Abstract
Lung cancer has the second highest incidence and highest mortality compared to all other cancers. Polycyclic aromatic hydrocarbon (PAH) molecules belong to a class of compounds that are present in tobacco smoke, diesel exhausts, smoked foods, as well as particulate matter (PM). PAH-derived reactive metabolites are significant contributors to lung cancer development. The formation of these reactive metabolites entails metabolism of the parent PAHs by cytochrome P4501A1/1B1 (CYP1A1/1B1) and epoxide hydrolase enzymes. These reactive metabolites then react with DNA to form DNA adducts, which contribute to key gene mutations, such as the tumor suppressor gene, p53 and are linked to pulmonary carcinogenesis. PAH exposure also leads to upregulation of CYP1A1 transcription by binding to the aryl hydrocarbon receptor (AHR) and eliciting transcription of the CYP1A1 promoter, which comprises specific xenobiotic-responsive element (XREs). While hepatic and pulmonary CYP1A1/1B1 metabolize PAHs to DNA-reactive metabolites, the hepatic CYP1A2, however, may protect against lung tumor development by suppressing both liver and lung CYP1A1 enzymes. Further analysis of these enzymes has shown that PAH-exposure also induces sustained transcription of CYP1A1, which is independent of the persistence of the parent PAH. CYP1A2 enzyme plays an important role in the sustained induction of hepatic CYP1A1. PAH exposure may further contribute to pulmonary carcinogenesis by producing epigenetic alterations. DNA methylation, histone modification, long interspersed nuclear element (LINE-1) activation, and non-coding RNA, specifically microRNA (miRNA) alterations may all be induced by PAH exposure. The relationship between PAH-induced enzymatic reactive metabolite formation and epigenetic alterations is a key area of research that warrants further exploration. Investigation into the potential interplay between these two mechanisms may lead to further understanding of the mechanisms of PAH carcinogenesis. These mechanisms will be crucial for the development of effective targeted therapies and early diagnostic tools.
Keywords: Pulmonary carcinogenesis, polycyclic aromatic hydrocarbon (PAH), Cytochrome P450 (CYP1) enzyme, epigenetics, lung cancer
Introduction
The importance of understanding the mechanisms contributing to lung cancer cannot be overstated. Lung cancer has the second highest incidence in both men and women, and has the highest mortality rate in both men and women [1]. Although research into lung cancer continues, it still remains to be the cause for the highest mortality among all cancers, in part, because of the lack of effective and easy screening methods available [1]. Currently, high risk factors are the basis for screening, and the only way to screen for the cancer is with low-dose computed tomography (LDCT) [2]. Because of the expense, inconvenience, and radiation exposure of a routine LDCT, this screening method is insufficient and limiting. Continued research into newer and better screening methods needs to be of the utmost priority, and many of these methods are reliant upon the understanding of the mechanisms behind pulmonary carcinogenesis.
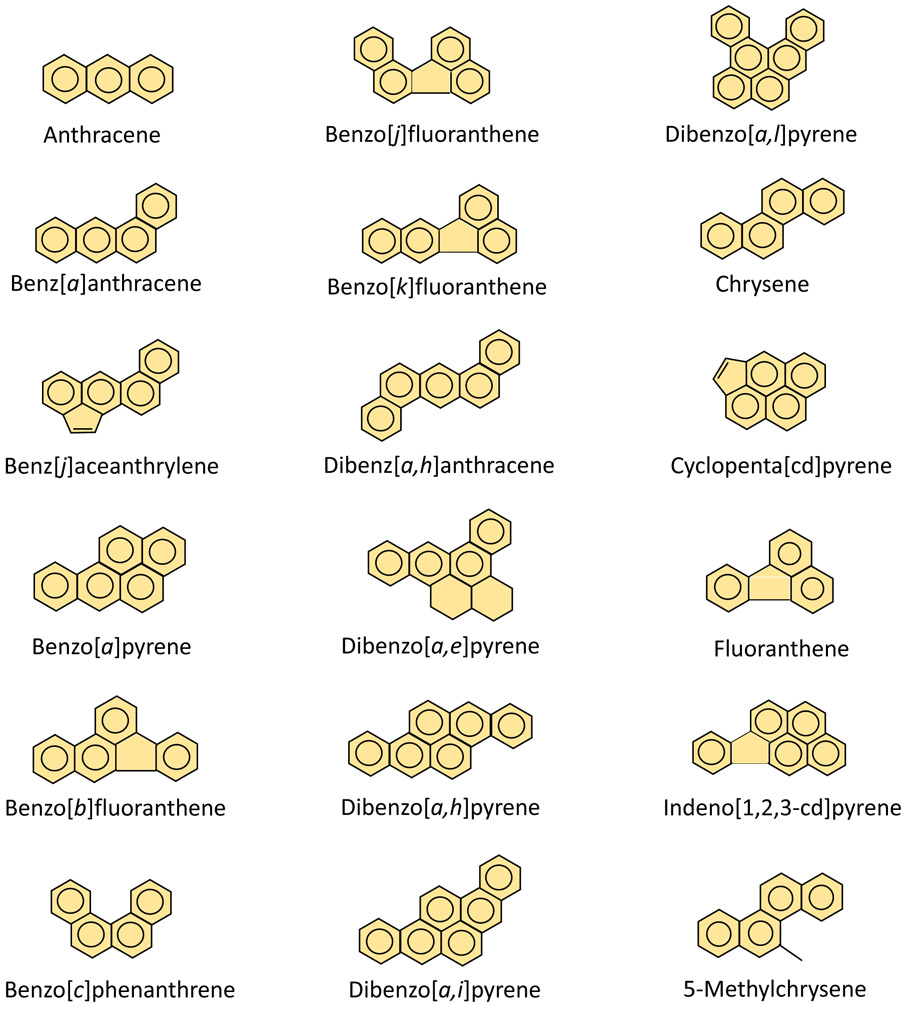
About 90% of lung cancer cases are associated with tobacco smoking, although outdoor air pollution, secondhand smoke, inhalation of incomplete indoor/outdoor combustion of coal and wood may also contribute to lung cancer as well [3, 4]. Polycyclic aromatic hydrocarbons (PAHs) are significant components in tobacco smoke and air pollution and are defined as a group of chemicals containing two or more fused benzene rings, but with no heteroatoms [5]. Examples of PAHs are benzo[a]pyrene (BP), dibenzo[a,h]anthracene (DBA), 3-methylcholanthrene (MC), 5-methylchrysene, and 7,12-dimethylbenz[a]anthracene [5]. The structures of the various molecules that are classified as PAHs are shown in Figure 1. One of the most notable PAHs is BP due to its high carcinogenic potential [6]. BP is a result of incomplete combustion of organic material and can be found in tobacco smoke, diesel or other types of motor vehicle exhaust, industrial byproducts, and forest fires [7]. Another notable PAH due to its potency is MC.
Figure 1.
Molecular structures of various PAH molecules. This class of molecules is defined by having two or more fused benzene rings and are able to diffuse across a plasma membrane due to their lipid solubility.
Inhaled PAH exposure enhances lung cancer risk in humans [8-11]. Smokers may also ingest large quantities of PAHs by swallowing the smoke, allowing it to enter the gastrointestinal tract [12-14]. Significant amounts of PAHs are also found in grilled, barbecued, or smoked meat products [15-20]. A recent study has found a direct link between dietary PAH exposure and lung cancer incidence, except for one recent study from China [21]. This review will discuss the mechanisms by which PAHs induce cancer, as it may lead to more targeted treatments for lung cancer, which has poor prognosis and leads to billions of dollars spent on this devastating disease.
PAHs and Carcinogenesis
Because of their lipophilic nature, PAHs are able to diffuse across the plasma membrane of a cell. Three major pathways have been identified in PAH metabolism, each utilizing a different set of enzymes. The pathway that is the focus of this review is the CYP1A1/CYP1B1 and epoxide hydrolase pathway. The second pathway uses the CYP peroxidase enzyme for metabolism, and the third uses aldo-keto reductase [4, 22, 23]. CYP1A2, which is liver-specific, has a protective role against carcinogenesis.
Role of CYP1A1/CYP1B1 and Epoxide Hydrolase Pathway in DNA adduct Formation
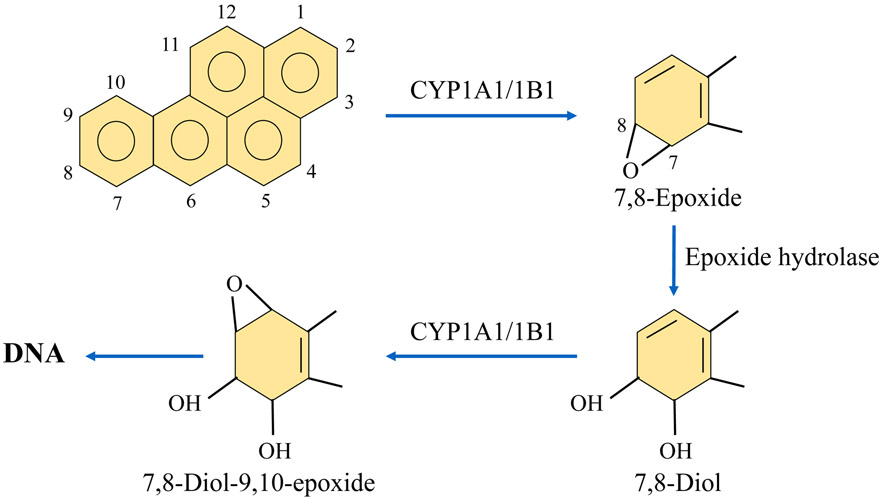
While PAHs themselves are not directly carcinogenic, they are considered procarcinogens because metabolism of these molecules results in carcinogenic metabolites that lead to DNA adduct formation. For example, BP is a PAH that is metabolized by CYP1A1/1B1 enzymes to form 7,8-epoxide. From here, 7, 8-epoxide is further metabolized by epoxide-hydrolase to form 7, 8-diol, and then again by CYP1A1/1B1 to form 7, 8-diol-9, 10-epoxide (BPDE), which is the ultimate carcinogenic metabolite of BP. BPDE reacts with N2 of guanosine to form a DNA adduct [24-26] (Figure 2).
Figure 2.
Mechanisms of DNA adduct formation by PAHs. BP is metabolized by CYP1A1 and CYP1B1 enzymes to form 7,8-epoxide metabolite. This product is then metabolized by epoxide hydrolase to produce 7,8-diol. After this step, CYP1A1/1B1 metabolizes 7, 8-diol to form the ultimate carcinogenic metabolite, 7, 8-diol-9, 10-epoxide. This metabolite reacts with N2 of guanosine to form DNA adducts, which contribute to carcinogenesis.
DNA adducts represent a critical event in the initiation of carcinogenesis [27, 28]. Several studies have linked DNA adduct formation to the modification of specific genes implicated in animal and human cancers [29, 30]. DNA adduct formation has been shown to be sequence-specific, and cause mutations in the tumor suppressor gene p53 and KRAS oncogene [31-35]. The p53 gene acts like breaks on the cellular replication process and causes cell cycle arrest to prevent errors from propagating and, thus prevents tumors from developing [36]. DNA adducts are formed on the gene encoding p53 resulting in mutations and malfunction of the gene, which allows cells to propagate with less regulation [31-35]. Denissenko, et al. demonstrated that in smokers, these DNA adducts are located on codons 157, 248, and 273 on the p53 gene, and mutations of these codons showed a correlation with human lung cancer [31]. Therefore, the DNA adduct formation produced by CYP1A1/CYP1B1 metabolism of PAH molecules may initiate carcinogenesis by mutating the p53 gene. Once initiated, carcinogenesis must undergo promotion and progression in order to result in cancer tumor formation. Based on this analysis, adduct formation seems to be the main mechanism in which p53 is mutated in lung cancer, and these studies provide direct evidence that link cigarette smoking to lung cancer [32].
An important byproduct of PAH metabolism is the formation of reactive oxygen species (ROS). The oxidation-reduction balance in cells are maintained by a balance between oxidative and anti-oxidative reactions, also referred to as redox cycling. When the balance is disrupted, ROS formation is increased and the cell experiences oxidative stress. PAH disrupts this balance by binding with the aryl hydrocarbon receptor (AHR), resulting in the upregulation of CYP1 enzyme transcription [37], as well as other genes of the AHR gene battery. This could result in increased production of ROS, which in turn contributes to DNA damage [38, 39]. The resulting DNA adducts and damage from PAH-AHR-mediated induction of CYP1 enzymes increases the risk for tumor development. In addition to playing a role in DNA adduct formation and tumorigenesis, AHR also contributes to downstream inflammation, cell proliferation, and loss of cell-cell adhesion, which may further result in cancer progression [10].
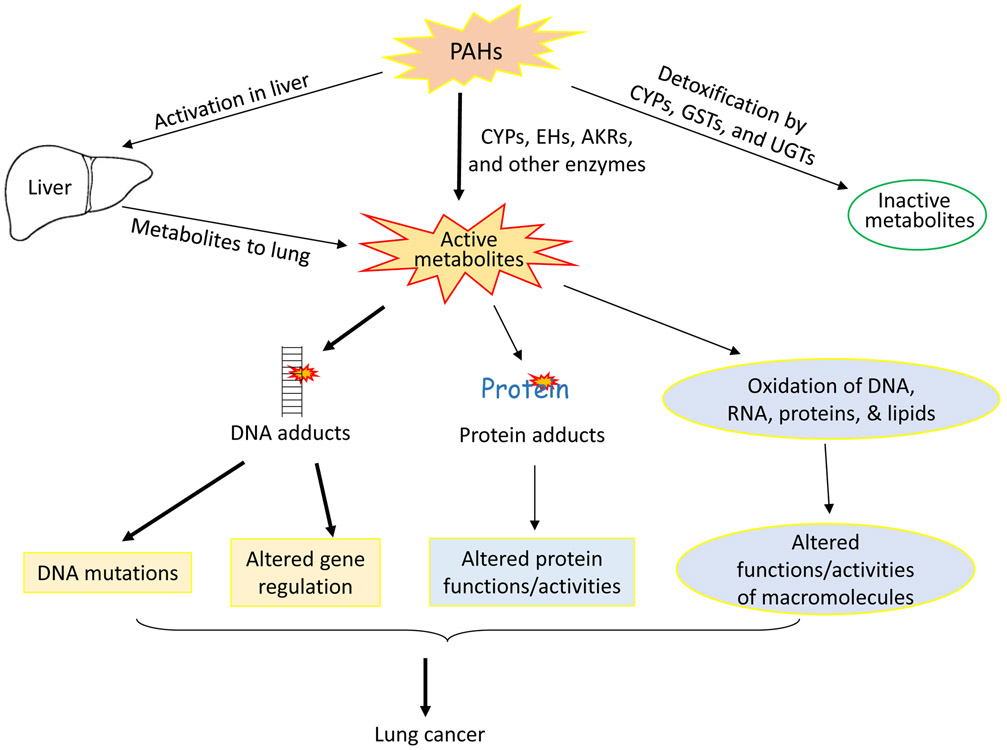
In response to increased ROS formation and resulting oxidative stress, the cell upregulates the transcription of antioxidant, phase II enzymes [40]. Activation of the transcription factor nuclear factor erythroid 2-related factor 2 (Nrf2) helps regulate this response [41-45]. In its inactivated form, Nrf2 is bound to a Keap1 complex, which signals for degradation of Nrf2 [46]. However, oxidative stress from PAH metabolism triggers Nrf2 dissociation from the Keap1 complex [47, 48]. Once released, Nrf2 is able to migrate to the nucleus where it activates transcription of antioxidant enzymes [43]. One of the main types of enzymes targeted in this pathway is the NAD(P)H quinone oxidoreductase (NQO1 and NQO2) enzymes [49]. In both the BP and MC metabolic pathways, NQO1 and NQO2 enzymes prevent production of harmful semiquinones that can form DNA adducts by metabolizing quinones into hydroquinones instead [50, 51]. UDP-glucuronosyltransferase (UGT) and glutathione S-transferase (GST) enzymes have also been shown to detoxify the harmful metabolite [50, 52, 53]. If these antioxidant, protective enzymes are not sufficient to compensate for the ROS formation, oxidative stress will still occur in the cell and enhance carcinogenesis potential [54]. Figure 3 summarizes the different mechanisms in which PAH may contribute to carcinogenesis.
Figure 3.
PAH exposure and resultant metabolism may produce inactive metabolites via the CYP, UDP-glucuronosyltransferase (UGT), or glutathione S-transferase (GST) pathways. Active metabolites may also be produced during PAH metabolism when metabolized by CYP1-epoxide hydrolase pathway or the aldo-keto reductase pathway. Production of these metabolites occurs in the liver as well, which can contribute to the effects seen in the lungs. These active metabolites can then form DNA adducts, resulting in DNA mutations and altered gene regulation. They can also form protein adducts, which directly alters protein function. Finally, the active metabolites may oxidize DNA, RNA, proteins, and lipids, contributing to altered function of macromolecules. All three of these alterations lead to lung cancer.
Role of PAH-AHR Complex in CYP1A1/CYP1B1 Transcription
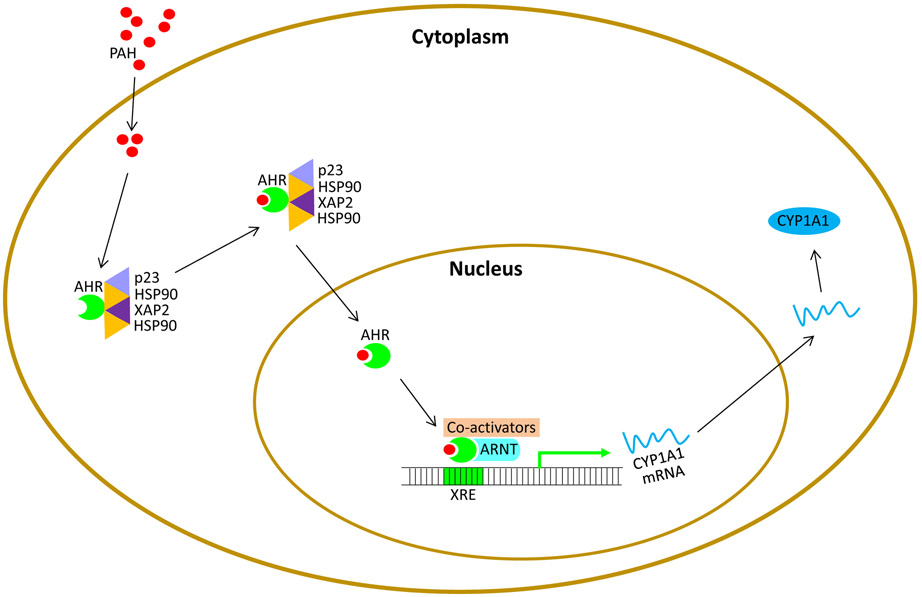
The basic mechanism by which PAH results in carcinogenesis begins when PAH enters the cell and binds to an AHR complex, which also consists of HSP90, XAP2, and p23 [55]. The AHR plays crucial roles in many species by binding to numerous exogenous and endogenous ligands. In humans, AHR has the highest levels in the liver, adipose tissue, and bronchioles. Studies of non-small cell lung cancer samples demonstrated increased levels of AhR mRNA with a positive correlation to CYP1A1 expression in cases of adenocarcinomas [56]. Once bound, the PAH-AHR complex sheds the other molecules from the complex and is able to translocate to the nucleus. This mechanism is facilitated by phosphorylation of protein kinases C sites adjacent to the nuclear localization sequences (56-58). In the nucleus, the AHR and an aryl hydrocarbon receptor nuclear translocator (ARNT) dimerize, and the PAH-AHR-ARNT complex bind the DNA-recognition site xenobiotic-responsive element (XRE) via a bHLH-PAS motif [55, 57, 58]. This binding of the DNA triggers the upregulation of CYP1A1, CYP1A2, and CYP1B1 transcription [57, 59-61]. The upregulation of the CYPI enzymes increases the metabolism of PAH into carcinogenic products in the sarcoplasmic reticulum. The CYP1A1 promoter in the non-induced state remains silent, as histone modifiers such as histone deacetylase (HDAC) and DNA methyltransferase (DNMT) are bound to the promoter. When the PAH-AHR-ARNT bind to the CYP1A1 promoter, the HDAC/DNMT complex dissociates, and transcription ensues. Upon binding to CYP1A1 promoter, the PAH-AHR-ARNT complex also recruits a number of co-activators, which alter the chromatin structure into a more accessible configuration via histone acetyltransferases and histone methyltransferase. In addition, other factors are recruited that include Kinases IKKa, MSK1, and MSK2, and coactivators, such as SP1, NCOA1, NCOA3, NCOA4, P300, BRCA1 tumor suppressor protein, and general transcription protein IIB, which is required for transcription initiation by RNA polymerase II (56-58). Figure 4 demonstrates the steps described in this pathway.
Figure 4.
Once the body is exposed to PAHs, the PAHs will enter the cytoplasm to react with the aryl hydrocarbon receptor (AHR) complex that also consists of p23, XAP2, and two HSP90 molecules. The PAH-AHR complex then dissociates from the other molecules and migrates to the nucleus. Dimerization occurs between AHR and ARNT, which allows AHR to bind to xenobiotic-responsive element DNA binding site. Once bound, co-activators are drawn to the site and help induce histone modifiers, histone deacetylase (HDAC) and DNA methyltransferase (DNMT), which were originally bound to the CYP1A1 promoter site. These histone modifiers dissociate, and CYP1A1 transcription occurs.
Like most pathways, this pathway is also self-regulated through a negative feedback-type system. In addition to upregulating CYP1 transcription, the activated AHR complex can also bind to the aryl hydrocarbon receptor repressor (Ahrr) gene [62]. Due to structural similarities between AHR and AHRR, AHRR is able to bind ARNT and occupy XRE target sites [63]. Therefore, AHRR acts as a competitive inhibitor of AHR, rendering CYP1 transcription inactive [63].
Differential Roles of CYP1 Enzymes to Pulmonary Carcinogenicity
CYP1 enzymes are important phase I enzymes in the breakdown of exogenous xenobiotic molecules as well as important endogenous molecules [64]. Epoxide products and ROS byproducts are common results of CYP1 metabolism. Based on this mechanism, CYP1A1 enzymes are key carcinogenic enzymes. However, results from recent studies suggest that the hepatic CYP1A2 enzyme may actually protect pulmonary cells against ROS and harmful DNA adduct formation [37]. This mechanism is not fully understood and requires further investigation. CYP1B1is mainly localized in the lungs and appears to promote carcinogenesis via mechanisms similar to CYP1A1 enzymes [4].
Carcinogenic Role of CYP1A1 in PAH Metabolism
Based on the mechanism discussed in the section “PAHs and carcinogenesis”, CYP1A1 enzymes play a significant carcinogenic role in PAH metabolism. These enzymes bioactivate PAHs into reactive metabolites that induce mutagenic DNA adducts, which can lead to cancer. Past studies have investigated the role of CYP1A1 in PAH bioactivation; however, the individual roles of each CYP1A enzymes are still unknown. In a recently published paper [38], we tested the hypothesis that mice lacking the genes for Cyp1a1 or Cyp1a2 will display altered susceptibilities to PAH-induced pulmonary carcinogenesis [37]. Wild-type (WT), Cyp1a1-null, and Cypla2-null male and female mice were treated with MC (a single dose of MC, i.p. 40 μmol/kg) for cancer initiation and tumor formation studies. In WT mice, CYP1A1 and 1A2 expression was induced by MC. Cyp1a1(−/−) and Cyp1a2(−/−) mice treated with PAHs displayed a compensatory pattern, where knocking out 1 Cypla gene led to increased expression of the other. Cyp1a1(−/−) mice were resistant to DNA adduct and tumor formation, whereas Cyp1a2(−/−) mice displayed increased levels of both. UALCAN analysis revealed that lung adenocarcinoma patients with high levels of CYP1A2 expression survive significantly better than patients with low/medium expression [38].
The reactive metabolites formed from CYP1A1 metabolism of PAH molecules results in DNA adduct formation and, eventual, pulmonary carcinogenesis. DNA adduct formation can mutate important tumor suppressor molecules, such as the p53 protein, resulting in tumor progression [31]. In addition to stimulating CYP1A1-mediated metabolism, PAH also upregulates CYP1A1 transcription by binding to AHR and activating the XRE DNA-recognition site. This important carcinogenic pathway involves several molecules that could be targeted to slow or inhibit lung cancer progression. For example, reduction of CYP1A1 expression via a CYP1A1 inhibitor, such as (E)-3-(3,4,5-trimethoxyphenyl)-1-(pyridin-4-yl)prop-2-en-1-one, could potentially offer an eventual treatment method for mitigating PAH-induced pulmonary carcinogenesis [65].
Further studies are needed to determine the effects that specific polymorphic CYP1A1 enzymes have on a patient’s risk for lung cancer development. One study found an association between CYP1A1 MspI polymorphism and an increased risk of lung cancer development in the Chinese population [66]. Other studies showed an association in the variant enzyme CYP1A1 rs4646903 with increased risk of lung cancer in smokers, specifically [67]. Non-smokers did not have an association between this enzyme and the risk of lung cancer development. As individualized medicine becomes more of a reality in today’s world, the relationship between CYP1A1 polymorphisms and lung cancer need to be further analyzed in order to develop effective treatment options for individual patients.
Protective Role of CYP1A2 in PAH Metabolism
CYP1A2 enzymes are found in high concentrations in the liver. Similar to CYP1A1 enzymes in the lungs, CYP1A2 enzymes metabolize PAHs. However, while CYP1A1 enzymes are carcinogenic, results from new studies are indicating that CYP1A2 enzyme may play a protective role against PAH-mediated carcinogenesis. In our recent study, WT, Cyp1a1−/−, and Cyp1a2−/− mice, were treated with MC [68], and DNA adducts in the lungs and liver were measured in all three groups. Figure 5 shows the results from this study. The most interesting part of this study was an increase in DNA adduct formation in the lungs in Cyp1a2-null mice even though CYP1A2 enzyme is located in the liver, but not the lungs [68]. These results indicate an important interrelationship between PAH metabolism via CYP1A2 enzyme in the liver, and DNA adduct formation in the lungs. Figure 5 exemplifies the anti-carcinogen effects hepatic CYP1A2 enzymes seem to have on pulmonary carcinogenesis. Furthermore, studies have found that humans who have a less active variant of CYP1A2 display an increased risk of developing lung cancer, again insinuating the protective role of hepatic CYP1A2 on pulmonary carcinogenesis [69, 70]. Interestingly, our studies of Cyp1a2−/− mice found upregulation of CYP1A1 enzymes in the lungs when compared to WT mice [68]. This result indicates that levels of hepatic CYP1A2 enzyme may impact levels of pulmonary CYP1A1 enzymes.
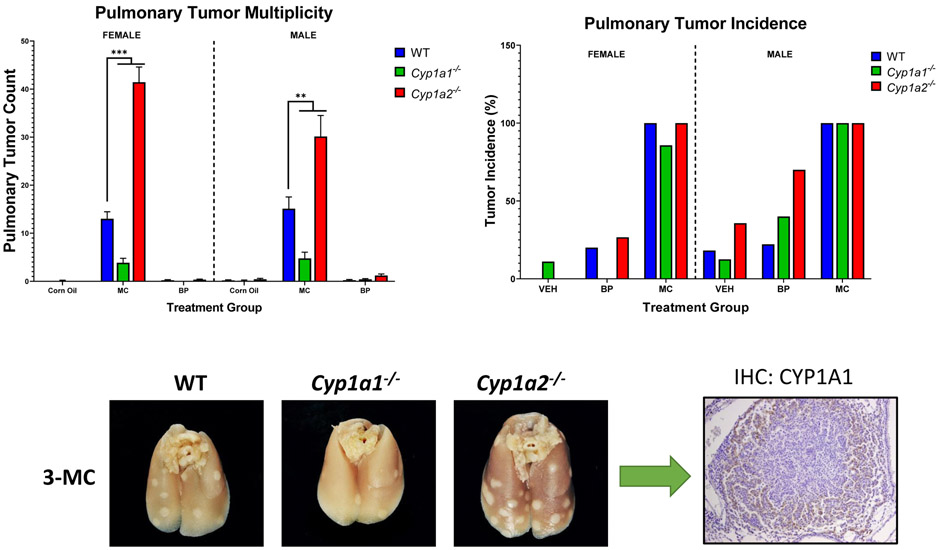
Figure 5.
Differential roles of CYP1A1 and 1A2 in PAH carcinogenesis. Compared to WT mice, Cyp1a1(−/−) showed a significantly lower pulmonary tumor count following exposure to 3-methylcholanthrene (3-MC). Cyp1a2(−/−) mice demonstrated a significantly higher pulmonary tumor count compared to WT mice. These results strongly indicate a protective function of hepatic CYP1A2 enzymes against pulmonary carcinogenesis. This protective mechanism may occur due to CYP1A2-induced suppression of CYP1A1 enzyme following PAH exposure. In Cyp1a2(−/−) mice, CYP1A1 is not suppressed, allowing for increased DNA adduct formation and resulting lung tumor development.
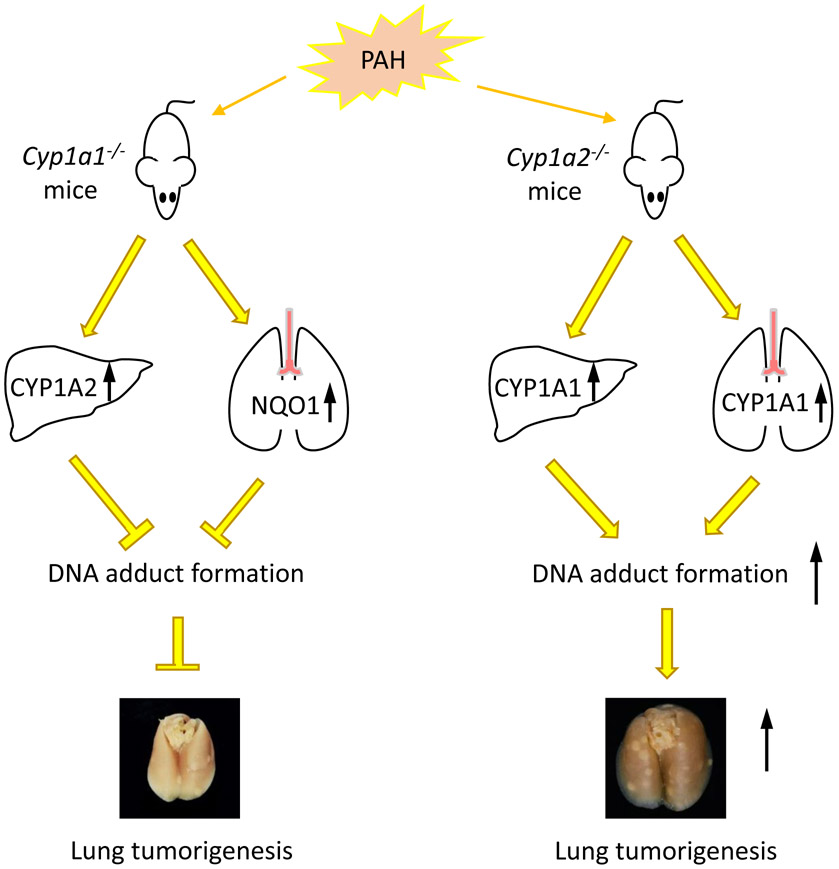
Upon analysis of TCGA RNA expression in lung cancer patients, Grady, et al. found that patients with lung adenocarcinoma had a higher chance of survival with higher levels of CYP1A2 expression compared to patients with low or medium expression [37]. Analysis of sex differences showed that the high levels of CYP1A2 imparted a survival benefit to males and much less so to females [37]. Further study of this possible interconnected nature is crucial to understanding the role CYP1A2 enzymes and hepatic metabolism of PAHs may play in pulmonary tumor formation. Figure 6 summarizes the biochemical changes that occur in Cyp1a1 and 1a2-null mice when exposed to PAH.
Figure 6.
PAH exposure in Cyp1a1-null mice causes upregulation of hepatic CYP1A2 enzyme and attenuation of hepatic DNA adduct formation. In the lungs, NQO1 enzyme is upregulated. Upon analysis, Cyp1a1-null mice demonstrated reduced lung tumor formation compared to WT mice. In Cyp1a2-null mice, PAH exposure caused an upregulation of CYP1A1 enzyme in both the liver and the lungs, which increases DNA adduct formation. These mice showed increased lung tumor formation compared to WT mice.
Upregulation of hepatic CYP1A2 enzymes may offer a potential therapeutic target for lung cancer
Recently, a myriad number of studies have been undertaken with a goal to prevent cancer using chemicals such as drugs, herbal preparations, and from dietary sources (e.g., green tea) [71-75]. While this area of research is getting great attention, the approach to use specific CYP1A2 inducers has not been explored, and this will be a logical extension of the proposed mechanism. In fact, there are many drugs such as olanzapine, a CYP1A2 inducer in humans, which is being used to treat psychiatric disorders [76, 77]. It is highly possible that the proposed research could lead to novel therapeutic approaches to prevent/treat lung cancer in humans. However, the use of CYP1A2 inducers must be used with caution because CYP1A2 enzymes may also activate aromatic amines, which could contribute to other cancers, including prostate and breast cancer [78, 79]. The balance between the protective role of CYP1A2 in pulmonary carcinogenesis and its contributory role in other cancers must fully be understood before CYP1A2 inducers can be utilized as a potential lung cancer treatment.
Carcinogenic Role of CYP1B1 in PAH Metabolism
CYP1B1 enzyme is found in a variety of tissues and it plays a similar role as CYP1A1 enzyme in PAH metabolism [80-82]. The production of reactive metabolites and DNA adduct formation by CYP1B1-mediated metabolism of PAHs indicate that CYP1B1 is also carcinogenic. However, CYP1A1 and CYP1B1 differ slightly with regards to their products from BP metabolism. While they both produce BP-cis and trans-7,8-dihydrodiol isomers, CYP1B1, in conjunction with epoxide hydrolase, produces BP-7,8-diol at a higher rate than CYP1A1, and CYP1A1 more preferentially produces BP diol-epoxides [4]. Animal studies of Cyp1b1−/− mice demonstrated reduced tumor production in these mice when compared to WT mice with prolonged exposure of MC [68]. Furthermore, Cyp1a1/1a2−/− mice showed increased tumor formation and Cyp1a1/1a2/1b1−/− mice showed decreased tumor formation after MC exposure when compared to WT mice [68]. These results provide a significant indication of the importance of CYP1B1 in augmenting the pulmonary carcinogenesis process. Therefore, CYP1A1 should not be the only enzyme potentially targeted for therapy in lung cancer. Concentrations of CYP1A2 and CYP1B1 enzymes may also need to be evaluated and altered to enhance the efficacy of treatment. Similar to CYP1A1 enzymes, certain CYP1B1 polymorphic enzymes also may augment the risk of developing lung cancer [83, 84]. CYP1B1 enzymes must be considered in conjunction with CYP1A1/2 enzymes when analyzing genetic risk factors and developing new therapeutic strategies for lung cancer treatment.
Another important aspect of the CYP1B1 metabolic mechanism is its connection to estrogen levels and the potential for estrogen-mediated sex differences [85]. CYP1B1 is a key enzyme in the metabolism of 17β-estradiol and, therefore, is expressed in higher levels in tissues connected to estrogen metabolism, such as mammary glands, the uterus, and ovaries [85]. Although our studies indicate a possibility of sex differences in CYP1B1 expression and resulting differences in pulmonary carcinogenesis via PAH metabolism, more experimentation of these sex differences are required to understand its significance [68].
Sustained Induction of CYP1A1 Enzymes
Studies from our laboratories have shown intriguing results in regard to a mechanism that elicits induces sustained induction of CYP1A1 enzyme following PAH exposure in that the induction continues to occur even after the PAH molecule has been dispelled from the body. These results indicate that biomolecular changes occur that may induce transcription of the enzyme even after PAH has been removed from the physiological environment.
Our research has found that MC exposure leads to sustained induction of CYP1A1 by mechanisms other than persistence of the drug. We reported earlier that 4 daily administrations of MC or DBA lead to induction of ethoxyresorufin O-deethylase (EROD) activity, CYP1A1 apoprotein, and mRNA expression for several weeks after MC withdrawal [86, 87]. MC undergoes extensive hepatic biotransformation to water-soluble metabolites, which are excreted into bile [88, 89]. Our discovery that MC elicits a sustained induction of CYP1A1 for as many as 45 days after cessation of treatment led to the hypothesis that MC elicits this persistent induction by mechanisms other than persistence of the drug [90, 91]. Our studies showed rapid elimination of MC from the body, and after 15 days, when high CYP1A1 activities are expressed, the intra-hepatic concentration of the parent compound was 270 pmol/g, a concentration which we have shown to be insufficient to induce CYP1A1 in liver [90]. These results support our hypothesis that the phenomenon of persistent induction by MC occurs by mechanisms other than persistence of the drug and that novel molecular mechanisms mediate this effect. We hypothesize that MC elicits sustained induction of CYP1A1 by persistent increase in the transcription of the CYP1A1 gene [90-92].
Role of CYP1A2 in the Persistent Induction of CYP1A1 by MC
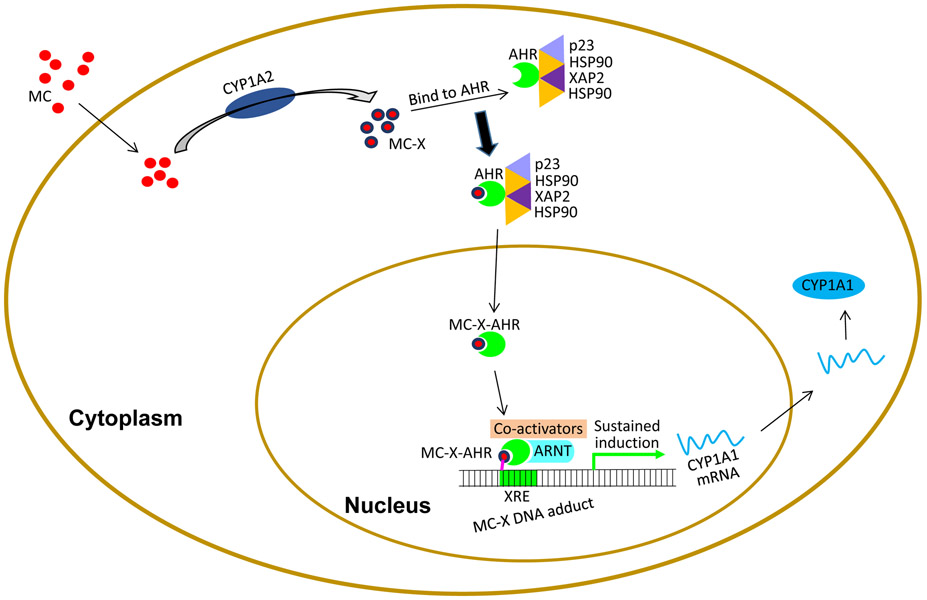
• We reported earlier the phenomenon of persistent induction of hepatic and pulmonary CYP1A1 by MC in C57BL/6J mice [92]. When we exposed WT and Cyp1a2-null mice to MC, the extent of initial nduction of hepatic CYP1A1 was much higher in the Cyp1a2-null mice, suggesting that CYP1A2 plays a regulatory role in the suppression of CYP1A1 induction by MC. While persistence of induction in the liver declined at a faster rate than WT mice at later time points, it was enhanced in the lung. Moreover, we observed increased tumorigenesis in the Cyp1a2-null mice exposed to MC. We hypothesize that CYP1A2-derived MC-X contributes to sustained induction via sequence-specific DNA adduct formation on the CYP1A1 promoter [93]. We posit that in the non-induced state, CYP1A1 is silenced by binding of HDAC and DNMT to CYP1A1 promoter. When cells are exposed to MC after the initial induction of CYP1A1 and 1A2 occurs, CYP1A2 converts MC to MC-X, which binds to the AHR, enters the nucleus. The MC-X-AHR complex then binds to ARNT, and being electrophilic, the MC-X binds covalently to XREs on the CYP1A1 promoter. This binding prevents binding of HDAC and DNMT to the promoter, thereby maintaining sustained CYP1A1 induction (Figure 7).
Figure 7.
Role of CYP1A2 in the sustained CYP1A1 induction. We propose that CYP1A2 may do this by metabolizing MC to MC-X, which binds to the AHR complex. After dissociation, the MC-X-AHR complex binds to ARNT at the XRE binding site and forms a MC-X DNA adduct. We posit that the MC-X binds covalently to DNA, and prevents binding of DNMT and HDAC that keep the promoter in silenced state, leading to sustained CYP1A1 induction.
These results reinforce the possible importance of hepatic enzyme driven PAH metabolism in pulmonary carcinogenesis. The fact that persistent CYP1A1 induction is attenuated in liver of Cyp1a2-null mice could be due to absence of formation of MC-X in the liver. In the lungs, it is possible that another CYP isoform (e.g., CYP1B1) catalyzes the formation of MC-X, thereby leading to persistent CYP1A1 induction in lung in the Cyp1a2-null mice.
Significance of Persistent CYP1A1 Induction in Lung
Our preliminary studies showing increased expression of CYP1A1 in pulmonary tumor cells, compared to normal cells several months after MC treatment, strongly suggest that the phenomenon of persistent CYP1A1 induction plays a mechanistic role in lung tumorigenesis by PAHs. The studies of Shervington, et al. [94], identifying a novel association between CYP1A1 and telomerase in A549 cells suggest that CYP1A1 siRNA can be an effective treatment for lung cancer [95]. We reported earlier that MC causes sustained induction of CYP1A1 in mouse lung [92]. We also observed that MC elicits long-term induction of pulmonary CYP1A1 in mice lacking the gene for CYP1A2 [92], a phenomenon that may be of mechanistic relevance to MC-mediated lung tumorigenesis. Furthermore, we observed that MC causes sustained induction of phase II enzymes in rats [96]. These results hold great significance since we have seen that in lung cancer patients who quit smoking, it takes 3 months for CYP1A1 levels to return to normal [97, 98]. This mechanism may have an effect on continued pulmonary tumorigenesis even after cessation of smoking. Thus, determination of the mechanisms of persistent induction and the biological implications of this phenomenon will enable development of rational strategies for the prevention and treatment of cancers caused by environmental chemicals.
Role of Epigenetics Alterations on PAH- Carcinogenesis
During PAH exposure, the carcinogenic affects from the CYP1A1/CYP1B1 pathway may be compounded by PAH-induced epigenetic alterations. As is the case with most mechanisms of carcinogenesis development, one primary mechanism is not sufficient to elicit carcinogenesis. Other carcinogenetic mechanisms, such as epigenetic alterations, must also occur concurrently. Broadly, epigenetics entails alterations in DNA methylation and histone modification via acetylation and deacetylation. DNA methylation prevents transcription of DNA segments, often turning off tumor suppressor genes. Another epigenetic alteration can occur via non-coding RNA, which binds and downregulates its target gene [99]. Tumor progression occurs when the downregulated target gene is a tumor suppressor gene. Epigenetic gene silencing can be propagated through cell cycling and replication until an accumulation of these mutations results in cancer induction.
PAH-Induced DNA Methylation
PAH exposure affects the levels of DNA methylation of AHRR and Coagulation Factor II Thrombin Receptor-Like 3 (F2RL3) genes in the blood. The hypomethylation of these genes occurs at CpG sites. As previously described in the section “Role of PAH in CYP1A1/CYP1B1 transcription”, AHRR competitively inhibits AHR from binding with ARNT and triggering CYP1 enzyme transcription. Hypomethylation of AHRR may inactivate it, thus promoting CYP1 transcription and subsequent DNA adduct formation and pulmonary carcinogenesis. Interestingly, F2RL3 plays a crucial role in thrombin-mediated platelet activation in the coagulation system [100]. Although the mechanistic connection between PAH exposure and F2RL3 hypomethylation is not fully understood, it is hypothesized that PAH triggers an inflammatory response in peripheral blood cells, which includes Coagulation F2RL3 [100, 101]. Studies have found that F2RL3 and AHRR methylation levels in the blood have a high sensitivity and selectivity for lung cancer incidence and mortality [102, 103]. These results indicate that peripheral blood levels of F2RL3 and AHRR methylation may be a promising biomarker that could potentially be used for early lung cancer diagnosis.
Other DNA methylation pathways have also been shown to occur that may promote pulmonary carcinogenesis. BP diol epoxide (BPDE)-exposed human bronchial epithelial (HBE) cells display an increase in the expression of DNA methyltransferases as well as the Cadherin 13 (CDH13) [104]. CDH13 is an anti-oncogene; therefore, downregulation of this gene via hypermethylation of its promoter region may contribute to pulmonary carcinogenesis [105]. Furthermore, Li, et al. found that certain single nucleotide polymorphisms (SNPs) near the CDH13 CpG promoter sites may increase the risk of lung cancer development, reinforcing the idea of an interplay between genetic and epigenetic factors [105].
Another mechanism in which DNA methylation may promote lung cancer is via the p16 tumor suppressor pathway. HBE cells exposed to BP displayed hypermethylation at the CpG cites of p16 genes, thereby inhibiting p16 gene expression, which prevents cell cycle arrest and apoptosis [106]. Based on the proposed mechanism by Yang, et al., p16 hypermethylation may be induced by the DNA damage caused by CYP1A1-produced DNA adducts binding to the p53 CpG promoter regions [106]. Although this proposed mechanism has not yet been confirmed, it further suggests an important connection between the genetic and epigenetic alterations caused by PAH exposure and warrants further experimentation. BP-induced p16 inhibition is an additional mechanism that may significantly promote tumor formation and progression. Furthermore, understanding these mechanisms may offer potential biomarkers that could be targeted for diagnosis and therapeutic treatment.
Histone Modifications
Histone acetylation via histone acetyltransferases (HATs) reduces histone protein affinity, thereby, opening the previously tightly bound structure so that DNA transcription may occur [107]. Alterations in HATs and HDACs levels may disrupt DNA transcription, resulting in carcinogenesis. Studies with mice have found that cigarette smoke exposure to be implicated in downregulating HDAC2 due to prolonged oxidative stress exposure [108, 109]. One proposed mechanism is that lipid peroxidation caused by oxidative stress from components of cigarette smoke covalently modify the HDAC2 protein, resulting in decreased activity [108, 110]. The decreased HDAC2 protein would result in an imbalance between HATs and HDACs, leading to increased acetylation of histones and inflammatory gene transcription [111]. More studies need to be conducted to examine the exact mechanism in which this downregulation of HDAC2 may contribute to the epigenetic component of pulmonary carcinogenesis.
Another significant epigenetic alteration due to PAH exposure is the di-methylation of Lysine 79 in histone H3 (H3K79). One study found that PAH exposure in Chinese coke oven workers resulted in decreased H3K79me2 and increased DNA damage [112]. Studies have proposed the mechanism behind this relationship may be due to BP-mediated suppression of H3K79 methyltransferase DOT1 expression, resulting in decreased methylation of H3K79. Decreased levels of H3K79me2 may increase cellular sensitivity to gamma-irradiation, which would further propagate chromosomal instability and carcinogenesis [113]. Other studies have proposed that H3K79me2 binds to 53DP1 and enhances double-strand break (DSB) repair in DNA [114], [115]. Furthermore, methylated H3K79 may also promote DNA repair by augmenting nuclear excision repair of DNA lesions created by ultraviolet exposure [116]. Both of these mechanisms indicate the importance of H3K79me2 in DNA repair. Therefore, reduction of H3K79me2 levels due to PAH exposure will increase errors in DNA replication, resulting in carcinogenesis promotion.
Histone modification can occur in a multitude of mechanisms due to PAH and tobacco smoke exposure. Altering the balance between HATs and DHACs allows histone acetylation of inflammatory DNA strands and subsequent transcription to be upregulated. Moreover, decreased methylation of histones directly may also inhibit DNA repair. These mechanisms may play important contributory roles to pulmonary carcinogenesis and require further exploration as potential targets of cancer diagnosis and treatment.
BP-Induced LINE-1 Retrotransposition
Long interspersed nuclear elements (LINEs) represent a class of transposable elements in the human genome. The LINE family is composed of LINE-1, LINE-2, and LINE-3, and contribute to a fifth of the human genome; however, LINE-1 (L1) is the only member of the family that can still be activated by demethylation [117]. L1 retrotransposition requires activation of its two open reading frames, ORF1 and ORF2 [118]. ORF2 encodes a reverse transcriptase and endonuclease that allows reverse transcription of L1 into cDNA, which can be inserted into the genome resulting in instability [119]. Regulatory region L1 insertions, homologous recombination, and L1-induced chromosomal deletions are all mechanisms by which L1 may cause genomic instability and cancer progression [120]. Studies have found that BP may further promote carcinogenesis by inducing sustained activation of L1 [121, 122]. Previous work has shown that BP reactivates LINE-1 in bronchial epithelial cells through displacement of nucleosome remodeling and deacetylase (NuRD) corepressor complexes, and interfering with retinoblastoma-regulated epigenetic signaling [74]. It is not clearly known whether LINE-1 in coordination with other genes within its regulatory network contributes to the in vivo genotoxic response to BP. Hassanin, et al. recently showed that intratracheal instillation of ORFeusLSL mice with BP alone or in combination with adenovirus (adeno)-CRE recombinase is genotoxic to the lung and associated with activation of the human LINE-1 transgene present in these mice [121]. LINE-1 reactivation modulated the expression of genes involved in oncogenic signaling, and these responses were most pronounced in female mice compared with males and synergized by adeno-CRE recombinase. This is the first report linking LINE-1 and genes within its oncogenic regulatory network with early sexually dimorphic responses of the lung in vivo [121].
Effects of PAH on Non-Coding RNA
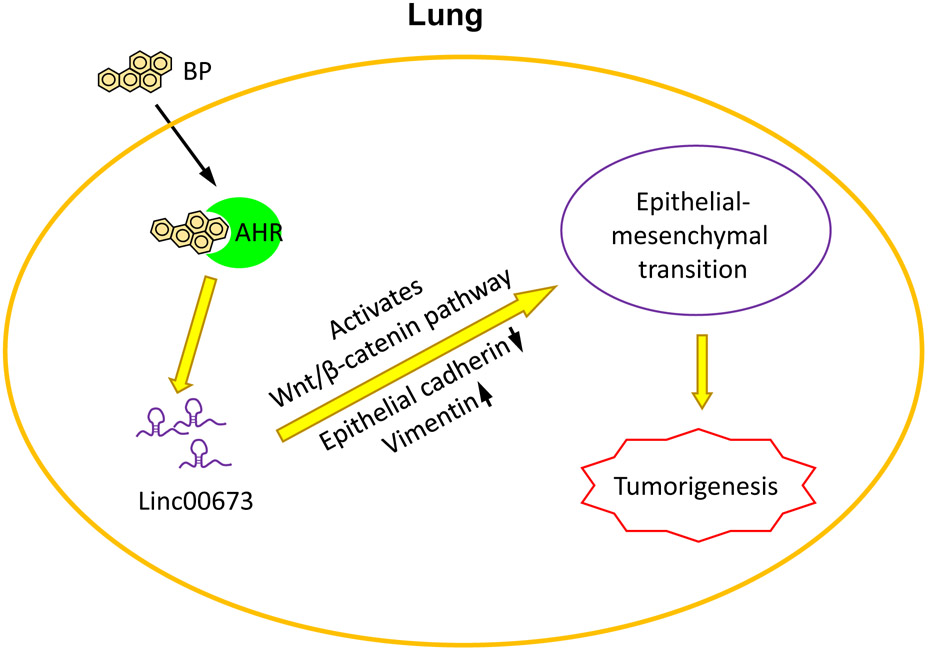
Changes in non-coding RNA, or microRNA (miRNA), have been correlated with changes in tumor suppressor gene and oncogene levels. Based on the results of various studies, exposure of PAH and its metabolites on HBE cells triggered both carcinogenic and compensatory mechanisms via non-coding RNA, which may contribute to pulmonary carcinogenesis. Some key oncogenic non-coding RNA that were examined in these experiments were miR-22 and miR-494, which decrease the tumor suppressor gene PTEN, miR-106a, which prevents cell cycle arrest and apoptosis, and miR-638, which may suppress BRCA1 proteins [123]. Studies found that HBE cells exposed to anti-benzo[a]pyrene-trans-7,8-diol-9,10-epoxide had increased levels of these four non-coding RNA segments. Another mechanism discovered to promote lung cancer following BP exposure is by upregulation of the non-coding RNA linc00678 [124]. Upregulation of linc00678 increased transition of epithelial cells to mesenchymal cells, thus promoting lung cancer development. Figure 8 demonstrates this mechanism as an example in which PAH induces non-coding RNA.
Figure 8.
Role of non-coding RNA in lung cancer. One epigenetic mechanism altered by BP exposure is non-coding RNA alterations. For example, BP binds to AHR, which in turn upregulates the non-coding RNA linc00673. This upregulation promotes epithelial-mesenchymal transition, resulting in lung cancer. This is just one example of the epigenetic alterations caused by PAH exposure.
On the other hand, compensatory mechanisms via non-coding RNA in response to the oncogenic changes also occurred. These HBE cells contained miR-622 and miR-506, which suppressed malignant transformation by decreasing KRAS and NRAS proteins as well as NRAS mRNA levels [125, 126]. In another study, BP-treated human bronchial epithelial cells showed a positive correlation with miR-34c and phosphorylated p53 proteins, which decreased BP carcinogenesis [127]. These changes seem to be occurring as a compensatory mechanism in response to BP-induced cellular alterations, but further investigation of these non-coding RNA changes and their possible impact on pulmonary carcinogenesis require further investigation.
Future Research Needs
The areas of research relating to PAH-induced pulmonary carcinogenesis are numerous. However, there are few key areas that warrant further studies. Based on the promising results from our studies, one of the first mechanisms that needs more dedicated research is the protective role hepatic CYP1A2 enzymes may play in pulmonary carcinogenesis. This interconnected pathway between enzyme metabolism in the liver and tumor progression in the lungs may offer tremendous insight into other such pathways. This pathway may also revolutionize cancer treatment strategies by targeting organs other than the cancerous organ. Because of the regulatory mechanism CYP1A2 enzymes have on CYP1A1 enzymes, the CYP1A2 enzyme expression could potentially be utilized to downregulate or control CYP1A1 enzyme expression and possibly control cancer progression. Animal studies will continue to be a key tool for understanding this mechanism and for eventually utilizing the mechanism to develop alternative treatment strategies.
The second area of research that will have a significant impact on our understanding and treatment of lung cancer is the polymorphic variations of key enzymes in pulmonary tumorigenesis. Increased risk of lung cancer has been linked to not only CYP1A1 and CYP1B1 enzymes but also GSTM1, GSTT1, GSTP1, AKR, microsomal epoxide hydrolase (EPHX1), and NQO1 [4]. These polymorphisms may also affect epigenetic alterations that could further increase the risk of cancer development and progression. As individualized medicine continues to become more of a reality in medicine, genomic risk factors may start to play a role in early diagnosis and treatment strategy selection. It is imperative to continue this genetic exploration in order to keep up with the rapidly changing medical landscape.
Finally, various PAH mixtures are required to develop a structured scientific research. because BP makes up about 51-64% of the total carcinogenic potential, current studies on PAH molecules and their effects on pulmonary carcinogenesis often utilize BP as representative of the PAH class of molecules [7]. However, many more of these molecules have been identified as having carcinogenic effects and warrant further analysis, namely acenapthene, benzo[b]fluoranthene, benzo[j]fluoranthene, benzo[k]fluoranthene, chrysene, diben[a,h]anthracene, dibenzo[a,l]pyrene, and indeno[1,2,3-c,d]pyrene [7]. Furthermore, PAHs often occur in mixtures, rather than individually. The kinetics, reactivity, enzymatic induction, and carcinogenic potential may alter depending on the exact composition of the different mixtures. One study of lung cancer patients in Romania found that the mixture of PAHs contained BP, benz[a]anthracene, anthracene, fluoranthene, benzo[b]fluoranthene, and benzo[k]fluoranthene [128]. This study demonstrated how diverse PAH exposures resulting in lung cancer can be. By incorporating these mixtures into studies, we would emulate the real-world exposure and mechanisms more accurately. Cataloging various PAH mixtures and, subsequently, using these mixtures in animal models or basic science studies could reveal important differences in properties these mixtures may have, such as carcinogenic potential and kinematics.
Finally, as highlighted in Figure 9, CYP1 enzymes and epigenetic alterations both play a role in PAH-mediated pulmonary carcinogenesis. While many studies have been conducted to understand these mechanisms separately, the interplay between these two systems may hold tremendous insight into cancer development and requires further exploration. Their effects may compound to increase the risk of tumor progression. Furthermore, identification and understanding of any causal or sequential relationship between the two mechanisms may illuminate more effective treatment methods. Integrating these key pathways that have been previously studied separately will be imperative as cancer research continues to progress.
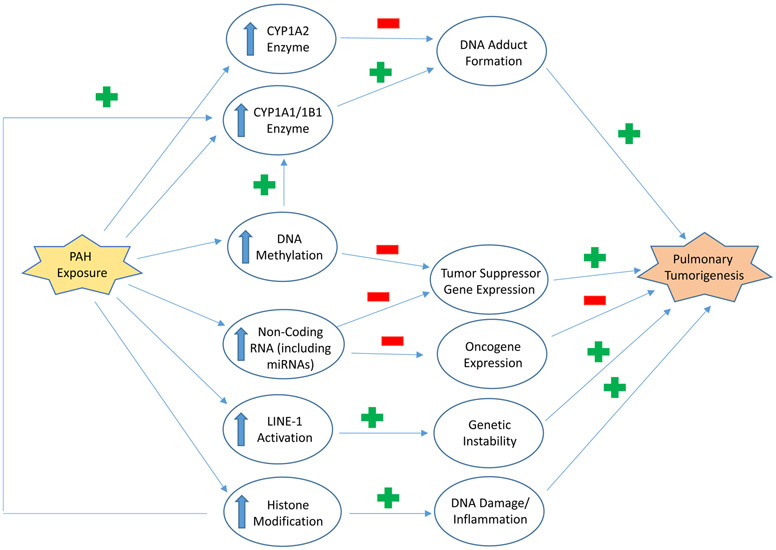
Figure 9.
PAH may contribute to pulmonary carcinogenesis through a multitude of mechanisms. One mechanism is through the upregulation of CYP1A1/CYP1B1 enzymes, which form DNA adducts on key gene sequences, such as p53. These mutations result in lung tumor formation. CYP1A2 is also upregulated by PAH exposure but has a protective mechanism against carcinogenesis. PAH exposure also induces epigenetic changes of DNA methylation and non-coding RNAs, including microRNA (miRNA). These epigenetic mechanisms downregulate tumor suppressor gene expression, thereby promoting pulmonary tumorigenesis. Non-coding enzymes can also inhibit oncogene expression, which serves as a protective mechanism and prevents tumor formation. Long interspersed nuclear element (LINE) activation is also induced by PAH exposure. Due to activation, LINE-1 insertions cause genetic instability and tumor development. PAHs have has been linked to histone modification, which induces proteins that promote DNA damage and inflammation. Histone modification also occurs during upregulation of CYP1A1 transcription caused by the PAH-AHR complex and further contribute to pulmonary carcinogenesis via DNA adduct formation.
Summary and Conclusions
The mechanisms behind PAH-induced pulmonary carcinogenesis may contain important implications for improvements in lung cancer screening, diagnosis, and treatment. PAH exposure occurs in everyday life through tobacco smoke exposure, consumption of charcoal-smoked meats, and air pollutant inhalation. PAHs are metabolized hrough three key pathways: (1) CYP1A1/CYP1B1 and epoxide hydrolase pathway, (2) CYP peroxidase pathway, and (3) aldo-keto reductase. This review analyzed the significance of the CYP1A1/CYP1B1 pathway and the contribution of these enzymes to pulmonary carcinogenesis.
Both CYP1A1 and CYP1B1 are found throughout the body, especially in the lungs. During the metabolism of PAH molecules, CYP1A1/1B1 enzymes produce reactive metabolites that react to form DNA adducts, which can contribute to tumor progression. One important protein mutated from these DNA adducts is p53, which prevent cell cycle arrest and apoptosis. Furthermore, PAH upregulates CYP1A and CYP1B1 transcription by binding to AHR, which promotes XRE DNA binding. Human studies demonstrate upregulation of these enzymes in pulmonary tumor cells, and animal studies show how deficiencies in these enzymes reduce tumor progression. These findings indicate the importance CYP1A1 and CYP1B1 enzymes may have in development of new lung cancer treatment options. On the other hand, hepatic CYP1A2 enzymes seem to play a protective role by reducing both pulmonary and hepatic CYP1A1 expression, thus, preventing DNA adduct formation and protecting against pulmonary carcinogenesis.
New studies indicate a revolutionary mechanism in which CYP1A1 enzymes undergo sustained induction following PAH exposure and subsequent removal. The implications for this persistent induction of the enzymes may hold clinical relevance since CYP1A1 enzyme levels remain high up to 3 months following smoking cessation. This time frame allows for further DNA adduct formation and resulting carcinogenesis. CYP1A2 may further play a protective role against lung cancer progression by suppressing this sustained induction. These results highlight the importance of regulating these enzymes even after removing the PAH exposure in order to prevent any further tumor progression.
The PAH-induced epigenetic alterations have also been shown to contribute to pulmonary carcinogenesis. The CYP1 enzyme transcription seems to be upregulated by hypomethylation of AHRR at CpG sites. Tumor suppressor proteins CDH13 and p16 may be suppressed via PAH-induced hypermethylation. PAH exposure seems to further contribute to epigenetic alterations by disrupting the balance between HATs and HDACs, which can promote DNA damage and inflammation. BP exposure has also been shown to promote LINE-1 activation, which leads to genetic instability via DNA insertions. Another epigenetic factor that may enhance lung tumor progression is through non-coding RNA levels, which inhibit key tumor suppressor genes. These epigenetic changes may compound with CYP1 enzymatic DNA adduct formation to further enhance pulmonary carcinogenesis triggered by PAH exposure.
The importance of improving lung cancer diagnosis and treatment is significant. Because lung cancer has the second highest incidence and has the leading mortality rate of all cancers, new screening methods and therapy options should be at the forefront of medical research. Consideration of PAH exposure and its underlying mechanisms may be critical to achieving these advancements.
Other areas that require further research include the significance of polymorphic enzymes on increased lung cancer risk as well as the kinematic differences of PAH mixtures. Finally, as highlighted in this review, the relationship between DNA adduct formation induced by CYP1-dependent metabolism and epigenetic alterations may provide a compounding mechanism in which pulmonary carcinogenesis can occur. Further investigation into how these two mechanisms relate may give great insight into lung cancer progression. Metabolic enzyme regulation and epigenetic biomarker utilization are only a few of the many promising mechanisms that could be utilized in the development of these tools and therapies.
Acknowledgements:
This work was supported in part by USPHS grants 5R01ES 009132, R01HL129794, 1R01ES029382, and 1P42 ES0327725, and RP190279 from Cancer Prevention Research Institute of Texas (CPRIT) to BM.
Abbreviations
- BP
Benzo(a)pyrene
- LINE
Long interspersed nuclear element
- AHR
Ah receptor
- ARNT
Ah receptor nuclear translocator
- PAH
Polycyclic aromatic hydrocarbon
- CYP
Cytochrome P450
- XRE
Xemobiotic regulatory enzymes
- LDCT
Low-dose computed tomography
- DBA
Dibenzo[a,h]anthracene
- MC
3-methylcholanthrene
- WT
Wild type
- ROS
Reactive Oxygen Speces
- Nrf2
Nuclear factor erythroid 2-related factor 2
- NQO
NAD(P)H quinone oxidoreductase
- UGT
UDP-glucuronosyltransferase
- GST
Glutathione S-transferase
- HDAC
Histone deacetylase
- DNMT
DNA methyltransferase
- AHHR
Aryl hydrocarbon receptor repressor
References
- 1.Wong MCS, et al. , Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep, 2017. 7(1): p. 14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Who Should Be Screened for Lung Cancer? 2021. March 11, 2021; Available from: https://www.cdc.gov/cancer/lung/basic_info/screening.htm. [Google Scholar]
- 3.Reid BC, et al. , Research opportunities for cancer associated with indoor air pollution from solid-fuel combustion. Environ Health Perspect, 2012. 120(11): p. 1495–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moorthy B, Chu C, and Carlin DJ, Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol Sci, 2015. 145(1): p. 5–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agency for Toxic Substances and Disease Registry (ATSDR), P.H.S., Polycyclic Aromatic Hydrocarbons, December 1990, U.S.D.o.H.a.H.S. U.S. Public Health Service, Atlanta, GA, Editor. December, 1990. [Google Scholar]
- 6.Choi H, H. R, Komulainen H, and Delgado Saborit JM, Polycyclic aromatic hydrocarbons. WHO Guidelines for Indoor Air Quality: Selected Pollutants, 2010. 6. [Google Scholar]
- 7.Group IW, BENZO[a]PYRENE, in Chemical Agents and Related Occupations. 2012, Lyon (FR): International Agency for Research on Cancer. [Google Scholar]
- 8.Eom SY, et al. , Polycyclic aromatic hydrocarbon-induced oxidative stress, antioxidant capacity, and the risk of lung cancer: a pilot nested case-control study. Anticancer Res, 2013. 33(8): p. 3089–97. [PubMed] [Google Scholar]
- 9.Osgood RS, et al. , Polycyclic aromatic hydrocarbon-induced signaling events relevant to inflammation and tumorigenesis in lung cells are dependent on molecular structure. PLoS One, 2013. 8(6): p. e65150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsay JJ, et al. , Aryl hydrocarbon receptor and lung cancer. Anticancer Res, 2013. 33(4): p. 1247–56. [PMC free article] [PubMed] [Google Scholar]
- 11.DeMarini DM, Genotoxicity of tobacco smoke and tobacco smoke condensate: a review. Mutat Res, 2004. 567(2-3): p. 447–74. [DOI] [PubMed] [Google Scholar]
- 12.Nebert DW, et al. , Oral Benzo[a]pyrene: Understanding Pharmacokinetics, Detoxication and Consequences--Cyp1 Knockout Mouse Lines as a Paradigm. Mol Pharmacol, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jarup L, Hazards of heavy metal contamination. Br Med Bull, 2003. 68: p. 167–82. [DOI] [PubMed] [Google Scholar]
- 14.Rozman KKK, D. C, Absorption, distribution and excretion of toxicants, in Casarett and Doul´s Toxicology: The Basic Science of Poisons, Klaassen CD, Editor. 2007, McGraw-Hill: New York. p. 107–132. [Google Scholar]
- 15.Simko P, Determination of polycyclic aromatic hydrocarbons in smoked meat products and smoke flavouring food additives. J Chromatogr B Analyt Technol Biomed Life Sci, 2002. 770(1-2): p. 3–18. [DOI] [PubMed] [Google Scholar]
- 16.Mottier P, Parisod V, and Turesky RJ, Quantitative determination of polycyclic aromatic hydrocarbons in barbecued meat sausages by gas chromatography coupled to mass spectrometry. J Agric Food Chem, 2000. 48(4): p. 1160–6. [DOI] [PubMed] [Google Scholar]
- 17.Sinha R, et al. , Pan-fried meat containing high levels of heterocyclic aromatic amines but low levels of polycyclic aromatic hydrocarbons induces cytochrome P4501A2 activity in humans. Cancer Res, 1994. 54(23): p. 6154–9. [PubMed] [Google Scholar]
- 18.Hansen AM, Olsen IL, and Poulsen OM, Polycyclic aromatic hydrocarbons in air samples of meat smokehouses. Sci Total Environ, 1992. 126(1-2): p. 17–26. [DOI] [PubMed] [Google Scholar]
- 19.Larsson BK, et al. , Polycyclic aromatic hydrocarbons in grilled food. J Agric Food Chem, 1983. 31(4): p. 867–73. [DOI] [PubMed] [Google Scholar]
- 20.Masuda Y, Mori K, and Kuratsune M, Polycyclic aromatic hydrocarbons in common Japanese foods. I. Broiled fish, roasted barley, shoyu, and caramel. Gann, 1966. 57(2): p. 133–42. [PubMed] [Google Scholar]
- 21.Cai Y, et al. , Dietary exposure estimates of 16 polycyclic aromatic hydrocarbons (PAHs) in Xuanwei and Fuyuan, counties in a high lung cancer incidence area in China. J Environ Monit, 2012. 14(3): p. 886–92. [DOI] [PubMed] [Google Scholar]
- 22.Jiang H, et al. , Competing roles of cytochrome P450 1A1/1B1 and aldo-keto reductase 1A1 in the metabolic activation of (+/−)-7,8-dihydroxy-7,8-dihydro-benzo[a]pyrene in human bronchoalveolar cell extracts. Chem Res Toxicol, 2005. 18(2): p. 365–74. [DOI] [PubMed] [Google Scholar]
- 23.Stiborova M, Rupertova M, and Frei E, Cytochrome P450- and peroxidase-mediated oxidation of anticancer alkaloid ellipticine dictates its anti-tumor efficiency. Biochim Biophys Acta, 2011. 1814(1): p. 175–85. [DOI] [PubMed] [Google Scholar]
- 24.Gelboin HV, Benzo[alpha]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol Rev, 1980. 60(4): p. 1107–66. [DOI] [PubMed] [Google Scholar]
- 25.Meehan T, Straub K, and Calvin M, Benzo[alpha]pyrene diol epoxide covalently binds to deoxyguanosine and deoxyadenosine in DNA. Nature, 1977. 269(5630): p. 725–7. [DOI] [PubMed] [Google Scholar]
- 26.Meehan T and Straub K, Double-stranded DNA steroselectively binds benzo(a)pyrene diol epoxides. Nature, 1979. 277(5695): p. 410–2. [DOI] [PubMed] [Google Scholar]
- 27.Guengerich FP, Roles of cytochrome P-450 enzymes in chemical carcinogenesis and cancer chemotherapy. Cancer Res, 1988. 48(11): p. 2946–54. [PubMed] [Google Scholar]
- 28.Hemminki K, DNA adducts, mutations and cancer. Carcinogenesis, 1993. 14(10): p. 2007–12. [DOI] [PubMed] [Google Scholar]
- 29.Chaturvedi S and Lakshman MK, Site-specific modification of the human N-ras proto-oncogene with each diol epoxide metabolite of benzo[a]pyrene and thermal denaturation studies of the adducted duplexes. Carcinogenesis, 1996. 17(12): p. 2747–52. [DOI] [PubMed] [Google Scholar]
- 30.Wei D, Maher VM, and McCormick JJ, Effect of nuclear environment on the distribution of benzo[a]pyrene diol epoxide-induced adducts in the HPRT gene of human fibroblasts. Carcinogenesis, 1996. 17(12): p. 2695–701. [DOI] [PubMed] [Google Scholar]
- 31.Denissenko MF, et al. , Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science, 1996. 274(5286): p. 430–2. [DOI] [PubMed] [Google Scholar]
- 32.Pfeifer GP and Besaratinia A, Mutational spectra of human cancer. Hum Genet, 2009. 125(5-6): p. 493–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Geacintov NE, et al. , NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol, 1997. 10(2): p. 111–46. [DOI] [PubMed] [Google Scholar]
- 34.Rojas M, et al. , High benzo[a]pyrene diol-epoxide DNA adduct levels in lung and blood cells from individuals with combined CYP1A1 MspI/Msp-GSTM1*0/*0 genotypes. Pharmacogenetics, 1998. 8(2): p. 109–18. [PubMed] [Google Scholar]
- 35.Conney AH, Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res, 1982. 42(12): p. 4875–917. [PubMed] [Google Scholar]
- 36.Bennett WP, et al. , Molecular epidemiology of human cancer risk: gene-environment interactions and p53 mutation spectrum in human lung cancer. J Pathol, 1999. 187(1): p. 8–18. [DOI] [PubMed] [Google Scholar]
- 37.Gastelum G, et al. , Polycyclic Aromatic Hydrocarbon-induced Pulmonary Carcinogenesis in Cytochrome P450 (CYP)1A1- and 1A2-Null Mice: Roles of CYP1A1 and CYP1A2. Toxicol Sci, 2020. 177(2): p. 347–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kwack SJ and Lee BM, Correlation between DNA or protein adducts and benzo[a]pyrene diol epoxide I-triglyceride adduct detected in vitro and in vivo. Carcinogenesis, 2000. 21(4): p. 629–32. [DOI] [PubMed] [Google Scholar]
- 39.Moorthy B, et al. , The atherogen 3-methylcholanthrene induces multiple DNA adducts in mouse aortic smooth muscle cells: role of cytochrome P4501B1. Cardiovasc Res, 2002. 53(4): p. 1002–9. [DOI] [PubMed] [Google Scholar]
- 40.Shen G and Kong AN, Nrf2 plays an important role in coordinated regulation of Phase II drug metabolism enzymes and Phase III drug transporters. Biopharm Drug Dispos, 2009. 30(7): p. 345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cho HY, et al. , Role of NRF2 in protection against hyperoxic lung injury in mice. Am J Respir Cell Mol Biol, 2002. 26(2): p. 175–82. [DOI] [PubMed] [Google Scholar]
- 42.Reddy NM, et al. , PI3K-AKT Signaling via Nrf2 Protects against Hyperoxia-Induced Acute Lung Injury, but Promotes Inflammation Post-Injury Independent of Nrf2 in Mice. PLoS One, 2015. 10(6): p. e0129676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma Q, Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol, 2013. 53: p. 401–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawasaki Y, et al. , Clinicopathological significance of nuclear factor (erythroid-2)-related factor 2 (Nrf2) expression in gastric cancer. BMC Cancer, 2015. 15: p. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aleksunes LM and Klaassen CD, Coordinated regulation of hepatic phase I and II drug-metabolizing genes and transporters using AhR-, CAR-, PXR-, PPARalpha-, and Nrf2-null mice. Drug Metab Dispos, 2012. 40(7): p. 1366–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang DD, et al. , Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol, 2004. 24(24): p. 10941–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nguyen PM, et al. , Benzo[a]pyrene increases the Nrf2 content by downregulating the Keap1 message. Toxicol Sci, 2010. 116(2): p. 549–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kobayashi A, et al. , Oxidative and electrophilic stresses activate Nrf2 through inhibition of ubiquitination activity of Keap1. Mol Cell Biol, 2006. 26(1): p. 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S, et al. , Omeprazole induces NAD(P)H quinone oxidoreductase 1 via aryl hydrocarbon receptor-independent mechanisms: Role of the transcription factor nuclear factor erythroid 2-related factor 2. Biochem Biophys Res Commun, 2015. 467(2): p. 282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bock KW, Ah receptor- and Nrf2-gene battery members: modulators of quinone-mediated oxidative and endoplasmic reticulum stress. Biochem Pharmacol, 2012. 83(7): p. 833–8. [DOI] [PubMed] [Google Scholar]
- 51.Kwon J, et al. , Assurance of mitochondrial integrity and mammalian longevity by the p62-Keap1-Nrf2-Nqo1 cascade. EMBO Rep, 2012. 13(2): p. 150–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yeager RL, et al. , Introducing the "TCDD-inducible AhR-Nrf2 gene battery". Toxicol Sci, 2009. 111(2): p. 238–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu KC, Cui JY, and Klaassen CD, Effect of graded Nrf2 activation on phase-I and -II drug metabolizing enzymes and transporters in mouse liver. PLoS One, 2012. 7(7): p. e39006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aoki Y, et al. , Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol Appl Pharmacol, 2001. 173(3): p. 154–60. [DOI] [PubMed] [Google Scholar]
- 55.Hankinson O, The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol, 1995. 35: p. 307–40. [DOI] [PubMed] [Google Scholar]
- 56.Oyama T, et al. , Cytochrome P450 expression (CYP) in non-small cell lung cancer. Front Biosci, 2007. 12: p. 2299–308. [DOI] [PubMed] [Google Scholar]
- 57.Mimura J and Fujii-Kuriyama Y, Functional role of AhR in the expression of toxic effects by TCDD. Biochim Biophys Acta, 2003. 1619(3): p. 263–8. [DOI] [PubMed] [Google Scholar]
- 58.DeGroot DE and Denison MS, Nucleotide specificity of DNA binding of the aryl hydrocarbon receptor:ARNT complex is unaffected by ligand structure. Toxicol Sci, 2014. 137(1): p. 102–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujisawa-Sehara A, et al. , Characterization of xenobiotic responsive elements upstream from the drug-metabolizing cytochrome P-450c gene: a similarity to glucocorticoid regulatory elements. Nucleic Acids Res, 1987. 15(10): p. 4179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen ES and Whitlock JP Jr., Protein-DNA interactions at a dioxin-responsive enhancer. Mutational analysis of the DNA-binding site for the liganded Ah receptor. J Biol Chem, 1992. 267(10): p. 6815–9. [PubMed] [Google Scholar]
- 61.Zhang L, et al. , Characterization of the mouse Cyp1B1 gene. Identification of an enhancer region that directs aryl hydrocarbon receptor-mediated constitutive and induced expression. J Biol Chem, 1998. 273(9): p. 5174–83. [DOI] [PubMed] [Google Scholar]
- 62.Baba T, et al. , Structure and expression of the Ah receptor repressor gene. J Biol Chem, 2001. 276(35): p. 33101–10. [DOI] [PubMed] [Google Scholar]
- 63.Mimura J, et al. , Identification of a novel mechanism of regulation of Ah (dioxin) receptor function. Genes Dev, 1999. 13(1): p. 20–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guengerich FP, Cytochrome p450 and chemical toxicology. Chem Res Toxicol, 2008. 21(1): p. 70–83. [DOI] [PubMed] [Google Scholar]
- 65.Horley NJ, et al. , (E)-3-(3,4,5-Trimethoxyphenyl)-1-(pyridin-4-yl)prop-2-en-1-one, a heterocyclic chalcone is a potent and selective CYP1A1 inhibitor and cancer chemopreventive agent. Bioorg Med Chem Lett, 2017. 27(24): p. 5409–5414. [DOI] [PubMed] [Google Scholar]
- 66.Wang X, Yue K, and Hao LR, CYP1A1 MspI polymorphism and susceptibility to lung cancer in the Chinese population: an updated meta-analysis and review. Int J Clin Exp Med, 2015. 8(8): p. 11905–12. [PMC free article] [PubMed] [Google Scholar]
- 67.Wang J, et al. , Genetic predisposition to lung cancer: comprehensive literature integration, meta-analysis, and multiple evidence assessment of candidate-gene association studies. Sci Rep, 2017. 7(1): p. 8371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gastelum GM, Role of Cytochrome P4501 (CYP)1 Enzymes in Polycyclic Aromatic Hydrocarbon (PAH)-Induced Pulmonary Carcinogenesis, in The Graduate School of Biomedical Sciences 2020, Baylor College of Medicine [Google Scholar]
- 69.Guessous I, et al. , Caffeine intake and CYP1A2 variants associated with high caffeine intake protect non-smokers from hypertension. Hum Mol Genet, 2012. 21(14): p. 3283–92. [DOI] [PubMed] [Google Scholar]
- 70.Bu Z-B, et al. , Four Polymorphisms in the Cytochrome P450 1A2 (CYP1A2) Gene and Lung Cancer Risk: a Meta-analysis. Asian Pacific Journal of Cancer Prevention, 2014. 15(14): p. 5673–5679. [DOI] [PubMed] [Google Scholar]
- 71.Ma Q and Lu AY, CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos, 2007. 35(7): p. 1009–16. [DOI] [PubMed] [Google Scholar]
- 72.Eggler AL, Gay KA, and Mesecar AD, Molecular mechanisms of natural products in chemoprevention: induction of cytoprotective enzymes by Nrf2. Mol Nutr Food Res, 2008. 52 Suppl 1: p. S84–94. [DOI] [PubMed] [Google Scholar]
- 73.Brausi M, Rizzi F, and Bettuzzi S, Chemoprevention of human prostate cancer by green tea catechins: two years later. A follow-up update. Eur Urol, 2008. 54(2): p. 472–3. [DOI] [PubMed] [Google Scholar]
- 74.Ramos S, Cancer chemoprevention and chemotherapy: dietary polyphenols and signalling pathways. Mol Nutr Food Res, 2008. 52(5): p. 507–26. [DOI] [PubMed] [Google Scholar]
- 75.Surh YJ and Chun KS, Cancer chemopreventive effects of curcumin. Adv Exp Med Biol, 2007. 595: p. 149–72. [DOI] [PubMed] [Google Scholar]
- 76.Laika B, et al. , Pharmacogenetics and olanzapine treatment: CYP1A2*1F and serotonergic polymorphisms influence therapeutic outcome. Pharmacogenomics J, 2010. 10(1): p. 20–9. [DOI] [PubMed] [Google Scholar]
- 77.Urichuk L, et al. , Metabolism of atypical antipsychotics: involvement of cytochrome p450 enzymes and relevance for drug-drug interactions. Curr Drug Metab, 2008. 9(5): p. 410–8. [DOI] [PubMed] [Google Scholar]
- 78.Ayari I, et al. , Role of CYP1A2 polymorphisms in breast cancer risk in women. Mol Med Rep, 2013. 7(1): p. 280–6. [DOI] [PubMed] [Google Scholar]
- 79.Koda Masahide, I. M, Yamano Yuko, Lu Xi, Katoh Takahiko Association between NAT2, CYP1A1, and CYP1A2 genotypes, heterocyclic aromatic amines, and prostate cancer risk: a case control study in Japan. Environmental Health and Preventative Medicine, 2017. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Spivack SD, et al. , CYP1B1 expression in human lung. Drug Metab Dispos, 2001. 29(6): p. 916–22. [PubMed] [Google Scholar]
- 81.Larsen MC, et al. , Characterization of CYP1B1 and CYP1A1 expression in human mammary epithelial cells: role of the aryl hydrocarbon receptor in polycyclic aromatic hydrocarbon metabolism. Cancer Res, 1998. 58(11): p. 2366–74. [PubMed] [Google Scholar]
- 82.Murray GI, et al. , Tumor-specific expression of cytochrome P450 CYP1B1. Cancer Res, 1997. 57(14): p. 3026–31. [PubMed] [Google Scholar]
- 83.Lao X, et al. , Association of CYP1B1 Leu432Val polymorphism and lung cancer risk: an updated meta-analysis. Lung, 2014. 192(5): p. 739–48. [DOI] [PubMed] [Google Scholar]
- 84.Liu C, et al. , Genetic polymorphisms and lung cancer risk: Evidence from meta-analyses and genome-wide association studies. Lung Cancer, 2017. 113: p. 18–29. [DOI] [PubMed] [Google Scholar]
- 85.Tsuchiya Y, et al. , Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res, 2004. 64(9): p. 3119–25. [DOI] [PubMed] [Google Scholar]
- 86.Moorthy B, et al. , 3-Methylcholanthrene-inducible liver cytochrome(s) P450 in female Sprague-Dawley rats: possible link between P450 turnover and formation of DNA adducts and I-compounds. Carcinogenesis, 1993. 14(5): p. 879–86. [DOI] [PubMed] [Google Scholar]
- 87.Moorthy B, Sriram P, and Randerath K, Chemical structure- and time-dependent effects of polycyclic aromatic hydrocarbon-type inducers on rat liver cytochrome P450, DNA adducts, and I-compounds. Fundam Appl Toxicol, 1994. 22(4): p. 549–60. [DOI] [PubMed] [Google Scholar]
- 88.Bresnick E, et al. , Administration of 3-methylcholanthrene to rats increases the specific hybridizable mRNA coding for cytochrome P-450c. Proc Natl Acad Sci U S A, 1981. 78(7): p. 4083–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Takahashi G, Shah H, and Weinhouse S, Metabolism of [3H]-methylcholanthrene in the perfused rat liver. Cancer Res, 1977. 37(2): p. 369–75. [PubMed] [Google Scholar]
- 90.Moorthy B, Persistent expression of 3-methylcholanthrene-inducible cytochromes P4501A in rat hepatic and extrahepatic tissues. J Pharmacol Exp Ther, 2000. 294(1): p. 313–22. [PubMed] [Google Scholar]
- 91.Kondraganti SR, et al. , Effects of 3-methylcholanthrene on gene expression profiling in the rat using cDNA microarray analyses. Chem Res Toxicol, 2005. 18(11): p. 1634–41. [DOI] [PubMed] [Google Scholar]
- 92.Kondraganti SR, Jiang W, and Moorthy B, Differential regulation of expression of hepatic and pulmonary cytochrome P4501A enzymes by 3-methylcholanthrene in mice lacking the CYP1A2 gene. J Pharmacol Exp Ther, 2002. 303(3): p. 945–51. [DOI] [PubMed] [Google Scholar]
- 93.Moorthy B, et al. , 3-Methylcholanthrene elicits DNA adduct formation in the CYP1A1 promoter region and attenuates reporter gene expression in rat H4IIE cells. Biochem Biophys Res Commun, 2007. 354(4): p. 1071–7. [DOI] [PubMed] [Google Scholar]
- 94.Shervington A, et al. , Identification of a novel co-transcription of P450/1A1 with telomerase in A549. Gene, 2007. 388(1–2): p. 110–6. [DOI] [PubMed] [Google Scholar]
- 95.Mohammed K and Shervington A, Can CYP1A1 siRNA be an effective treatment for lung cancer? Cell Mol Biol Lett, 2008. 13(2): p. 240–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kondraganti SR, et al. , Persistent induction of hepatic and pulmonary phase II enzymes by 3-methylcholanthrene in rats. Toxicol Sci, 2008. 102(2): p. 337–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Petruzzelli S, et al. , Long-lasting effects of tobacco smoking on pulmonary drug-metabolizing enzymes: a case-control study on lung cancer patients. Cancer Res, 1988. 48(16): p. 4695–700. [PubMed] [Google Scholar]
- 98.Geneste O, et al. , Comparison of pulmonary DNA adduct levels, measured by 32P-postlabelling and aryl hydrocarbon hydroxylase activity in lung parenchyma of smokers and ex-smokers. Carcinogenesis, 1991. 12(7): p. 1301–5. [DOI] [PubMed] [Google Scholar]
- 99.Iqbal MA, et al. , MicroRNA in lung cancer: role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med, 2019. 70: p. 3–20. [DOI] [PubMed] [Google Scholar]
- 100.Rajkumar R, et al. , Genomewide RNA expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol, 2010. 298(4): p. H1235–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alhamdow A, et al. , DNA methylation of the cancer-related genes F2RL3 and AHRR is associated with occupational exposure to polycyclic aromatic hydrocarbons. Carcinogenesis, 2018. 39(7): p. 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang Y, et al. , F2RL3 methylation, lung cancer incidence and mortality. Int J Cancer, 2015. 137(7): p. 1739–48. [DOI] [PubMed] [Google Scholar]
- 103.Fasanelli F, et al. , Hypomethylation of smoking-related genes is associated with future lung cancer in four prospective cohorts. Nat Commun, 2015. 6: p. 10192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Damiani LA, et al. , Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res, 2008. 68(21): p. 9005–14. [DOI] [PubMed] [Google Scholar]
- 105.Li Y, et al. , Genetic variation in CDH13 gene was associated with non-small cell lung cancer (NSCLC): A population-based case-control study. Oncotarget, 2018. 9(1): p. 881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang P, et al. , CpG site-specific hypermethylation of p16INK4alpha in peripheral blood lymphocytes of PAH-exposed workers. Cancer Epidemiol Biomarkers Prev, 2012. 21(1): p. 182–90. [DOI] [PubMed] [Google Scholar]
- 107.Li J, et al. , Particulate matter-induced epigenetic changes and lung cancer. Clin Respir J, 2017. 11(5): p. 539–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Marwick JA, et al. , Cigarette smoke alters chromatin remodeling and induces proinflammatory genes in rat lungs. Am J Respir Cell Mol Biol, 2004. 31(6): p. 633–42. [DOI] [PubMed] [Google Scholar]
- 109.Adenuga D, et al. , Histone deacetylase 2 is phosphorylated, ubiquitinated, and degraded by cigarette smoke. Am J Respir Cell Mol Biol, 2009. 40(4): p. 464–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Klingbeil EC, et al. , Polycyclic aromatic hydrocarbons, tobacco smoke, and epigenetic remodeling in asthma. Immunol Res, 2014. 58(2–3): p. 369–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ito K, et al. , Cigarette smoking reduces histone deacetylase 2 expression, enhances cytokine expression, and inhibits glucocorticoid actions in alveolar macrophages. FASEB J, 2001. 15(6): p. 1110–2. [PubMed] [Google Scholar]
- 112.Zhang Z, et al. , Global H3K79 di-methylation mediates DNA damage response to PAH exposure in Chinese coke oven workers. Environ Pollut, 2021. 268(Pt B): p. 115956. [DOI] [PubMed] [Google Scholar]
- 113.Lin YH, et al. , Global reduction of the epigenetic H3K79 methylation mark and increased chromosomal instability in CALM-AF10-positive leukemias. Blood, 2009. 114(3): p. 651–8. [DOI] [PubMed] [Google Scholar]
- 114.Lazzaro F, et al. , Histone methyltransferase Dot1 and Rad9 inhibit single-stranded DNA accumulation at DSBs and uncapped telomeres. EMBO J, 2008. 27(10): p. 1502–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huyen Y, et al. , Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks. Nature, 2004. 432(7015): p. 406–11. [DOI] [PubMed] [Google Scholar]
- 116.Bostelman LJ, et al. , Methylation of histone H3 lysine-79 by Dot1p plays multiple roles in the response to UV damage in Saccharomyces cerevisiae. DNA Repair (Amst), 2007. 6(3): p. 383–95. [DOI] [PubMed] [Google Scholar]
- 117.Lander ES, et al. , Initial sequencing and analysis of the human genome. Nature, 2001. 409(6822): p. 860–921. [DOI] [PubMed] [Google Scholar]
- 118.Mathias SL, et al. , Reverse transcriptase encoded by a human transposable element. Science, 1991. 254(5039): p. 1808–10. [DOI] [PubMed] [Google Scholar]
- 119.Symer DE C. C S. ST C. EM; Cost GJ; Parmigiani G; Boeke JD, Human L1 Retrotransposition Is Associated with Genetic Instability In Vivo. Cell, 2002. 110: p. 327–338. [DOI] [PubMed] [Google Scholar]
- 120.Ponomaryova AA, et al. , Aberrant Methylation of LINE-1 Transposable Elements: A Search for Cancer Biomarkers. Cells, 2020. 9(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hassanin AAI, et al. , Lung genotoxicity of benzo(a)pyrene in vivo involves reactivation of LINE-1 retrotransposon and early reprogramming of oncogenic regulatory networks. Am J Physiol Lung Cell Mol Physiol, 2019. 317(6): p. L816–L822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Stribinskis V and Ramos KS, Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res, 2006. 66(5): p. 2616–20. [DOI] [PubMed] [Google Scholar]
- 123.Chappell G, et al. , Epigenetic alterations induced by genotoxic occupational and environmental human chemical carcinogens: A systematic literature review. Mutat Res Rev Mutat Res, 2016. 768: p. 27–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu Y, et al. , Benzo(a)pyrene regulated A549 cell migration, invasion and epithelial-mesenchymal transition by up-regulating long non-coding RNA linc00673. Toxicol Lett, 2020. 320: p. 37–45. [DOI] [PubMed] [Google Scholar]
- 125.Han Z, et al. , MicroRNA-622 functions as a tumor suppressor by targeting K-Ras and enhancing the anticarcinogenic effect of resveratrol. Carcinogenesis, 2012. 33(1): p. 131–9. [DOI] [PubMed] [Google Scholar]
- 126.Zhao Y, et al. , The role of miR-506 in transformed 16HBE cells induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide. Toxicol Lett, 2011. 205(3): p. 320–6. [DOI] [PubMed] [Google Scholar]
- 127.Han Z, et al. , Cell cycle changes mediated by the p53/miR-34c axis are involved in the malignant transformation of human bronchial epithelial cells by benzo[a]pyrene. Toxicol Lett, 2014. 225(2): p. 275–84. [DOI] [PubMed] [Google Scholar]
- 128.Cioroiu BI, et al. , Polycyclic aromatic hydrocarbons in lung tissue of patients with pulmonary cancer from Romania. Influence according as demographic status and ABO phenotypes. Chemosphere, 2013. 92(5): p. 504–11. [DOI] [PubMed] [Google Scholar]