Abstract
Flowering plants contain a large number of cyclin families, each containing multiple members, most of which have not been characterized to date. Here, we analyzed the role of the B1 subclass of mitotic cyclins in cell cycle control during Arabidopsis development. While we reveal CYCB1;5 to be a pseudogene, the remaining four members were found to be expressed in dividing cells. Mutant analyses showed a complex pattern of overlapping, development‐specific requirements of B1‐type cyclins with CYCB1;2 playing a central role. The double mutant cycb1;1 cycb1;2 is severely compromised in growth, yet viable beyond the seedling stage, hence representing a unique opportunity to study the function of B1‐type cyclin activity at the organismic level. Immunolocalization of microtubules in cycb1;1 cycb1;2 and treating mutants with the microtubule drug oryzalin revealed a key role of B1‐type cyclins in orchestrating mitotic microtubule networks. Subsequently, we identified the GAMMA‐TUBULIN COMPLEX PROTEIN 3‐INTERACTING PROTEIN 1 (GIP1/MOZART) as an in vitro substrate of B1‐type cyclin complexes and further genetic analyses support a potential role in the regulation of GIP1 by CYCB1s.
Keywords: CDK, CYCB1, endosperm, microtubule nucleation, mitosis
Subject Categories: Cell Cycle, Plant Biology
Studying multiple mutant combinations, this study reveals the role of B1‐type cyclins in plant development and microtubule organization.

Introduction
A highly elaborated control system guides cells through mitosis during which chromosomes are separated and distributed to the newly forming daughter cells. Cyclin‐dependent kinase (CDK)‐cyclin complexes stand in the center of this control system (Morgan, 1997; Lindqvist et al, 2009). In animals, Cdk1 together with B‐type cyclins are an essential part of the so‐called mitosis promoting factor (MPF) complex that phosphorylates a plethora of mitotic substrates including nuclear structure proteins, such as Lamin A and B, and chromosome segregation proteins, such as the spindle assembly factor TPX2 (Blethrow et al, 2008). MPF activity is kept low prior to mitotic entry by excluding Cyclin B1 (CycB1) from the nucleus (Hagting et al, 1998; Toyoshima et al, 1998; Yang et al, 1998). In addition, Cdk1‐cyclin B complexes are inhibited by phosphorylation on two inhibitory residues, Thr14 and Tyr15 (or the homologous amino acids in the P‐loop of the respective Cdk) by the action of Wee1 and/or Myt1 kinases (O’Farrell, 2001). After a threshold concentration of Cdk1‐CycB1 is reached, CycB1 accumulates in the nucleus and the Cdk‐CycB1 complex becomes activated by a group of dual‐specificity Cdc25 phosphatases that remove the inhibitory phosphorylation from the P‐loop of the kinase. Due to a negative feedback wiring with Wee1 and a positive feedback with Cdc25 (Tyson & Novak, 2001), Cdk1‐cyclin activity levels rise rapidly and promote entry and progression through mitosis, including the separation of the duplicated centrosomes (spindle pole body in yeast) as a key step to generate a bipolar spindle (Lacey et al, 1999; Haase et al, 2001). Finally, to complete mitosis and promote cytokinesis, Cdk1‐cyclin B levels have to drop. This is accomplished by the degradation of cyclin B mediated by the Anaphase Promoting Complex/Cyclosome (APC/C) (Nakayama & Nakayama, 2005).
While a wealth of information about the execution of mitosis exists in animals and yeast, information is still scarce in plants. Notably, flowering plants appear to regulate mitosis differently from yeast and animals. First of all, flowering plants do not contain centrosomes and it is still not fully understood how the mitotic spindle is organized, although many microtubule‐regulating components are conserved (Yamada & Goshima, 2017). Next, Arabidopsis WEE1 kinase was shown to prevent premature cell differentiation in S phase after DNA damage rather than functioning in mitotic control (De Schutter et al, 2007; Cools et al, 2011). Moreover, Arabidopsis does not contain a functional Cdc25 homolog, thus one of the most central control loops of the animal cell cycle is absent at least in this plant (Dissmeyer et al, 2009, 2010).
Another difference in mitotic regulation between plants and animals appears at the level of cyclins. In animals, D‐type cyclins control entry into the S phase (G1 cyclins), while cyclin A controls the S phase as well as early mitotic events, and B‐type cyclins control mitosis (Riabowol et al, 1989; Furuno et al, 1999). In contrast, functional studies and expression analyses have revealed that members of all three cyclin classes, that is, cyclin A, B, and D, are involved in the control of mitosis in plants (Schnittger et al, 2002; Menges et al, 2005; Dewitte et al, 2007; Boudolf et al, 2009; Vanneste et al, 2011). While there are only a few members in each cyclin family in metazoans, plant cyclin families are large, which makes functional studies challenging. For instance, as opposed to 3 B‐type cyclins in mammals (CycB1, CycB2, and CycB3) and 2 in Drosophila (CycB1 and CycB3), there are 11 predicted B‐type cyclins divided into three subgroups (B1, B2, and B3) in Arabidopsis that are all equally distant from animal B‐type cyclins, that is, Arabidopsis B1‐type cyclins are closer related to B2 and B3 from Arabidopsis than to any B‐type cyclin from animals (Doerner et al, 1996; Vandepoele et al, 2002; Wang et al, 2004). This classification is currently only based on sequence similarities and for B‐type cyclins, as for most other cyclins in plants, the biological role is far from being understood.
Here, we present a functional analysis of the largest class of B‐type cyclins in Arabidopsis, that is, the five‐member B1 group. We reveal a central role for CYCB1;2 that is backed up by one or more of the other B1‐type cyclins in a tissue‐dependent manner. Unlike CycB1 mutants in mouse (Brandeis et al, 1998), Arabidopsis cycb1;1 cycb1;2 double mutants are viable, presenting a unique opportunity to study cyclin B function at an organismic level. This allowed us to reveal the organization of mitotic microtubules as the main function of B1‐type cyclins in Arabidopsis, a finding supported by in vitro kinase assays that indicated that GIP1/MOZART, a key factor of microtubule organization, is a substrate of CDK‐CYCB1 complexes.
Results
CYCB1;1, CYCB1;2, and CYCB1;3 are redundantly required for endosperm proliferation
To start the characterization of B1‐type cyclins, we first determined their expression pattern. To this end, we used previously generated promoter reporter lines comprising GFP fused to the N‐terminal part of the respective cyclin (CYCB1;1 to CYCB1;4), including the destruction box (Weimer et al, 2016). For CYCB1;5, since different annotations exist for this gene, we generated three different reporter constructs, each reaching to the different predicted transcriptional start sites. One upstream of the first ATG, one upstream of the second ATG, and the third one including the first and second upstream regions. However, in none of these CYCB1;5 reporter lines a signal could be detected. Therefore, we next analyzed the expression of CYCB1;5 by qRT–PCR. Sequencing of the amplified products showed the existence of many different CYCB1;5 cDNAs that exhibited exon skipping, intron retention, and use of internal polyadenylation sites (Fig EV1, EV2, EV3, EV4, EV5), consistent with the lack of reliable transcriptional support for CYCB1;5 in public depositories (The Arabidopsis Information Resource, TAIR). Taken together with data from previous studies (Bulankova et al, 2013) and the fact that many Arabidopsis accessions have accumulated several point mutations and even deletions in CYCB1;5 (The Arabidopsis Information Resource, TAIR), we concluded that CYCB1;5 is a pseudogene. In the following study, we therefore concentrated on the analysis of CYCB1;1 through CYCB1;4.
Figure EV1. CYCB1;5 is a pseudogene.
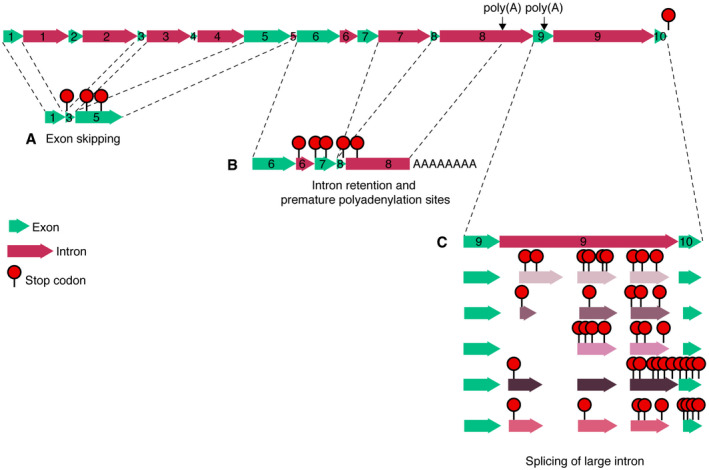
-
A–CThe predicted gene structure of CYCB1;5, including the observed cDNAs with exon skipping (A), intron retention and premature polyadenylation sites (B), and alternative splicing of a large intron (C).
Figure EV2. CYCB1;1, CYCB1;2, and CYCB1;3 but not CYCB1;4 are expressed during seed development.
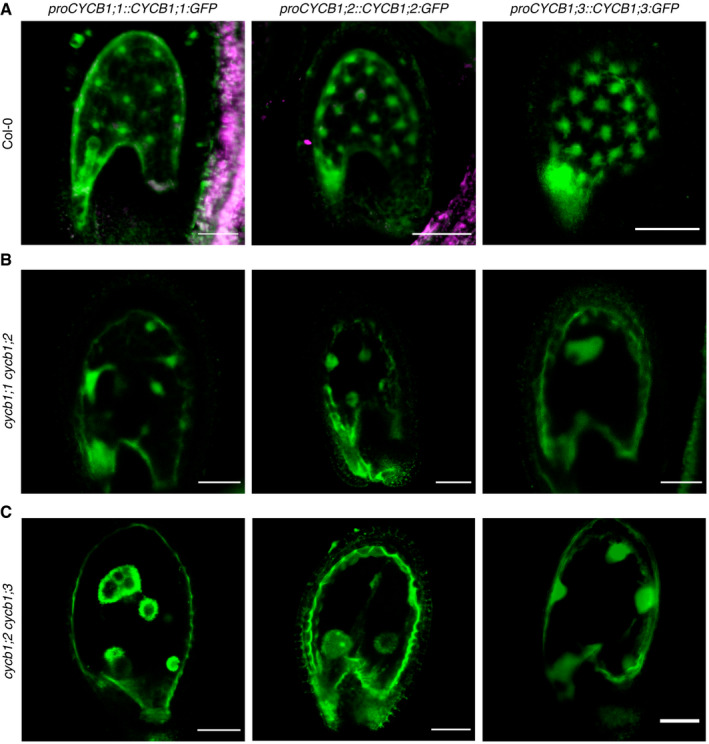
-
A–CConfocal microscope pictures of seeds expressing either proCYCB1;1:GFP, proCYCB1;2:GFP or proCYCB1;3:GFP in Col‐0 (A), cycb1;1 cycb1;2 (B), or cycb1;2 cycb1;3 (C). Scale bars: 30 μm.
Figure EV3. Oryzalin root growth assays across time in cycb1 single mutants.
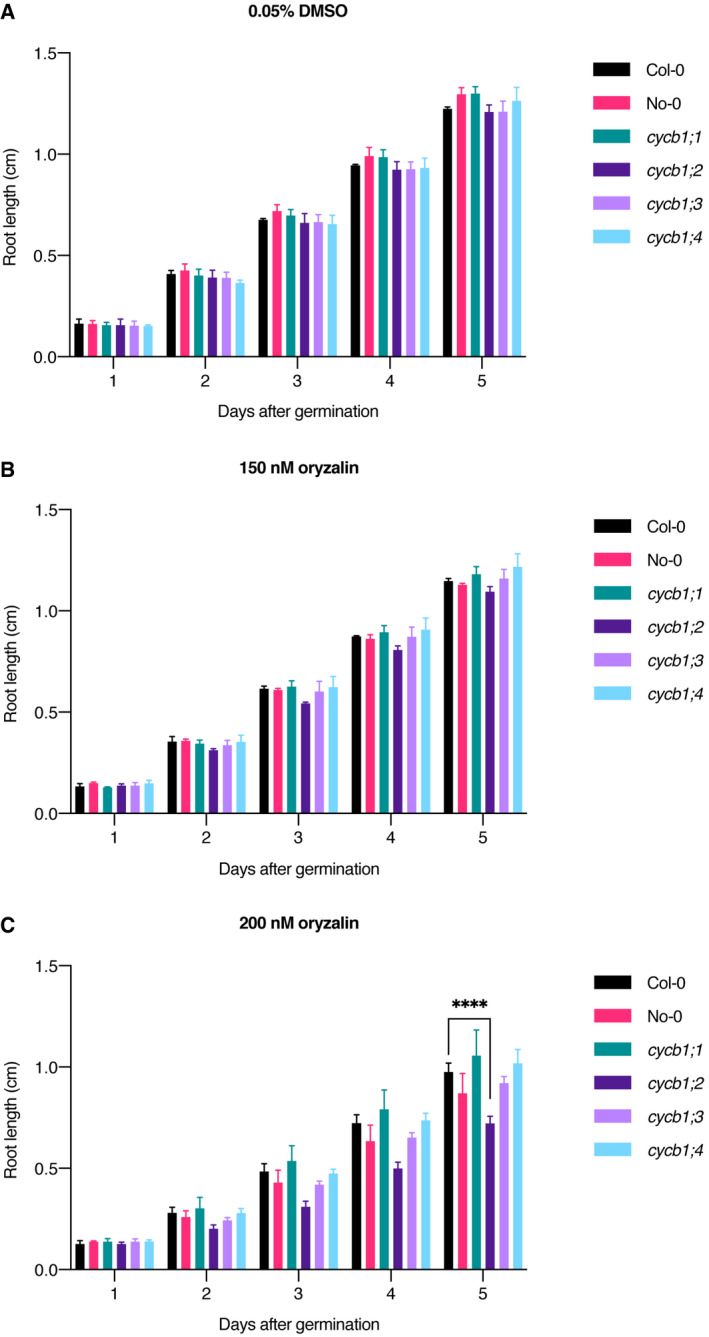
-
A–CQuantification of root length in a control condition (A), 150 nM oryzalin (B) and 200 nM oryzalin (C). Graphs show mean ± SD of three biological replicates with at least 10 plants per genotype per replicate. Asterisks indicate a significant difference in root length in a two‐way ANOVA followed by Tukey’s multiple comparisons test (****P < 0.0001).
Figure EV4. Oryzalin root growth assays across time in cycb1 double mutants.
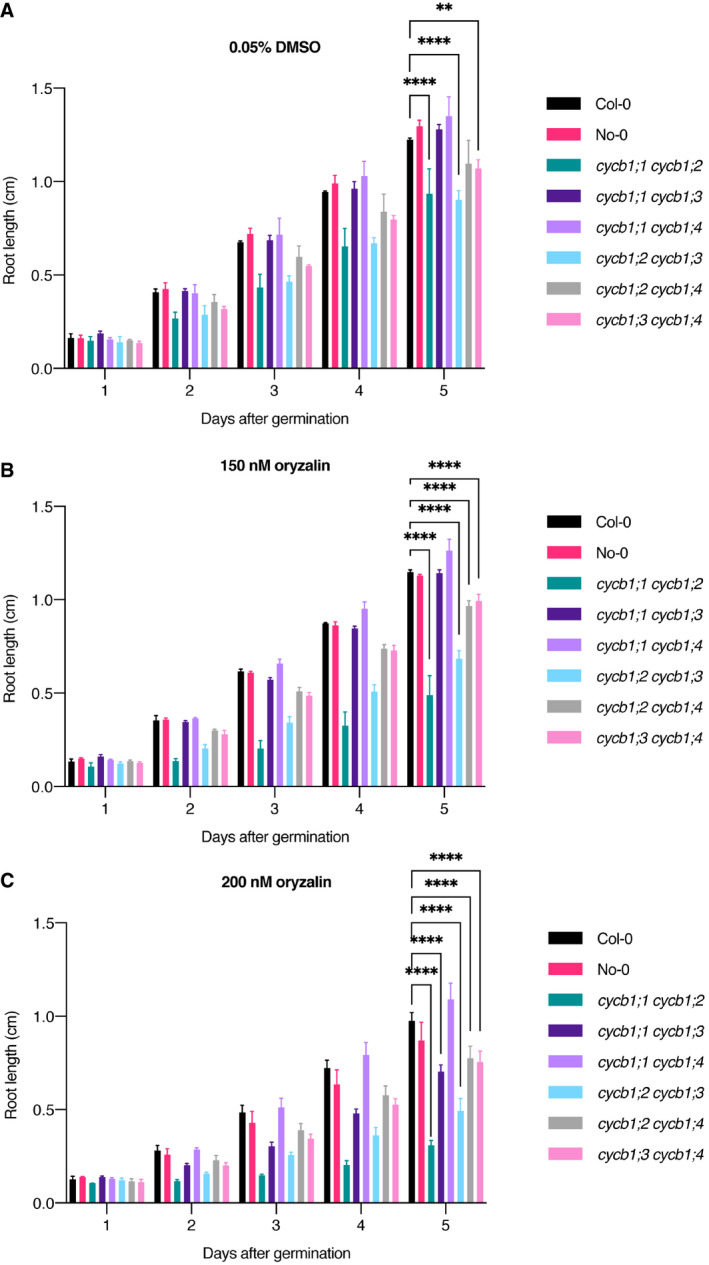
-
A–CQuantification of root length in a control condition (A), 150 nM oryzalin (B), and 200 nM oryzalin (C). Graphs show mean ± SD of three biological replicates with at least 10 plants per genotype per replicate. Asterisks indicate a significant difference in root length in a two‐way ANOVA followed by Tukey’s multiple comparisons test (**P < 0.01 and ****P < 0.0001).
Figure EV5. Ploidy analysis of young seedlings of the single and double mutants.
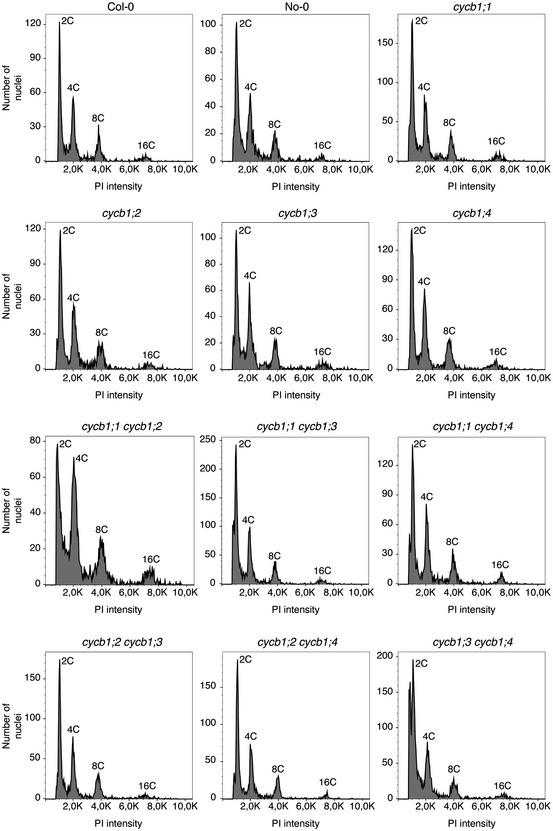
Flow cytometrical quantification of the different nuclear ploidies, as indicated by propidium iodide (PI) intensity. Single (top rows) and double (bottom rows) mutants have been analyzed. The individual genotypes are indicated on the top of each graph. Each peak has been labeled according to the expected nuclear content (2C, 4C, 8C or 16C).
The expression of the four B1‐type cyclins has been previously described in a patchy pattern in regions with high cell proliferation activity, such as in the roots (Weimer et al, 2016). Hence, these cyclins seem to be true mitotically expressed genes. Furthermore, previous genome‐wide expression studies have detected that the transcripts of all three B1‐type cyclins CYCB1;1, CYCB1;2, and CYCB1;3 are overrepresented in the developing endosperm (Day et al, 2008). In agreement, we found that the promoter reporter constructs for CYCB1;1, CYCB1;2, and CYCB1;3 but not CYCB1;4 were expressed during endosperm development (Fig EV2A).
To assess the individual biological role of B1‐type cyclins, we then analyzed previously isolated null mutants for all four B1‐type cyclins (Weimer et al, 2016). However, none of the single mutants showed an obvious deviation from the wild type under normal growth conditions, as for instance seen in root growth (Fig 1A) and seed viability (Fig 1F) in comparison to the wild type. This finding was consistent with former observations (Weimer et al, 2016). Since CYCB1;1, CYCB1;2, and CYCB1;3 reporters have similar enrichment in the proliferating endosperm (Fig EV2A), and CYCB1;1, CYCB1;2, CYCB1;3, and CYCB1;4 have a similar expression pattern in the roots, we reasoned that the B1‐type cyclins might control mitotic divisions redundantly. Therefore, we also generated and analyzed all six possible double mutant combinations. The growth of the cycb1;1 cycb1;2 double mutant was severely reduced (Fig 1B–D; for detailed characterization see below), while the size and morphology of the other double mutants were at a first look indistinguishable from the wild type.
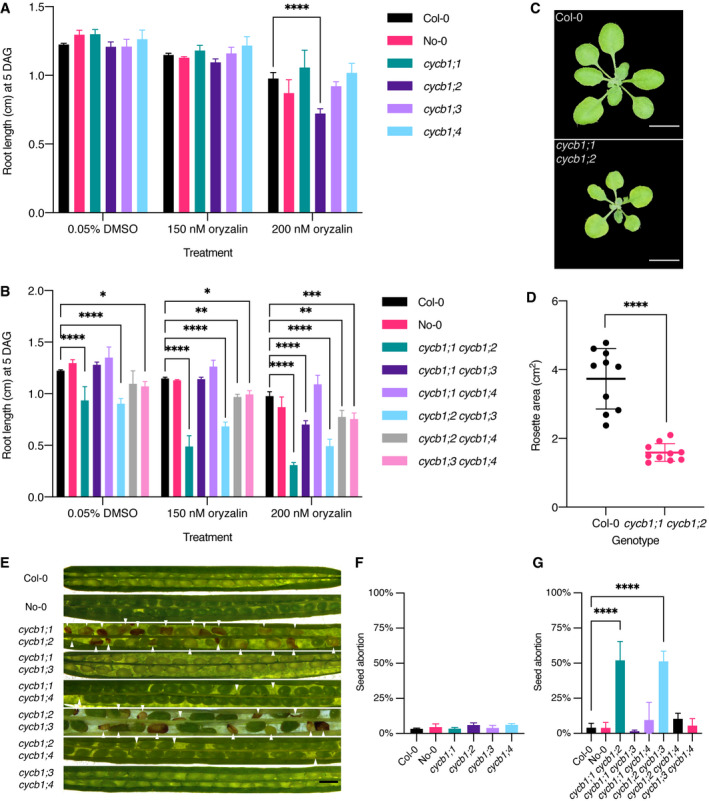
Figure 1. cycb1;1 cycb1;2 mutants show decreased root growth and shoot development as well as higher seed abortion.
-
A, BQuantification of oryzalin root growth assays in single (A) and double (B) mutants. DAG, days after germination. Graphs show mean ± SD of three biological replicates with at least 10 plants per genotype per replicate. Asterisks indicate a significant difference in root length in a two‐way ANOVA followed by Tukey’s multiple comparisons test (*P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001).
-
CRosette pictures of 20‐day‐old Col‐0 and cycb1;1 cycb1;2. Scale bar: 1 cm.
-
DQuantification of the rosette area using total leaf surface in Col‐0 and cycb1;1 cycb1;2. Graph represents the single rosette area values and the horizontal lines indicate the mean value ± SD, n = 10 plants per genotype. Asterisks indicate a significant difference in rosette area using an unpaired t‐test, t = 7.421, df = 18 (****P < 0.0001).
-
ESilique pictures of cycb1 double mutant combinations. White arrowheads indicate aborted ovules and seeds. Scale bars: 500 μm.
-
F, GQuantification of aborted seeds in single (F) and double (G) mutants. Graphs represent the average seed abortion rate per plant ± SD of three biological replicates, n = 550–1,029 seeds analyzed per genotype. Asterisks indicate significant differences in seed abortion rate in an ordinary one‐way ANOVA test, followed by a Dunnett’s multiple comparisons test (****P < 0.0001).
An analysis of the siliques in the double mutants revealed that cycb1;1 cycb1;3, cycb1;1 cycb1;4, and cycb1;3 cycb1;4 did not have a reduced seed set. In contrast, cycb1;1 cycb1;2 and cycb1;2 cycb1;3 had on average approximately half of the seeds aborted (Fig 1E and G; 52.0% ± 13.5%, n = 3 biological replicates, 550 seeds in total, for cycb1;1 cycb1;2 and 51.3% ± 7.2%, n = 3 biological replicates, 769 seeds in total, for cycb1;2 cycb1;3 vs. 4.0% ± 3.2%, n = 3 biological replicates, 821 seeds in total, for wild type, Col‐0; P < 0.0001 for both comparisons). The appearance of the aborted seeds varied in size and color (Fig 1E). Some seeds lacked the typical green color of a maturating embryo and looked transparent while others appeared brown and shriveled; in some cases, unfertilized or aborted ovules were visible.
To investigate the cause of this seed abortion, we collected, fixed and cleared seeds 3 days after pollination (DAP) (Fig 2). Since endosperm nuclei exhibit a strong autofluorescence, we were able to assess seed development quantitatively by using confocal laser scanning microscopy. In wild‐type Arabidopsis seeds, a fertilized central cell will undergo seven to eight cycles of free nuclear divisions leading to an estimated total of more than 200 nuclei in the endosperm of 3‐day‐old seeds (Boisnard‐Lorig et al, 2001). In our analysis, the general morphology of the seeds at the single mutant level seemed unchanged in comparison to the wild type (Fig 2A). However, counting the number of endosperm nuclei 3 DAP in these seeds displayed a strong reduction in endosperm divisions in the cycb1;2 mutant (Fig 2C; 94.1 ± 55.4 endosperm nuclei per seed, n = 30) in comparison to the wild type (Col‐0, 175.2 ± 43.6, n = 30; P < 0.0001).
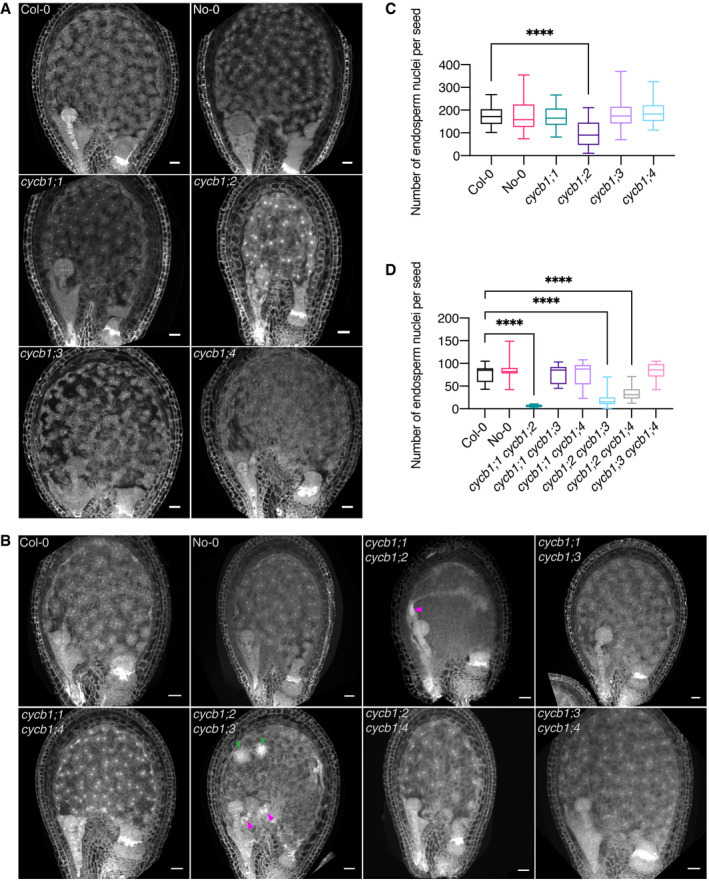
Figure 2. CYCB1 mutations delay endosperm proliferation.
-
A, BConfocal microscopy images of seeds 3 DAP. Endosperm and embryo morphology in cycb1 single (A) and double (B) mutants. Magenta arrowheads indicate enlarged endosperm nuclei, while green arrowheads indicate atypical agglomerates of endosperm nuclei. Scale bars: 25 μm.
-
C, DQuantification of endosperm nuclei in cycb1 single (C) and double (D) mutants. Boxes and whiskers represent min to max values with the median indicated as a central horizontal line, n = 26–30 seeds per genotype. Asterisks show significant differences in the number of endosperm nuclei per seed in a Kruskal–Wallis test followed by Dunn’s multiple comparisons test (****P < 0.0001).
At the double mutant level, the general morphology of cycb1;1 cycb1;2 and cycb1;2 cycb1;3 seeds appeared abnormal 3 DAP (Fig 2B). The endosperm nuclei number at this time point was dramatically reduced with only 6 ± 3 in cycb1;1 cycb1;2 (n = 26) and 28 ± 9 in cycb1;2 cycb1;3 (n = 30) in relation to 77 ± 19.3 in the wild type (Fig 2D; Col‐0, n = 30; P < 0.0001 for both comparisons). Moreover, the nuclei appeared to be extremely enlarged in cycb1;1 cycb1;2 and cycb1;2 cycb1;3 (Fig 2B; magenta arrowheads), and in cycb1;2 cycb1;3 atypical agglomerates of micro‐sized nuclei were seen (Fig 2B; green arrowheads). The strong accumulation of a reporter gene when expressed from the CYCB1;1, CYCB1;2, and CYCB1;3 promoters in the seeds of these two double mutants consistently revealed enlarged nuclei which appeared, on the basis of reporter activity, to be halted at G2/M (Fig EV2B and C). The double mutant cycb1;2 cycb1;4 also showed a decrease in endosperm nuclei number (33.4 ± 12.87, n = 30) in relation to the wild type (P < 0.0001), yet not as extensive as in the cycb1;1 cycb1;2 and cycb1;2 cycb1;3 double mutants and no major morphological abnormalities were identified, which could be explained by our observation that CYCB1;4 was never expressed in the developing endosperm, consistent with the non‐enrichment of the transcript in this tissue as shown in Day et al, 2008, and therefore CYCB1;4 might not play a major role in endosperm divisions.
Taken together, we conclude that CYCB1;2 is of major importance for the free nuclear divisions during endosperm development and acts redundantly with CYCB1;1 and CYCB1;3.
CYCB1;1, CYCB1;2, and CYCB1;4 together control female gametophyte development
Up to this point, we did not find a clear role for CYCB1;4, suggesting even higher levels of redundancy among the B1‐type cyclins or, alternatively, an overlapping function with other B‐type cyclins in Arabidopsis. To clarify the relative contribution of the four CYCB1 genes to development, we decided to investigate the role of the CYCB1 group in detail by first constructing the triple cycb1;1 −/− cycb1;3 −/− cycb1;4 −/− mutant. Notably, this triple mutant was not different from the wild type as, for instance, judged by overall growth, seed viability (Fig 3D), pollen development, and pollen viability (Fig 4C and E). This finding further underlined the paramount role of CYCB1;2 among the B1‐type cyclins. This result also indicated that CYCB1;4, if functionally relevant, may have a redundant role with either one or both of the two pairs CYCB1;1 CYCB1;2 and CYCB1;2 CYCB1;3. To test this, we generated the triple and quadruple mutant combinations cycb1;1 −/− cycb1;2 +/− cycb1;4 −/− and cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/−. While overall growth of the triple and quadruple mutant combinations (note that at least one B1 gene is not homozygous mutant in these combinations) was similar to the wild‐type, we found a strong reduction in fertility as siliques contained approximately 43% (± 0.4%, n = 3 biological replicates, 500 seeds; P < 0.0001) and 48% (± 2.1%, n = 3 biological replicates, 579 seeds; P < 0.0001) of aborting or unfertilized ovules and/or aborting seeds for cycb1;1 −/− cycb1;2 +/− cycb1;4 −/− and cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/−, respectively, in comparison to the wild type (Fig 3D and E; 0.8% ± 0.9%, n = 3 biological replicates, 487 seeds). This abortion rate suggested a female gametophytic defect and we, therefore, analyzed embryo sac development in the mutants.
Figure 3. Embryo sac development is controlled by CYCB1 members.
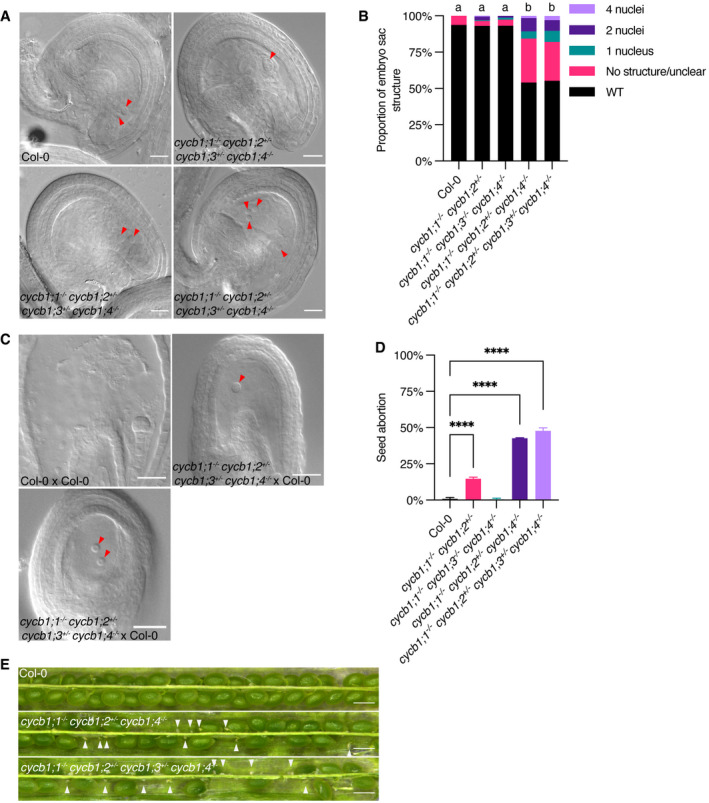
- DIC images of abnormal embryo sacs in cycb1 mutant combinations. Red arrowheads indicate the visible nuclei in Col‐0 embryo sacs (central and egg cells) and the corresponding structures in the quadruple cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutant. Scale bars: 20 μm.
- Quantification of the different abnormal embryo sac structures in cycb1 mutant combinations (n = 202–459 embryo sacs per genotype). Different letters indicate significant differences in the proportion of abnormal embryo sacs in a Chi‐squared test followed by the Marascuilo procedure to identify significant pairwise comparisons. WT, wild type.
- DIC images of embryo sacs 3 DAP with wild‐type pollen (female × male). Red arrowheads indicate the visible embryo sac nuclei in the crosses with the quadruple cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutant as a female donor, while the control Col‐0 × Col‐0 cross exhibits a developing embryo. Scale bars: 20 μm.
- Quantification of seed abortion in different cycb1 mutant combinations. Graph represents the average seed abortion rate per plant ± SD of three biological replicates, n = 463–579 seeds analyzed per genotype. Asterisks indicate significant differences in seed abortion rate in an ordinary one‐way ANOVA test, followed by a Dunnett’s multiple comparisons test (****P < 0.0001).
- Silique pictures of cycb1 triple and quadruple mutants. White arrowheads indicate early aborted ovules. Scale bars: 500 μm.
Figure 4. Pollen development is affected by mutations in the CYCB1 group.
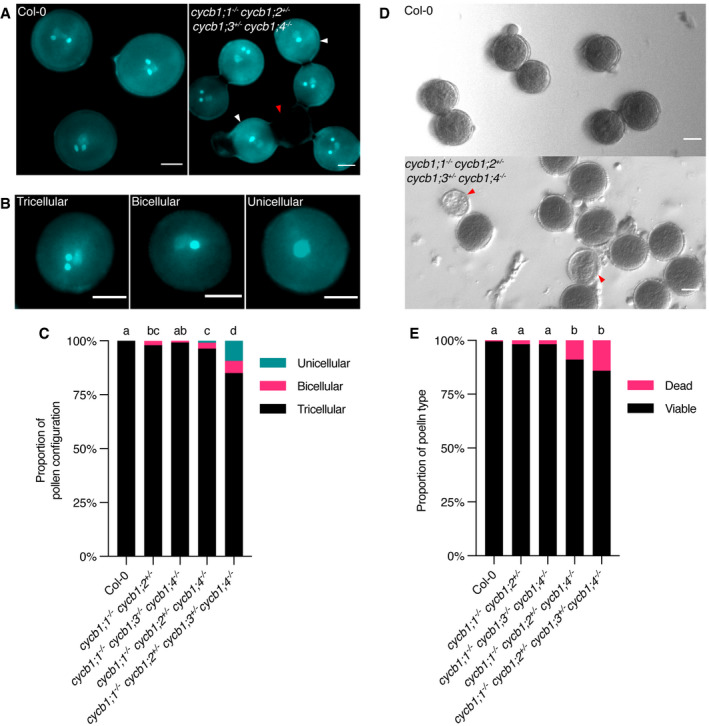
-
A, BDAPI staining of pollen in cycb1 mutants, including pollen configurations found in cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutants (B). Scale bars: 5 μm.
-
CQuantification of DAPI‐stained pollen configurations in different cycb1 mutant combinations, n = 420–616 pollen grains per genotype. Different letters indicate significant differences in the proportion of abnormal pollen (uni‐ and bicellular) in a Chi‐squared test followed by the Marascuilo procedure to identify significant pairwise comparisons.
-
DAlexander staining of mature pollen indicating pollen viability. Scale bars: 5 μm.
-
EQuantification of Alexander stained pollen viability, n = 403–498 pollen grains per genotype. Different letters indicate significant differences in the proportion of dead pollen in a Chi‐squared test followed by the Marascuilo procedure to identify significant pairwise comparisons.
Data information: Red arrowheads indicate dead pollen, while white arrowheads indicate bicellular pollen.
In wild‐type Arabidopsis plants, an embryo sac develops from a megaspore that is released after meiosis (Drews & Yadegari, 2002). Every megaspore undergoes three rounds of nuclear divisions resulting in an eight‐celled embryo sac that subsequently cellularizes. The two centrally located polar nuclei then fuse to generate the central cell nucleus while the three antipodal cells that lay at the opposite side of the egg cell undergo programmed cell death, resulting in a four‐celled mature embryo sac that consists of a large, homodiploid central cell and an egg cell (red arrowheads; Fig 3A, Col‐0) and two synergids that flank the egg cell (not shown). While this stereotypic wild‐type developmental pattern was not significantly altered in cycb1;1 −/− cycb1;2 +/− double mutant combinations, consistent with the full transmission of the mutant allele through the female gametophyte (Table 1), we found embryo sacs from cycb1;1 −/− cycb1;2 +/− cycb1;4 −/− and cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutant combinations with only one, two, or four nuclei that did not show any sign of cellularization (Fig 3A); in addition, fuzzy embryo sacs were present in 30 and 27% of the cases, respectively, likely indicating degenerating tissues, which is consistent with an early arrest of gametophytic development. In total, 46% (n = 459 embryo sacs analyzed) and 44.7% (n = 445) of embryo sacs from plants of the cycb1;1 −/− cycb1;2 +/− cycb1;4 −/− and cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− combinations, respectively, were abnormal, in comparison to 6.2% (n = 210) in the wild type (P < 0.0001) and 6.8% in the cycb1;1 −/− cycb1;3 −/− cycb1;4 −/− triple mutant (Fig 3B; P < 0.0001). The observation that the triple cycb1;1 −/− cycb1;2 +/− cycb1;4 −/− and quadruple cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutants displayed a similar number of mutant embryo sacs (Fig 3B) suggests that CYCB1;3 is not required, at least at this triple mutant level, for the divisions of the female gametophyte.
Table 1.
Transmission efficiency of the cycb1;1 and cycb1;3 mutant alleles in reciprocal crosses of a triple mutant with wild type.
| Parental genotypes (female x male) | Mutant allele | Number of seeds (n) | |
|---|---|---|---|
| cycb1;1 | cycb1;3 | ||
| cycb1;1 +/− cycb1;2 −/− cycb1;3 +/− × Col‐0 | 92% | 98% | 94 |
| Col‐0 × cycb1;1 +/− cycb1;2 −/− cycb1;3 +/− | 106% | 100% | 90 |
A transmission efficiency of 100% indicates full transmission of the mutant allele; that is, 50% of the genotyped F1 seedlings are heterozygous. A transmission efficiency may be higher than 100% if one of the two alleles is transmitted in a reduced manner, conversely making the other allele overrepresented. A z‐test for one proportion was performed to test whether the observed transmission frequencies differ from expected values and the significance level was corrected for multiple comparisons using Bonferroni. Asterisks indicate significant differences from expected values. Primers used for genotyping are listed in Table EV1.
To assess the functionality of these embryo sacs, we pollinated the quadruple mutant combination with pollen from wild‐type plants (Fig 3C). Supporting a female gametophytic defect, we observed a similar proportion of unfertilized and/or arrested embryo sacs in these crosses 3 DAP (44.3%, n = 684 seeds) in comparison to control crosses in which wild‐type plants were used as a female parent fertilized with wild‐type pollen (0.7% arrested embryo sacs, n = 597). In reciprocal control crosses with pollen from mutant plants onto stigmas of wild‐type plants, embryo and endosperm were formed in the developing seeds (seed abortion = 2.9%, n = 902 seeds). Thus, CYCB1;4, next to CYCB1;1 and CYCB1;2 appears to be required for embryo sac development. This was corroborated by analyzing the transmission of the cycb1;2 and cycb1;3 mutant alleles in reciprocal crosses of wild‐type plants with the quadruple cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutant. As expected, transmission of cycb1;2 was abolished through the female gametophyte (0%) and the efficiency in transmission was clearly reduced through the male gametophyte (70.8%) (Table 2), while the cycb1;3 allele could be transmitted without an obvious reduction in efficiency through the females (92.7%).
Table 2.
Transmission efficiency of the cycb1;2 and cycb1;3 mutant alleles in reciprocal crosses of a quadruple mutant with wild type.
| Parental genotypes (female × male) | Mutant allele | Number of seeds (n) | |
|---|---|---|---|
| cycb1;2 | cycb1;3 | ||
| cycb1;1 −/− cycb1;2+/− cycb1;3 +/− cycb1;4 −/− × Col‐0 | 0%**** | 92.7%**** | 192 |
| Col‐0 × cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− | 70.84%**** | 30.2%**** | 192 |
A transmission efficiency of 100% indicates full transmission of the mutant allele, that is, 50% of the genotyped F1 seedlings are heterozygous. A transmission efficiency may be higher than 100% if one of the two alleles is transmitted in a reduced manner, conversely making the other allele overrepresented. A z‐test for one proportion was performed to test if the observed transmission frequencies differ from expected values and the significance level was corrected for multiple comparisons using Bonferroni. Asterisks indicate significant differences from expected values (****P < 0.0001). Primers used for genotyping are listed in Table EV1.
Interestingly, the requirement of the B1‐type cyclins was different on the male side. Pollen develops from microspores through two consecutive divisions resulting in a tricellular grain that harbors two sperms next to one vegetative cell (McCormick, 2004) (Fig 4). We observed that both the cycb1;2 as well as the cycb1;3 mutant alleles were not fully transmitted through pollen in a cross of cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− with wild type plants (Table 2; 70.8 and 30.2% transmission efficiency, respectively), indicating that all four B1‐type cyclins contribute to the mitotic divisions of the developing pollen grain. Consistent with the reduced transmission, we also found pollen grains in mature anthers of cycb1;1 −/− cycb1;2 +/− cycb1;4 −/− and cycb1;1 −/− cycb1;2 +/− cycb1;3 +/− cycb1;4 −/− mutant combinations that comprised, instead of three, only two or sometimes even one cell (Fig 4A–C). Accordingly, differential staining of aborted and non‐aborted pollen showed an increased pollen abortion in the triple and quadruple mutants (Fig 4D and E) to 8.9% (n = 403 pollen grains analyzed) and 14.1% (n = 467), respectively, in relation to the wild type (Col‐0, 0.5%, n = 404; P < 0.0001).
Taken together, CYCB1;2 is also the most important B1‐type cyclin during gametophyte development. CYCB1;3 appears to have only a minor role during female gametophyte development where instead CYCB1;4 acts together with CYCB1;1 and CYCB1;2. Remarkably, after fertilization the requirement changes, as presented above, and CYCB1;3 instead of CYCB1;4 is necessary for endosperm development.
Root growth under microtubule‐destabilizing conditions underlines the redundant role of CYCB1;1, CYCB1;2, and CYCB1;3 in regulating the cytoskeleton
Considering the severe reduction of growth in cycb1;1 cycb1;2 mutants (Fig 1; see above) and to explore the role of CYCB1;2 and the other B1‐type cyclins in controlling the microtubule cytoskeleton, we next analyzed the growth of B1‐type cyclin mutants on medium containing the microtubule poison oryzalin. The rationale is that a minor defect in the mutants could become more prominent if the microtubule cytoskeleton is already slightly compromised. To that end, we compared the root growth of cycb1 single and double mutants in ½ MS medium containing 150 or 200 nM oryzalin (Fig 1A and B, respectively; Figs EV3A–C and EV4A–C). As shown above, under control conditions (0.05% DMSO), the single cycb1 mutants had similar root growth to the wild type (Fig 1A). Once oryzalin was applied at 200 nM, cycb1;2 grew significantly less (0.7 ± 0.03 cm, n = 3 biological replicates with at least 10 plants each) when compared to the wild type (Col‐0, 1.0 ± 0.04 cm, n = 3; P < 0.0001).
At the double mutant level, some combinations already showed a significantly shorter root even in control conditions (Fig 1B), such as cycb1;1 cycb1;2 (0.9 ± 0.1 cm, n = 3; P < 0.0001) and cycb1;2 cycb1;3 (0.9 ± 0.05 cm, n = 3; P < 0.0001) in comparison to the wild type (Col‐0, 1.2 ± 0.009 cm, n = 3). When 150 nM oryzalin was applied, the growth of Col‐0 was reduced by approximately 6%, while cycb1;1 cycb1;2 had a reduction of almost 50% in growth and cycb1;2 cycb1;3 had a reduction of around 24%. As the concentration of oryzalin increased to 200 nM oryzalin, the difference in growth between Col‐0 and other double mutants, such as cycb1;2 cycb1;4 and cycb1;3 cycb1;4, became more pronounced. However, most strikingly, cycb1;1 cycb1;3 mutants, which so far had shown no specific phenotype and no reduction in shoot or root growth, grew significantly shorter at 200 nM oryzalin (0.7 ± 0.04 cm, n = 3) in comparison to the wild type (Col‐0, 1.0 ± 0.04 cm, n = 3; P < 0.0001).
Collectively, a major function of all four B1‐type cyclins seems to be regulating the microtubule cytoskeleton.
CYCB1;1 and CYCB1;2 control the organization of different microtubule arrays in the roots
Following our finding that cycb1;1 cycb1;2 and cycb1;2 cycb1;3 mutants have a severe reduction in root growth both in control and especially in microtubule‐depolymerizing conditions, we performed whole mount immunolocalization studies against α‐tubulin and KNOLLE, which is a G2/M and cell plate marker (Figs 5 and 6). By analyzing mitotic divisions in the roots of these double mutants, a more detailed picture of cell division appeared.
Figure 5. The double cycb1;1 cycb1;2 mutant has abnormal microtubule arrays.
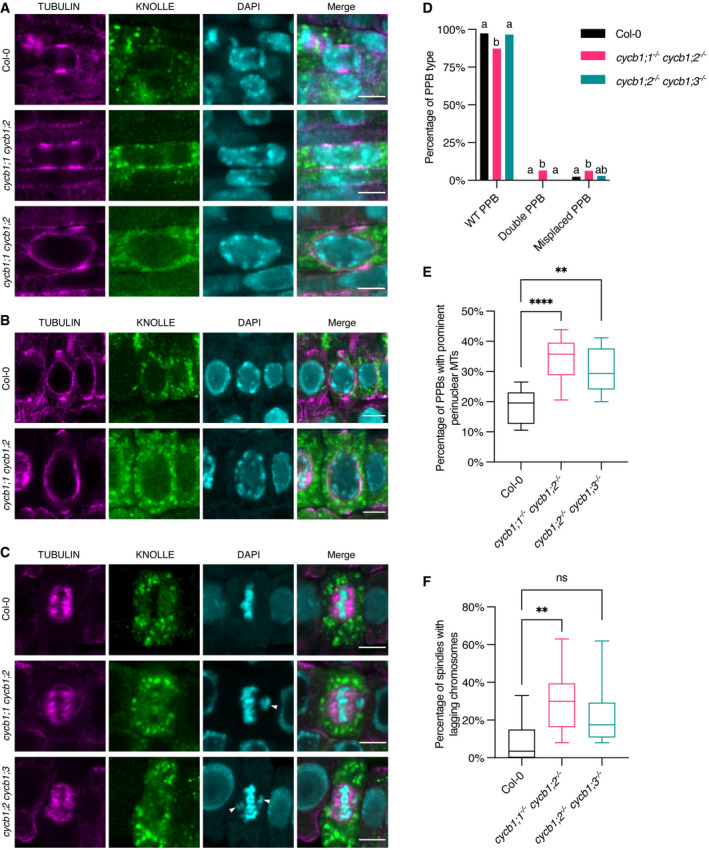
-
A–CCo‐immunolocalization against tubulin (magenta) and KNOLLE (green) in root meristematic cells. Nuclei were counterstained with DAPI for the DNA (cyan). White arrowheads indicate laggards in the metaphase stage. Scale bars: 5 μm.
-
DQuantification of wild‐type (WT), double, and misplaced PPBs. Different letters indicate significant differences in the proportions of the different arrays per category in a Chi‐squared test followed by the Marascuilo procedure to identify significant pairwise comparisons. Ten roots were analyzed per genotype.
-
EQuantification of PPBs with prominent perinuclear microtubules (MTs). Boxes and whiskers represent min to max values with the median indicated as a central horizontal line, n = 10 roots per genotype. Asterisks show significant differences in the percentage of PPBs with prominent perinuclear microtubules per root in an ANOVA test followed by Dunnett’s multiple comparisons test (**P < 0.01 and ****P < 0.0001).
-
FQuantification of spindles with lagging chromosomes. Boxes and whiskers represent min to max values with the median indicated as a central horizontal line, n = 10 roots per genotype. Asterisks show significant differences in the percentage of spindles with lagging chromosomes per root in a Kruskal–Wallis test followed by Dunn’s multiple comparisons test (**P < 0.01 and ns, non‐significant).
Source data are available online for this figure.
Figure 6. Both cycb1;1 cycb1;2 and cycb1;2 cycb1;3 have abnormal phragmoplasts and extended spindle stages.
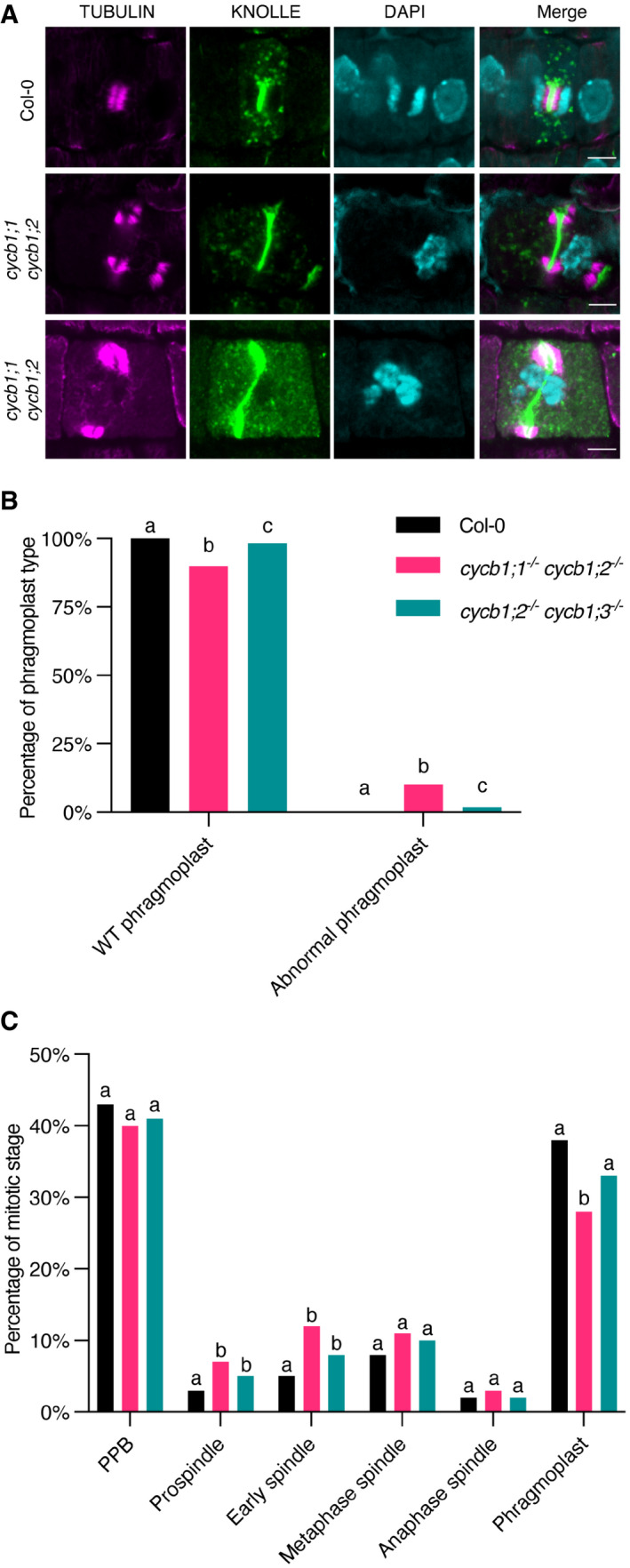
- Co‐immunolocalization against tubulin (magenta) and KNOLLE (green) in root meristematic cells. Nuclei were counterstained with DAPI for the DNA (cyan). Scale bars: 5 μm.
- Quantification of wild‐type (WT) and abnormal phragmoplasts. Different letters indicate significant differences in the proportions of the different arrays per category in a Chi‐squared test followed by the Marascuilo procedure to identify significant pairwise comparisons. Ten roots were analyzed per genotype.
- Quantification of the different mitotic stages in roots of the different genotypes. Different letters indicate significant differences in the proportions of the different arrays per category in a Chi‐squared test followed by the Marascuilo procedure to identify significant pairwise comparisons. Ten roots were analyzed per genotype.
Source data are available online for this figure.
Preceding a mitotic division, a band of microtubules that encircles the nucleus at the equatorial plane, the so‐called preprophase band (PPB), will form at the site of the future division plane. The PPB functions as a positional cue and anchoring site for proteins involved in the cell division site determination and by that contributes to the robustness of cell divisions (Schaefer et al, 2017). Following nuclear envelope breakdown, the barrel‐shaped acentrosomal spindle that is responsible for separating the chromosomes forms. After proper bipolar kinetochore‐microtubule attachments occur and enough tension is sensed, sister chromatids are pulled toward opposing poles. Next, the phragmoplast, which is a bipolar microtubule structure that expands in time toward the cell cortex, forms. The phragmoplast is a scaffold for cell wall formation on which vesicles are transported toward the microtubule‐devoid midzone, where a growing cell plate is located.
As the cell progresses from G2 toward mitosis, the nuclear surface becomes a prominent site of microtubule nucleation. In cycb1;1 cycb1;2 and cycb1;2 cycb1;3 mutants, at the PPB stage, we observed an increase in perinuclear microtubules, with an average of 18.3% PPBs with prominent microtubules in Col‐0 versus 34 and 30% in cycb1;1 cycb1;2 and cycb1;2 cycb1;3, respectively (Fig 5B and E). Either cycb1 mutations induce an early accumulation of perinuclear microtubules or else the “mature” PPB stage, that is, with perinuclear microtubules, lasts longer in the cycb1 mutants, and cells have trouble progressing into mitosis. In the stele and pericycle cells of the cycb1;1 cycb1;2 double mutant, we also observed cells harboring double PPBs and cells with misplaced PPBs, that is, PPBs that did not align properly at the equatorial plane of the nucleus. These double and misplaced PPBs were rarely seen in Col‐0 wild‐type plants; we found 6.50% double PPBs in cycb1;1 cycb1;2 in comparison to 0.22% in wild type (P < 0.0001) and 6.21% misplaced PPBs in comparison to 2.38% in wild type (P = 0.009; Fig 5A and D).
At the spindle stage, irregular chromosome configurations were observed in cycb1;1 cycb1;2, with a significantly larger number of metaphase and anaphase spindles with chromosome laggards (Fig 5C and F). Although the number of spindles with lagging chromosomes in cycb1;2 cycb1;3 was not significantly larger than in the wild type, the impairment seen in those mutants in chromosome alignment was much more severe than that of the wild‐type plants, that is, chromosomes were seen far away from the metaphase plate. Finally, abnormal phragmoplasts were observed in the two double mutant combinations, including fragmented phragmoplasts, deformed phragmoplasts around abnormally shaped nuclei, and daughter cells with incompletely separated nuclei (P < 0.0001; Fig 6A and B). These abnormal phragmoplasts were likely a consequence of the irregular chromosome alignment and segregation seen in metaphase and anaphase. In short, all microtubule arrays were affected in the double mutants to a smaller or larger degree.
Next, we analyzed the proportion of cells at PPB, spindle, and phragmoplast stages per root (Fig 6C). A significantly larger proportion of cells in both prospindle and early spindle stages were observed in cycb1;1 cycb1;2 and cycb1;2 cycb1;3 mutants (P < 0.0001), which indicates that these stages are delayed in those mutants. The phragmoplast stage is proportionally shorter in the cycb1;1 cycb1;2 mutant, although this could be a direct consequence of the extended spindle stage since, if the proportions of some stages increase, the others decrease automatically. Accordingly, a flow cytometrical analysis revealed that cycb1;1 cycb1;2 mutants have a higher proportion of 4C, 8C, and 16C nuclei in comparison to the wild type (Fig EV5), which is an indication that these mutants have longer G2 and/or M phases. An increase in polyploid cells could have two, not mutually excluding, reasons. First, a failure to undergo cytokinesis. Second, a compromised division program leading to premature exit from proliferation and entry into differentiation accompanied by endoreplication. In addition, broader peaks were observed, suggesting the formation of aneuploidies as a result of irregular mitotic divisions in this genotype.
In summary, CYCB1;1 and CYCB1;2 seem to be both redundantly required for robust root mitotic divisions under normal conditions, with CYCB1;3 playing a secondary role.
The CYCB1 group forms active complexes mainly together with CDKB2;2 and can phosphorylate a MAP
Previous studies have shown that all B1‐type cyclins can interact with all five major cell cycle CDKs from Arabidopsis, that is, CDKA;1, CDKB1;1, CDKB1;2, CDKB2;1, and CDKB2;2 (Van Leene et al, 2010). However, when we assessed the biochemical activity of all four B1‐type cyclins with CDKA;1, CDKB1;1, and CDKB2;2, as representative members of the major cell cycle CDKs, in comparative in vitro kinase assays against Histone H1, a well‐known, generic CDK substrate (Harashima & Schnittger, 2012), a more complex pattern appeared (Fig 7A and C). As a general principle, all four B1‐type cyclins build the most active complexes with CDKB2;2, which is strictly expressed in mitosis. CYCB1;1 and CYCB1;4 showed overall the highest activity levels with CDKB2;2, followed by CYCB1;2 with CDKB2;2, while the CYCB1;3‐CDKB2;2 pair was the least active among the CYCB1‐CDKB2 complexes. Although much less than in complex with CDKB2;2, CYCB1;1, CYCB1;2, and CYCB1;4 could also phosphorylate Histone H1 together with CDKB1;1, but little to no activity was found in complexes with CDKA;1. In contrast, we could not detect any activity of CYCB1;3‐CDKB1;1 complexes, while CYCB1;3 with CDKA;1 was almost as active as CYCB1;3‐CDKB2;2 pairs.
Figure 7. CYCB1‐CDKB2;2 complexes are the most active and are able to phosphorylate a MT‐nucleation factor.
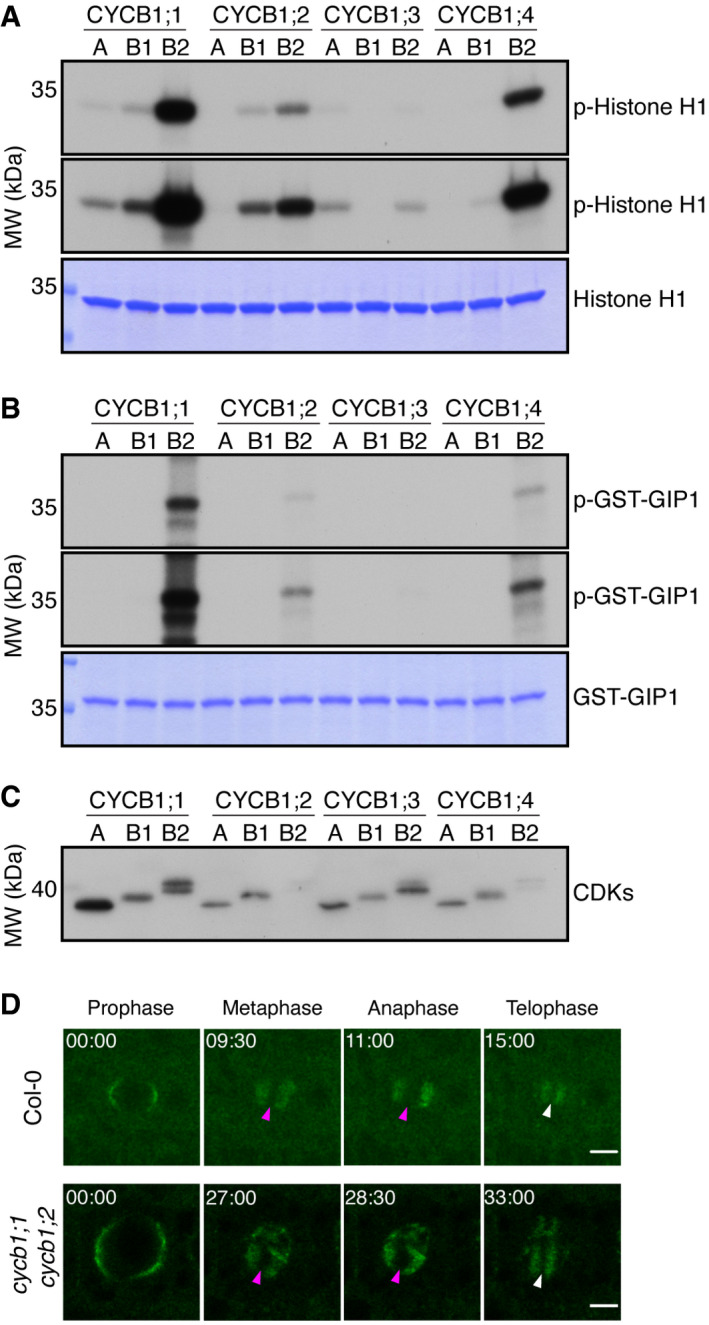
- Kinase assays against Histone H1. Top and middle panels indicate shorter and longer exposures, respectively, of the same kinase assays. Bottom panel is a CBB staining of Histone H1 showing equal loading of the protein. A: CDKA;1, B1: CDKB1;1, B2: CDKB2;2.
- Kinase assays against GIP1. Top and middle panels indicate shorter and longer exposures, respectively, of the same kinase assays. Bottom panel is a CBB staining of GIP1 showing equal loading of the protein. A: CDKA;1, B1: CDKB1;1, B2: CDKB2;2.
- Western blot against StrepIII‐tagged proteins to show loaded amounts of the CDKs. A: CDKA;1, B1: CDKB1;1, B2: CDKB2;2. Double CDKB2;2 bands are likely due to a truncation of the expressed protein.
- Time lapse of confocal microscope pictures of root meristematic cells tagged with GFP‐GIP1 in Col‐0 (top panel) and cycb1;1 cycb1;2 (bottom panel). GIP1 localizes at the nuclear polar caps, followed by co‐localization with microtubules at the spindle and phragmoplast arrays. In cycb1;1 cycb1;2 double mutants, GIP1 exhibited an abnormal localization, being found at the spindle (magenta arrowheads) and phragmoplast (white arrowheads) midzones, which are normally devoid of the protein. Scale bars: 5 μm.
The abnormal microtubule pattern observed in cycb1;1 cycb1;2 mutants was reminiscent of the defects observed in microtubule binding and organizing protein mutants, such as in gip1 gip2 double mutants (Janski et al, 2012; Nakamura et al, 2012), which are homologs of MOZART1 in animals. The gip1 gip2 double knockdown mutant displays growth defects, sterility, defective microtubule arrays, and spindles with irregular polarity, which is linked to chromosome laggards in metaphase and anaphase and aneuploidy (Janski et al, 2012). Additionally, the γ‐tubulin tubg1 tubg2 mutants display similar aberrant female and male gametophytes, with abnormal embryo sacs and reduced pollen nuclei number (Pastuglia et al, 2006). This suggested that CDK complexes containing B1‐type cyclins might phosphorylate the GIPs and/or other microtubule‐organizing proteins. Indeed, GIP1 but not GIP2 contains a consensus CDK phosphorylation site at position T67. Therefore, we expressed GIP1 in bacteria and subjected it to in vitro kinase assays with all four CYCB1 members each paired with either CDKA;1, CDKB1;1, or CDKB2;2. High activity levels against GIP1 were found for CYCB1;1, CYCB1;2, and CYCB1;4 (Fig 7B and C). However, these B1‐type cyclins phosphorylated GIP1 only in combination with CDKB2;2, highlighting the importance of both the cyclin and the CDK partner for substrate recognition in plants and further emphasizing B2‐type CDKs as the most important partners of the cyclin B1 group.
Following the finding that GIP1 is phosphorylated by CYCB1‐CDKB2;2 complexes, we decided to generate triple gip1 cycb1;1 cycb1;2 and gip2 cycb1;1 cycb1;2 mutants. However, we were never able to isolate gip2 cycb1;1 cycb1;2 mutants (Table 3). To address whether the missing triple mutant was due to a gametophytic and/or embryonic defect, we performed reciprocal crosses with gip1 −/− cycb1;1 −/− cycb1;2 +/− and gip2 −/− cycb1;1 −/− cycb1;2 +/− as male and female donors with Col‐0 (Table 4). With the exception of a reduced transmission efficiency of cycb1;2 through the female gametophyte of approximately 65% in gip2 −/− cycb1;1 −/− cycb1;2 +/− crosses with the wild type, we observed that both gip1 cycb1;1 cycb1;2 and gip2 cycb1;1 cycb1;2 gametes were largely viable and transmitted through both the female and male sides. This indicated that the triple gip2 −/− cycb1;1 −/− cycb1;2 −/− mutation is embryo lethal.
Table 3.
Distortion of cycb1;2 segregation in a gip2 −/− cycb1;1 −/− cycb1;2 +/− background.
| Genotype (selfed) | Genotype of progeny | Number of seeds (n) | ||
|---|---|---|---|---|
| cycb1;2 +/+ | cycb1;2 +/− | cycb1;2 −/− | ||
| cycb1;1 −/− cycb1;2 +/− | 25.26% | 65.26% | 9.47%** | 95 |
| gip1 −/− cycb1;1 −/− cycb1;2 +/− | 31.25% | 64.58% | 4.17%**** | 96 |
| gip2 −/− cycb1;1 −/− cycb1;2 +/− | 34.55% | 65.44% | 0%**** | 191 |
| Expected Mendelian | 25% | 50% | 25% | ‐ |
The expected Mendelian segregation reflects the proportion of F1 seedlings with the respective genotypes if the mutant alleles promote no deleterious effects. A z‐test for one proportion was performed to test if the observed homozygous mutant frequencies differ from the expected Mendelian value and the significance level was corrected for multiple comparisons using Bonferroni. Asterisks indicate significant differences from the expected value (25%) (**P < 0.01, ****P < 0.0001). Primers used for genotyping are listed in Table EV1.
Table 4.
Transmission efficiency of the cycb1;2 mutant allele in reciprocal crosses of gip1 −/− cycb1;1 −/− cycb1;2 +/− and gip2 −/− cycb1;1 −/− cycb1;2 +/− with wild type.
| Parental genotypes (female × male) | Allele | Number of seeds (n) | |
|---|---|---|---|
| Wild‐type | cycb1;2 | ||
| Col‐0 × gip1 −/− cycb1;1 −/− cycb1;2 +/− | 93.62% | 106.38% | 94 |
| Col‐0 × gip2 −/− cycb1;1 −/− cycb1;2 +/− | 111.36% | 88.64%**** | 88 |
| gip1 −/− cycb1;1 −/− cycb1;2 +/− × Col‐0 | 112.36% | 87.64%**** | 89 |
| gip2 −/− cycb1;1 −/− cycb1;2 +/− × Col‐0 | 134.78% | 65.22%**** | 92 |
A transmission efficiency of 100% indicates full transmission of the mutant allele, that is, 50% of the genotyped F1 seedlings are heterozygous. A transmission efficiency may be higher than 100% if one of the two alleles is transmitted in a reduced manner, conversely making the other allele overrepresented. A z‐test for one proportion was performed to test if the observed transmission frequencies differ from expected values and the significance level was corrected for multiple comparisons using Bonferroni. Asterisks indicate significant differences from expected values (****P < 0.0001). Primers used for genotyping are listed in Table EV1.
Based on the results of our segregation analysis and reciprocal crosses (Tables 3 and 4), the assumption that GIP1 and GIP2 are completely interchangeable is challenged. It seems likely that GIP1 but not GIP2 is regulated by a CYCB1‐dependent process. Thus, we generated a 2,849‐bp genomic GFP‐GIP1 reporter in order to follow protein localization in the cycb1;1 cycb1;2 mutant background (Fig 7D). We reasoned that, if GIP1 is indeed modulated by the CYCB1‐CDK complexes, protein localization would be impaired in a cycb1 mutant background. A cause–consequence relationship, however, is hard to establish, since defective microtubule structures as a consequence of other faulty pathways might also result in mislocalization of GIP1.
GIP1 is a microtubule nucleation factor and mainly localizes to microtubule minus ends across mitosis. At prophase, it localizes at the nuclear surface. At metaphase and anaphase, it is directed to the two spindle poles, co‐localizing with microtubule minus ends. At telophase, it localizes at the two opposing sides of the phragmoplast, directing microtubule nucleation toward the midzone. With some degree of variation between divisions, GFP‐GIP1 localization differed greatly in cycb1;1 cycb1;2 mutants in comparison to the wild type. In some cases, GFP‐GIP1 was found to remain in the spindle midzone (Fig 7D; magenta arrowheads) during metaphase in abnormal mitotic divisions. The resulting phragmoplast, which is normally devoid of GIP1 in its midzone (Fig 7D; white arrowheads), also contained remaining GIP1. The duration of these abnormal mitotic divisions in the double mutant was also around double the time of the wild‐type divisions (Fig 7D). Thus, we conclude that B1‐type cyclins and in particular CYCB1;2 might control microtubule organization through the regulation of GIP1 and more likely through several additional substrates.
Discussion
Angiosperms have undergone an extensive expansion of the cyclin family in comparison to yeast and mammals, containing for instance a total of 10 different cyclin families with more than 50 protein‐encoding cyclin genes in Arabidopsis (Wang et al, 2004; Jia et al, 2014). For most of these genes, functional information is still lacking. Although genetic dissection of such an enlarged number of cyclin members may be more challenging and require the construction of multiple mutant combinations, it also provides an opportunity to study the function of these cyclins in compromised, yet viable mutant combinations of redundantly acting genes. In contrast, mutants for the CycB1 in mice, for example, are not viable and die in utero making its analysis, especially at the developmental level, challenging (Brandeis et al, 1998). Here, we have functionally dissected the group of B1‐type cyclins and created various double and multiple mutant combinations. In particular the combination of cycb1;1 and cycb1;2 proved to be a valuable tool to study the function of this class of cyclins.
Endosperm—a demanding structure
During plant development, many different cell cycle programs are executed (Jakoby & Schnittger, 2004). One of the most particular proliferation modes are the free nuclear divisions during endosperm proliferation (Berger et al, 2006). Despite its importance for seed growth and embryo nutrition, there is currently very little known about the cell cycle machinery that drives these free nuclear divisions. Laser dissection microscopy‐based transcriptional profiling of Arabidopsis endosperm revealed that B1‐type cyclins are among the most prominently expressed cell cycle regulators in this tissue (Day et al, 2008). Consistently, we found that nuclear divisions are reduced in cycb1;2 single mutants and aberrant mitotic divisions appear in cycb1;1 cycb1;2 and cycb1;2 cycb1;3 double mutants. Correspondingly, seed development defects were reported as an effect of silencing cyclin B1 expression in rice (Guo et al, 2010).
Strikingly, the phenotypes of cycb1;1 cycb1;2 and cycb1;2 cycb1;3 double mutant endosperm closely resemble the defects seen in mutants for ENDOSPERM DEFECTIVE 1 (EDE1), a plant‐specific microtubule‐binding protein (Pignocchi et al, 2009). EDE1 contains short CDK consensus phosphorylation sites (S/T‐P) and so far has not been identified in CDK substrate searches in Arabidopsis (Pusch et al, 2012; Harashima et al, 2016). However, EDE1 could be phosphorylated by human Cdk complexes in in vitro kinase assays and it is known that short CDK consensus sites are sufficient to be phosphorylated by CDK/cyclin complexes (Ubersax et al, 2003; Pignocchi & Doonan, 2011). Interestingly, many cytoskeletal components are highly expressed in proliferating endosperm tissue and the free nuclear divisions might be very sensitive to alterations in cytoskeleton function, providing a possible reason why these divisions are apparently more sensitive to the loss of CYCB1 function (Day et al, 2009). Endosperm development in Arabidopsis might thus advance as a model system to study cell biological questions. However, endosperm is difficult to access, since it is buried in maternal structures, such as the seed coat and the silique. Therefore, morphological analyses always require mechanical preparation steps. In this light, the identification of cycb1;1 cycb1;2 homozygous double mutants represents a unique tool to investigate the control of mitosis in roots or other much more easily accessible plant tissues than gametophytes.
B1‐type cyclins and the control of microtubule nucleation
Microtubules nucleate from ring‐shaped complexes that contain γ‐tubulin, and a family of related proteins called γ‐tubulin complex proteins (GCPs). The composition of γ‐tubulin ring complexes (γTURC) varies between organisms: budding yeast contains only the γ‐tubulin small complex (γTUSC), with two molecules of γ‐tubulin, and one each of GCP2 and GCP3 (Vinh et al, 2002). On the other hand, animal nucleating complexes are made of multiple copies of the γTUSC plus GCP4, GCP5, and GCP6, as well as other non‐GCP constituents, such as GIP1/MOZART1, MOZART2, and NEDD1, which is a localization factor (Tovey & Conduit, 2018). In plants, γ‐tubulin complexes contain all GCP subunits, the GIP1/MOZART1 protein, and a NEDD1 homolog (Lee & Liu, 2019).
The dynamic assembly and disassembly of the microtubule network generally runs in parallel with the cell cycle and, for example, even strong defects in microtubule arrays, for example, lack of the PPB formation (Schaefer et al, 2017), do not block the cell cycle. However, rearrangements of the microtubule cytoskeleton in plant cells are obviously coupled with the cell cycle. Specific microtubule arrays accompany each stage of the cell cycle, either in interphase (the interphase cortical microtubule array), in pre‐mitosis (the PPB), or in mitosis (spindle and phragmoplast). Moreover, several observations indicate a tight—at least temporal—coordination of both cycles. For instance, the PPB is formed in the late G2‐prophase in somatic tissues. Rapid PPB dismantling precisely coincides with nuclear envelope breakdown and entry into metaphase. Prospindle and spindle formation also takes place at precise stages of the cell cycle. Likewise, the phragmoplast is precisely initiated at telophase from remnants of the spindle. However, very little is currently known about the molecular mechanisms of how this coordination is achieved. Interestingly, several cell cycle regulators including CDKA;1 have been identified at microtubule arrays such as the PPB, spindle, and phragmoplast (Boruc et al, 2010). Our finding that GIP1/MOZART1 is phosphorylated by CDKB2‐CYCB1 complexes offers a potential mechanism of how the cell cycle might orchestrate microtubule assembly. Interestingly, double PPBs and asymmetric PPBs, as we report here to be present in cycb1;1 cycb1;2 mutants, have also been described in a gip1 gip2 double knockdown mutant previously, further strengthening that CYCB1 controls the cytoskeleton via regulation of the γTURC complex components (Janski et al, 2012). The exact mechanism by which this regulation happens, including if phosphorylation of GIP1 at T67 plays an important role and if there is indeed a differential phosphorylation regulation of GIP1 and GIP2, remains to be explored.
Moreover, other factors of the γTURC have also been found to be phosphorylated in animals and yeast. For instance, all core units of the γTURC (γ‐tubulin, GCP2‐GCP6, GCP‐WD, and GCP8/MOZART2), but GIP1/MOZART1, have been found to be phosphorylated in mammals (Teixidó‐Travesa et al, 2012). In particular, CDKs were shown to phosphorylate γTURC components including γ‐tubulin and others in yeast and Nedd1 in humans (Zhang et al, 2009; Keck et al, 2011). However, the functional importance of these phosphorylation sites is not understood and an analysis of microtubule dynamics in animals is complicated due to the lethality of core cell cycle regulators such as Cdk1 or CycB1 (Brandeis et al, 1998; Santamaría et al, 2007).
In plants, all of the core γTURC components (GIP1, GCP2, GCP3, GCP4, GCP5a, GCP5b, NEDD1, and γ‐tubulin 1 as well as γ‐tubulin 2), but GIP2, have at least one CDK consensus phosphorylation site, and for NEDD1 and GCP4 as well as GCP5a a phosphorylated Ser/Thr in a consensus CDK site has been deposited in the PhosPhAt database (http://phosphat.uni‐hohenheim.de). In addition, CYCB1;3 has been found to bind to GCP3 and γ‐tubulin 1 (Van Leene et al, 2010). Thus, the regulation of the γTURC complex by CYCB1s likely goes even beyond the phosphorylation of GIP1 reported here.
The CYCB1 group has an evolutionarily conserved role in microtubule networks
Mammalian CycB1 is mainly cytoplasmic in interphase, rapidly accumulates in the nucleus at the end of prophase, and associates with the mitotic apparatus in the course of mitosis, that is, chromatin, microtubules, kinetochores, and centrosomes (Toyoshima et al, 1998; Yang et al, 1998; Hagting et al, 1999; Bentley et al, 2007). Loss of the CycB1 function in mice results in very early embryo lethality (Brandeis et al, 1998). In contrast, mammalian cyclin B2 (CycB2) localizes mostly to the Golgi apparatus in both interphase and metaphase and CycB2 knockout mice are viable (Jackman et al, 1995; Brandeis et al, 1998; Draviam et al, 2001). However, knocking down both CycB1 and CycB2 in HeLa cells showed a redundant function for both cyclins (Soni et al, 2008). Cyclin B3 is only poorly expressed in mitotic cells, but its mRNA is readily observed in both male and female meiosis (Lozano et al, 2002; Nguyen et al, 2002).
Interestingly, CycB1 in mammals localizes to the outer plate of the kinetochore at prometaphase and later on to the spindle poles following microtubule attachment to kinetochores (Bentley et al, 2007; Chen et al, 2008). Reduction of CycB1 by the use of RNA interference results in the irregular attachment of kinetochores to microtubules, chromosome alignment defects and delays anaphase (Chen et al, 2008), which is reminiscent of the chromosome alignment and segregation problems in addition to the extended spindle stages we found in cycb1;1 cycb1;2 mutants.
In contrast to many other eukaryotes, the setup of interphasic and mitotic microtubule networks in flowering plants is not driven by microtubule‐organizing centers containing centrioles/basal bodies. Instead, it has been proposed that mitotic microtubule networks nucleate from chromatin. Consistent with a role in microtubule nucleation, CYCB1;1 and CYCB1;2 have been found to be present mainly at chromatin during mitosis, while CYCB1;3 is localized to both chromatin and cytoplasm and CYCB1;4 is localized mainly in the cytoplasm as well as in the region of the cytoplasm that co‐localizes with the mitotic spindle (Bulankova et al, 2013). Thus, although the CYCB1 group in Arabidopsis appears from a general point of view to regulate the mitotic microtubule network similarly to CycB1 in mammals, the localization of B1‐type cyclins is different in both species, indicating that the work of B‐type cyclins in different species is differently distributed among its members.
Remarkably, and in contrast to CycB1 localization in mammals, CYCB1;1, CYCB1;2, and CYCB1;3 were not found at the mitotic spindle. We cannot rule out at the moment that B1‐type cyclins do not have a function in further organizing the mitotic spindle. However, it seems likely that other, yet to be characterized subgroups of mitotic cyclins, in particular the B2 and B3 group, might play a key role here especially since a recent analysis of CYCB3;1 found that this cyclin is localized to the spindle, at least in meiosis (Sofroni et al, 2020). With this, it will be exciting to have eventually a complete view on B‐type cyclin function in Arabidopsis.
Materials and Methods
Plant material and growth conditions
The accessions Columbia (Col‐0) and Nossen (No‐0) were used as wild type controls. The single cycb1;1, cycb1;2, cycb1;3, and cycb1;4 mutants were previously described and characterized (Weimer et al, 2016). The cycb1;3 T‐DNA insertion is in a No‐0 background. The gip1 (GABI_213D01) and gip2 (FLAG_36406) mutants were also previously characterized (Janski et al, 2012). Genotyping primers are listed in Table EV1.
Arabidopsis thaliana seeds were sown on half‐strength (½) Murashige and Skoog (MS) medium (basal salt mixture, Duchefa Biochemie) containing 0.5% sucrose and 0.8% plant agar (Duchefa Biochemie) at pH 5.8. Seeds were either sterilized with chlorine gas or by liquid sterilization. For the liquid sterilization, a 2% bleach, 0.05% Triton X‐100 solution was applied for 5 min, followed by three washing steps with sterile distilled water and the addition of 0.05% agarose. Stratification of the seeds on plates was performed at 4°C for 2–3 days in the dark. Plants were initially grown in vitro at 22°C in a 16‐h light regime and subsequently transferred to soil with a 16‐h light/21°C and 8‐h/18°C dark regime with 60% humidity.
Quantitative PCR
Total RNA was isolated from plant tissues using the RNeasy Plant Mini Kit (Qiagen). Dnase (TaKaRa) treatment was performed to avoid DNA contamination and RNA concentration was measured using a Nanodrop ND‐1000 instrument. A total of 3.5 µg of total RNA was reverse transcribed using SuperScript® III reverse transcriptase kit (Invitrogen). An additional step of Rnase H treatment at 37°C for 20 min was performed to eliminate the remaining RNA. The cDNA was further purified and concentrated by using QIAquick PCR Purification Kit (Qiagen) and the concentration was determined by Nanodrop ND‐1000 instrument. Finally, using cDNA as the template, qPCR was performed on a Light‐cycler LC480 instrument (Roche) as per the manufacturer’s instructions.
Plasmid construction and plant transformation
To analyze the expression of the CYCB1 group in seeds, we used previously generated promoter reporter lines for CYCB1;1 to CYCB1;4 fused at the N‐terminus to GFP (Weimer et al, 2016).
To generate a PROGIP1:GFP:GIP1 construct, a 2,849‐bp genomic region including the native promoter and terminator was amplified by PCR and integrated into a pENTR‐D‐TOPO vector. A SmaI restriction site was introduced before the first ATG codon of the GIP1 CDS. After linearization of the construct by restriction digest with SmaI, a ligation with GFP was performed, followed by LR reaction with the destination vector pGWB501. The constructs were transformed in Arabidopsis thaliana by floral dipping.
Flow cytometry assay
Ten 7‐day‐old seedlings per genotype were chopped with a new razorblade in homogenization buffer (45 mM MgCl2, 20 mM MOPS, 30 mM sodium citrate, 0.1% Triton X‐100, pH 7.0), followed by filtration through a 15‐μm nylon mesh. After that, propidium iodide (Sigma) and Rnase A (Sigma) were added to final concentrations of 50 μg/ml and 10 μg/ml, respectively. Samples were left on ice for 5 min and then analyzed in a S3e Cell Sorter (Bio‐Rad) with a laser excitation at 488 nm. The scatterplots were analyzed and processed using the FlowJo software.
Endosperm nuclei proliferation analysis
Flower buds were initially emasculated before the visible maturation and release of pollen. Emasculated flowers were then hand pollinated with pollen from the same genotype after 2–3 days. After 3 days, siliques were dissected and fixed in a solution of 4% glutaraldehyde in 12.5 mM cacodylate buffer, pH 6.8, followed by vacuum application for 20 min and storage at 4°C overnight. The following day, individual seeds were mounted on microscope slides containing a clearing 1:8:2 glycerol:chloral hydrate:water solution and stored at 4°C overnight. Imaging was performed with a Zeiss LSM 780 or 880 confocal microscope with excitation at 488 nm and detection between 498 and 586 nm and Z‐stacks were analyzed using the Fiji software.
Pollen staining
To identify single nuclei in mature pollen, pollen grains were released into a DAPI staining solution (2.5 μg/ml DAPI, 0.01% Tween, 5% dimethyl sulfoxide, 50 mM Na phosphate buffer, pH 7.2) and incubated at 4°C overnight. Pollen viability was analyzed by mounting pollen as previously described (Alexander, 1969). Imaging was performed with a Zeiss Axioimager.
Embryo sac analysis
Mature ovules and developing seeds were prepared from siliques before and 3 days after fertilization, respectively, mounted on microscope slides on a clearing 1:8:2 glycerol:chloral hydrate:distilled water solution and kept at 4°C overnight before analysis as previously described (Nowack et al, 2006). Differential Interference Contrast (DIC) imaging was performed on a Zeiss Axioimager.
Microtubule cytoskeleton dynamics in roots
Meristematic cell divisions in the root were observed in 5–7 day‐old seedlings under a layer of ½ MS medium using a Leica TCS SP8 inverted confocal microscope.
Immunostaining
Roots of 4‐day‐old Arabidopsis seedlings were fixed in 4% paraformaldehyde and 0.1% Triton X‐100 in ½ MTSB buffer (25 mM PIPES, 2.5 mM MgSO4, 2.5 mM EGTA, pH 6.9) for 1 h under vacuum, then rinsed in PBS 1X for 10 min. Samples were then permeabilized in ethanol for 10 min and rehydrated in PBS for 10 min. Cell walls were digested using the following buffer for 1 h: 2 mM MES pH 5, 0.20% driselase and 0.15% macerozyme. Tissues were hybridized overnight at room temperature with the B‐5‐1‐2 monoclonal anti‐α‐tubulin (Sigma) and the anti‐KNOLLE antibody (kind gift of G. Jürgens, University of Tübingen, Germany; Lauber et al, 1997). The next day, tissues were washed for 15 min in PBS, 50 mM glycine, incubated with secondary antibodies (Alexa Fluor 555 goat anti‐rabbit for KNOLLE antibody and Alexa Fluor 488 goat anti‐mouse for the tubulin antibody) overnight and washed again in PBS, 50 mM glycine, and DAPI 20 ng/ml. Tissues were mounted in VECTASHIELD and DAPI and viewed using an SP8 confocal laser microscope (Leica Microsystems). Samples were excited sequentially at 405 nm (DAPI), 488 nm (@TUB/Alexa Fluor 488), and 561 nm (@KNOLLE/Alexa Fluor 555), with an emission band of 420–450 nm (DAPI), 495–545 nm (Alexa Fluor 488), and 560–610 nm (Alexa Fluor 555) using a PMT for DAPI imaging, and hybrid detectors for MT and KNOLLE imaging. All stacks have been imaged using the same zoom (x 1.60) with a pixel size xyz of 200 nm × 200 nm × 500 nm. KNOLLE is particularly useful when mutants do not form a PPB to unambiguously identify cells at the G2/M stage, although this was not the case in our study.
A blind counting was set up to count mitotic MT arrays seen in 10 roots of each genotype. The “Cell counter” plugin was used to count the occurrence of MT arrays within each root stack (https://imagej.nih.gov/ij/plugins/cell‐counter.html).
Protein expression and purification and in vitro kinase assays
Purified GIP1 was kindly donated by Nicolas Baumberger (IBMP, Strasbourg). GIP1 CDS was cloned into the pGEX‐2TK vector (GE Healthcare; courtesy of Etienne Herzog) and transformed into the BL21(DE3) E. coli strain. An overnight culture cultivated at 37°C was used to inoculate an expression culture at an OD600 of 0.1. The expression culture was grown at 37°C and 250 rpm until it reached an OD600 of 0.6. Afterwards, 0.5mM IPTG was added and the growth continued at 37°C for 6 h. Cells were collected by centrifugation at 5,000 g for 15 min and the pellet resuspended in 50 mM Tris pH 8, 300 mM NaCl, 5% glycerol, 5 mM EDTA, 0.1% Tween 20. Cells were lyzed by sonication and the lysate clarified by centrifugation at 10,000 g for 20 min at 4°C. GST‐GIP was purified by passage onto a glutathione‐sepharose GSTrap HP 1ml column (GE Healthcare) with 50 mM Tris pH 7.5, 10 mM MgCl2, 100 mM NaCl as an equilibration/washing buffer and 50 mM Tris pH 7.5, 10 mM MgCl2, 100 mM NaCl plus 10 mM reduced glutathione as an elution buffer. Elution fractions were analyzed on polyacrylamide gel and concentrated by ultrafiltration before being frozen and stored at −80°C. Histone H10 was purchased from NEB. In vitro kinase assays were performed as described previously (Harashima & Schnittger, 2012).
Oryzalin root growth assays
Plants were sown on ½ MS medium containing either 0.05% DMSO as a control or oryzalin. 100 mM oryzalin stock solutions were prepared in DMSO and stored at −20°C and further diluted to a final concentration of 150 nM or 200 nM for the root assays. Root growth was recorded daily up until 5 days after germination, when plates were scanned, and root length was subsequently measured using the Fiji software. Three biological replicates with at least 10 plants per genotype were performed. The mean root length of each individual experiment was determined and again averaged.
Statistical analysis
The employed statistical tests are indicated in the figure legends. Statistical tests were performed using the GraphPad Prism 9 software and the XLSTAT plugin for Microsoft Excel. The distribution of the measured values was tested beforehand, for example, by the Anderson–Darling test. If the distribution was significantly different from a normal distribution, a non‐parametric test was employed. Significance levels are P ≥ 0.05 (ns), P < 0.05 (*), P < 0.01 (**), P < 0.001 (***), P < 0.0001 (****). In the case of the Chi‐squared test followed by the Marascuilo procedure, significant pairwise comparisons are indicated by letters.
Author contributions
MM, XZ, MP, FR, HH, SK, MH, and AS designed the research; MM, XZ, MP, KB, FR, MKo, HH, SK, MKu, and PB performed the experiments; MM, XZ, and MP performed the statistical analysis; MM, XZ, MP, KB, FR, MKo, HH, SK, MKu, PB, MH, KR, DB, and AS analyzed and discussed the data; DB, KR, and AS provided material and reagents, MM and AS wrote the article; MM, XZ, MP, FR, MKo, HH, SK, MKu, PB, MH, KR, DB, and AS revised and approved the article.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Expanded View Figures PDF
Table EV1
Source Data for Figure 5
Source Data for Figure 6
Acknowledgments
This work was supported by a grant from the Ohsumi Frontier Science Foundation to S.K., a fellowship from the International Graduate School in Genetics and Functional Genomics, University of Cologne to M.Ku., by the Ministry of Education, Youth, and Sports of the Czech Republic, European Regional Development Fund‐Project REMAP (grant CZ.02.1.01/0.0/0.0/15_003/0000479) to K.R., by the Human Frontier Science Program to D.B. and A.S., and by a grant from the Deutsche Forschungsgemeinschaft (SCHN 736/2‐2) to A.S. Open access funding enabled and organized by Projekt DEAL.
EMBO reports (2022) 23: e53995.
Data availability
This study includes no data deposited in external repositories.
References
- Alexander MP (1969) Differential staining of aborted and nonaborted pollen. Biotech Histochem 44: 117–122 [DOI] [PubMed] [Google Scholar]
- Bentley AM, Normand G, Hoyt J, King RW (2007) Distinct sequence elements of Cyclin B1 promote localization to chromatin, centrosomes, and kinetochores during mitosis. Mol Biol Cell 18: 4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Grini PE, Schnittger A (2006) Endosperm: an integrator of seed growth and development. Curr Opin Plant Biol 9: 664–670 [DOI] [PubMed] [Google Scholar]
- Blethrow JD, Glavy JS, Morgan DO, Shokat KM (2008) Covalent capture of kinase‐specific phosphopeptides reveals Cdk1‐cyclin B substrates. Proc Natl Acad Sci U S A 105: 1442–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisnard‐Lorig C, Colon‐Carmona A, Bauch M, Hodge S, Doerner P, Bancharel E, Dumas C, Haseloff J, Berger F (2001) Dynamic analyses of the expression of the histone: YFP fusion protein in Arabidopsis show that syncytial endosperm is divided in mitotic domains. Plant Cell 13: 495–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J, Mylle E, Duda M, de Clercq R, Rombauts S, Geelen D, Hilson P, Inzé D, van Damme D, Russinova E (2010) Systematic localization of the Arabidopsis core cell cycle proteins reveals novel cell division complexes. Plant Physiol 152: 553–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudolf V, Lammens T, Boruc J, Van Leene J, Van Den Daele H, Maes S, Van Isterdael G, Russinova E, Kondorosi E, Witters E et al (2009) CDKB1;1 forms a functional complex with CYCA2;3 to suppress endocycle onset. Plant Physiol 150: 1482–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandeis M, Rosewell I, Carrington M, Crompton T, Jacobs MA, Kirk J, Gannon J, Hunt T (1998) Cyclin B2‐null mice develop normally and are fertile whereas cyclin B1‐null mice die in utero. Proc Natl Acad Sci U S A 95: 4344–4349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulankova P, Akimcheva S, Fellner N, Riha K (2013) Identification of Arabidopsis meiotic cyclins reveals functional diversification among plant cyclin genes. PLoS Genet 9: e1003508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Zhang X, Jiang Q, Clarke PR, Zhang C (2008) Cyclin B1 is localized to unattached kinetochores and contributes to efficient microtubule attachment and proper chromosome alignment during mitosis. Cell Res 18: 268–280 [DOI] [PubMed] [Google Scholar]
- Cools T, Iantcheva A, Weimer AK, Boens S, Takahashi N, Maes S, van den Daele H, van Isterdael G, Schnittger A, de Veylder L (2011) The Arabidopsis thaliana checkpoint kinase WEE1 protects against premature vascular differentiation during replication stress. Plant Cell 23: 1435–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC (2008) Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiol 148: 1964–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Müller S, Macknight RC (2009) Identification of cytoskeleton‐associated genes expressed during Arabidopsis syncytial endosperm development. Plant Signal Behav 4: 883–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Scofield S, Alcasabas AA, Maughan SC, Menges M, Braun N, Collins C, Nieuwland J, Prinsen E, Sundaresan V et al (2007) Arabidopsis CYCD3 D‐type cyclins link cell proliferation and endocycles and are rate‐limiting for cytokinin responses. Proc Natl Acad Sci U S A 104: 14537–14542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, Pusch S, de Schutter K, Kamei CLA, Nowack MK, Novak B, Duan GL, Zhu YG, de Veylder L et al (2009) Control of cell proliferation, organ growth, and DNA damage response operate independently of dephosphorylation of the Arabidopsis Cdk1 Homolog CDKA;1. Plant Cell 21: 3641–3654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N, Weimer AK, de Veylder L, Novak B, Schnittger A (2010) The regulatory network of cell cycle progression is fundamentally different in plants versus yeast or metazoans. Plant Signal Behav 5: 1613–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerner P, Jørgenser JE, You R, Steppuhn J, Lamb C (1996) Control of root growth and development by cyclin expression. Nature 380: 520–523 [DOI] [PubMed] [Google Scholar]
- Draviam VM, Orrechia S, Lowe M, Pardi R, Pines J (2001) The localization of human cyclins B1 and B2 determines CDK1 substrate specificity and neither enzyme requires MEK to disassemble the Golgi apparatus. J Cell Biol 152: 945–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drews GN, Yadegari R (2002) Development and function of the angiosperm female gametophyte. Annu Rev Genet 36: 99–124 [DOI] [PubMed] [Google Scholar]
- Furuno N, Den EN, Pines J (1999) Human cyclin A is required for mitosis until mid prophase. J Cell Biol 147: 295–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Wang F, Song J, Sun W, Zhang XS (2010) The expression of Orysa;CycB1;1 is essential for endosperm formation and causes embryo enlargement in rice. Planta 231: 293–303 [DOI] [PubMed] [Google Scholar]
- Haase SB, Winey M, Reed SI (2001) Multi‐step control of spindle pole body duplication by cyclin‐dependent kinase. Nat Cell Biol 3: 38–42 [DOI] [PubMed] [Google Scholar]
- Hagting A, Jackman M, Simpson K, Pines J (1999) Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation‐dependent nuclear import signal. Curr Biol 9: 680–689 [DOI] [PubMed] [Google Scholar]
- Hagting A, Karlsson C, Clute P, Jackman M, Pines J (1998) MPF localization is controlled by nuclear export. EMBO J 17: 4127–4138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Dissmeyer N, Hammann P, Nomura Y, Kramer K, Nakagami H, Schnittger A (2016) Modulation of plant growth in vivo and identification of kinase substrates using an analog‐sensitive variant of CYCLIN‐DEPENDENT KINASE A;1. BMC Plant Biol 16: 1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harashima H, Schnittger A (2012) Robust reconstitution of active cell‐cycle control complexes from co‐expressed proteins in bacteria. Plant Methods 8: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman M, Firth M, Pines J (1995) Human cyclins B1 and B2 are localized to strikingly different structures: B1 to microtubules, B2 primarily to the Golgi apparatus. EMBO J 14: 1646–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoby M, Schnittger A (2004) Cell cycle and differentiation. Curr Opin Plant Biol 7: 661–669 [DOI] [PubMed] [Google Scholar]
- Janski N, Masoud K, Batzenschlager M, Herzog E, Evrard JL, Houlné G, Bourge M, Chabouté ME, Schmit AC (2012) The GCP3‐interacting proteins GIP1 and GIP2 are required for γ‐tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell 24: 1171–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia RD, Guo CC, Xu GX, Shan HY, Kong HZ (2014) Evolution of the cyclin gene family in plants. J Syst Evol 52: 651–659 [Google Scholar]
- Keck JM, Jones MH, Wong CCL, Binkley J, Chen D, Jaspersen SL, Holinger EP, Xu T, Niepel M, Rout MP et al (2011) A cell cycle phosphoproteome of the yeast centrosome. Science 332: 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T (1999) Cyclin‐dependent kinase control of centrosome duplication. Proc Natl Acad Sci U S A 96: 2817–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber MH, Waizenegger I, Steinmann T, Schwarz H, Mayer U, Hwang I, Lukowitz W, Jürgens G (1997) The Arabidopsis KNOLLE protein is a cytokinesis‐specific syntaxin. J Cell Biol 139: 1485–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YRJ, Liu B (2019) Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells. New Phytol 222: 1705–1718 [DOI] [PubMed] [Google Scholar]
- Van Leene J, Hollunder J, Eeckhout D, Persiau G, Van De Slijke E, Stals H, Van Isterdael G, Verkest A, Neirynck S, Buffel Y, et al (2010) Targeted interactomics reveals a complex core cell cycle machinery in Arabidopsis thaliana . Mol Syst Biol 6: 397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A, Rodríguez‐Bravo V, Medema RH (2009) The decision to enter mitosis: feedback and redundancy in the mitotic entry network. J Cell Biol 185: 193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano JC, Schatt P, Peaucellier G, Picard A, Perret E, Arnould C (2002) Molecular cloning, gene localization, and structure of human cyclin B3. Biochem Biophys Res Commun 291: 406–413 [DOI] [PubMed] [Google Scholar]
- McCormick S (2004) Control of male gametophyte development. Plant Cell 16: 142–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menges M, De Jager SM, Gruissem W, Murray JAH (2005) Global analysis of the core cell cycle regulators of Arabidopsis identifies novel genes, reveals multiple and highly specific profiles of expression and provides a coherent model for plant cell cycle control. Plant J 41: 546–566 [DOI] [PubMed] [Google Scholar]
- Morgan DO (1997) Cyclin‐dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13: 261–291 [DOI] [PubMed] [Google Scholar]
- Nakamura M, Yagi N, Kato T, Fujita S, Kawashima N, Ehrhardt DW, Hashimoto T (2012) Arabidopsis GCP3‐interacting protein 1/MOZART 1 is an integral component of the γ‐tubulin‐containing microtubule nucleating complex. Plant J 71: 216–225 [DOI] [PubMed] [Google Scholar]
- Nakayama KI, Nakayama K (2005) Regulation of the cell cycle by SCF‐type ubiquitin ligases. Semin Cell Dev Biol 16: 323–333 [DOI] [PubMed] [Google Scholar]
- Nguyen TB, Manova K, Capodieci P, Lindon C, Bottega S, Wang XY, Refik‐Rogers J, Pines J, Wolgemuth DJ, Koff A (2002) Characterization and expression of mammalian cyclin B3, a prepachytene meiotic cyclin. J Biol Chem 277: 41960–41969 [DOI] [PubMed] [Google Scholar]
- Nowack MK, Grini PE, Jakoby MJ, Lafos M, Koncz C, Schnittger A (2006) A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet 38: 63–67 [DOI] [PubMed] [Google Scholar]
- O’Farrell PH (2001) Triggering the all‐or‐nothing switch into mitosis. Trends Cell Biol 11: 512–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastuglia M, Azimzadeh J, Goussot M, Camilleri C, Belcram K, Evrard JL, Schmit AC, Guerche P, Bouchez D (2006) γ‐tubulin is essential for microtubule organization and development of Arabidopsis . Plant Cell 18: 1412–1425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Doonan JH (2011) Interaction of a 14‐3‐3 protein with the plant microtubule‐associated protein EDE1. Ann Bot 107: 1103–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignocchi C, Minns GE, Nesi N, Koumproglou R, Kitsios G, Benning C, Lloyd CW, Doonan JH, Hills MJ (2009) Endosperm Defective1 is a novel microtubule‐associated protein essential for seed development in Arabidopsis . Plant Cell 21: 90–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusch S, Harashima H, Schnittger A (2012) Identification of kinase substrates by bimolecular complementation assays. Plant J 70: 348–356 [DOI] [PubMed] [Google Scholar]
- Riabowol K, Draetta G, Brizuela L, Vandre D, Beach D (1989) The cdc2 kinase is a nuclear protein that is essential for mitosis in mammalian cells. Cell 57: 393–401 [DOI] [PubMed] [Google Scholar]
- Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M (2007) Cdk1 is sufficient to drive the mammalian cell cycle. Nature 448: 811–815 [DOI] [PubMed] [Google Scholar]
- Schaefer E, Belcram K, Uyttewaal M, Duroc Y, Goussot M, Legland D, Laruelle E, De Tauzia‐Moreau ML, Pastuglia M, Bouchez D (2017) The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356: 186–189 [DOI] [PubMed] [Google Scholar]
- Schnittger A, Schöbinger U, Bouyer D, Weinl C, Stierhof YD, Hülskamp M (2002) Ectopic D‐type cyclin expression induces not only DNA replication but also cell division in Arabidopsis trichomes. Proc Natl Acad Sci U S A 99: 6410–6415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K, Joubès J, Cools T, Verkest A, Corellou F, Babiychuk E, Van Der Schueren E, Beeckman T, Kushnir ST, Inzé D et al (2007) Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofroni K, Takatsuka H, Yang C, Dissmeyer N, Komaki S, Hamamura Y, Böttger L, Umeda M, Schnittger A (2020) CDKD‐dependent activation of CDKA;1 controls microtubule dynamics and cytokinesis during meiosis. J Cell Biol 219: 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni DV, Sramkoski RM, Lam M, Stefan T, Jacobberger JW (2008) Cyclin B1 is rate limiting but not essential for mitotic entry and progression in mammalian somatic cells. Cell Cycle 7: 1285–1300 [DOI] [PubMed] [Google Scholar]
- Teixidó‐Travesa N, Roig J, Lüders J (2012) The where, when and how of microtubule nucleation ‐ one ring to rule them all. J Cell Sci 125: 4445–4456 [DOI] [PubMed] [Google Scholar]
- Tovey CA, Conduit PT (2018) Microtubule nucleation by γ‐tubulin complexes and beyond. Essays Biochem 62: 765–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E (1998) Nuclear export of cyclin B1 and its possible role in the DNA damage‐induced G2 checkpoint. EMBO J 17: 2728–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson JJ, Novak B (2001) Regulation of the eukaryotic cell cycle: molecular antagonism, hysteresis, and irreversible transitions. J Theor Biol 210: 249–263 [DOI] [PubMed] [Google Scholar]
- Ubersax JA, Woodbury EL, Quang PN, Paraz M, Blethrow JD, Shah K, Shokat KM, Morgan DO (2003) Targets of the cyclin‐dependent kinase Cdk1. Nature 425: 859–864 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Raes J, De Veylder L, Rouzé P, Rombauts S, Inzé D (2002) Genome‐wide analysis of core cell cycle genes in Arabidopsis . Plant Cell 14: 903–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Coppens F, Lee E, Donner TJ, Xie Z, Van Isterdael G, Dhondt S, De Winter F, De Rybel B, Vuylsteke M et al (2011) Developmental regulation of CYCA2s contributes to tissue‐specific proliferation in Arabidopsis . EMBO J 30: 3430–3441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinh DBN, Kern JW, Hancock WO, Howard J, Davis TN (2002) Reconstitution and characterization of budding yeast γ‐tubulin complex. Mol Biol Cell 13: 1144–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Kong H, Sun Y, Zhang X, Zhang W, Altman N, DePamphilis CW, Ma H (2004) Genome‐wide analysis of the cyclin family in Arabidopsis and comparative phylogenetic analysis of plant cyclin‐like proteins. Plant Physiol 135: 1084–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer AK, Biedermann S, Harashima H, Roodbarkelari F, Takahashi N, Foreman J, Guan Y, Pochon G, Heese M, Van Damme D et al (2016) The plant‐specific CDKB 1‐ CYCB 1 complex mediates homologous recombination repair in Arabidopsis . EMBO J 35: 2068–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Goshima G (2017) Mitotic spindle assembly in land plants: molecules and mechanisms. Biology 6: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Bardes ESG, Moore JD, Brennan J, Powers MA, Kornbluth S (1998) Control of Cyclin B1 localization through regulated binding of the nuclear export factor CRM1. Genes Dev 12: 2131–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen Q, Feng J, Hou J, Yang F, Liu J, Jiang Q, Zhang C (2009) Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the γTuRC to the centrosome. J Cell Sci 122: 2240–2251 [DOI] [PubMed] [Google Scholar]


