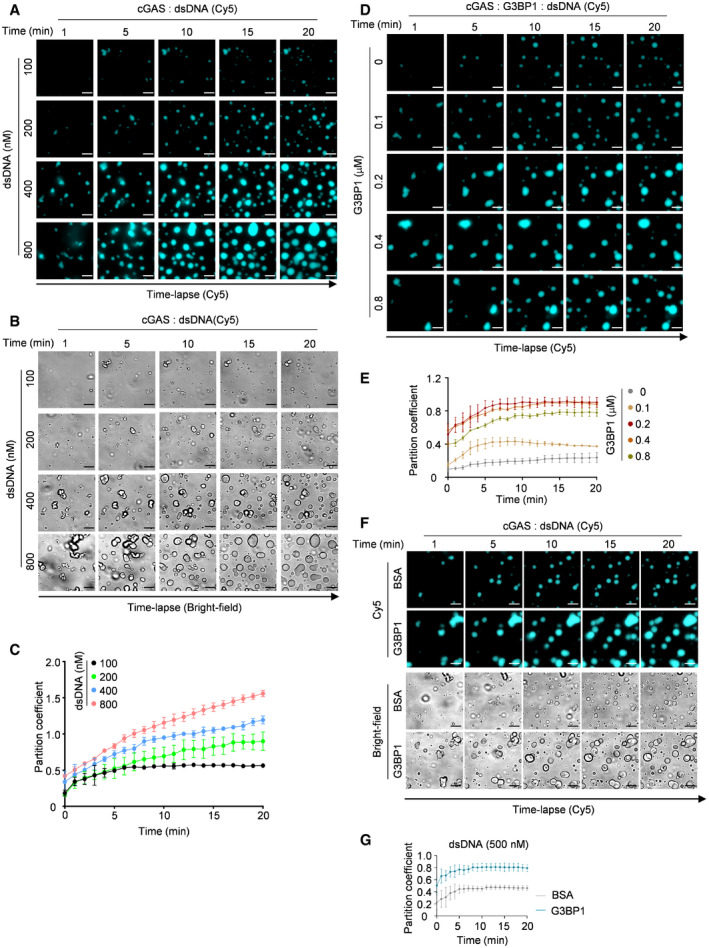
-
A, BTime‐lapse imaging of droplets formed by recombinant cGAS (10 μM) and indicated amounts of Cy5‐labeled dsDNA.
-
CQuantitative analysis of (A). n = 3 biological replicates.
-
DTime‐lapse imaging of cGAS‐DNA droplets in the presence of the indicated amounts of G3BP1. 10 μM cGAS and 200 nM Cy5‐labeled dsDNA were used.
-
EQuantitative analysis of (D). n = 3 biological replicates.
-
FTime‐lapse imaging of cGAS‐DNA condensates in the presence of heparin affinity chromatography purified G3BP1. 10 μM cGAS and 500 nM Cy5‐labeled dsDNA were used.
-
GQuantitative analysis of (F). n = 3 biological replicates.
Data information: Representative images are shown (A, B, D and F), scale bars, 10 μm. Error bars, mean with s.d. of (C, E and G). The partition coefficient was calculated as the total fluorescence intensity of droplets/bulk fluorescence intensity of background (C, E and G).
Source data are available online for this figure.
Figure 4. G3BP1 promotes DNA‐induced LLPS and activation of cGAS.
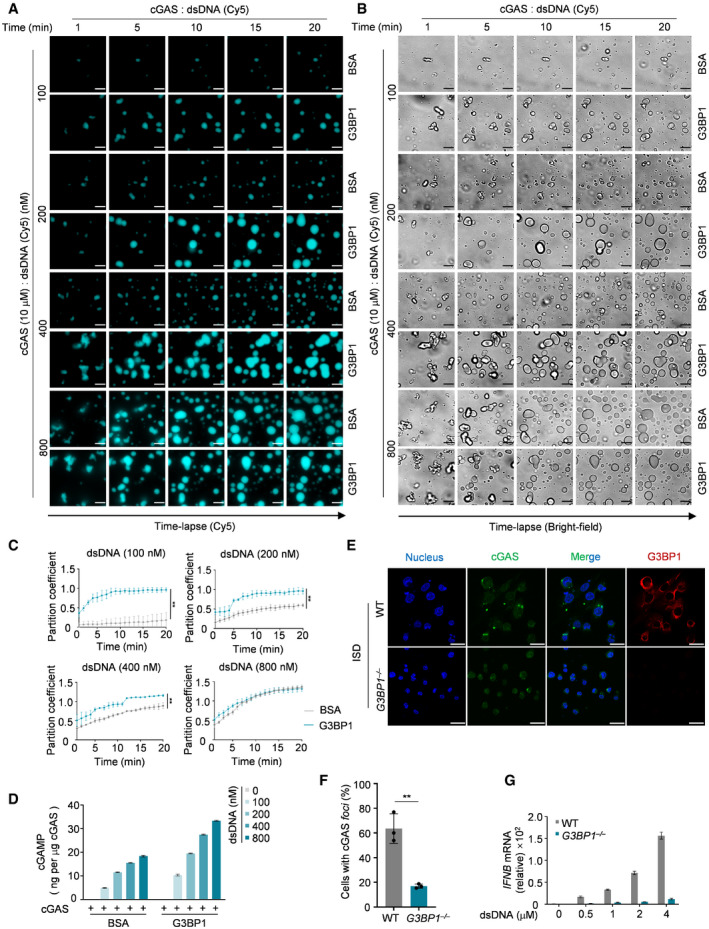
-
A, BRecombinant cGAS (10 μM) and the indicated amounts of Cy5‐labeled dsDNA were incubated, with or without G3BP1 (2 μM). The DNA‐induced cGAS‐DNA liquid droplets were analyzed with time‐lapse imaging.
-
CQuantitative analysis of (A). The partition coefficient was calculated as the total fluorescence intensity of droplets/bulk fluorescence intensity of background. n = 3 biological replicates.
-
DcGAMP production assays. Recombinant cGAS (10 μM) was incubated with the indicated amounts of dsDNA, with or without G3BP1 (10 μM). The produced cGAMP was quantitatively analyzed by LC‐MS/MRM. n = 3 technical replicates.
-
E, FImmunofluorescent staining of cGAS in WT and G3BP1 −/− U937 cells were transfected with ISD (2 μg ml−1) for 3 h (E). The percentage of cells with cGAS foci was quantified, n = 3 biological replicates (F). Hoechst (blue), nuclear staining.
-
GqPCR analysis of IFNB mRNA levels in WT and G3BP1 −/− U937 cells transfected with dsDNA as indicated. n = 3 technical replicates.
Data information: Representative images are shown (A, B and E). Error bars, mean with s.d. (C, D, F and G), **P < 0.01, two‐tailed t‐test. Scale bars, 10 μm (A and B), 20 μm (E). ISD, interferon stimulatory DNA. See also Fig EV4.
Full‐length G3BP1 binds to cGAS and promotes LLPS of cGAS
G3BP1 contains three domains/motifs named NTF2‐like, RRM and RGG (Fig 5A; Tourriere et al, 2003; Sanders et al, 2020). We purified domain deleted and truncated variants of G3BP1 (Fig 5B) and found that none of the G3BP1 regions was dispensable for its interaction with cGAS (Fig 5C). Next, we examined the function of each G3BP1 domain/motif in cGAS LLPS, and we found that only full‐length G3BP1 promoted DNA‐induced LLPS of cGAS (Figs 5D and E, and EV5A, Movies EV9). By measuring the clarity of the cGAS‐G3BP1 variants mixture, we found that only full‐length G3BP1 triggered the opacification of the cGAS solution (Fig EV5B–D). Together with the analysis of cGAMP production (Fig 5F), these data suggest that a multivalent interaction between G3BP1 and cGAS was important for G3BP1 to promote cGAS LLPS and activation.
Figure 5. Full‐length G3BP1 binds to cGAS and promotes LLPS of cGAS.
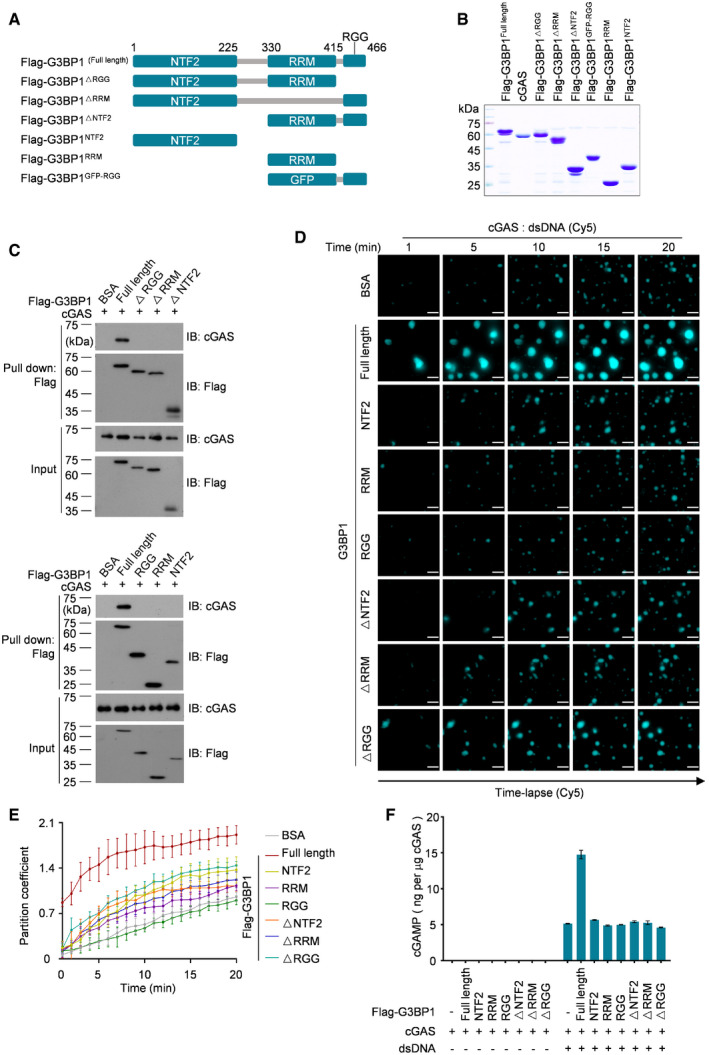
-
ASchematic drawing of domain architecture and truncations of human G3BP1.
-
BCoomassie blue staining of purified cGAS, full length and truncated G3BP1 recombinant proteins.
-
CcGAS‐G3BP1 interaction analysis. Full‐length and truncated G3BP1 was pulled‐down using anti‐Flag M2 beads. The cGAS‐G3BP1 interaction was further analyzed with immunoblotting (IB). 2 μg recombinant full‐length cGAS and 2 μg each of truncated G3BP1 were used in the assay.
-
D, ETime‐lapse imaging of DNA‐induced LLPS of cGAS, in the presence of full length or truncated G3BP1 (2 μM) (D). Partition coefficient of D was calculated as the total fluorescence intensity of droplets / bulk fluorescence intensity of background (E). n = 3 biological replicates.10 μM cGAS and 100 nM dsDNA were used in the assay.
-
FcGAMP production assays. Recombinant cGAS (10 μM) and full length or truncated G3BP1 (10 μM each) were incubated, in the presence or absence of dsDNA (200 nM). The produced cGAMP was quantitatively analyzed by LC‐MS/MRM. n = 3 technical replicates.
Data information: Representative images are shown (D). Error bars, mean with s.d. (E and F). Scale bars, 10 μm (D). See also Fig EV5.
Source data are available online for this figure.
Figure EV5. Full‐length G3BP1 binds to cGAS and promotes LLPS of cGAS.
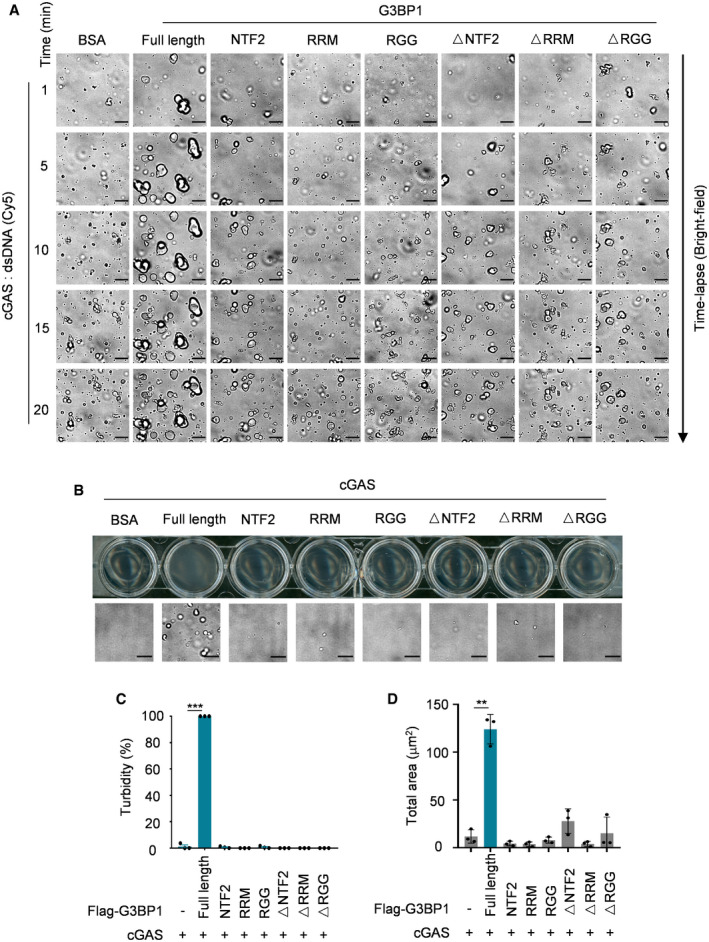
- Bright‐field microscope images of Fig 5D.
- Turbidity (top) and bright‐field microscope images (bottom) of indicated groups. 10 μM cGAS, 5 μM full length or truncated G3BP1 were used in the assay.
- Quantitative analysis of (B) (top) were measured by absorbance at 600 nm. n = 3 biological replicates.
- Quantitative analysis of total droplets area of (B) (bottom). n = 3 biological replicates.
Data information: Representative images are shown (A, B), scale bars, 10 μm. Error bars, mean with s.d. (C and D), **P < 0.05, ***P < 0.001, two‐tailed t‐test.
Source data are available online for this figure.
Zn2+ promotes the condensation and DNA‐induced LLPS of cGAS
It has been reported that zinc ions strongly promote cGAS activity (Du & Chen, 2018). We therefore incubated recombinant cGAS with KCl, MgCl2, or ZnCl2 and found that only ZnCl2 robustly induced the formation of cGAS condensates (Fig 6A). Further, the presence of ZnCl2 significantly enhanced cGAS activation (Fig 6B) and DNA‐induced LLPS of cGAS (Fig 6C and D). Interestingly, G3BP1 and ZnCl2 showed additive effects in promoting cGAS activation and LLPS in response to DNA (Fig 6E and F), suggesting that G3BP1 and ZnCl2 regulate LLPS of cGAS by different mechanisms. Consistent with the in vitro data, ZnCl2 treatment remarkably led to the formation of cGAS foci in cells (Fig 6G). Therefore, both G3BP1‐ and ZnCl2‐mediated primary condensates of cGAS seem to be important to prime cGAS for further activation by DNA.
Figure 6. Zn2+ promotes condensation and the DNA‐induced LLPS of cGAS.
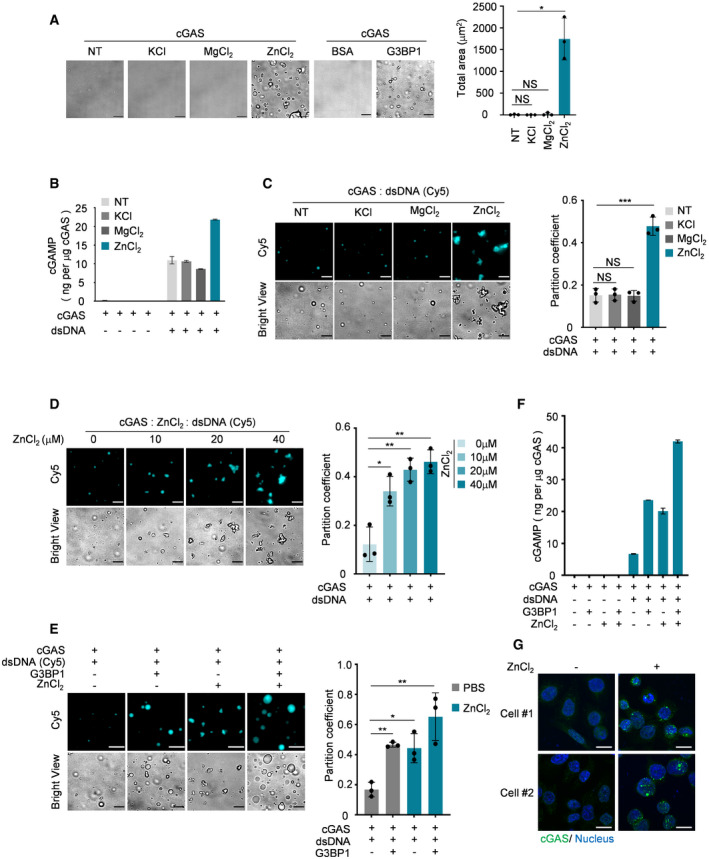
-
ABright‐field microscope images of indicated groups (left) and a quantitative analysis of total area of droplets (right). n = 3 biological replicates. 10 μM cGAS, 5 μM G3BP1 and 50 μM KCl, 50 μM MgCl2, 50 μM ZnCl2 were used.
-
BcGAMP production assays. Recombinant cGAS (10 μM) and indicated ions (100 μM) were incubated with or without dsDNA (100 nM). The production of cGAMP was analyzed by LC‐MS/MRM. n = 3 technical replicates.
-
CFluorescent images and analysis of DNA‐induced cGAS LLPS in the presence of the indicated reagent. n = 3 biological replicates. 10 μM cGAS, 50 μM of each ion and 100 nM dsDNA were used in these assays.
-
DFluorescent images and analysis of DNA‐induced LLPS of cGAS in the presence of different amounts of ZnCl2. n = 3 biological replicates. 10 μM cGAS and 100 nM dsDNA were used in the assay.
-
EFluorescent images and analysis of DNA‐induced LLPS of cGAS in the presence of G3BP1 (2 μM) and/or ZnCl2 (20 μM) as indicated. n = 3 biological replicates.
-
FcGAMP production assays. Recombinant cGAS (10 μM), G3BP1(10 μM) and ZnCl2 (100 μM) were incubated with or without dsDNA (100 nM) as indicated. The produced cGAMP was quantitatively analyzed by LC‐MS/MRM. n = 3 technical replicates.
-
GImmunofluorescence staining of cGAS in HeLa cells stimulated with ZnCl2 (100 μM). The images were acquired by ZEISS LSM 880 Confocal Microscopy.
Data information: Representative images are shown (A, C–E and G). Scale bars, 10 μm. The partition coefficient was calculated as the total fluorescence intensity of droplets/bulk fluorescence intensity of background (C–E). Hoechst (blue), nuclear staining. Error bars, mean with s.d. (A‐F), *P < 0.05, **P < 0.01, ***P < 0.001, two‐tailed t‐test. NS, non‐significant; NT, non‐treated.
EGCG inhibits G3BP1‐promoted cGAS phase condensation and activation
Epigallocatechin gallate (EGCG) is a natural compound isolated from green tea leaves (Cao & Cao, 1999; Singh et al, 2011). As an inhibitor of G3BP1, EGCG is known to block cGAS activation through inhibition of G3BP1 (Liu et al, 2019). We thus reasoned that EGCG may disrupt G3BP1‐promoted cGAS LLPS. To test this possibility, we added EGCG to the cGAS‐G3BP1 reaction and found that this chemical strongly reduced the formation of the cGAS‐G3BP1 condensates (Fig 7A). In accordance with our recent report (Liu et al, 2019), we confirmed the regions in G3BP1 that are critical for EGCG‐binding (Fig 7B and C). Further, EGCG significantly inhibited G3BP1‐indcued cGAS LLPS upon DNA treatment (Fig 7D). Consistently, EGCG abolished G3BP1‐facilitated cGAS activation in in vitro cGAMP synthesis assays (Fig 7E) and disrupted the interaction between cGAS and G3BP1 (Fig 7F). In line with our findings that EGCG inhibited G3BP1‐promoted cGAS LLPS, it also blocked DNA‐induced cGAS activation and the production of IFN (Fig 7G–I). EGCG seemed to suppress cGAS activation in the DNA‐cGAS‐IFN pathway through selectively targeting G3BP1 since it could no longer inhibit HT‐DNA‐stimulated IFNB mRNA expression in G3BP1‐null cells (Fig 7H). Taken together, our results indicate that by engaging cGAS in a primary condensation state, G3BP1 primes cGAS for efficient activation in response to cytoplasmic DNA (Fig 8).
Figure 7. EGCG inhibits G3BP1‐promoted cGAS phase condensation and activation.
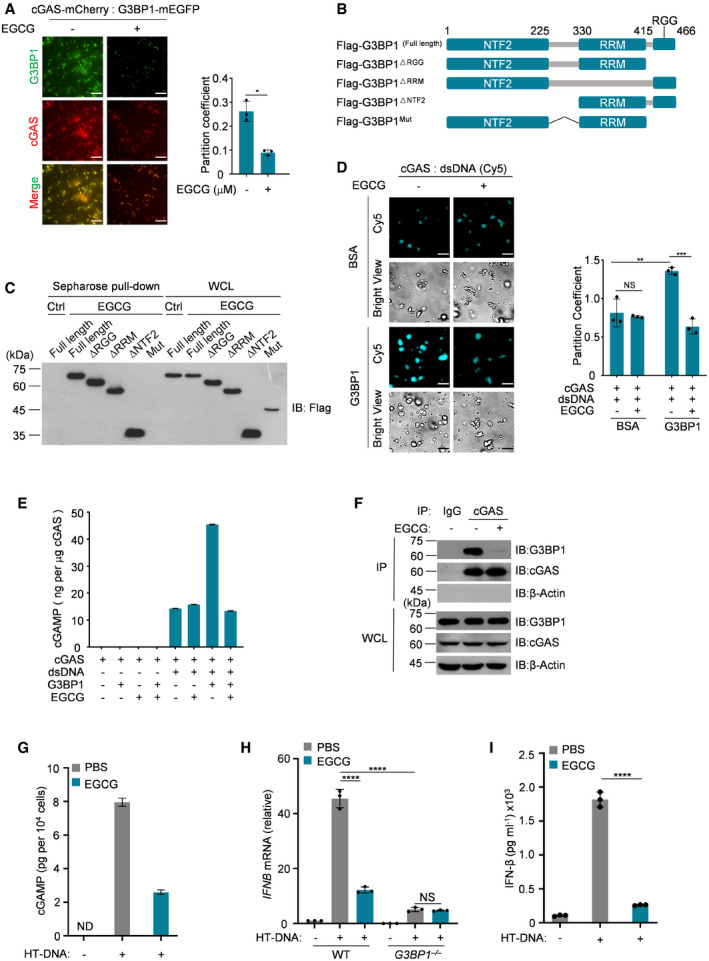
-
AFluorescent images of cGAS‐G3BP1 droplets in the presence or absence of 10 μM EGCG (left). The partition coefficient was calculated as the total fluorescence intensity of droplets/bulk fluorescence intensity of background (right), n = 3 biological replicates. 45 μM cGAS‐mCherry and 10 μM G3BP1‐mEGFP were used in these assays.
-
BSchematic drawing of the full‐length and truncations of G3BP1.
-
CEGCG‐G3BP1 interaction analysis. EGCG was pulled‐down using EGCG conjugated sepharose beads. The EGCG‐G3BP1 interaction was further analyzed by immunoblotting.
-
DFluorescent images of DNA‐induced LLPS of cGAS, with or without G3BP1, in the absence or presence of 10 μM EGCG (left). The partition coefficient was calculated as in (A), n = 3 biological replicates. 10 μM cGAS, 5 μM G3BP1, and 100 nM dsDNA were used in these assays.
-
EcGAMP production assays. Recombinant cGAS (10 μM) and G3BP1 (10 μM) were incubated, in the presence or absence of dsDNA (200 nM), with or without EGCG (20 μM). The produced cGAMP was quantitatively analyzed by LC‐MS/MRM. n = 3 technical replicates.
-
FcGAS‐G3BP1 interaction analysis. cGAS was precipitated using anti‐cGAS antibodies in U937 cells treated with EGCG as indicated. IgG served as control. The cGAS‐G3BP1 interaction was further analyzed by immunoblotting.
-
GcGAMP production assays. U937 cells were untreated (left) or treated (right) with EGCG (20 μM), followed by HT‐DNA transfection. The produced cGAMP was quantitatively analyzed by LC‐MS/MRM. ND, not detected. n = 3 technical replicates.
-
H, IU937 cells were untreated (left) or treated (right) with EGCG (20 μM), followed by HT‐DNA transfection. qPCR analysis of IFNB mRNA expression in cells (H) and ELISA analysis of secreted IFN‐β (I) were performed. n = 3 biological replicates. NS, non‐significant.
Data information: Representative images are shown (A, C, D and F). Scale bars, 10 μm (A and D). Error bars, mean with s.d. (A, D, E and G–I), *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, two‐tailed t‐test. NS, non‐significant; IB, immunoblotting; WCL, whole cell lysate; Ctrl, control; IP, immunoprecipitation; WT, wild type.
Source data are available online for this figure.
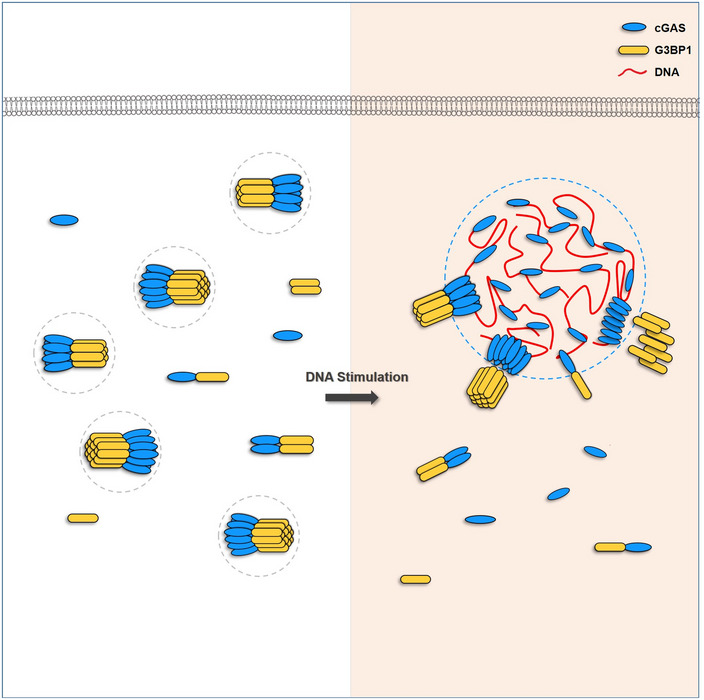
Figure 8. Schematic drawing of G3BP1 engaging cGAS in a primary condensation state to enable a rapid response to DNA.
Discussion
Liquid–liquid phase separation (LLPS) mediates the formation of membrane‐less cellular compartments that govern a variety of biological procedures (Alberti et al, 2019). Upon infection, the viral DNA can be introduced into the cytoplasm of the host cell, triggering cGAS‐DNA condensation via LLPS (Du & Chen, 2018). In these condensates, cGAS and DNA are highly concentrated, thus becoming microreactors for cGAMP production. We previously reported that G3BP1 promotes the DNA‐binding and activation of cGAS (Liu et al, 2019). In the current study, we further reveal the molecular mechanism underlying G3BP1‐mediated promotion of cGAS activation. We found that G3BP1 primes cGAS activation by engaging a primary condensed state of cGAS in resting cells. While these condensates were not as dynamic as DNA‐cGAS condensates, the formation of these cGAS‐G3BP1 condensates was important for efficient LLPS of cGAS and expeditious cGAS activation in response to DNA. By showing a G3BP1‐mediated regulation of cGAS LLPS, our study provides important insights to further understand the precise regulation of cGAS activation by phase separation.
Using high‐resolution microscopy, we showed that the particle‐like morphological characteristics of cGAS in cytoplasm was markedly weakened in G3BP1 −/− cells, in which cGAS‐mediated IFN production was also suppressed. Because G3BP1 deficiency did not obviously affect the localization and expression levels of cGAS, our data collectively indicate that dampened cGAS activation in the absence of G3BP1 is mainly due to the disorganization of the primary condensation state of cGAS. In line with a recent study showing that DNA treatment led to nucleus‐to‐cytosol translocation (Sun et al, 2021), our data also suggested that DNA‐triggered cGAS activation may occur mainly in the cytoplasm.
A growing number of publications described roles of nuclear cGAS and the mechanisms underlying cGAS inhibition in the nucleus (Xia et al, 2018; Boyer et al, 2020; Guey et al, 2020; Kujirai et al, 2020; Michalski et al, 2020; Pathare et al, 2020; Zhao et al, 2020). Here, we found that cGAS also exhibits particle‐like morphological characteristics in the nucleus. This is most likely regulated independently of G3BP1, because G3BP1 is predominantly localized in the cytoplasm. Future studies need to investigate whether LLPS is involved in the regulation of cGAS activation also in the nucleus. A recent publication showed that a circular RNA, named cia‐cGAS, binds and prevents the activation of cGAS in the nucleus (Xia et al, 2018). RNAs were previously reported to form liquid droplets with cGAS, however, cGAS was not activated by RNAs (Du & Chen, 2018). In our study, we also observed the same phenomena. It is possible that the formation of RNA‐cGAS condensates contributes to cGAS inhibition in the nucleus. Moreover, we showed that treatment with DNA, but not with RNA, led to the dissociation of G3BP1 from cGAS. This interesting observation may help to understand the deeper mechanisms by which cGAS distinguishes DNA and RNA.
Under physiological conditions, both infection‐ and cellular damage–derived cytosolic DNAs would be very limited. The efficient detection of small amounts of DNA is a challenge for cGAS to execute its function in eliciting a timely immune response against infections and damage. While cGAS can directly bind to DNA (Li et al, 2013b; Sun et al, 2013), several mechanisms have been discovered to illustrate how cGAS‐DNA binding is further regulated in cells. HMGB and TFAM have been found to prearrange DNA as ladder‐like structures to facilitate cGAS–DNA binding (Andreeva et al, 2017). PQBP1 is a critical coreceptor of cGAS and promotes the recognition of HIV‐1 viral DNA by cGAS (Yoh et al, 2015). IFI16 can cooperate with cGAS to detect nuclear viral DNA (Orzalli et al, 2015; Almine et al, 2017; Jonsson et al, 2017), and PKR has been recently shown to be necessary for intracellular DNA sensing by cGAS (Hu et al, 2019). In addition, several post‐translational modifications have also been reported to promote cGAS function (Hu et al, 2016; Liu et al, 2018; Seo et al, 2018; Song et al, 2020). Our study revealed an important mechanism, by which G3BP1 pre‐organizes cGAS proteins as condensates through phase separation. Interestingly, zinc ions that are known to enhance cGAS activity (Du & Chen, 2018) also induced the condensation of cGAS. Thus, the formation of a primary condensation state of cGAS is critical for efficient DNA detection and cGAS activation.
Recent advances highlight the pivotal role of G3BP1 in the formation of SGs. In particular, G3BP1 functions as a tunable switch that drives SG formation by LLPS (Guillen‐Boixet et al, 2020; Sanders et al, 2020; Yang et al, 2020). SGs are one prominent type of cellular granules that are assembled in response to numerous stresses, including viral infection (Emara & Brinton, 2007; Onomoto et al, 2014; McCormick & Khaperskyy, 2017) and intracellular RNA responses (Yoo et al, 2014; Reineke & Lloyd, 2015; Kim et al, 2019). In our study, we further revealed the critical role of G3BP1 in regulating cytosolic DNA‐sensing pathways. G3BP1 seems to be a critical regulator that orchestrates a spectrum of stress responses, including cytosolic DNA‐induced stress. G3BP1 may carry out such functions by mediating the formation of different condensates through LLPS. In the case of cGAS, G3BP1 mediates a primary cGAS‐G3BP1 phase. While this step is not enough to activate cGAS, it could efficiently promote the formation of the secondary cGAS‐DNA phase, in which cGAS is then activated by DNA. Such a process would ensure the quick and robust activation of cGAS, as well as subsequent downstream immune responses. Fully understanding these molecular events would provide new insights in designing strategies to modulate cGAS‐mediated signaling, thus treating cGAS‐related diseases.
Materials and Methods
Antibodies and reagents
Anti‐Flag (F3165) was from Sigma‐Aldrich; anti‐human cGAS (15102) was from Cell Signaling Technology; anti‐β‐Actin (sc‐47778) was from Santa Cruz Biotechnology; anti‐G3BP1 (H00010146‐M01) was from Abnova; anti‐IRF3 (ab68481) was from Abcam; anti‐human cGAS and anti‐human GAPDH antibodies for immunoblotting were prepared in our laboratory. The cGAS antibody was generated by immunizing rabbits with human cGAS (amino acids 1–160); the GAPDH antibody was generated by immunizing rabbits with human GAPDH protein. Anti‐Rabbit IgG (H + L) Highly Cross‐Adsorbed Secondary Antibody, Alexa Fluor 488‐(A‐11034) and 546‐(A‐11030) conjugated secondary antibody were from Thermo Fisher.
U937 (CRL‐1593.2) and HeLa (CCL‐2) cell lines were from ATCC. E. coli DH5α (S101‐02), and E. coli BL21(DE3) (S106‐02), Ni‐NTA (G106‐100), isopropyl‐β‐D‐thiogalactopyranoside (IPTG) (VA20321‐100), spectinomycin (VA28361), and bovine serum albumin (BSA) were from Genstar. PBS (C10010500), HT‐DNA (D6898), Anti‐Flag M2 affinity beads (a2220) and Triton X‐100 (X100) were from Sigma‐Aldrich. Tube* protein purification (29924), TRIzol (15596018), ATP Solution (R0441), and GTP Solution (R0461) were from ThermoFisher Scientific. Phenylmethylsulfonyl fluoride (PMSF) (PA116‐01) was from Biomed. Cocktail (B14001) was from Bimake. Dithiothreitol (DTT) (A620058‐0005) and cover glass (F518112) were from Sangon biotech. SYBR Green Master Mix (A25778) was from ABI. PrimeScript RT reagent (RR037A) was from TaKaRa. Epigallocatechin gallate (EGCG) (S2250) was from Selleck. 2’3’‐cGAMP (tlrl‐cga23) and lipofectamine 2000 (11668019) were from Invitrogen. Amicon Ultra‐0.5 Centrifugal Filter Unit (ufc5050bk) was from Millipore. Frosted glass (ZLI‐9354) was from ZSGB‐BIO. (CNBr)‐activated Sepharose 4B (GE17‐0430‐01) was from GE healthcare. Nonidet P‐40 (AR0107) was from BOSTER. EDTA (MC011.1) and EGTA (MC012) were from MACGENE. Phorbol‐12‐myristate‐13‐acetate (PMA) (524400) was from Merck. Sheep serum (ZLI‐9056) was from ZSGB. Human IFN‐β ELISA kit (EHC026b.96.2) was from NEOBIOSCIENCE.
The HSV‐1 60mer oligonucleotide (5’‐TAAGACACGATGCGATAAAATCTGTTTGTAAAATTTATTAAGGGTACAAATTGCCCTAGC‐3’), ssRNA and dsRNA (5’ CGCGACGUGCUCGUACGUGGCUUUGGAGACUCCGUGGAGGAGGUCUUAUCAGAGGCACGU‐3’) and ISD (5’‐TACAGATCTACTAGTGATCTATGACTGATCTGTACATGATCTACA‐3’) was synthesized from Tsingke Biological Technology.
Plasmids
pCDF‐cGAS, pCDF‐flag‐G3BP1FL, pCDF‐flag‐G3BP1RGG, pCDF‐flag‐G3BP1RRM, pCDF‐flag‐G3BP1NTF2, pCDF‐flag‐G3BP1▵NTF2, pCDF‐flag‐G3BP1▵RRM, pCDF‐flag‐G3BP1▵RGG, pCDH‐flag‐G3BP1FL, pCDH‐flag‐G3BP1RGG, pCDH‐flag‐G3BP1RRM, pCDH‐flag‐G3BP1NTF2, pCDH‐flag‐G3BP1▵RRM, pCDH‐flag‐G3BP1▵RGG, pCDH‐flag‐G3BP1▵NTF2 and pCDH‐flag‐G3BP1Mut were prepared in our laboratory before (Liu et al, 2019).
We subcloned the coding sequence of cGAS into pET28a(+) vector for recombinant protein purification and fused with an mCherry tag at C‐terminus (connected with a 3×GGGGS linker). The coding sequence of G3BP1 was subcloned into pET28a(+) vector and fused with an mEGFP tag at C‐terminus (connected with a 3×GGGGS linker).
Cell culture and transfection
HeLa and Hs27 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% (vol/vol) fetal bovine serum (FBS), 2 mM l‐glutamine, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin. U937 cells were cultured in RPMI‐1640 medium containing 10% FBS, 2 mM l‐glutamine, 100 U ml−1 penicillin and 100 mg ml−1 streptomycin. U937 cells were differentiated with 0.1 μM PMA for 36 h before transfection or other treatment. HeLa, Hs27 and U937 cells were obtained from ATCC. G3BP1‐deficient U937 and HeLa cells were prepared in our laboratory (Liu et al, 2019). All cell lines were tested to be mycoplasma free by PCR. Transfection of interferon stimulatory DNA (ISD), double strain DNA (dsDNA) and Herring testes DNA (HT‐DNA) were performed with Lipofectamine 2000 at final concentrations as indicated.
Protein expression and purification
The protein‐coding plasmids were expressed in E. coli BL21 (DE3) strain before purification. Cells were grown in LB medium overnight with 50 μg ml−1 spectinomycin at 37°C, 220 rpm. The overnight cultured bacteria were inoculated into fresh LB medium containing 50 μg ml−1 spectinomycin with the ratio of 1:100 for further culturing at 37°C, 220 rpm. When the OD600 reached 0.6–1.0, 0.5 mM IPTG was added into the medium for further culturing for 16 h at 16°C, 180 rpm. The cells were then harvested, washed with PBS, and purified with Ni‐NTA Sefinose (TM) Resin (Settled Resin) according to the manufacturer’s protocol (Genstar). Heparin affinity chromatography was used to further purify recombinant cGAS and G3BP1 proteins to remove nuclei acids contamination.
Quantification of cGAMP in cells
WT or G3BP1 −/− U937 cells were seeded into 12‐well plates and differentiated with PMA for 36 h, followed by transfection of 2 μg ml−1 ISD for 3 h. The samples were harvested with extraction solvent (50:50:100 (v:v:v) methanol‐acetonitrile‐ddH2O) as described previously (Wu et al, 2013).
The quantification of cGAMP in cells were then performed as previously described (Dai et al, 2019). The LC–MS/MS system consisted of an ekspert ultraLC 110‐XL system (SHIMAZU) and a SIL‐30AD triple quadropole mass spectrometer (SHIMAZU). The Qtrap6500 triple quadropole mass spectrometer was operated in positive ionization mode for MRM analysis of cGAMP. The source conditions were set as follows: ionspray voltage was 5.5 kV, ion source temperature was 550°C, curtain gas was 20 (arbitrary units), collision gas was Medium (CAD), and the dwell time for cGAMP was 100 ms. The optimized ion transitions were: cGAMP m/z 675 → 524; m/z 675 → 506; m/z 675 → 136. The parameters, entrance potential (EP), declustering potential (DP), collision energy (CE), and collision exit potential (CXP), were set as 7 V, 90 V, 29 V, and 18 V, respectively.
In vitro cGAMP synthesis assay
Recombinant cGAS protein was incubated with dsDNA in reaction buffer (20 mM HEPES, pH 7.5, 5 mM MgCl2, 2 mM ATP, 2 mM GTP) at 37°C for 1 h, with or without recombinant human G3BP1 or different ions. The samples were harvested by extraction solvent (40:40:20 (v:v:v) methanol‐acetonitrile‐sample) and centrifuged at 4°C, 16,000 g, for 10 min (Wu et al, 2013). Supernatants were then collected and lyophilized by gentle stream of nitrogen gas. The dried extracts were resuspended in ammonium acetate buffer (10 mM ammonium acetate, 0.05% acetate), vortexed. After another centrifugation at 4°C, 16,000 g, for 10 min, the supernatants were used for cGAMP quantification by LC‐MS/MRM analysis.
RNA isolation and quantitative PCR (qPCR)
Cells were collected and total RNA was extracted with TRIzol reagent. 500 ng RNA were used to perform the reverse transcription with PrimeScript RT reagent kit. To analysis relative mRNA levels, qPCR was performed with SYBR Green Master Mix on an ABI StepOnePlus Real‐Time PCR System. Data were analyzed with StepOne Plus software. The primers were synthesized from Sango Biotech. Human GAPDH mRNA levels were used for normalization.
Human IFNB: forward: 5'‐AGGACAGGATGAACTTTGAC‐3', reverse: 5'‐TGATAGACATTAGCCAGGAG‐3'.
Human GAPDH: forward: 5'‐GAGTCAACGGATTTGGTCGT‐3', reverse: 5'‐TTGATTTTGGAGGGATCTCG‐3'.
Fluorescence recovery after photobleaching
mCherry‐tagged cGAS protein was used to LLPS assays in vitro. Confocal microscope (LSM 880) was used to acquire the images at room temperature. Selected regions were photobleached at 561 nm, and the fluorescence intensity were recorded every 1 s afterwards. The fluorescence intensity was normalized to the initial intensity before bleaching.
Phase condensation of cGAS
Cyclic GMP‐AMP synthase proteins were diluted in PBS; dsDNA was added into the reaction system, mixed, and observed in a glass‐bottom cell culture dish (NEST, 801002) for microscopic observation and image acquirement.
For assays with G3BP1 and zinc ion, G3BP1 or zinc ion was pre‐mixed with cGAS in PBS before dsDNA was added. The mixture was observed in the glass‐bottom cell culture dishes for microscopic observation and image acquirement. For EGCG inhibition experiment, indicated concentration of G3BP1 and EGCG were pre‐incubated before cGAS was added.
Turbidity analysis
Recombinant cGAS protein, G3BP1 protein, and dsDNA were diluted in PBS on ice, mixed, and incubated in 0.6 ml centrifuge tube for 5 min. The mixtures were transferred into Clear Standard Module Plate (Thermo Scientific Nunc, 469949). The images were obtained by a regular scanner. The turbidity was measured by the absorbance at 600 nm using SpectraMax i3 (Molecular Devices).
Immunofluorescence
Wild‐type and G3BP1‐deficient U937 cells were seeded on the coverslips in 24‐well plates and differentiated with PMA for 36 h before dsDNA transfection. HeLa cells were seeded on the coverslips in 24‐well plates, followed by ZnCl2 (100 μM) treatment for 3 h. Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 0.3% Triton X‐100 for 10 min and blocked in 5% sheep serum for 1 h. Cells were incubated with primary antibodies, anti‐cGAS and anti‐G3BP1, overnight at 4°C. Alexa Fluor 488‐ and 546‐conjugated secondary antibodies were incubated for 1 h at room temperature. Images were acquired using Leica TCS SP8 or ZEISS LSM 880 Confocal Microscopy. Cytosolic dsDNA staining was performed as described previously (Lam et al, 2014; Shen et al, 2015).
Pull‐down and immunoprecipitation assay
Recombinant cGAS protein (2 μg) and different truncation mutants of G3BP1 with Flag tag (2 μg) were mixed in a buffer (20 mM Tris‐HCl, pH 7.5, 0.5% Nonidet P‐40, 10 mM NaCl, 3 mM EDTA and 3 mM EGTA) containing complete protease inhibitor cocktail (Roche 04693132001) at 4°C for 1 h. Pull‐down assay was performed with anti‐flag M2 beads according to the manufacturer’s instruction (Sigma‐Aldrich) and analyzed with immunoblotting.
As previously described, EGCG pull‐down assays were performed (Shim et al, 2010). Briefly, cyanogen bromide (CNBr)–activated Sepharose 4B was conjugated with EGCG. Cells were lysed with lysis buffer (20 mM Tris–HCl, pH 7.5; 0.5% Nonidet P‐40; 250 mM NaCl; 3 mM EDTA and 3 mM EGTA) containing complete protease inhibitor cocktail, PMSF and DTT, for 1 h, followed by centrifugation at 20,000 g for 20 min at 4°C. EGCG‐conjugated Sepharose 4B was incubated with cell lysates at 4°C for 3 h. The beads were then washed five times with lysis buffer. EGCG‐bound proteins were analyzed with immunoblotting.
For immunoprecipitation, cells were lysed with lysis buffer described above. Cell lysates were centrifuged at 20,000 g for 20 min at 4°C. The cell lysates were then incubated with anti‐cGAS antibody at 4°C for 12 h. The beads were then washed five times with lysis buffer. Whole cell lysis or immunoprecipitants were analyzed with immunoblotting.
ELISA
PMA‐differentiated U937 cells were seeded into 12‐well plates at a density of 5 × 105 cells per well and treated as indicated. The secreted interferon in cell culture medium was analyzed with ELISA kit according to the manufacturer’s instruction (NEOBIOSCIENCE).
RNA interference
Hs27 cells were transfected with siRNAs using RNAiMax (Thermo) transfection reagent for 48 h. The siRNAs used were as following: si‐G3BP1 (5′‐GGTCCGTCTGAATGTCGAA‐3′).
Statistical analysis
To determine the partition coefficients of indicated groups, at least three independent microscopy images were calculated. For each image, the fluorescence intensity was acquired by Volocity. Partition coefficient was calculated as the total fluorescence intensity of droplets divided by the bulk fluorescence intensity of background.
The statistical analysis was performed with GraphPad Prism 8.0. Data are presented as mean with s.d. A standard two‐tailed unpaired Student’s t‐test was used for statistical analysis of two groups.
Author contributions
Tao Li and W‐HL supervised the project. Tao Li, MZ, TX, J‐QX, and L‐HY designed the experiments. MZ, TX, J‐QX, L‐HY, JP, J‐YL, L‐MS, and MW performed cell experiments, qPCR analysis, and western blotting. TX, KW, Teng Li, and XX acquired images using ZEISS LSM 880 Confocal Microscopy and Lecia TCS SP8 Microscopy. MZ, J‐YL, L‐MS, HC, X‐WL, and WX performed the LLPS and in vitro cGAMP synthesis assays. JM, NW, KH, TZ, and A‐LL performed cGAMP detection by LC‐MS/MRM. MZ, J‐QX, Tingting Li, and Q‐YH performed pull down assays and the immunoprecipitation. MZ, TX, J‐QX, X‐MZ, W‐HL, and Tao Li analyzed the data. MZ, TX, J‐QX, and Tao Li wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Movie EV8
Movie EV9
Source Data for Expanded View
Source Data for Figure 2
Source Data for Figure 5
Source Data for Figure 7
Acknowledgments
The authors sincerely thank C. Widmann (University of Lausanne) for providing the HA‐G3BP1 plasmid, J. W. Chin (Cambridge) for providing the pCDF PylT‐1 plasmid, J. U. Jung (University of Southern California) for providing the cGAS cDNA construct, and M. Yang (Tsinghua University) for help with protein purification. They are grateful to Z.G. Liu (National Institutes of Health) for helpful discussion and critical reading of the manuscript. This work was supported by grants from the National Key Research and Development Program of China (No. 2020YFA0707702 to Tao Li and No. 2020YFA0707703 to T.Z.) and China National Natural Science Foundation (No. 81925017 and No. 81771708 to Tao Li, No. 81872153 to T.Z.).
EMBO reports (2022) 23: e53166.
See also: MP Gantier (January 2022)
Contributor Information
Wei‐Hua Li, Email: whli@ncba.ac.cn.
Tao Li, Email: tli@ncba.ac.cn.
Data availability
No primary datasets have been deposited.
References
- Ablasser A, Goldeck M, Cavlar T, Deimling T, Witte G, Rohl I, Hopfner KP, Ludwig J, Hornung V (2013) cGAS produces a 2'‐5'‐linked cyclic dinucleotide second messenger that activates STING. Nature 498: 380–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn J, Barber GN (2014) Self‐DNA, STING‐dependent signaling and the origins of autoinflammatory disease. Curr Opin Immunol 31: 121–126 [DOI] [PubMed] [Google Scholar]
- Aicardi J, Goutieres F (1984) A progressive familial encephalopathy in infancy with calcifications of the basal ganglia and chronic cerebrospinal fluid lymphocytosis. Ann Neurol 15: 49–54 [DOI] [PubMed] [Google Scholar]
- Alberti S, Gladfelter A, Mittag T (2019) Considerations and challenges in studying liquid‐liquid phase separation and biomolecular condensates. Cell 176: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almine JF, O’Hare CAJ, Dunphy G, Haga IR, Naik RJ, Atrih A, Connolly DJ, Taylor J, Kelsall IR, Bowie AG et al (2017) IFI16 and cGAS cooperate in the activation of STING during DNA sensing in human keratinocytes. Nat Commun 8: 14392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreeva L, Hiller B, Kostrewa D, Lässig C, de Oliveira Mann CC, Jan Drexler D, Maiser A, Gaidt M, Leonhardt H, Hornung V et al (2017) cGAS senses long and HMGB/TFAM‐bound U‐turn DNA by forming protein‐DNA ladders. Nature 549: 394–398 [DOI] [PubMed] [Google Scholar]
- Barbalat R, Ewald SE, Mouchess ML, Barton GM (2011) Nucleic acid recognition by the innate immune system. Annu Rev Immunol 29: 185–214 [DOI] [PubMed] [Google Scholar]
- Boeynaems S, Alberti S, Fawzi NL, Mittag T, Polymenidou M, Rousseau F, Schymkowitz J, Shorter J, Wolozin B, Van Den Bosch L et al (2018) Protein phase separation: a new phase in cell biology. Trends Cell Biol 28: 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH et al (2017) Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell 65: 1044–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie A (2012) The STING in the tail for cytosolic DNA‐dependent activation of IRF3. Sci Signal 5: pe9 [DOI] [PubMed] [Google Scholar]
- Boyer JA, Spangler CJ, Strauss JD, Cesmat AP, Liu P, McGinty RK, Zhang Q (2020) Structural basis of nucleosome‐dependent cGAS inhibition. Science 370: 450–454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R (2009) Eukaryotic stress granules: the ins and outs of translation. Mol Cell 36: 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Cao R (1999) Angiogenesis inhibited by drinking tea. Nature 398: 381 [DOI] [PubMed] [Google Scholar]
- Chong PA, Vernon RM, Forman‐Kay JD (2018) RGG/RG motif regions in RNA binding and phase separation. J Mol Biol 430: 4650–4665 [DOI] [PubMed] [Google Scholar]
- Crow YJ, Manel N (2015) Aicardi‐Goutieres syndrome and the type I interferonopathies. Nat Rev Immunol 15: 429–440 [DOI] [PubMed] [Google Scholar]
- Dai J, Huang Y‐J, He X, Zhao M, Wang X, Liu Z‐S, Xue W, Cai H, Zhan X‐Y, Huang S‐Y et al (2019) Acetylation blocks cGAS activity and inhibits self‐DNA‐induced autoimmunity. Cell 176: 1447–1460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du M, Chen ZJ (2018) DNA‐induced liquid phase condensation of cGAS activates innate immune signaling. Science 361: 704–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emara MM, Brinton MA (2007) Interaction of TIA‐1/TIAR with West Nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc Natl Acad Sci USA 104: 9041–9046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei J, Jadaliha M, Harmon TS, Li ITS, Hua B, Hao Q, Holehouse AS, Reyer M, Sun Q, Freier SM et al (2017) Quantitative analysis of multilayer organization of proteins and RNA in nuclear speckles at super resolution. J Cell Sci 130: 4180–4192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feric M, Vaidya N, Harmon TS, Mitrea DM, Zhu L, Richardson TM, Kriwacki RW, Pappu RV, Brangwynne CP (2016) Coexisting liquid phases underlie nucleolar subcompartments. Cell 165: 1686–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Nakagawa S, Hirose T, Bond CS (2018) Paraspeckles: where long noncoding RNA meets phase separation. Trends Biochem Sci 43: 124–135 [DOI] [PubMed] [Google Scholar]
- Franzmann TM, Alberti S (2019) Protein phase separation as a stress survival strategy. Cold Spring Harb Perspect Biol 11: a034058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzmann TM, Jahnel M, Pozniakovsky A, Mahamid J, Holehouse AS, Nüske E, Richter D, Baumeister W, Grill SW, Pappu RV et al (2018) Phase separation of a yeast prion protein promotes cellular fitness. Science 359: eaao5654 [DOI] [PubMed] [Google Scholar]
- Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y, May AI, Knorr RL, Suzuki K, Ohsumi Y et al (2020) Phase separation organizes the site of autophagosome formation. Nature 578: 301–305 [DOI] [PubMed] [Google Scholar]
- Gao PU, Ascano M, Wu Y, Barchet W, Gaffney B, Zillinger T, Serganov A, Liu Y, Jones R, Hartmann G et al (2013a) Cyclic [G(2',5')pA(3',5')p] is the metazoan second messenger produced by DNA‐activated cyclic GMP‐AMP synthase. Cell 153: 1094–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao PU, Ascano M, Zillinger T, Wang W, Dai P, Serganov A, Gaffney B, Shuman S, Jones R, Deng L et al (2013b) Structure‐function analysis of STING activation by c[G(2',5')pA(3',5')p] and targeting by antiviral DMXAA. Cell 154: 748–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guey B, Wischnewski M, Decout A, Makasheva K, Kaynak M, Sakar MS, Fierz B, Ablasser A (2020) BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 369: 823–828 [DOI] [PubMed] [Google Scholar]
- Guillen‐Boixet J, Kopach A, Holehouse AS, Wittmann S, Jahnel M, Schlussler R, Kim K, Trussina I, Wang J, Mateju D et al (2020) RNA‐induced conformational switching and clustering of G3BP drive stress granule assembly by condensation. Cell 181: 346–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurtler C, Bowie AG (2013) Innate immune detection of microbial nucleic acids. Trends Microbiol 21: 413–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu MM, Yang Q, Xie XQ, Liao CY, Lin H, Liu TT, Yin L, Shu HB (2016) Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45: 555–569 [DOI] [PubMed] [Google Scholar]
- Hu S, Sun H, Yin L, Li J, Mei S, Xu F, Wu C, Liu X, Zhao F, Zhang DI et al (2019) PKR‐dependent cytosolic cGAS foci are necessary for intracellular DNA sensing. Sci Signal 12: eaav7934 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN (2008) STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455: 674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wang S, Huang Y, He X, Cui H, Zhu X, Zheng Y (2015) Phase transition of spindle‐associated protein regulate spindle apparatus assembly. Cell 163: 108–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jønsson KL, Laustsen A, Krapp C, Skipper KA, Thavachelvam K, Hotter D, Egedal JH, Kjolby M, Mohammadi P, Prabakaran T et al (2017) IFI16 is required for DNA sensing in human macrophages by promoting production and function of cGAMP. Nat Commun 8: 14391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassiotis G, Stoye JP (2016) Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol 16: 207–219 [DOI] [PubMed] [Google Scholar]
- Kim SS, Sze L, Liu C, Lam KP (2019) The stress granule protein G3BP1 binds viral dsRNA and RIG‐I to enhance interferon‐beta response. J Biol Chem 294: 6430–6438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S, Maharana S, Mateju D, Malinovska L, Nuske E, Poser I, Richter D, Alberti S (2015) Promiscuous interactions and protein disaggregases determine the material state of stress‐inducible RNP granules. Elife 4: e06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kujirai T, Zierhut C, Takizawa Y, Kim R, Negishi L, Uruma N, Hirai S, Funabiki H, Kurumizaka H (2020) Structural basis for the inhibition of cGAS by nucleosomes. Science 370: 455–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam AR, Le Bert N, Ho SSW, Shen YJ, Tang MLF, Xiong GM, Croxford JL, Koo CX, Ishii KJ, Akira S et al (2014) RAE1 ligands for the NKG2D receptor are regulated by STING‐dependent DNA sensor pathways in lymphoma. Cancer Res 74: 2193–2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K‐H, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A et al (2016) C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane‐less organelles. Cell 167: 774–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Shu C, Yi G, Chaton CT, Shelton CL, Diao J, Zuo X, Kao CC, Herr AB, Li P (2013a) Cyclic GMP‐AMP synthase is activated by double‐stranded DNA‐induced oligomerization. Immunity 39: 1019–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XD, Wu J, Gao D, Wang H, Sun L, Chen ZJ (2013b) Pivotal roles of cGAS‐cGAMP signaling in antiviral defense and immune adjuvant effects. Science 341: 1390–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisnevskaia L, Murphy G, Isenberg D (2014) Systemic lupus erythematosus. Lancet 384: 1878–1888 [DOI] [PubMed] [Google Scholar]
- Liu Z‐S, Cai H, Xue W, Wang M, Xia T, Li W‐J, Xing J‐Q, Zhao M, Huang Y‐J, Chen S et al (2019) G3BP1 promotes DNA binding and activation of cGAS. Nat Immunol 20: 18–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZS, Zhang ZY, Cai H, Zhao M, Mao J, Dai J, Xia T, Zhang XM, Li T (2018) RINCK‐mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell Biosci 8: 35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin N, Tian L, Spencer D, Coutable‐Pennarun A, Anderson JLR, Mann S (2019) Photoswitchable phase separation and oligonucleotide trafficking in DNA coacervate microdroplets. Angew Chem Int Ed Engl 58: 14594–14598 [DOI] [PubMed] [Google Scholar]
- McCormick C, Khaperskyy DA (2017) Translation inhibition and stress granules in the antiviral immune response. Nat Rev Immunol 17: 647–660 [DOI] [PubMed] [Google Scholar]
- Michalski S, de Oliveira Mann CC, Stafford CA, Witte G, Bartho J, Lammens K, Hornung V, Hopfner KP (2020) Structural basis for sequestration and autoinhibition of cGAS by chromatin. Nature 587: 678–682 [DOI] [PubMed] [Google Scholar]
- Mitrea DM, Chandra B, Ferrolino MC, Gibbs EB, Tolbert M, White MR, Kriwacki RW (2018) Methods for physical characterization of phase‐separated bodies and membrane‐less organelles. J Mol Biol 430: 4773–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, Taylor JP (2015) Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA (2013) Immunology. Sensing the dark side of DNA. Science 339: 763–764 [DOI] [PubMed] [Google Scholar]
- Onomoto K, Yoneyama M, Fung G, Kato H, Fujita T (2014) Antiviral innate immunity and stress granule responses. Trends Immunol 35: 420–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orzalli MH, Broekema NM, Diner BA, Hancks DC, Elde NC, Cristea IM, Knipe DM (2015) cGAS‐mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc Natl Acad Sci USA 112: E1773–E1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathare GR, Decout A, Gluck S, Cavadini S, Makasheva K, Hovius R, Kempf G, Weiss J, Kozicka Z, Guey B et al (2020) Structural mechanism of cGAS inhibition by the nucleosome. Nature 587: 668–672 [DOI] [PubMed] [Google Scholar]
- Protter DSW, Parker R (2016) Principles and properties of stress granules. Trends Cell Biol 26: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke LC, Lloyd RE (2015) The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J Virol 89: 2575–2589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riback JA, Katanski CD, Kear‐Scott JL, Pilipenko EV, Rojek AE, Sosnick TR, Drummond DA (2017) Stress‐triggered phase separation is an adaptive, evolutionarily tuned response. Cell 168: 1028–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders DW, Kedersha N, Lee DSW, Strom AR, Drake V, Riback JA, Bracha D, Eeftens JM, Iwanicki A, Wang A et al (2020) Competing protein‐RNA interaction networks control multiphase intracellular organization. Cell 181: 306–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU (2018) TRIM56‐mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun 9: 613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen YJ, Le Bert N, Chitre AA, Koo CX, Nga XH, Ho SS, Khatoo M, Tan NY, Ishii KJ, Gasser S (2015) Genome‐derived cytosolic DNA mediates type I interferon‐dependent rejection of B cell lymphoma cells. Cell Rep 11: 460–473 [DOI] [PubMed] [Google Scholar]
- Shim JH, Su ZY, Chae JI, Kim DJ, Zhu F, Ma WY, Bode AM, Yang CS, Dong Z (2010) Epigallocatechin gallate suppresses lung cancer cell growth through Ras‐GTPase‐activating protein SH3 domain‐binding protein 1. Cancer Prev Res (Phila) 3: 670–679 [DOI] [PubMed] [Google Scholar]
- Singh BN, Shankar S, Srivastava RK (2011) Green tea catechin, epigallocatechin‐3‐gallate (EGCG): mechanisms, perspectives and clinical applications. Biochem Pharmacol 82: 1807–1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song ZM, Lin H, Yi XM, Guo W, Hu MM, Shu HB (2020) KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc Natl Acad Sci USA 117: 21568–21575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R (2006) Recognition of cytosolic DNA activates an IRF3‐dependent innate immune response. Immunity 24: 93–103 [DOI] [PubMed] [Google Scholar]
- Sun H, Huang YU, Mei S, Xu F, Liu X, Zhao F, Yin L, Zhang DI, Wei L, Wu C et al (2021) A nuclear export signal is required for cGAS to sense cytosolic DNA. Cell Rep 34: 108586 [DOI] [PubMed] [Google Scholar]
- Sun L, Wu J, Du F, Chen X, Chen ZJ (2013) Cyclic GMP‐AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339: 786–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Li Y, Chen L, Chen H, You F, Zhou X, Zhou Y, Zhai Z, Chen D, Jiang Z (2009) ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc Natl Acad Sci USA 106: 8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Akira S (2010) Pattern recognition receptors and inflammation. Cell 140: 805–820 [DOI] [PubMed] [Google Scholar]
- Tourriere H, Chebli K, Zekri L, Courselaud B, Blanchard JM, Bertrand E, Tazi J (2003) The RasGAP‐associated endoribonuclease G3BP assembles stress granules. J Cell Biol 160: 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Watson RO, Bell SL, MacDuff DA, Kimmey JM, Diner EJ, Olivas J, Vance RE, Stallings CL, Virgin HW, Cox JS (2015) The cytosolic sensor cGAS detects Mycobacterium tuberculosis DNA to induce type I interferons and activate autophagy. Cell Host Microbe 17: 811–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Cheng YY, Yin H, Song XN, Li WW, Zhou XX, Zhao LP, Tian LJ, Han JC, Yu HQ (2013) Oxygen promotes biofilm formation of Shewanella putrefaciens CN32 through a diguanylate cyclase and an adhesin. Sci Rep 3: 1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32: 461–488 [DOI] [PubMed] [Google Scholar]
- Xia P, Wang S, Ye B, Du Y, Li C, Xiong Z, Qu Y, Fan Z (2018) A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS‐mediated exhaustion. Immunity 48: 688–701 [DOI] [PubMed] [Google Scholar]
- Xie W, Lama L, Adura C, Tomita D, Glickman JF, Tuschl T, Patel DJ (2019) Human cGAS catalytic domain has an additional DNA‐binding interface that enhances enzymatic activity and liquid‐phase condensation. Proc Natl Acad Sci USA 116: 11946–11955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Mathieu C, Kolaitis R‐M, Zhang P, Messing J, Yurtsever U, Yang Z, Wu J, Li Y, Pan Q et al (2020) G3BP1 Is a tunable switch that triggers phase separation to assemble stress granules. Cell 181: 325–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda S, Tsuchiya H, Kaiho AI, Guo Q, Ikeuchi K, Endo A, Arai N, Ohtake F, Murata S, Inada T et al (2020) Stress‐ and ubiquitylation‐dependent phase separation of the proteasome. Nature 578: 296–300 [DOI] [PubMed] [Google Scholar]
- Yoh S, Schneider M, Seifried J, Soonthornvacharin S, Akleh R, Olivieri K, De Jesus P, Ruan C, de Castro E, Ruiz P et al (2015) PQBP1 is a proximal sensor of the cGAS‐dependent innate response to HIV‐1. Cell 161: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo J‐S, Takahasi K, Ng CS, Ouda R, Onomoto K, Yoneyama M, Lai JC, Lattmann S, Nagamine Y, Matsui T et al (2014) DHX36 enhances RIG‐I signaling by facilitating PKR‐mediated antiviral stress granule formation. PLoS Pathog 10: e1004012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youn JY, Dyakov BJA, Zhang J, Knight JDR, Vernon RM, Forman‐Kay JD, Gingras AC (2019) Properties of stress granule and P‐body proteomes. Mol Cell 76: 286–294 [DOI] [PubMed] [Google Scholar]
- Zhao B, Xu P, Rowlett CM, Jing T, Shinde O, Lei Y, West AP, Liu WR, Li P (2020) The molecular basis of tight nuclear tethering and inactivation of cGAS. Nature 587: 673–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong BO, Yang Y, Li S, Wang Y‐Y, Li Y, Diao F, Lei C, He X, Zhang LU, Tien PO et al (2008) The adaptor protein MITA links virus‐sensing receptors to IRF3 transcription factor activation. Immunity 29: 538–550 [DOI] [PubMed] [Google Scholar]


