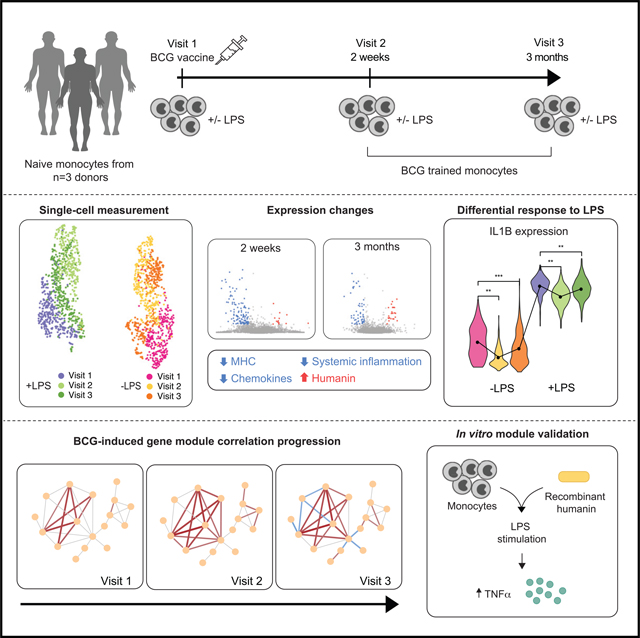
SUMMARY
Bacillus Calmette-Guérin (BCG) vaccine is one of the most widely used vaccines worldwide. In addition to protection against tuberculosis, BCG confers a degree of non-specific protection against other infections by enhancing secondary immune responses to heterologous pathogens, termed “trained immunity.” To better understand BCG-induced immune reprogramming, we perform single-cell transcriptomic measurements before and after BCG vaccination using secondary immune stimulation with bacterial lipopolysaccharide (LPS). We find that BCG reduces systemic inflammation and identify 75 genes with altered LPS responses, including inflammatory mediators such as CCL3 and CCL4 that have a heightened response. Co-expression analysis reveals that gene modules containing these cytokines lose coordination after BCG. Other modules exhibit increased coordination, including several humanin nuclear isoforms that we confirm induce trained immunity in vitro. Our results link in vivo BCG administration to single-cell transcriptomic changes, validated in human genetics experiments, and highlight genes that are putatively responsible for non-specific protective effects of BCG.
In brief
Kong et al. use single-cell transcriptomics to characterize the gene expression changes involved in the altered immune response in monocytes after BCG vaccination. The authors show that BCG reduces systemic inflammation and identify a set of genes implicated in the vaccine’s non-specific protective effects.
Graphical Abstract
INTRODUCTION
As the world becomes more interconnected, preventing/mitigating global health crises such as the ongoing coronavirus disease 2019 (COVID-19) pandemic is fast becoming a centerpiece of public policy. The Bacillus Calmette-Guérin (BCG) vaccine, which is nearly a century old, has garnered renewed attention in this regard. BCG was originally developed to prevent infection by Mycobacterium tuberculosis (TB), although it also has protective effects against other heterologous pathogens, including viruses (Matsumoto et al., 1983; O’Neill and Netea, 2020; Starr et al., 1976), and is reported to decrease susceptibility to a range of respiratory tract infections (Giamarellos-Bourboulis et al., 2020; Nemes et al., 2018; O’Neill and Netea, 2020). BCG is one of the most widely used vaccines globally, although its use in developed countries has declined over the past decades due to lower incidence of TB (Zwerling et al., 2011). This decline in BCG vaccination rates has been hypothesized as a factor leading to greater mortality from COVID-19 in certain countries (Gursel and Gursel, 2020).
Immune memory has typically been associated solely with the adaptive immune system. However, a growing body of literature has shown that some cells of the innate immune system have adaptive properties. In particular, vaccination by BCG induces an epigenetic-based reprogramming of myeloid cells and their bone marrow progenitors (Cirovic et al., 2020) that boosts the subsequent response mounted against secondary infections that may be unrelated to Mycobacteria (Kleinnijenhuis et al., 2012; Netea et al., 2016), an effect termed “trained immunity” (Netea et al., 2020). The heightened antimicrobial response includes enhanced production of inflammatory cytokines, such as interleukin (IL)1β or necrosis factor alpha (TNF-α), and IL6 (Jing et al., 1990). Epigenetic changes include the methylation and acetylation of histones, enhancing chromatin accessibility and enabling greater expression of antimicrobial genes (Netea et al., 2016).
BCG-induced trained immunity has been shown to confer protection against models of controlled human infection (Benn et al., 2013), including the yellow fever vaccine virus (Arts et al., 2018) and malaria (Walk et al., 2019). Heightened cytokine responses have also been observed for BCG-trained monocytes challenged with lipopolysaccharide (LPS) and Candida albicans (Arts et al., 2018). Because the non-specific protective effects of BCG vaccination can last up to 5 years (Nankabirwa et al., 2015), BCG can serve as a promising stopgap strategy against emerging pathogens until a proper vaccine can be developed.
To better understand the non-specific protective effects of BCG-induced trained immunity, we require deeper understanding of the mechanisms underlying these effects, along with the specific transcriptional modifications mediating the altered innate immune response. Here, we performed single-cell RNA-sequencing (scRNA-seq) in monocytes before and after administering BCG both in vivo and in vitro to characterize these modifications at the single-cell level.
RESULTS
BCG vaccination reduces the inflammatory response
We collected monocytes from three healthy donors during three visits. On the first visit, blood collection was performed before BCG vaccination. Follow-up samples were collected 2 weeks and 3 months after the initial visit (Figure 1A). No infections or adverse reactions occurred between vaccination and follow-up visits. Monocytes were either incubated in RPMI medium (negative control) or stimulated for 4 h with TLR4 agonist LPS, a microbial ligand from the Gram-negative bacteria Escherichia coli (unrelated to mycobacteria) to assess non-specific immune response differences induced by BCG. LPS-induced changes were greater than differences between donors, causing scRNA-seq expression profiles from the 1,710 collected monocytes to primarily cluster by LPS treatment rather than donor (Figure 1B).
Figure 1. Study design.
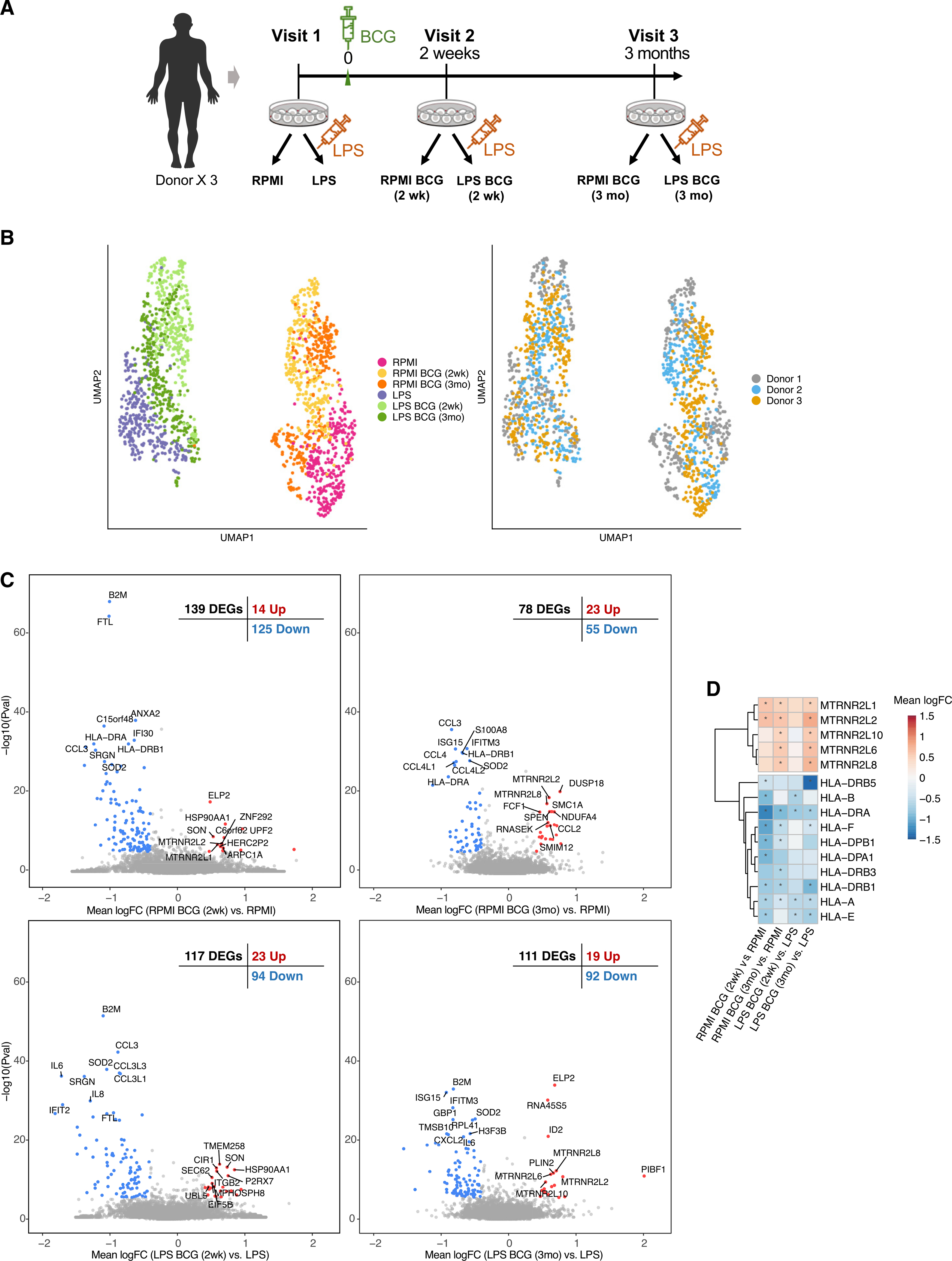
(A) Single-cell RNA-seq was performed on monocytes from 3 donors over 3 visits each. Donors were vaccinated after blood was collected at the first visit. For each collection, monocytes were exposed to RPMI or LPS (10 ng/mL) for 4 h.
(B) UMAP visualization of 1710 monocytes colored by condition (left) and by donor (right).
(C) Volcano plots comparing expression after BCG training to before training in both RPMI and LPS conditions. DEGs are colored (blue and red). The top 10 DEGs per direction are labeled.
(D) Heatmap of mean logFC of DEGs in the MHC and humanin-like nuclear isoforms in all four comparisons. Stars indicate the gene is differentially expressed in that comparison (STAR Methods).
Trained immunity persists for at least 3 months after vaccination (Kleinnijenhuis et al., 2012), at which time BCG has been eliminated from the local injection site, and the transcriptional and immunological profile can be considered to be due to the memory-like response induced by trained immunity. Although we found more differential expression 2 weeks after vaccination (139 differentially expressed genes [DEGs], false discovery rate [FDR] <0.05) (Figure 1C; Table S1), some important differences persisted up to 3 months (78 DEGs). Interestingly, at both time points, the majority of DEGs were downregulated (125 down and 14 up at 2 weeks, 55 down and 23 up at 3 months), which was also observed in the LPS-stimulated cells (Figure 1C). These genes were enriched in immune-related pathways (Table S2), suggesting that BCG vaccination reduces systemic inflammation, consistent with recent observations (Koeken et al., 2020). An earlier microarray-based study also only found a downregulation trend among DEGs, which included enrichment for IL6 receptor activity (Schreiber et al., 2010).
Specific downregulated systems in monocytes after BCG vaccination, before ex vivo LPS restimulation, included components of the major histocompatibility complex (MHC). Most prominent were HLA-A and HLA-DRA (downregulated in all conditions); other components included B2M (one of the most prominently downregulated genes in 3 conditions) (Figure 1D), HLA-E, HLA-F, HLA-DPB1, HLA-DRB1, and HLA-DRB5, suggesting that BCG training influences the interaction of antigen-presenting cells with T cells. We also found a number of downregulated chemokines, including CCL3, CCL4, and associated genes (CCL3L1, CCL3L3, CCL4L1, and CCL4L2), suggesting that BCG affects the inter-cellular immune signaling program. CCL2 showed the opposite trend and was upregulated at 3 months. Supporting the above, concordant changes in the CCL family were also observed, particularly CCL3 and associated genes, in a bulk RNA-seq dataset with similar design (Arts et al., 2018) (Figure S1). Several interferon-induced antiviral genes were also consistently downregulated, including GBP1, IFITM1, IFITM2, and IFITM3. H3F3B, the predominant form of histone H3 in non-dividing cells, was downregulated at all time points after training, suggesting an interaction between BCG training and cell-cycle regulation in monocytes. Among the smaller subset of genes that were upregulated after training, we found a set of humanin-like nuclear isoforms (MTRNR2L*) (Figure 1D), particularly in cells incubated in RPMI, and these were more prominent 3 months after training compared to 2 weeks. Humanin has cytoprotective and anti-apoptotic properties, and reduced expression of its nuclear isoforms is associated with a number of diseases (Bik-Multanowski et al., 2015). A similar trend for these genes was also observed in bulk RNA-seq data (Arts et al., 2018) (Figure S1).
Several genes identified as DEGs 2 weeks after BCG vaccination, but not at 3 months, may be involved in mediating the BCG-induced transcriptional alterations. Contrary to the majority of DEGs at 2 weeks, several of these were upregulated, most notably SON, a post-transcriptional gene regulator (Ahn et al., 2013), and HSP90AA1, a central gene regulator that induces epigenetic modifications (Ahn et al., 2013; Khurana and Bhattacharyya, 2015), suggesting the epigenetic changes involved in BCG training are in part facilitated by this chaperone. Of note, IL1β was the only cytokine differentially expressed at 2 weeks but not 3 months. Transcription factors including ATF5, CEBPB, REL (downregulated), HCFC2, and MTDH (upregulated) were differentially expressed at 2 weeks in RPMI, but not other conditions, and may therefore be part of the training process. Another important immune regulatory gene significantly downregulated at different time points was both components of the calprotectin heterodimer, S100A8 and S100A9.
We leveraged the single-cell resolution data to investigate heterogeneity in the transcriptional programs by attempting to partition the monocytes into distinct clusters in both RPMI and LPS conditions (Figure 2A). In RPMI, some population structure can be observed, with monocytes at baseline and 2 weeks being most different from each other, and monocytes 3 months after training containing a mixture of these two groups. This suggests that at 2 weeks, most monocytes have been impacted by BCG training. Although transcription returns to normal in circulating monocytes over the course of later weeks, their capacity to respond to restimulation is changed through epigenetic rewiring of their progenitors in the bone marrow (Cirovic et al., 2020).
Figure 2. BCG alters the response to LPS.
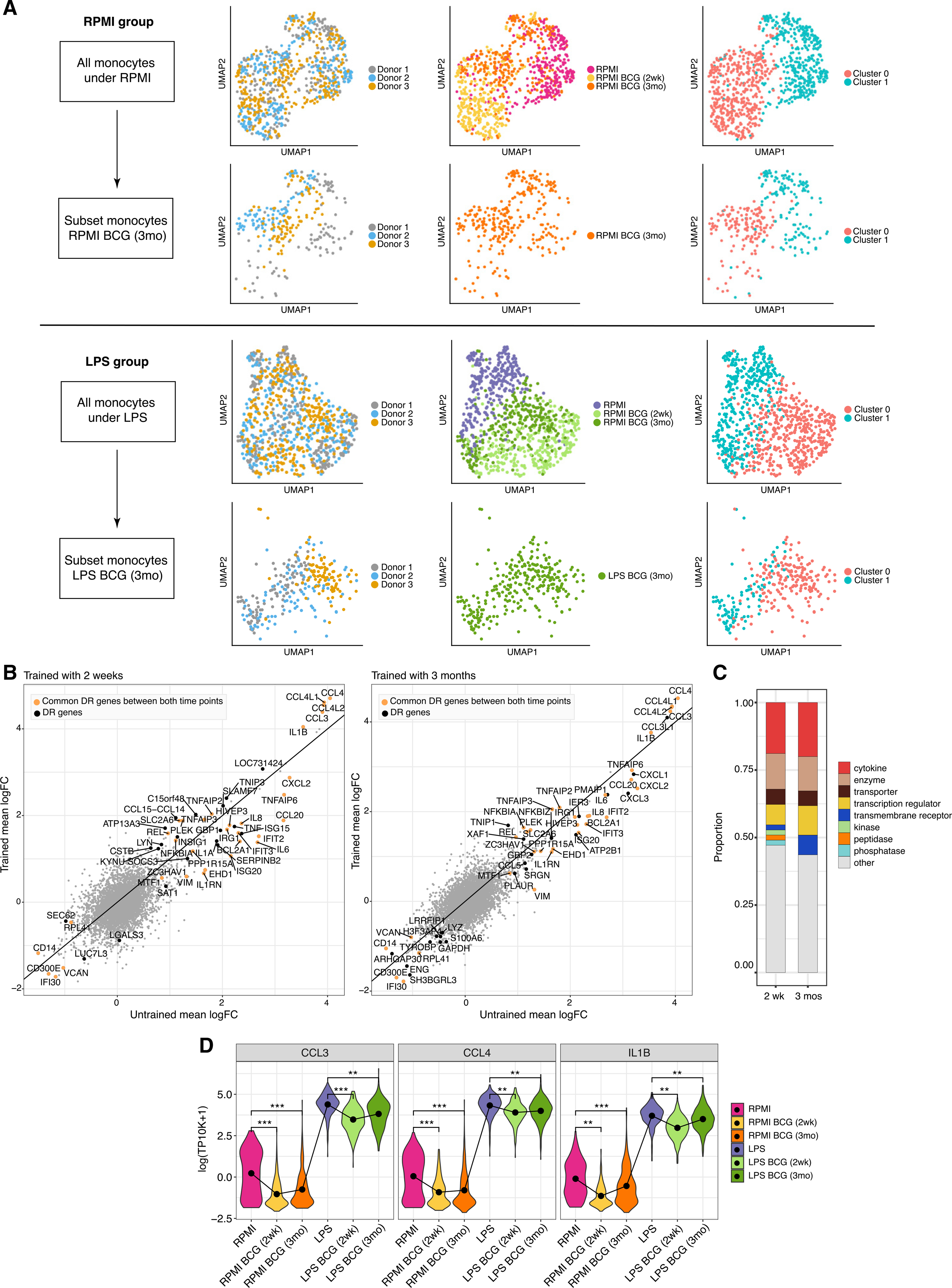
(A) UMAP visualization of expression profiles in RPMI (top rows) and LPS (bottom rows) colored by donor, time point, and identified cluster. The second row in each condition is subset to the 3-month data.
(B) Mean logFC of genes between RPMI and LPS conditions before and after BCG training. Colored points (yellow and black) are DR genes; yellow DR genes are common between both time points. The solid line denotes no difference.
(C) Distribution of gene types of DR genes.
(D) Violin plots of select DR genes (minimum adjusted p value among 3 donors; ***p < 10-15, **p < 0.01; STAR Methods) (all 75 profiles in Figure S2B).
BCG also altered the monocytes’ transcriptional response to LPS stimulation (Figure 2B). These included 53 differentially responding genes (DR genes) (STAR Methods) at 2 weeks and 55 DR genes at 3 months, with 33 genes in common. Larger fold-change differences were observed 2 weeks after training compared to 3 months. DR genes were enriched in cytokines and other signaling genes and depleted in transmembrane receptors (Figure 2C) compared to BCG-related DEGs (Figure S2A). Consistent with this, DR genes were strongly enriched in immune-related Gene Ontology (GO) terms such as Defense Response, Response to Cytokines, and Inflammatory Response (Table S2). Enriched transcription factor motifs included BACH2 and nuclear factor κB (NF-κB) after BCG-induced training. Because both are broad transcription factors associated with immune response, this suggests that additional factors are involved in DR gene regulation. At both time points, the genes with the largest LPS response (e.g., CCL3 and CCL4) had increased relative response after training, due largely to the reduction in baseline expression (Figure 2D).
BCG alters single-cell level correlations between genes
Having single cell-level transcriptomic profiles allows us to further examine BCG- and LPS-induced changes in correlations between genes. Between DR genes, we found more highly correlated gene pairs exist in the LPS conditions (Figures 3A-3D), and correlations among remaining DR genes tend to increase (Figure 3, insets), consistent with these cells mounting a coordinated immune response to the presence of LPS. However, at all time points, BCG-induced trained immunity reduced the correlations between the genes with the highest degree of coordination. Similar results were also obtained in vitro (Figure S3A).
Figure 3. BCG training affects correlations among DR genes.
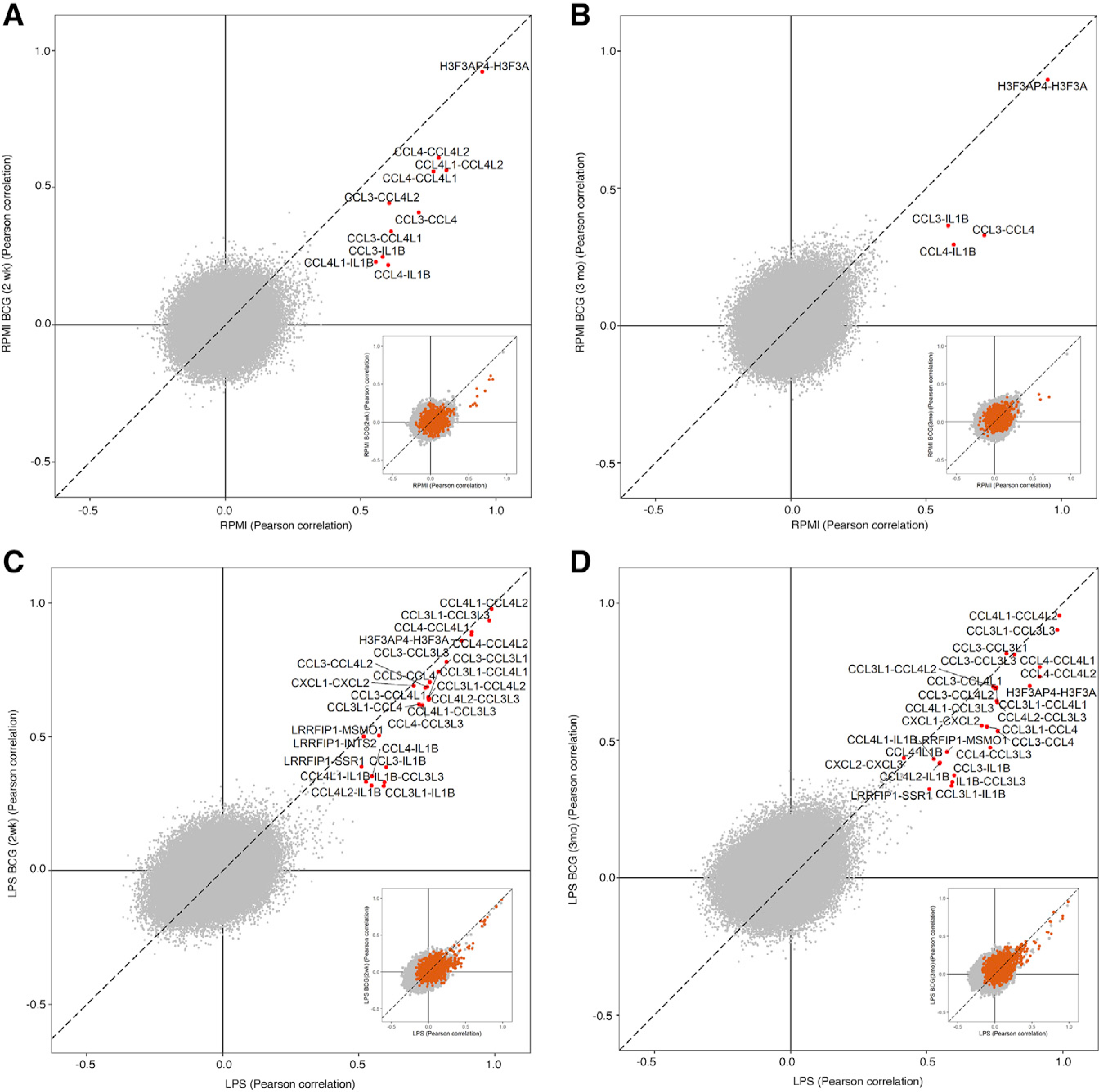
(A–D) Change in Pearson correlation between pairs of genes from before BCG training (x axis) and after (y axis), where one gene in the pair is a DR gene. Highlighted gene pairs (red) are those at least 0.6 away from (0,0). Inset scatterplots highlight DR-DR gene pairs, showing a general increase in correlation between DR genes in the LPS conditions (C and D).
The strongest associations observed here were mostly between DR genes. Non-DR genes with significant correlations to DR genes included MSMO1, SSR1, CCL3L3, and INTS2 (FDR <0.05). The strongest consistently correlated gene pair involving a DR gene was H3F3AP4-H3F3A. H3F3AP4 expression was downregulated by LPS stimulation 3 months after BCG training, but not before, suggesting that H3F3AP4 and H3F3A, a histone H3 variant, play a role in epigenetic reprogramming during BCG training.
Examining single-cell-level correlations between the BCG-implicated cytokines IL1β, IL6, and TNF-α and the DR genes (Figure S3B), we found that IL1β in particular is strongly correlated with several of the DR chemokines before training, including CCL3, CCL4, and CXCL1–3. After training, these associations weaken or disappear, suggesting that transcriptional reprogramming during BCG training has weakened the sensitivity of these genes to these proinflammatory signals. These associations tend to be stronger with LPS stimulation, although a reduction in correlation due to training is also observed in this condition. IL6 and TNF-α follow a similar pattern of association with DR chemokines.
Correlated gene modules affected by BCG
We next studied alterations to correlated gene modules among all genes (Figure 4) rather than only the DR genes above. Several gene modules were consistent across conditions. One module, consisting of DR genes including IL1β and CCL*, exhibits a reduction in correlation after training consistent with the decrease in correlation among DR genes observed earlier. Another larger module that exhibited the same pattern includes cell-cycle-related genes such as ORC4 (Figure 4). More tightly coupled modules after training included another module of cell-cycle regulators SPDYE*, and a larger module including several of the humanin-like nuclear isoforms and an antiviral Zinc finger ZNF793. Examining expression distributions of these modules did not reveal a clear connection to the population structure in Figure 2A. This larger module includes negative associations only significant 3 months after BCG training.
Figure 4. BCG alters the gene correlation network.
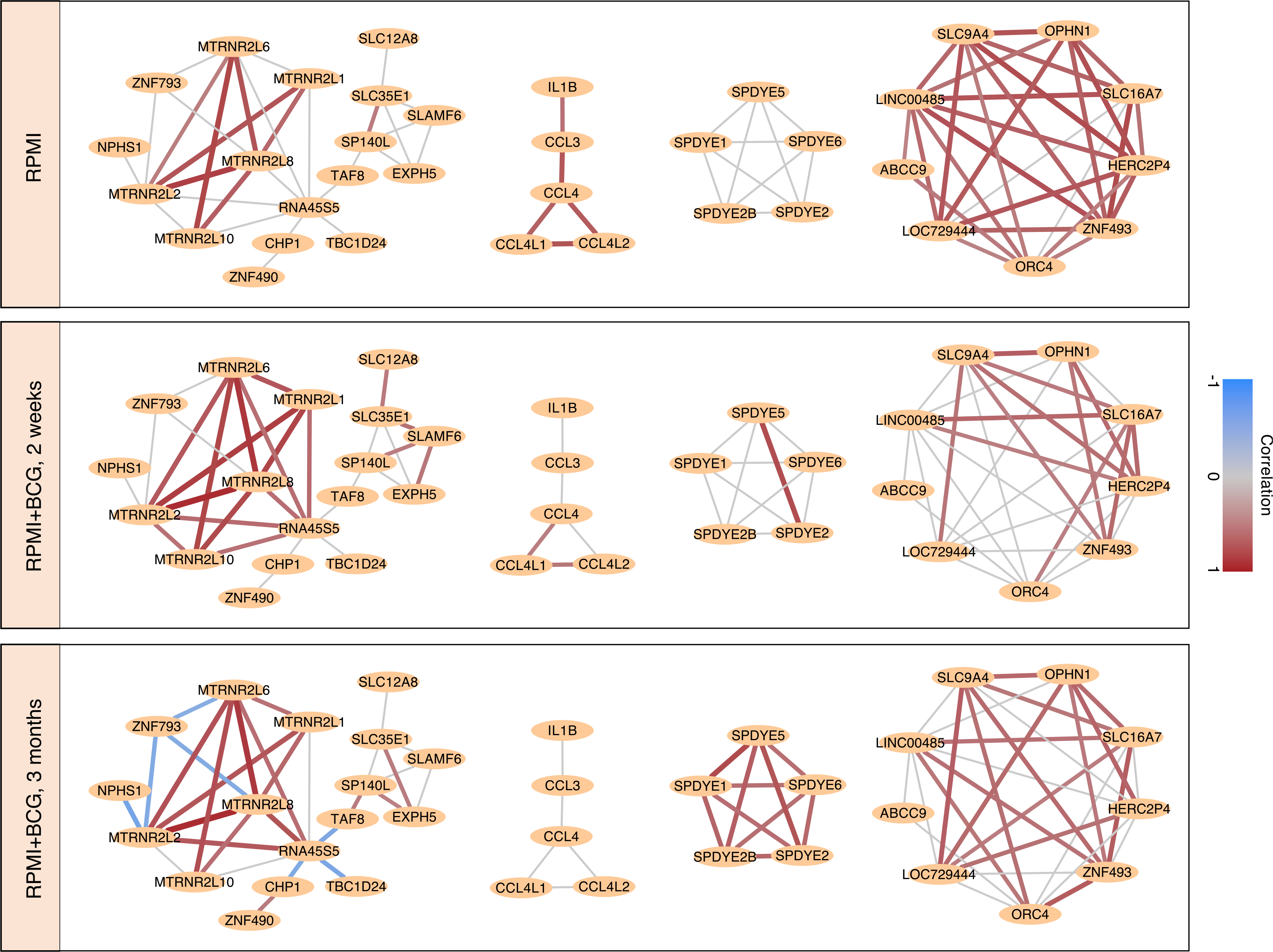
Pearson correlation network built from gene pairs with significantly correlated expression (FDR < 0.05; see STAR Methods) across cells at baseline, 2 weeks, and 3 months after BCG training, in the RPMI condition. A red (blue) connection implies that cells with higher expression of one gene in the pair have higher (lower) expression of the other gene. Gray edges are not significant at that time point. Selected gene modules are shown. These networks after stimulation by LPS can be found in Figure S4.
Many of these modules were also recovered after exposure to LPS. The IL1β/CCL* module grows considerably as these genes mount a coordinated response to the stimulus (Figure S4). The humanin module also reappears, although gathers a different set of negatively associated genes 3 months after BCG training. Other larger modules included the aforementioned ORC4 module and two modules not observed without LPS. Of note, the SPDYE* module, strongly correlated in RPMI, is lost in the LPS condition.
Genetic validation using expression quantitative trait locus that affect in vitro and in vivo BCG-induced trained immunity
To validate the relevance of the genes identified above after BCG-induced trained immunity, we tested whether common genetic polymorphisms (minor allele frequency [MAF] >5%) in genes that showed a significant differential expression at the single-cell level affect in vitro BCG-induced trained immunity responses. For this, we performed expression quantitative trait locus (eQTL) mapping using genetic data and in vitro cytokine production from adherent monocytes isolated from 267 healthy individuals, from the 300BCG cohort in the Human Functional Genomics study (https://www.humanfunctionalgenomics.org). Monocytes were stimulated for 24 h with RPMI control medium or BCG and were restimulated on day 6 with either LPS or control medium for 24 h. We identified common genetic polymorphisms within a window of 100 kb around several of the DEGs identified earlier that significantly influence the fold change of IL6 or TNF-α cytokine production (p < 0.001) (Figure 5A; Table S3). These genes include the nuclear-encoded humanin isoforms MTRNR2L1 and MTRNR2L6, the latter of which affected both TNF-α and IL6. H3F3AP4, CEBPB, and CCL3/4 were also significantly associated with TNF-α. This genetic association indicates that these genes play a role in BCG-induced trained immunity.
Figure 5. Genetic validation.
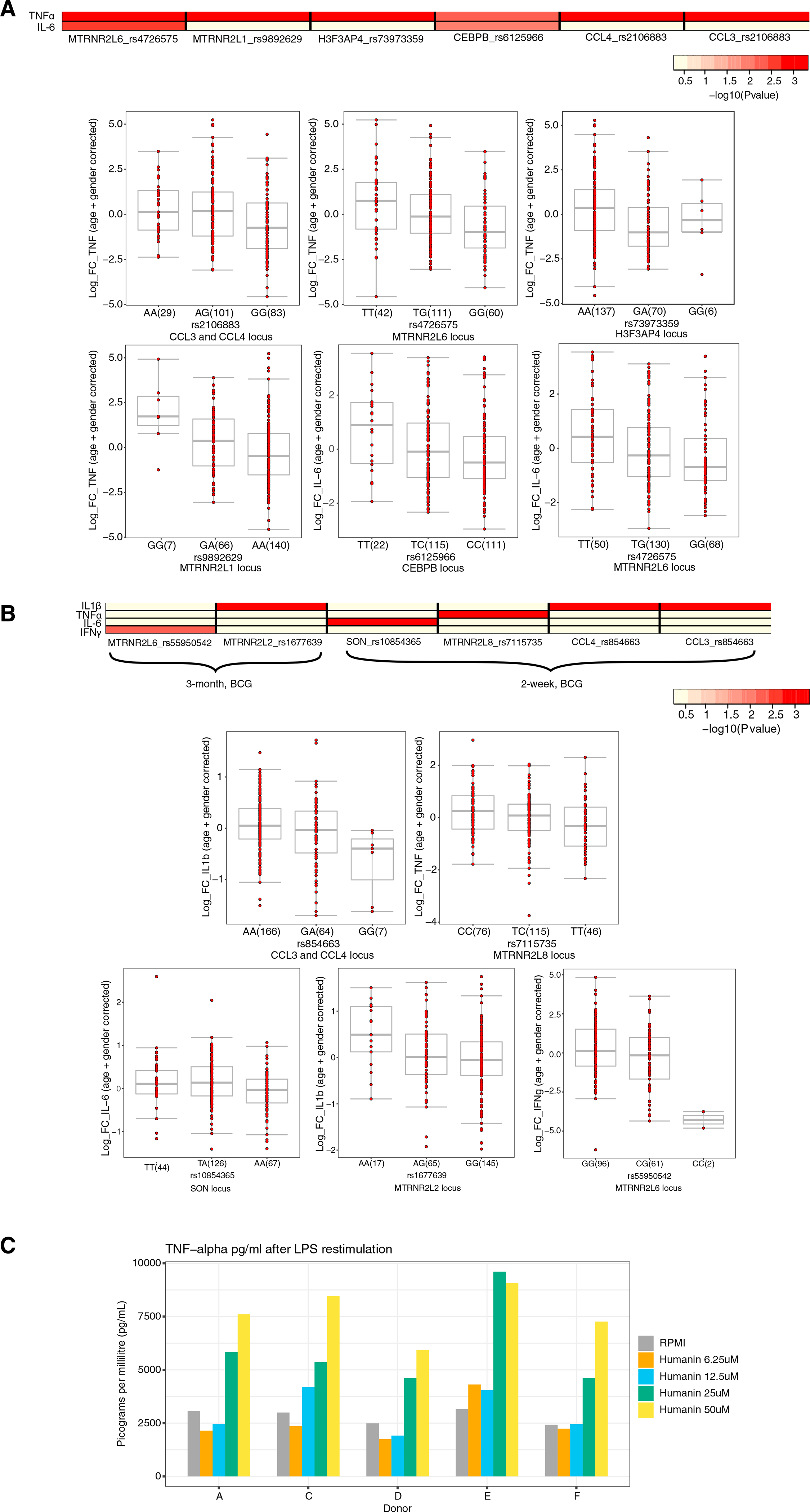
(A) Heatmap of the p values of association between SNPs mapped within a window of 100 kb around genes that were significantly differentially expressed at a single-cell level and the magnitude of cytokine production capacity by monocytes trained with BCG in vitro isolated from 300BCG cohort. Boxplots show the genotype-stratified cytokine changes from monocytes trained with BCG. Boxplots show median, upper, and lower quartiles, and whiskers extend to the most extreme point less than 1.5 times the interquartile range from the box.
(B) Similar to (A), although with S. aureus stimulated PBMCs isolated from healthy volunteers (300BCG) 2 weeks and 3 months after BCG vaccination. A linear regression model was used with age and sex as covariates to identify associations between SNPs and cytokine log fold-changes. A detailed description of the QTL mapping using the in vitro and ex vivo trained immunity responses is provided in the STAR Methods section “Genetic validation using the 300BCG cohort.” Boxplots show median, upper, and lower quartiles, and whiskers extend to the most extreme point less than 1.5 times the interquartile range from the box.
(C) TNF-α expression after stimulation with LPS of monocytes that were incubated with varying concentrations of recombinant humanin.
In addition to the in vitro cytokine production, we further investigated whether the same genetic polymorphisms affect the in vivo trained immunity response upon S. aureus restimulation at 2 weeks and 3 months after BCG vaccination from 296 healthy individuals of the 300BCG cohort. Genetic polymorphisms in a window of 100 kb around our genes of interest significantly affect the fold change of IL1β, TNF-α, IFNγ, and IL6 upon ex vivo exposure to S. aureus (p < 0.001) (Figure 5B; Table S3). Common genes with the in vitro experiment included MTRNR2L6, although this affected only IFNγ here, as well as CCL3/4 that was associated with IL1β. Other genes included MTRNR2L2, MTRNR2L8, and SON. This further supports an important role for these genes in the induction of BCG-induced trained immunity.
Finally, to further validate the role of humanin for the modulation of trained immunity, we initiated a series of functional validation experiments in an in vitro model of trained immunity (STAR Methods). Humanin pre-exposure significantly increased TNF-α production capacity compared to naive monocytes (Figure 5C), indicating induction of trained immunity, validating the conclusions of our transcriptional studies.
DISCUSSION
We investigated the single-cell transcriptional signature of BCG-induced trained immunity in circulating monocytes, with LPS as a secondary immune challenge. We observed a general reduction in expression of inflammatory markers after BCG, while after restimulation, BCG vaccination had a mixed effect on the transcriptional response to LPS, with genes responding with increased or decreased expression compared to baseline. Increased fold changes, in particular for a set of cytokines with the strongest transcriptional response to LPS, were driven primarily by a reduction in baseline expression rather than increased response to LPS. This is an important observation, in order not to confuse this for an increased overall response, when in fact many of these genes display a decreased expression both at baseline and after restimulated expression.
Earlier studies by our group and others have reported that BCG induces an increased responsiveness of myeloid cells to re-stimulation. Although this can be observed here for some inflammation-related genes, the single-cell analysis paints a more complex and important picture in which transcription of a significant number of genes was also downregulated by BCG vaccination, consistent with a recent study showing reduction in systemic inflammation by BCG vaccination in humans (Koeken et al., 2020). The mechanisms driving these modulatory effects of BCG remain to be studied in detail, but one hypothesis could involve the induction of autophagy (Buffen et al., 2014), which is well-known to modulate inflammatory processes (Buffen et al., 2014; Netea-Maier et al., 2016). The molecular mechanisms behind the increased transcription of some chemokines such as CCL2 after BCG, while other genes involved in inflammation are downregulated, remains an interesting topic for future studies. The specificity of CCL2 as a chemoattractant for monocytes is an interesting observation and may contribute to the biological effects of BCG.
In total, we identified 75 genes with an altered response to LPS due to BCG training (i.e., with a differential response to LPS). Although these tended to be related to the immune response, this group also included several genes affecting methylation such as HSP90AA1 (Khurana and Bhattacharyya, 2015) and H3F3B, consistent with epigenetic reprogramming as the molecular mechanism underlying trained immunity. We found that single-cell-level expression of DR genes as a group became more correlated after training, suggesting that the monocytes are expressing a more coordinated response, particularly to LPS where the greatest correlations among DR genes were observed. However, at the same time, we observed a counterintuitive reduction in the correlation between the most correlated DR gene pairs. This reduction was observed in both RPMI and LPS conditions, in which the expression levels of these genes differed drastically, showing that this reduction cannot be attributed to the generally lower level of expression of these genes after BCG training. Combined, this implies that more genes are becoming involved in the coordinated immune response, although the expression program learned during induction of trained immunity is not the program originally executed by the BCG-naive monocytes. The stronger impact of BCG vaccination on transcriptional responses in the present study compared to earlier investigations (e.g., Arts et al., 2018) is likely due to methodological aspects: the earlier studies have performed bulk RNA-seq in which differences at the single-cell level are lost. The earlier timing of post-BCG transcriptional differences in the current study (day 14) compared to the later time point (day 30) in Arts et al. (2018) can also account for some differences.
We also found several highly correlated gene modules, the most prominent of which involved nuclear isoforms of humanin (MTRNR2L*). These genes were upregulated in both RPMI and LPS conditions after BCG training, polymorphisms in or near these genes were shown to affect the in vivo and in vitro trained immunity response, and in an in vitro model of trained immunity, we showed that these genes induce a trained immunity response. Humanin is a cytoprotective, anti-apoptotic factor, and reduced expression of a number of these nuclear isoforms have been associated with increased risk of poor immune outcomes such as type 1 diabetes (Hoang et al., 2010) and inflammation (Zapała et al., 2010; Zhao et al., 2013). Our results strongly suggest that these effects may be an important mediator of non-specific beneficial effects of BCG training. Of note, 4 of the 5 humanin nuclear isoforms identified here (MTRNR2L1, MTRNR2L6, MTRNR2L8, and MTRNR2L10) were recently reported together in a cluster of genes co-expressed with ACE2 specifically in COVID-19 patients (He et al., 2020). MTRNR2L1 and MTRNR2L8 were also upregulated in the respiratory tract in active COVID-19 patients (Zhou et al., 2020). This similarity raises the intriguing possibility that BCG may impart a protective effect against this and similar diseases.
In conclusion, in this study, we described at an unprecedented level of detail the transcriptional response of monocytes after BCG vaccination, which is crucial for understanding the mechanisms underlying trained immunity and heterologous protective effects of BCG. We identified the molecular substrate for the beneficial immunological effects of BCG that combines a lower inflammatory status in the circulation with increased responsiveness of the innate immune cells to reinfection. These data contribute to understanding trained immunity and they represent an important resource for improving future development of vaccines by incorporating trained immunity-inducing properties.
Limitations of study
The present study is limited by the inclusion of a relatively small number of available donors, introducing the risk of selection bias. Future studies are needed to assess the degree to which the findings observed here differ between people. The relatively small number of monocytes per donor limits the identification of heterogeneity in transcriptional programs exhibited by the monocytes. A greater number of sampled monocytes may enable the identification of additional monocyte subgroups which interact differently with BCG. Finally, this study focused on monocytes as a representative population in which trained immunity can be observed. Other mechanisms may underlie further alterations to the immune response relevant to the non-specific protective effects of BCG in other cell types.
STAR*METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ramnik J. Xavier (xavier@molbio.mgh.harvard.edu)
Materials availability
This study did not generate new unique reagents.
Data and code availability
The datasets generated during and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/): GSE184241.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Individuals from the 300BCG cohort were vaccinated with 0.1 mL of BCG (BCG vaccine strain Bulgaria; Intervax, Canada). Blood was collected before vaccination, 2 weeks and 3 months after vaccination. Written informed consent was received from participants prior to inclusion in the study. The study was approved by the Ethical Committee of Radboud University Nijmegen, the Netherlands (no. NL58553.091.16). Experiments were conducted according to the principles expressed in the Declaration of Helsinki.
METHOD DETAILS
Monocyte isolation and stimulation experiments
Venous blood from the cubital vein of volunteers from the 300BCG cohort was drawn into 10 mL EDTA tubes (Monoject). Peripheral blood mononuclear cell (PBMC) isolation was performed by density centrifugation of blood diluted 1:1 in pyrogen-free PBS over Ficoll-Paque (GE healthcare, UK). Cells were washed twice in PBS and suspended in RPMI culture medium (Roswell Park Memorial Institute medium, Invitrogen, CA, USA) supplemented with 50 mg/mL gentamicin (Centrafarm), 2 mM glutamax (GIBCO), and 1mM pyruvate (GIBCO). Next, monocytes were isolated from PBMCs by negative selection using a MACS system (Miltenyi Biotech, Bergisch Gladbach, Germany), according to the manufacturer’s instructions. 1 × 106 Monocytes were incubated either with culture medium (RPMI) only as a negative control or 10 ng/mL LPS for 4 hours at 37°C.
Single monocytes were sorted on the presence of a cell death marker (LIVE/DEAD® Fixable Violet Dead Cell Stain Kit, Life Technologies) in skirted-side 96-well PCR plates containing buffer TCL (QIAGEN) (TCL + 2% of 2M DTT). Plates were sealed and centrifuged (800 g, 1 min) upon sorting and kept at −80°C until lysate cleanup.
In vitro model of trained immunity in human monocytes
Monocytes were isolated from human healthy volunteers as described above and incubated either with culture medium only (RPMI) or 5 μg/mL BCG (BCG-Bulgaria, Intervax, Canada) for 24 hours at 37°C (all in the presence of 10% pooled human serum). After 24h, cells were washed once with warm PBS and incubated for 5 more days in culture medium with 10% human pooled serum. Medium was changed once on day 3. On day 6, cells were restimulated for 4 hours with either RPMI or LPS 10 ng/mL (serotype 055: B5; Sigma) and sorted as described earlier. In experiments in which cytokine production was measured, cells were stimulated on day 6 for 24 hours with either RPMI or LPS 10 ng/mL. After 24 hours of stimulation, supernatants were collected and stored at −20°C until assayed.
For the separate experiments with humanin, 6.25 to 50 μM of recombinant humanin was used to induce trained immunity in the first 24h of incubation. Human primary monocytes were isolated from blood and exposed to RPMI or increasing concentrations of recombinant humanin for 24h. Thereafter, the cells were washed, rested for another 5 days, and restimulated for 24h with LPS (10 ng/ml). On day 7, supernatants were collected and TNFα production was measured by ELISA.
Assessment of trained immunity responses in BCG-vaccinated volunteers
PBMCs were isolated from healthy volunteers as described above and stimulated ex vivo with 5 × 106 CFU/ml heat-killed Staphylococcus aureus before vaccination, and 2 weeks and 3 months after vaccination. Cytokine production was measured in 24 hours (IL1β, IL6 and TNFα) and 7 days (IFNγ) supernatants. The fold change in cytokine production (after vaccination compared to baseline) was used as a measurement of the magnitude of the trained immunity response.
Genetic validation using the 300BCG cohort
The 300BCG is a population-based cohort of healthy individuals of Western descent from the Human Functional Genomics Project. The 300BCG cohort consists of 325 adults from the Netherlands. DNA samples of individuals were genotyped using the commercially available SNP chip, Infinium Global Screening Array MD v1.0 from Illumina. optiCall 0.7.0 (Shah et al., 2012) with default settings was used for genotype calling. Genetic variants with a call rate ≤ 0.01 were excluded, as were variants with a Hardy-Weinberg equilibrium (HWE) ≤ 0.0001, and minor allele frequency (MAF) ≤ 0.001. The strands and variants identifiers were aligned to the 1000 Genomes reference panel using Genotype Harmonizer (Deelen et al., 2014). One sample was excluded from the pre-imputed dataset due to high relatedness. We then imputed the samples on the Michigan imputation server using the human reference consortium (HRC r1.1 2016) (McCarthy et al., 2016) as a reference panel. Data were phased using Eagle v2.3, and we filtered out genetic variants with an R2 < 0.3 for imputation quality. We identified and excluded 17 genetic outliers, and selected 4,296,841 SNPs with MAF 5% for follow-up QTL mapping.
Both genotype and cytokine data on in vitro trained immunity responses induced by BCG in monocytes was obtained for a total of 267 individuals. Three samples were excluded due to medication use (of which one was identified as a genetic outlier), and one sample due to onset of type 1 diabetes during the study. First, the fold change of cytokine production between trained and non-trained cells was taken as a measurement for the magnitude of the trained immunity response. Following quality check for cytokine distribution and after excluding genetic outliers, we mapped the log-transformed fold changes of cytokine production to genotype data using a linear regression model with age and sex as covariates to correct the distributions of fold change of cytokine production. Similar approach was followed to identify QTLs using the ex vivo cytokine production from PBMCs after BCG vaccination in healthy volunteers. Both genotype and cytokine data on ex vivo trained immunity responses were obtained for a total of 296 individuals. The fold change of cytokine production before versus after 2-week and 3-month BCG vaccination was log-transformed, and was mapped to genotype data using a linear regression model with age and sex as covariates. R-package Matrix-eQTL was used for QTL mapping. We used a cutoff of p < 9.99 × 10−3 to identify suggestive QTL associations that affect trained immunity response within a window of 100kb around our genes of interest.
Sample preparation and sequencing
Single cells were sorted as described above into 96 well plates containing 10ul TCL buffer (QIAGEN) with 2-Mercaptoethanol. RNA was isolated from the lysate with Agencourt RNA Clean Spri beads (Beckman Coulter). A reverse transcription reaction was performed with the RNA-bound Spri beads, a polydT oligo, 4 unique TSOs multiplexed per well, and Maxima Reverse Transcriptase (Thermo Fisher). Unique TSO were composed of a 5' Biotin, a truncated Illumina adaptor, a 7 base pair unique molecular identifier, and 3 riboguanosines at the 3' end. The 5' end of the polydT oligo was comprised of a 5' Biotin and a truncated Illumina adaptor. Oligo sequences can be found in Table S4. Following reverse transcription the reactions were treated with Exonuclease I (NEB) and then in-well whole transcriptome amplification (WTA) was performed with Herculase II (Agilent). The primers in the WTA targeted the two truncated Illumina adaptor sequences used in the TSO and in the polydT oligo from the RT reaction. After WTA all 96 reactions in a plate were pooled and purified over a single Zymoclean gel DNA recovery column (Zymo Research Corporation). The volume of DNA was reduced by using Agencourt AMPure XP (Beckman Coulter) beads after the column elution.
To generate a 5'DGE library, WTA product (0.6–1ng) was tagmented for 10 minutes using the Nextera XT Library Prep Kit (Illumina) followed by a 14–15 cycle PCR to add adapters and amplify the fragmented library. The 14–15 cycle PCR incorporates the provided Nextera XT i5 adapters (Illumina) and a custom universal primer that targets the TSO-labeled end and includes an Illumina P7 sequencing adaptor and 8 base pair sample barcode. The tagmentation reaction was cleaned with Agencourt AMPure XP beads (Beckman Coulter). Libraries from each of 96-well plates were multiplexed and sequenced with a 75-cycle NextSeq 500/550 kit (Illumina) by paired-end reads of 46 and 21 cycles and one adaptor read.
QUANTIFICATION AND STATISTICAL ANALYSIS
Single-cell data processing
After sequencing, BCL files were demultiplexed with bcl2fastq (v.2.17.1.14, using “–no-lane-splitting–mask-short-adapter-reads 10– minimum-trimmed-read-length 10”), then fastq files were aligned to the human genome (hg19) with Burrows-Wheeler Aligner (Li and Durbin, 2009) (“bwa aln -l 24”). Seurat (Stuart et al., 2019) (v.3.1.0) was used to process the reads. Briefly, cells that have unique gene counts less than 200 were filtered out, and genes that cannot be detected at least in 3 cells were removed.
Prior to differential expression analysis, we applied a centered log-ratio transform (Arts et al., 2018) to the Seurat-normalized expression values, since we observed that a large fraction of reads were frequently attributed to small numbers of genes. This adjustment mitigates the impact of the compositional nature of the data.
Single-cell clustering and UMAP visualization
Clustering and UMAP visualization were done by following the standard Seurat v3 integration workflow.
Differential expression analysis
Differential expression analysis was using the Seurat function “FindMarkers” with MAST method. The test was performed within the donor to avoid interpersonal differences. DEGs were genes with abs(mean(logFC)) > log(1.5) and min(adj.p.val) < 0.05. Here, the mean and the min are taken across donors.
Differential response analysis
Let logFCTrained be the log fold-change of a comparison between an LPS-stimulated expression profile and its paired RPMI measurement, at one of the time points after BCG training (either 2 weeks or 3 months). Similarly, let logFCUntrained be the corresponding measurement without BCG training. A gene is labeled as DR if its mean |mean(logFCTrained)| > log(2), |mean(logFCUntrained)| > log(2), | mean(logFCTrained) - mean(logFCUntrained)| > 0.2, and all 3 individual adj.p.val < 0.05 in either the trained or untrained conditions.
Enrichment analysis
GO terms and pathway enrichment analyses were performed by using R package fgsea (Korotkevich et al., 2021), fast preranked gene set enrichment analysis (GSEA): minSize = 15, maxSize = 500 and nperm = 100,000. Gene sets “c5.all.v7.1.symbols.gmt,” “c2.cp.v7.1.symbols.gmt” and “c3.tft.v7.3.symbols.gmt” were in used the analysis, and these gene sets were obtained from the MSigDB collections: https://www.gsea-msigdb.org/gsea/msigdb/collections.jsp.
Correlation analysis and correlation network
Gene to gene Pearson correlations were calculated using function corr.test in the R package psych. P values were adjusted using Benjamini-Hochberg for multiple hypothesis correction. Correlations were calculated within donors to avoid including interpersonal differences in the correlations. Only genes that have non-zero expression in at least 25% cells with a given condition were included in this analysis. The minimum correlation value among all three donors was used as the final correlation value for each gene pair. Similarly, we used the maximum of the FDR value among all donors. The correlation network was visualized with Cytoscape (ver 3.7.1) (Shannon et al., 2003) with the gene pairs that have significant correlation (FDR < 0.05) within a given condition.
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Biological samples | ||
|
| ||
| Monocytes from BCG vaccinated subjects | Radboud University Medical Center | 300BCG |
|
| ||
| Deposited data | ||
|
| ||
| Monocyte single-cell RNA sequencing | Gene Expression Omnibus | GEO: GSE184241 |
| Human genome (hg19) | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001405.13 |
| MSigDB pathways | Liberzon et al., 2015 | https://www.gsea-msigdb.org/gsea/msigdb/index.jsp |
|
| ||
| Software and algorithms | ||
|
| ||
| bcl2fastq (ver 2.17.1.14) | N/A | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html |
| Burrows-Wheeler Aligner(ver 0.7.17) | Li and Durbin, 2009 | http://bio-bwa.sourceforge.net |
| Seurat (ver 3.1.0) | Butler et al., 2018 | http://satijalab.org/seurat |
| fgsea (R package) | Korotkevich et al., 2021 | http://bioconductor.org/packages/release/bioc/html/fgsea.html |
| psych (R package) | Revelle, 2021 | https://cran.r-project.org/web/packages/psych/index.html |
| Cytoscape (ver 3.7.1) | Shannon et al., 2003 | https://cytoscape.org |
Highlights.
Single-cell transcriptomic monocyte profiling uncovers mechanisms of trained immunity
BCG-induced transcription changes are consistent with systemic inflammation reduction
75 genes show BCG-induced altered response to LPS stimulation
Gene coexpression identifies a humanin module that induces trained immunity in vitro
ACKNOWLEDGMENTS
This study was supported by an ERC Advanced Grant (833247 to M.G.N.), a Spinoza grant of the Netherlands Organization for Scientific Research (to M.G.N.), and funding from the NIH (RC2 DK114784 to R.J.X.).
Footnotes
DECLARATION OF INTERESTS
R.J.X. is co-founder of Celsius Therapeutics and Jnana Therapeutics. M.G.N. and L.A.B.J. are scientific founders of Trained Therapeutix and Discovery (TTxD).
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.110028.
REFERENCES
- Ahn EE-Y, Higashi T, Yan M, Matsuura S, Hickey CJ, Lo M-C, Shia W-J, DeKelver RC, and Zhang D-E (2013). SON protein regulates GATA-2 through transcriptional control of the microRNA 23a 27a 24–2 cluster. J. Biol. Chem. 288, 5381–5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts RJW, Moorlag SJCFM, Novakovic B, Li Y, Wang S-Y, Oosting M, Kumar V, Xavier RJ, Wijmenga C, Joosten LAB, et al. (2018). BCG Vaccination Protects against Experimental Viral Infection in Humans through the Induction of Cytokines Associated with Trained Immunity. Cell Host Microbe 23, 89–100.e5. [DOI] [PubMed] [Google Scholar]
- Benn CS, Netea MG, Selin LK, and Aaby P (2013). A small jab - a big effect: nonspecific immunomodulation by vaccines. Trends Immunol. 34, 431–439. [DOI] [PubMed] [Google Scholar]
- Bik-Multanowski M, Pietrzyk JJ, and Midro A (2015). MTRNR2L12: A Candidate Blood Marker of Early Alzheimer’s Disease-Like Dementia in Adults with Down Syndrome. J. Alzheimers Dis. 46, 145–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffen K, Oosting M, Quintin J, Ng A, Kleinnijenhuis J, Kumar V, van de Vosse E, Wijmenga C, van Crevel R, Oosterwijk E, et al. (2014). Autophagy controls BCG-induced trained immunity and the response to intravesical BCG therapy for bladder cancer. PLoS Pathog. 10, e1004485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirovic B, de Bree LCJ, Groh L, Blok BA, Chan J, van der Velden WJFM, Bremmers MEJ, van Crevel R, Händler K, Picelli S, et al. (2020). BCG Vaccination in Humans Elicits Trained Immunity via the Hematopoietic Progenitor Compartment. Cell Host Microbe 28, 322–334.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deelen P, Bonder MJ, van der Velde KJ, Westra H-J, Winder E, Hendriksen D, Franke L, and Swertz MA (2014). Genotype harmonizer: automatic strand alignment and format conversion for genotype data integration. BMC Res. Notes 7, 901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos-Bourboulis EJ, Tsilika M, Moorlag S, Antonakos N, Kotsaki A, Domínguez-Andreés J, Kyriazopoulou E, Gkavogianni T, Adami M-E, Damoraki G, et al. (2020). Activate: Randomized Clinical Trial of BCG Vaccination against Infection in the Elderly. Cell 183, 315–323.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gursel M, and Gursel I (2020). Is global BCG vaccination-induced trained immunity relevant to the progression of SARS-CoV-2 pandemic? Allergy 75, 1815–1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Mok TN, Yun L, He C, Li J, and Pan J (2020). Single-cell RNA sequencing analysis of human kidney reveals the presence of ACE2 receptor: A potential pathway of COVID-19 infection. Mol. Genet. Genomic Med 8, e1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang PT, Park P, Cobb LJ, Paharkova-Vatchkova V, Hakimi M, Cohen P, and Lee K-W (2010). The neurosurvival factor Humanin inhibits beta-cell apoptosis via signal transducer and activator of transcription 3 activation and delays and ameliorates diabetes in nonobese diabetic mice. Metabolism 59, 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing SQ, Spencer T, Miller K, Hopkins C, and Trowbridge IS (1990). Role of the human transferrin receptor cytoplasmic domain in endocytosis: localization of a specific signal sequence for internalization. J. Cell Biol. 110, 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana N, and Bhattacharyya S (2015). Hsp90, the concertmaster: tuning transcription. Front. Oncol. 5, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinnijenhuis J, Quintin J, Preijers F, Joosten LAB, Ifrim DC, Saeed S, Jacobs C, van Loenhout J, de Jong D, Stunnenberg HG, et al. (2012). Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc. Natl. Acad. Sci. USA 109, 17537–17542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koeken VA, de Bree LCJ, Mourits VP, Moorlag SJ, Walk J, Cirovic B, Arts RJ, Jaeger M, Dijkstra H, Lemmers H, et al. (2020). BCG vaccination in humans inhibits systemic inflammation in a sex-dependent manner. J. Clin. Invest. 130, 5591–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A (2021). Fast gene set enrichment analysis. bioRxiv 10.1101/060012. [DOI] [Google Scholar]
- Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdoóttir H, Ghandi M, Mesirov JP, and Tamayo P (2015). The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Otani T, Une T, Osada Y, Ogawa H, and Azuma I (1983). Stimulation of nonspecific resistance to infection induced by muramyl dipeptide analogs substituted in the gamma-carboxyl group and evaluation of N alpha-muramyl dipeptide-N epsilon-stearoyllysine. Infect. Immun. 39, 1029–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, et al. ; Haplotype Reference Consortium (2016). A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 48, 1279–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nankabirwa V, Tumwine JK, Mugaba PM, Tylleskär T, and Sommerfelt H; PROMISE- EBF Study Group (2015). Child survival and BCG vaccination: a community based prospective cohort study in Uganda. BMC Public Health 15, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemes E, Geldenhuys H, Rozot V, Rutkowski KT, Ratangee F, Bilek N, Mabwe S, Makhethe L, Erasmus M, Toefy A, et al. ; C-040–404 Study Team (2018). Prevention of M. tuberculosis Infection with H4:IC31 Vaccine or BCG Revaccination. N. Engl. J. Med. 379, 138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Joosten LAB, Latz E, Mills KHG, Natoli G, Stunnenberg HG, O’Neill LAJ, and Xavier RJ (2016). Trained immunity: A program of innate immune memory in health and disease. Science 352, aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea MG, Domínguez-Andreés J, Barreiro LB, Chavakis T, Divangahi M, Fuchs E, Joosten LAB, van der Meer JWM, Mhlanga MM, Mulder WJM, et al. (2020). Defining trained immunity and its role in health and disease. Nat. Rev. Immunol. 20, 375–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netea-Maier RT, Plantinga TS, van de Veerdonk FL, Smit JW, and Netea MG (2016). Modulation of inflammation by autophagy: Consequences for human disease. Autophagy 12, 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill LAJ, and Netea MG (2020). BCG-induced trained immunity: can it offer protection against COVID-19? Nat. Rev. Immunol. 20, 335–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revelle W (2021). psychTools: Tools to Accompany the ’psych; Package for Psychological Research. https://rdrr.io/cran/psychTools/.
- Schreiber F, Huo Z, Giemza R, Woodrow M, Fenner N, Stephens Z, Dougan G, Prideaux S, Castello-Branco LRR, and Lewis DJM (2010). An investigation of clinical and immunological events following repeated aerodigestive tract challenge infections with live Mycobacterium bovis Bacille Calmette Guérin. Vaccine 28, 5427–5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah TS, Liu JZ, Floyd JAB, Morris JA, Wirth N, Barrett JC, and Anderson CA (2012). optiCall: a robust genotype-calling algorithm for rare, low-frequency and common variants. Bioinformatics 28, 1598–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr SE, Visintine AM, Tomeh MO, and Nahmias AJ (1976). Effects of immunostimulants on resistance of newborn mice to herpes simplex type 2 infection. Proc. Soc. Exp. Biol. Med. 152, 57–60. [DOI] [PubMed] [Google Scholar]
- Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walk J, de Bree LCJ, Graumans W, Stoter R, van Gemert G-J, van de Vegte-Bolmer M, Teelen K, Hermsen CC, Arts RJW, Behet MC, et al. (2019). Outcomes of controlled human malaria infection after BCG vaccination. Nat. Commun. 10, 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapa1a B, Kaczyński Ł, Kieć-Wilk B, Staszel T, Knapp A, Thoresen GH, Wybrańska I, and Dembińska-Kieć A (2010). Humanins, the neuroprotective and cytoprotective peptides with antiapoptotic and anti-inflammatory properties. Pharmacol. Rep. 62, 767–777. [DOI] [PubMed] [Google Scholar]
- Zhao S-T, Zhao L, and Li J-H (2013). Neuroprotective Peptide humanin inhibits inflammatory response in astrocytes induced by lipopolysaccharide. Neurochem. Res. 38, 581–588. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, Guo L, Yang J, Wang C, Jiang S, et al. (2020). Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe 27, 883–890.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwerling A, Behr MA, Verma A, Brewer TF, Menzies D, and Pai M (2011). The BCG World Atlas: a database of global BCG vaccination policies and practices. PLoS Med. 8, e1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/): GSE184241.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


