Key Points
Question
Can common genetic variants associated with essential tremor (ET) be identified?
Findings
In this genome-wide association study and meta-analysis including genetic data on 483 054 individuals, 5 genome-wide significant loci were associated with risk of ET and common variants were associated with approximately 18% of ET heritability.
Meaning
Findings of this study may help identify new genes and inform ET biology.
This genome-wide association study identifies common variants associated with risk of essential tremor.
Abstract
Importance
Essential tremor (ET) is one of the most common movement disorders, affecting 5% of the general population older than 65 years. Common variants are thought to contribute toward susceptibility to ET, but no variants have been robustly identified.
Objective
To identify common genetic factors associated with risk of ET.
Design, Setting, and Participants
Case-control genome-wide association study. Inverse-variance meta-analysis was used to combine cohorts. Multicenter samples collected from European populations were collected from January 2010 to September 2019 as part of an ongoing study. Included patients were clinically diagnosed with or reported having ET. Control individuals were not diagnosed with or reported to have ET. Of 485 250 individuals, data for 483 054 passed data quality control and were used.
Main Outcomes and Measures
Genotypes of common variants associated with risk of ET.
Results
Of the 483 054 individuals included, there were 7177 with ET (3693 [51.46%] female; mean [SD] age, 62.66 [15.12] years), and 475 877 control individuals (253 785 [53.33%] female; mean [SD] age, 56.40 [17.6] years). Five independent genome-wide significant loci and were identified and were associated with approximately 18% of ET heritability. Functional analyses found significant enrichment in the cerebellar hemisphere, cerebellum, and axonogenesis pathways. Genetic correlation (r), which measures the degree of genetic overlap, revealed significant common variant overlap with Parkinson disease (r, 0.28; P = 2.38 × 10−8) and depression (r, 0.12; P = 9.78 × 10−4). A separate fine-mapping of transcriptome-wide association hits identified genes such as BACE2, LRRN2, DHRS13, and LINC00323 in disease-relevant brain regions, such as the cerebellum.
Conclusions and Relevance
The results of this genome-wide association study suggest that a portion of ET heritability can be explained by common genetic variation and can help identify new common genetic risk factors for ET.
Introduction
Essential tremor (ET) is a complex neurological disorder affecting 1% of the general population and up to 5% of individuals older than 65 years.1,2 ET is clinically characterized as a bilateral, largely symmetric kinetic or postural tremor,3 which can greatly decrease the quality of life and debilitate daily function. Previous studies have implicated the cerebellum as a putative region of interest for ET.4,5 Specifically, abnormalities of Purkinje cells have been observed in postmortem brain tissue obtained from individuals with ET.6 Several transcriptomic studies and imaging studies have also highlighted the importance of the cerebellum in ET.4,7,8
The genetic etiology of ET remains elusive, despite twin studies that have shown the trait to be heritable.9,10,11 For instance, 1 twin study indicated ET had a concordance of 69% to 93% in monozygotic twins and 27% to 29% in dizygotic twins.9 Studies in which individuals with ET in the same family were sequenced have also implicated specific genes.12,13,14 For instance, rare variants in FUS and TENM4 were found to segregate in large families but the lack of replication suggests they are potentially private variants.15,16 Past genome-wide association studies (GWAS) have also identified putative ET loci, but none of these loci were statistically significant at a genome-wide level, likely owing to the size of the cohorts examined.17,18 The loci from these GWAS implicated nearby genes, such as STK32B and LINGO1, for which subsequent replication studies were conducted.19,20,21,22,23 However, most of these GWAS loci had conflicting replication results and were conducted in smaller cohorts.
Here, we present a genome-wide meta-analysis identifying the first genome-wide significant loci for ET using a cohort of 7177 individuals with ET and 475 877 control individuals. We additionally identified novel loci, implicated tissue-relevant genes, and found a significant genetic overlap between Parkinson disease (PD) and ET. This report supported the heritable nature of ET and implicated new disease-relevant loci and genes.
Methods
Statistical Analyses
Sample Description
The meta-analysis included cohorts in North America and Europe, totaling 7177 individuals with ET and 475 877 control individuals. This population was further grouped into cohorts based on study cohort, chip, and time of genotyping as described in the eMethods in Supplement 1. Individual clinical diagnoses are described previously or in the eMethods in Supplement 1. The review board at the McGill University Health Center Research Ethics Board approved the study protocols (reference number: IRB00010120). Written informed consent was obtained from all participants.
Genotyping and Quality Control
The cohorts were genotyped and followed standardized quality control, imputation, and postimputation quality control. Samples were removed if there was greater than 2% missingness, autosomal heterozygous deviation (F < 0.2), or failed sex check. Low quality single-nucleotide variants (SNVs) were removed based on Hardy-Weinberg equilibrium (P > 1 × 10−6) and SNV missingness less than 0.02 after sample removal. Samples were mapped against the 1000 Genomes Project phase 3 reference panel after pruning and removing SNVs from high–linkage disequilibrium (LD) regions, and only individuals of inferred European ancestry were retained owing to low sample size for individuals of non-European ancestry. No relatedness filter was applied because a linear mixed model was subsequently used to account for relatedness. Imputation was done using the Sanger Imputation Server with Eagle version 2.3.5 and the Haplotype Reference Consortium Reference Panel version 1.1.24 Further details on cohort and quality control for the UK BioBank and 23andMe data sets are described in the eMethods in Supplement 1.
Genome-Wide Association
A bayesian linear mixed model was done using BOLT-LMM 2.3.4, including 20 principal components and sex as covariates to accelerate convergence.25 The noninfinitesimal model was used if there was an increase in power. Subsequently, the data were meta-analyzed using an inverse-variance–weighted fixed-effects model with METAL.26 Only markers with an effective sample size N = 4 / (1 / No. affected + 1/No. controls) > 70% were retained, leaving a total of 6 892 661 variants (eTable 12 in Supplement 9).27
SNV Heritability and Partitioning of Heritability
To determine SNV heritability on the liability scale, the slope of the LD Score regression (LDSC) was calculated with individuals of European ancestry from the 1000 Genomes Project.28 The effects of confounding factors were determined by assessing the deviation of the LDSC intercept from 1. Specifically, the ratio between the (intercept −1) divided by the (mean χ2 − 1) indicated confounding other than polygenicity.28 Heritability was partitioned by different tissue, cell, and functional sets using the LDSC.29 The 1000 Genomes Project cohort was used for LD and allele frequencies.
Genetic Correlation
Genetic correlation was calculated for ET and other GWAS traits using LD Hub.30 This platform uses the LDSC to broadly assess multiple traits with publicly available GWAS. Traits with an updated GWAS were replaced, as defined by a larger sample size and/or a more recently published GWAS. Only traits with European ancestry were retained and data with low relative z scores (as reported by LD Hub) were excluded. SNVs from the major histocompatibility complex region were removed for traits. One of any duplicate traits was retained, prioritizing the most recent study or largest sample size.
Conditional Analysis
To determine whether there were any genome-wide significant loci with multiple independent signals, genome-wide complex trait analysis–conditional and joint analysis was used.31 The program takes the ET summary statistics and conditions genome-wide significant lead SNVs while using the LD of a reference panel. Here, the raw genotyping data from control samples and the CARTaGENE cohort were used as the reference panel.32 A stepwise approach was used to condition the top independent SNVs (P < 5 × 10−8) and a minor allele frequency greater than 0.01.
Multitrait Analysis
To increase power, we did a multitrait analysis of GWAS (MTAG), using phenotypes with significant positive correlation (PD and depression).33,34,35 The MTAG program was used to conduct the analysis. MTAG jointly meta-analyzed summary statistics from PD and depression with ET to increase power to identify ET-specific associations. Increase in power was defined as (multitrait analysis GWAS mean χ2 −1) / (non–multitrait analysis GWAS mean χ2 −1) × 100%.
Multitrait Conditional Analysis
To assess the association between PD and ET, a multitrait conditional analysis was done using multitrait conditional and joint analysis, adjusting ET by PD. Summary statistics from Nalls et al34 were used. Multitrait conditional and joint analysis removed pleiotropic signal with PD from the ET GWAS. Typically, most pleiotropic loci should have reduced conditional effect sizes, but trait-specific effects would have larger conditional effects.
Gene-Based, Gene-Set, and Tissue-Set Enrichment Analyses
P values that quantify the genic associations and gene-set enrichment for ET were calculated using MAGMA version 1.08 as implemented in FUMA (https://fuma.ctglab.nl).36,37 Bonferroni correction was applied for the number of genes (N = 18 517) tested with a threshold of P = 2.70 × 10−6. Enrichment among GTEx version 8 was also done using FUMA, with a significance threshold of P = 9.26 × 10−4.
Transcriptome-Wide Association
To identify genes influenced by cis-eQTLs, a transcriptome-wide association study (TWAS) was done using FUSION. Brain imputation panels were used from the Genotype-Tissue Expression (GTEx) project and the CommonMind Consortium.38,39 The 1000 Genomes Project version 3 LD panel was used for TWAS. Bonferroni-adjusted P values less than .05 were considered transcriptome-wide significant. A brain omnibus test was done to test for effect across reference panels, which accounts for pairwise correlation between features. A Bonferroni threshold was used for the omnibus (0.05 / 7221) × (number of genes tested).
To address coregulation in TWAS, fine-mapping of causal gene sets (FOCUS) was used for genome-wide significant loci to model predicted expression correlations and assign a posterior probability for causality in the previously mentioned imputation panels.40 FOCUS identified genes for each TWAS signal to be included in a 90% credible set.
Gene-set analyses were done using GeneNetwork version 2.0 (https://genenetwork.nl), which leverages RNA-sequencing data (n = 31 499) to provide coregulated genes within each pathway.41 Genes meeting a Bonferroni-adjusted P value less than .05 were used. Agnostic analyses of pathways in databases such as Reactome and GO were used.
To assess potential colocalization between the top significant loci with eQTLs, FUMA was used to map eQTLs with the top significant loci. Data from GTEx 53 version 8 for brain tissue and the CommonMind Consortium were used. All SNV-gene pairs of cis-eQTLs that were nominally significant were included (P < .05). The sign of the original eQTL data indicates the direction of effect for the tested allele. The lead SNVs from the 5 genome-wide significant loci were compared with the eQTL data.
Phenome-Wide Association
To investigate whether any top loci were associated with other phenotypes, the SNVs were assessed on the genetics Open Targets Platform (http://genetics.opentargets.org) and a PheWeb for the UK BioBank imaging data (https://open.win.ox.ac.uk/ukbiobank/big40/pheweb/).
Bivariate Gaussian Mixture Modeling of Polygenicity
To determine the univariate estimate of non-null SNVs (polygenicity) and shared polygenicity with PD, MiXeR version 1.3 was used on the summary statistics of ET and PD.42 In the cross-trait analysis, MiXeR modeled additive genetic effects as a combination of the following components: SNVs not influencing either ET or PD, SNVs influencing only 1 of 2 traits, or SNVs influencing both traits. After fitting the model, the dice coefficient (a parameter that estimates the proportion of overlapping variants), was calculated.
Results
Genome-Wide Significant ET Risk Loci
For the GWAS, samples from 14 different clinical centers and 2 biobanks were included (eMethods in Supplement 1). The cohorts were divided by genotyping array, leading to a total of 4 genotyping cohorts. The first clinical cohort was genotyped on the Axiom Genome-Wide CEU 1 Array (Affymetrix). The second clinical cohort was genotyped on the Illumina GSA array. The 2 biobank cohorts were from 23andMe and the UK BioBank. Prior to analysis, stringent quality control was performed on the data and ancestrally predicted Europeans were retained based on the 1000 Genomes Project phase 3 reference panel. Independent cohorts were meta-analyzed using an inverse-variance–weighted fixed-effects model. Of the 483 054 samples included, there were 7177 individuals with ET (3693 [51.46%] female; mean [SD] age, 62.66 [15.12] years), and 475 877 control individuals (253 785 [53.33%] female; mean [SD] age, 56.40 [17.6] years). Subsequently, variants with an effective sample size greater than 70% of the full meta-analysis were retained, leaving 6 892 661 markers.
From the meta-analyzed GWAS, the heritability explained by SNVs, was estimated to be 0.1829 (standard error [SE], 0.0141) using LDSC. The LDSC intercept, which indicates the degree of inflation due to confounding, was 1.051 (SE, 0.00074), which suggests low levels of confounding. The attenuation ratio, which assesses the degree of inflation due to polygenicity instead of confounding, was 0.14 (SE, 0.036), suggesting most inflation was due to polygenicity (eFigure 1 in Supplement 1).28 The genomic inflation factor (λ1000) was also determined to be 1.01. Genetic correlation, which captures the degree of genetic overlap with another cohort or trait, was calculated between cohorts using LDSC. Across the cohorts, correlations were significant and positive, providing evidence that effects were consistent across cohort designs (eTable 1 in Supplement 1). The clinical cohorts had a genetic correlation of 0.88 (SE, 0.20; P = 1.86 × 10−5), and these clinical cohorts respectively had a genetic correlation of 0.96 ± 0.148 (P = 9.34 × 10−11) and 0.52 ± 0.14 (P = 1.21 × 10−4) with the 23andMe cohort (eTable 1 in Supplement 1). The lower genetic correlation of 0.52 was between the clinical samples genotyped on the Illumina Array and 23andMe. Owing to the small effective sample size of the UK Biobank cohort, pairwise genetic correlation was not calculated.
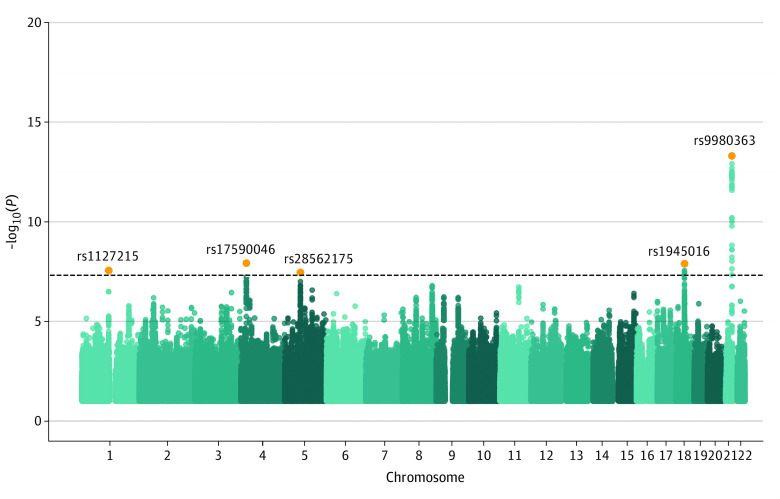
A total of 5 genome-wide significant loci (P < 5 × 10−8) were identified (Figure 1; Table). None of the of the top loci were found to be significantly heterogeneous (eTable 2 in Supplement 1). Furthermore, there were no additional independent secondary genome-wide significant signals found after conditioning on the lead SNVs iteratively.
Figure 1. Manhattan Plot of Genome-Wide Association Study of 7177 Individuals With Essential Tremor and 475 877 Control Individuals.
The x-axis shows chromosome number (chromosomes 1-22) and points are ordered by genomic position. The y-axis shows the statistical significance of each variant, represented as -log10(P). The black-dashed line indicates the genome-wide significance threshold P = 5 × 10-8). Lead SNPs are highlighted in orange.
Table. Lead Single-Nucleotide Variants (SNVs) for Genome-Wide Significant Loci in 7177 Individuals With Essential Tremor and 475 877 Control Individuals.
| CHR | Position (hg19) | rsID | Alleles (eff/ref) | Odds ratio (95% CI) | z Score | P value |
|---|---|---|---|---|---|---|
| 1 | 117532790 | rs1127215 | C/T | 1.09 (1.06 to 1.13) | 5.55 | 2.756 × 10−08 |
| 4 | 24362541 | rs17590046 | C/T | 0.89 (0.85 to 0.93) | −5.70 | 1.180 × 10−08 |
| 5 | 67827456 | rs28562175 | C/T | 0.92 (0.88 to 0.95) | −5.51 | 3.483 × 10−08 |
| 18 | 37207175 | rs1945016 | G/T | 1.10 (1.06 to 1.13) | 5.69 | 1.255 × 10−08 |
| 21 | 42520134 | rs9980363 | C/T | 1.16 (1.12 to 1.20) | 7.52 | 4.921 × 10−14 |
Abbreviations: CHR, chromosome; eff, effective allele; hg19, human genome version 19; ref, reference; rsID, reference SNV number.
Transcriptome-Wide Associations
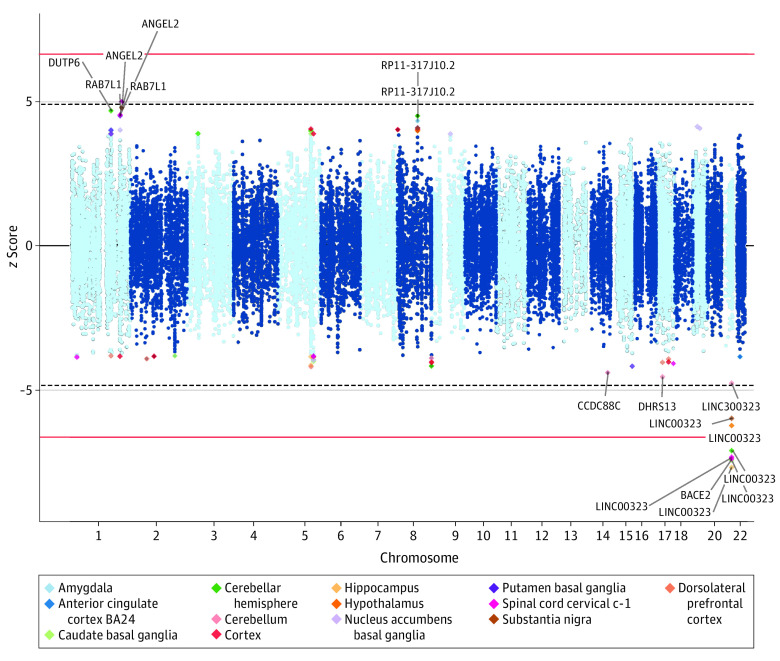
A TWAS was conducted using FUSION by leveraging brain data from the GTEx and the CommonMind Consortium.38,39,43 The TWAS identified genes that were predicted to have altered their expression due to ET-associated common variants. The Bonferroni-significant hits were BACE2 and LINC00323 (Figure 2; eTable 3 in Supplement 2). A brain omnibus test, which assesses the degree of shared signal across brain tissues, showed that BACE2, LINC00323, and ANGEL2 were significant after Bonferroni correction, suggesting effect across multiple brain tissue types (eTable 4 in Supplement 3).
Figure 2. Mirrored Manhattan Plot of Transcriptome-Wide Associations for Essential Tremor.
The red line indicates Bonferroni-significance threshold. The dashed blue line indicates the false-discovery rate threshold.
To prioritize the most genes, TWAS fine-mapping was done using FOCUS across the set of 90%-credible genes.40 FOCUS models the correlation among TWAS signals so that the likely causal gene(s) at genome-wide significant loci are prioritized. Across the genome-wide significant loci, BACE2 was prioritized with a posterior inclusion probability of 0.80 (z = −7.09) (eTable 5 in Supplement 4).
Additionally, the top significant loci were mapped to eQTLs derived from the GTEx 53 brain tissues and the CommonMind Consortium. The colocalization pointed toward three genes, PTGFRN, LINC00323, and BACE2. The PTGFRN gene was only significant in the cerebellum, whereas the latter 2 genes were implicated across multiple brain tissues (eTable 6 in Supplement 5).
Functional Enrichment of Genomic Regions
To identify patterns of heritability from the GWAS data, heritability was partitioned by function annotations using partitioned LD score regression.29 This analysis indicated that there were significant enrichments from SNVs in H3K9ac peaks, H3K27ac, and conserved regions (eTable 7 in Supplement 1).
Moreover, genome-wide analyses were done using MAGMA, which aggregates significance across loci into gene-level significance.36 The top gene set identified after enrichment was axonogenesis (Bonferroni P = 0.047) (eTable 8; eFigures 2 and 3 in Supplement 1). There was significant enrichment in the cerebellum (P = 6.3 × 10−5) and cerebellar hemisphere (P = 7.9 × 10−5) in GTEx 53 and overall enrichment in the brain (P = .001) (eFigure 4 in Supplement 1).37 There was no significant gene enrichment across the 29 different ages in the BrainSpan cohort (eFigure 5 in Supplement 1).44
Phenome-Wide Associations of Top Loci
For the genome-wide significant loci, a phenome-wide association study (pheWAS) was done to identify putatively relevant phenotypes. For the top BACE2 locus associated with ET (rs9980363), the pheWAS of brain imaging data showed it was a top significant association for white matter intracellular volume fraction in the left and right inferior cerebellar peduncle (β, 0.08; SE, 0.012; P = 1.2 × 10−12).
Genetic Correlation
To determine whether ET had a significant genome-wide genetic correlation with other diseases and traits, LD Hub and LDSC were used.28,30 After correcting for multiple testing, ET was shown to have a significant genetic correlation with PD (ρg, 0.28; SE, 0.051; P = 6.44 × 10−8) and depression (ρg, 0.12; SE, 0.04; P = 9.78 × 10−4) (eTable 9 in Supplement 6).
Association Between ET and PD
Considering the epidemiological implications and positive genetic correlation that have been reported between ET and PD, we sought to further dissect their genetic association. MiXer, a bivariate causal mixture model, was used to estimate the number of causally shared SNVs. It was found that there were 500 causally shared SNVs between PD and ET, with a total of 700 variants that influence ET and 4800 that influence PD.
To determine whether the genome-wide significant signals were pleiotropic for ET and PD, multitrait conditional and joint analysis using GWAS summary data was used to estimate the SNV effect size of the outcome trait (ET) after conditioning on exposure trait (PD).31 To do this, multitrait conditional and joint analysis takes the PD genome-wide significant loci to estimate the effect of exposure on ET, then undergoes a genome-wide conditioning with the estimated effect for the outcome trait.31 All the genome-wide significant ET loci remained significant after conditioning, suggesting no pleiotropy with PD for these loci (eTable 10 in Supplement 7).
To assess whether novel ET loci could be identified given the genetic correlation with PD, a multitrait association analysis was done using MTAG (multitrait analysis of genome-wide association studies).33 This method leverages the genetic correlation between traits to increase power for each respective phenotype. An additional genome-wide significant locus was identified through MTAG on chromosome 3 (lead SNV: rs703174), with an increase in power up to 8.5% (eTable 11 in Supplement 8).
Discussion
This genome-wide association study identified 5 genome-wide significant loci for ET, demonstrating the importance of common variants. One of the signals on chromosome 4 had nominal significance in a previous GWAS and was found to be consistent across the other cohorts included in this meta-analysis.17 The previous largest GWAS did not find any genome-wide significant loci that met the multiple-testing significance threshold but found 3 suggestive loci.24
For the chromosome 1 locus (rs1127215), the UK Biobank cohort did not have a consistent direction with the other cohorts. This may be because of bias from population biobanks, batch effects, and smaller case count. Interestingly, the UK Biobank cohort also had a low prevalence (approximately 0.06%), despite the expected prevalence of 1% to 5%. This may suggest an underreporting of ET in biobank surveys or a bias where there was decreased participation of patients with ET in the UK Biobank. Our study revealed multiple characteristics about the genetic architecture of ET. SNV-based heritability was found to be 18.29% on the liability scale, suggesting that a considerably large portion of heritability is explained by common variants. This is comparable with a variety of other brain-relevant disorders such as bipolar disorder, intracranial aneurysms, and PD.34,45,46 We also found this heritability to be enriched in histone markers such as H3K9ac and H3K27ac, which suggest future studies could investigate the importance of epigenetics for ET. We found an additional novel locus by leveraging the genetic overlap of PD with MTAG, which may suggest this locus is pleiotropic for the 2 phenotypes. Through gene-set enrichment analysis, we identified axonogenesis as an important cellular process for the disease, consistent with previous studies that implicate the importance of axons.20,47,48,49 Furthermore, we found significant associations between ET and the cerebellum, providing further evidence that ET may be a cerebellar disorder or reflective of neurons driving the signal due to high proportion of neurons in the cerebellum.50,51,52,53
A transcriptome-wide association study using brain cis-eQTL data from GTEx and the CommonMind Consortium found BACE2, LINC00323, and ANGEL2 to be transcriptome-wide significant for ET. Probabilistic fine mapping further prioritized BACE2 among the TWAS signals. The eQTL mapping also found converging evidence and implicated BACE2. The gene BACE2 encodes for a β-secretase homologue that is capable of cleaving amyloid β precursor protein resulting in the formation of amyloid-β protein, a major component in the pathogenesis of Alzheimer disease.54,55,56 Interestingly, a postmortem study has found increased levels of insoluble and soluble amyloid-β protein in the cerebellar and parietal cortex of patients with ET compared with control individuals and patients with PD.57
Moreover, ET was found to be significantly genetically correlated with depression and PD, suggesting common variant overlap. Previous studies have found that ET is associated with both self-reported depression and antidepressant medication use, concordant with the genetic correlation results.58 We conditioned ET by PD and found an attenuation of genome-wide association strength but that the top ET loci were still genome-wide significant. This result suggests that these loci are likely to be robustly associated with ET and not with PD.
Phenome-wide association analysis of these loci revealed that the BACE2 locus was associated with increased intracellular volume fraction, a marker of neuronal density, in the inferior cerebellar peduncles. The inferior cerebellar peduncles harbors the main afferent fibers of the cerebellum, channeling proprioceptive information from the spinal cord and brain stem nuclei to the cerebellar cortex.59 A recent volumetric analysis of magnetic resonance imaging brain scans found that the middle and inferior peduncles of the cerebellum of patients with ET displayed significant atrophy compared with healthy control individuals.60 In addition, stimulation of these afferent proprioceptive fibers was shown to be effective at reducing tremors in patients with ET.61 Interestingly, another study showed increased radial diffusivity, a parameter strongly associated with myelin abnormalities, in the inferior cerebellar peduncles of patients with ET.62 These results, paired with our findings of enrichment of genes in this region of the cerebellum and axonogenesis, highlight the potential implication of inferior cerebellar peduncles afferent fibers in ET pathophysiology.
Limitations
This study has limitations, including the lack of diverse ancestry. We did not have a large number of non-European samples, which prevented us from conducting multiancestral analyses. Moreover, there is a lack of deep phenotyping information for population-based biobanks such as 23andMe, which may lead to an increased number of diagnostic inaccuracies at the expense of increased power. However, there was still a correlation with the clinical cohorts, suggesting that the population-based biobanks are still capturing genetic signal.
Conclusions
This study identified 5 genome-wide significant loci for ET in a meta-analysis of 7177 individuals with ET and 475 877 control individuals, suggesting that approximately 18% of ET heritability might be explained by common variation. The meta-analysis implicated genes such as BACE2 and reinforced the importance of the cerebellum for the etiology of ET. The results also point toward approximately 30% shared common variant overlap with PD, and no genetic evidence for ET as a risk factor for PD.
eMethods
eFigure 1. QQ plot of GWAS meta-analysis
eFigure 2. QQ plot of ET gene-level association study
eFigure 3. Manhattan plot of ET gene-level association study
eFigure 4. Gene enrichment across GTEx 53 samples
eFigure 5. Gene enrichment across BrainSpan cohort
eTable 1. Genetic correlation between cohorts with Neff greater than 5000
eTable 2. Heterogeneity of the lead SNPs for ET GWAS
eTable 7. Significant partitioned heritability by functional annotation
eTable 8. Gene-level pathway enrichment hits for ET
eReferences
eTable 3. Significant transcriptome-wide association hits for ET
eTable 4. Significant brain omnibus transcriptome-wide association study
eTable 5. Finemapping ET transcriptome-wide association hits
eTable 6. Significant eQTL mapping of top ET loci
eTable 9. Genetic correlations with ET
eTable 10. ET GWAS conditioned by PD
eTable 11. Multi-trait analysis of ET, PD and Depression
eTable 12. Summary statistics
References
- 1.Hopfner F, Höglinger GU, Kuhlenbäumer G, et al. β-Adrenoreceptors and the risk of Parkinson’s disease. Lancet Neurol. 2020;19(3):247-254. doi: 10.1016/S1474-4422(19)30400-4 [DOI] [PubMed] [Google Scholar]
- 2.Louis ED, Ferreira JJ. How common is the most common adult movement disorder? update on the worldwide prevalence of essential tremor. Mov Disord. 2010;25(5):534-541. doi: 10.1002/mds.22838 [DOI] [PubMed] [Google Scholar]
- 3.Elble RJ. What is essential tremor? Curr Neurol Neurosci Rep. 2013;13(6):353. doi: 10.1007/s11910-013-0353-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axelrad JE, Louis ED, Honig LS, et al. Reduced Purkinje cell number in essential tremor: a postmortem study. Arch Neurol. 2008;65(1):101-107. doi: 10.1001/archneurol.2007.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liao C, Sarayloo F, Rochefort D, et al. Multiomics analyses identify genes and pathways relevant to essential tremor. Mov Disord. 2020;35(7):1153-1162. doi: 10.1002/mds.28031 [DOI] [PubMed] [Google Scholar]
- 6.Louis ED, Kerridge CA, Chatterjee D, et al. Contextualizing the pathology in the essential tremor cerebellar cortex: a patholog-omics approach. Acta Neuropathol. 2019;138(5):859-876. doi: 10.1007/s00401-019-02043-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharifi S, Nederveen AJ, Booij J, van Rootselaar AF. Neuroimaging essentials in essential tremor: a systematic review. Neuroimage Clin. 2014;5:217-231. doi: 10.1016/j.nicl.2014.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martuscello RT, Kerridge CA, Chatterjee D, et al. Gene expression analysis of the cerebellar cortex in essential tremor. Neurosci Lett. 2020;721:134540. doi: 10.1016/j.neulet.2019.134540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorenz D, Frederiksen H, Moises H, Kopper F, Deuschl G, Christensen K. High concordance for essential tremor in monozygotic twins of old age. Neurology. 2004;62(2):208-211. doi: 10.1212/01.WNL.0000103236.26934.41 [DOI] [PubMed] [Google Scholar]
- 10.Tanner CM, Goldman SM, Lyons KE, et al. Essential tremor in twins: an assessment of genetic vs environmental determinants of etiology. Neurology. 2001;57(8):1389-1391. doi: 10.1212/WNL.57.8.1389 [DOI] [PubMed] [Google Scholar]
- 11.Deng H, Wu S, Jankovic J. Essential tremor: genetic update. Expert Rev Mol Med. 2019;21:e8. doi: 10.1017/erm.2019.7 [DOI] [PubMed] [Google Scholar]
- 12.Sailani MR, Jahanbani F, Abbott CW, et al. Candidate variants in TUB are associated with familial tremor. PLoS Genet. 2020;16(9):e1009010. doi: 10.1371/journal.pgen.1009010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hor H, Francescatto L, Bartesaghi L, et al. Missense mutations in TENM4, a regulator of axon guidance and central myelination, cause essential tremor. Hum Mol Genet. 2015;24(20):5677-5686. doi: 10.1093/hmg/ddv281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merner ND, Girard SL, Catoire H, et al. Exome sequencing identifies FUS mutations as a cause of essential tremor. Am J Hum Genet. 2012;91(2):313-319. doi: 10.1016/j.ajhg.2012.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houle G, Schmouth J-F, Leblond CS, et al. Teneurin transmembrane protein 4 is not a cause for essential tremor in a Canadian population. Mov Disord. 2017;32(2):292-295. doi: 10.1002/mds.26753 [DOI] [PubMed] [Google Scholar]
- 16.Ortega-Cubero S, Lorenzo-Betancor O, Lorenzo E, et al. Fused in sarcoma (FUS) gene mutations are not a frequent cause of essential tremor in Europeans. Neurobiol Aging. 2013;34(10):2441.e9-2441.e11. doi: 10.1016/j.neurobiolaging.2013.04.024 [DOI] [PubMed] [Google Scholar]
- 17.Müller SH, Girard SL, Hopfner F, et al. Genome-wide association study in essential tremor identifies three new loci. Brain. 2016;139(Pt 12):3163-3169. doi: 10.1093/brain/aww242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefansson H, Steinberg S, Petursson H, et al. Variant in the sequence of the LINGO1 gene confers risk of essential tremor. Nat Genet. 2009;41(3):277-279. doi: 10.1038/ng.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Zhao Y, Zhou X, et al. Assessment of three new loci from genome-wide association study in essential tremor in Chinese population. Sci Rep. 2017;7(1):7981. doi: 10.1038/s41598-017-08863-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao C, Sarayloo F, Vuokila V, et al. Transcriptomic changes resulting from STK32B overexpression identify pathways potentially relevant to essential tremor. Front Genet. 2020;11:813. doi: 10.3389/fgene.2020.00813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bourassa CV, Rivière J-B, Dion PA, et al. LINGO1 variants in the French-Canadian population. PLoS ONE. 2011;6(1):e16254. doi: 10.1371/journal.pone.0016254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiménez-Jiménez FJ, García-Martín E, Lorenzo-Betancor O, Pastor P, Alonso-Navarro H, Agúndez JAG. LINGO1 and risk for essential tremor: results of a meta-analysis of rs9652490 and rs11856808. J Neurol Sci. 2012;317(1-2):52-57. doi: 10.1016/j.jns.2012.02.030 [DOI] [PubMed] [Google Scholar]
- 23.Vilariño-Güell C, Ross OA, Wider C, et al. LINGO1 rs9652490 is associated with essential tremor and Parkinson disease. Parkinsonism Relat Disord. 2010;16(2):109-111. doi: 10.1016/j.parkreldis.2009.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarthy S, Das S, Kretzschmar W, et al. ; Haplotype Reference Consortium . A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016;48(10):1279-1283. doi: 10.1038/ng.3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loh P-R, Tucker G, Bulik-Sullivan BK, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47(3):284-290. doi: 10.1038/ng.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190-2191. doi: 10.1093/bioinformatics/btq340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winkler TW, Day FR, Croteau-Chonka DC, et al. ; Genetic Investigation of Anthropometric Traits (GIANT) Consortium . Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9(5):1192-1212. doi: 10.1038/nprot.2014.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan BK, Loh P-R, Finucane HK, et al. ; Schizophrenia Working Group of the Psychiatric Genomics Consortium . LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47(3):291-295. doi: 10.1038/ng.3211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finucane HK, Bulik-Sullivan B, Gusev A, et al. ; ReproGen Consortium; Schizophrenia Working Group of the Psychiatric Genomics Consortium; RACI Consortium . Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47(11):1228-1235. doi: 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng J, Erzurumluoglu AM, Elsworth BL, et al. ; Early Genetics and Lifecourse Epidemiology (EAGLE) Eczema Consortium . LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics. 2017;33(2):272-279. doi: 10.1093/bioinformatics/btw613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu Z, Zheng Z, Zhang F, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat Commun. 2018;9(1):224. doi: 10.1038/s41467-017-02317-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Awadalla P, Boileau C, Payette Y, et al. ; CARTaGENE Project . Cohort profile of the CARTaGENE study: Quebec’s population-based biobank for public health and personalized genomics. Int J Epidemiol. 2013;42(5):1285-1299. doi: 10.1093/ije/dys160 [DOI] [PubMed] [Google Scholar]
- 33.Turley P, Walters RK, Maghzian O, et al. ; 23andMe Research Team; Social Science Genetic Association Consortium . Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat Genet. 2018;50(2):229-237. doi: 10.1038/s41588-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nalls MA, Blauwendraat C, Vallerga CL, et al. ; 23andMe Research Team; System Genomics of Parkinson’s Disease Consortium; International Parkinson’s Disease Genomics Consortium . Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019;18(12):1091-1102. doi: 10.1016/S1474-4422(19)30320-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Howard DM, Adams MJ, Clarke TK, et al. ; 23andMe Research Team; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium . Genome-wide meta-analysis of depression identifies 102 independent variants and highlights the importance of the prefrontal brain regions. Nat Neurosci. 2019;22(3):343-352. doi: 10.1038/s41593-018-0326-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11(4):e1004219. doi: 10.1371/journal.pcbi.1004219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. doi: 10.1038/s41467-017-01261-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Battle A, Brown CD, Engelhardt BE, Montgomery SB; GTEx Consortium; Laboratory, Data Analysis &Coordinating Center (LDACC)—Analysis Working Group; Statistical Methods groups—Analysis Working Group; Enhancing GTEx (eGTEx) groups; NIH Common Fund; NIH/NCI; NIH/NHGRI; NIH/NIMH; NIH/NIDA; Biospecimen Collection Source Site—NDRI; Biospecimen Collection Source Site—RPCI; Biospecimen Core Resource—VARI; Brain Bank Repository—University of Miami Brain Endowment Bank; Leidos Biomedical—Project Management; ELSI Study; Genome Browser Data Integration &Visualization—EBI; Genome Browser Data Integration &Visualization—UCSC Genomics Institute, University of California Santa Cruz; Lead analysts; Laboratory, Data Analysis &Coordinating Center (LDACC); NIH program management; Biospecimen collection; Pathology; eQTL manuscript working group . Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204-213. doi: 10.1038/nature24277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoffman GE, Bendl J, Voloudakis G, et al. CommonMind Consortium provides transcriptomic and epigenomic data for schizophrenia and bipolar disorder. Sci Data. 2019;6(1):180. doi: 10.1038/s41597-019-0183-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mancuso N, Freund MK, Johnson R, et al. Probabilistic fine-mapping of transcriptome-wide association studies. Nat Genet. 2019;51(4):675-682. doi: 10.1038/s41588-019-0367-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deelen P, van Dam S, Herkert JC, et al. Improving the diagnostic yield of exome- sequencing by predicting gene-phenotype associations using large-scale gene expression analysis. Nat Commun. 2019;10(1):2837. doi: 10.1038/s41467-019-10649-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frei O, Holland D, Smeland OB, et al. Bivariate causal mixture model quantifies polygenic overlap between complex traits beyond genetic correlation. Nat Commun. 2019;10(1):2417. doi: 10.1038/s41467-019-10310-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245-252. doi: 10.1038/ng.3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen EH, Overly CC, Jones AR. The Allen Human Brain Atlas: comprehensive gene expression mapping of the human brain. Trends Neurosci. 2012;35(12):711-714. doi: 10.1016/j.tins.2012.09.005 [DOI] [PubMed] [Google Scholar]
- 45.Mullins N, Forstner AJ, O’Connell KS, et al. Genome-wide association study of over 40,000 bipolar disorder cases provides novel biological insights. medRxiv. Preprint posted September 18, 2020. doi: 10.1101/2020.09.17.20187054 [DOI]
- 46.Bakker MK, van der Spek RAA, van Rheenen W, et al. ; HUNT All-In Stroke; China Kadoorie Biobank Collaborative Group; BioBank Japan Project Consortium; ICAN Study Group; CADISP Group; Genetics and Observational Subarachnoid Haemorrhage (GOSH) Study investigators; International Stroke Genetics Consortium (ISGC) . Genome-wide association study of intracranial aneurysms identifies 17 risk loci and genetic overlap with clinical risk factors. Nat Genet. 2020;52(12):1303-1313. doi: 10.1038/s41588-020-00725-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louis ED, Yi H, Erickson-Davis C, Vonsattel JPG, Faust PL. Structural study of Purkinje cell axonal torpedoes in essential tremor. Neurosci Lett. 2009;450(3):287-291. doi: 10.1016/j.neulet.2008.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gironell A. The GABA hypothesis in essential tremor: lights and shadows. Tremor Other Hyperkinet Mov (N Y). 2014;4:254. doi: 10.5334/tohm.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Louis ED, Faust PL. Essential tremor pathology: neurodegeneration and reorganization of neuronal connections. Nat Rev Neurol. 2020;16(2):69-83. doi: 10.1038/s41582-019-0302-1 [DOI] [PubMed] [Google Scholar]
- 50.Kuo SH, Tang G, Louis ED, et al. LINGO1 expression is increased in essential tremor cerebellum and is present in the basket cell pinceau. Acta Neuropathol. 2013;125(6):879-889. doi: 10.1007/s00401-013-1108-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paris-Robidas S, Brochu E, Sintes M, et al. Defective dentate nucleus GABA receptors in essential tremor. Brain. 2012;135(Pt 1):105-116. doi: 10.1093/brain/awr301 [DOI] [PubMed] [Google Scholar]
- 52.Deuschl G, Wenzelburger R, Löffler K, Raethjen J, Stolze H. Essential tremor and cerebellar dysfunction clinical and kinematic analysis of intention tremor. Brain. 2000;123(Pt 8):1568-1580. doi: 10.1093/brain/123.8.1568 [DOI] [PubMed] [Google Scholar]
- 53.Louis ED, Faust PL. Essential tremor: the most common form of cerebellar degeneration? Cerebellum Ataxias. 2020;7(1):12. doi: 10.1186/s40673-020-00121-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan R, Munzner JB, Shuck ME, Bienkowski MJ. BACE2 functions as an alternative α-secretase in cells. J Biol Chem. 2001;276(36):34019-34027. doi: 10.1074/jbc.M105583200 [DOI] [PubMed] [Google Scholar]
- 55.Azkona G, Amador-Arjona A, Obradors-Tarragó C, et al. Characterization of a mouse model overexpressing beta-site APP-cleaving enzyme 2 reveals a new role for BACE2. Genes Brain Behav. 2010;9(2):160-172. doi: 10.1111/j.1601-183X.2009.00538.x [DOI] [PubMed] [Google Scholar]
- 56.Solans A, Estivill X, de La Luna S. A new aspartyl protease on 21q22.3, BACE2, is highly similar to Alzheimer’s amyloid precursor protein β-secretase. Cytogenet Cell Genet. 2000;89(3-4):177-184. doi: 10.1159/000015608 [DOI] [PubMed] [Google Scholar]
- 57.Béliveau E, Tremblay C, Aubry-Lafontaine É, et al. Accumulation of amyloid-β in the cerebellar cortex of essential tremor patients. Neurobiol Dis. 2015;82:397-408. doi: 10.1016/j.nbd.2015.07.016 [DOI] [PubMed] [Google Scholar]
- 58.Louis ED, Benito-León J, Bermejo-Pareja F; Neurological Disorders in Central Spain (NEDICES) Study Group . Self-reported depression and anti-depressant medication use in essential tremor: cross-sectional and prospective analyses in a population-based study. Eur J Neurol. 2007;14(10):1138-1146. doi: 10.1111/j.1468-1331.2007.01923.x [DOI] [PubMed] [Google Scholar]
- 59.Perrini P, Tiezzi G, Castagna M, Vannozzi R. Three-dimensional microsurgical anatomy of cerebellar peduncles. Neurosurg Rev. 2013;36(2):215-224. doi: 10.1007/s10143-012-0417-y [DOI] [PubMed] [Google Scholar]
- 60.Prasad S, Pandey U, Saini J, Ingalhalikar M, Pal PK. Atrophy of cerebellar peduncles in essential tremor: a machine learning-based volumetric analysis. Eur Radiol. 2019;29(12):7037-7046. doi: 10.1007/s00330-019-06269-7 [DOI] [PubMed] [Google Scholar]
- 61.Schlaier J, Anthofer J, Steib K, et al. Deep brain stimulation for essential tremor: targeting the dentato-rubro-thalamic tract? Neuromodulation. 2015;18(2):105-112. doi: 10.1111/ner.12238 [DOI] [PubMed] [Google Scholar]
- 62.Juttukonda MR, Franco G, Englot DJ, et al. White matter differences between essential tremor and Parkinson disease. Neurology. 2019;92(1):e30-e39. doi: 10.1212/WNL.0000000000006694 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods
eFigure 1. QQ plot of GWAS meta-analysis
eFigure 2. QQ plot of ET gene-level association study
eFigure 3. Manhattan plot of ET gene-level association study
eFigure 4. Gene enrichment across GTEx 53 samples
eFigure 5. Gene enrichment across BrainSpan cohort
eTable 1. Genetic correlation between cohorts with Neff greater than 5000
eTable 2. Heterogeneity of the lead SNPs for ET GWAS
eTable 7. Significant partitioned heritability by functional annotation
eTable 8. Gene-level pathway enrichment hits for ET
eReferences
eTable 3. Significant transcriptome-wide association hits for ET
eTable 4. Significant brain omnibus transcriptome-wide association study
eTable 5. Finemapping ET transcriptome-wide association hits
eTable 6. Significant eQTL mapping of top ET loci
eTable 9. Genetic correlations with ET
eTable 10. ET GWAS conditioned by PD
eTable 11. Multi-trait analysis of ET, PD and Depression
eTable 12. Summary statistics