Abstract
Whole-brain mapping is an effective approach to investigate which brain areas are activated by the exploration of a novel environment. Previous studies analyzing neuronal activity promoted by novelty focused mostly on one specific area instead of the whole brain and measured activation using cfos immunohistochemistry. In this study, we utilized TRAP2 mice exposed to a novel and familiar environment to examine neuronal activity in exploratory, learning, and memory circuits. We analyzed the behavior of mice during environment exploration. Brain tissue was processed using tissue clarification and neurons active during exploration of an environment were mapped based on the cfos expression. Neuronal activity after each experience were quantified in regions of interest. We observed increased exploratory behavior in mice exposed to a novel environment in comparison to familiar (170.5s ± 6.47 vs. 112.5s ± 9.54, p=0.0001). Neuronal activity was significantly increased in the dentate gyrus (115.56 ± 53.84 vs. 32.24 ± 12.32, p=0.02) during the exploration of a novel environment. Moreover, examination of the remaining regions of interest showed some increase in the number of active neurons in the novel condition, however, those differences were not statistically significant. Brief exposure to a novel environment results in increased exploratory behavior and significant neuronal activity in the dentate gyrus.
1. Introduction
Whole-brain activity mapping can elucidate which brain regions are involved in the exploration of a novel environment. Recognition of novelty is a process dependent on intact memory (Montag-Sallaz et al., 1999; Snyder et al., 2008). Immediate early genes, such as cfos and Arc, have been used as a marker of active cells in the brain during seizures and behavior in several studies (Burnsed et al., 2019; Guzowski et al., 2005; Méndez-López et al., 2009; Montag-Sallaz et al., 1999; Naik et al., 2021; Radulovic et al., 1998; VanElzakker et al., 2008). Expression of cfos is low in physiologic conditions in resting animals but can be induced by external stimuli, which renders cfos useful for mapping brain regions involved in certain behaviors, including novel environment exploration.
Previous studies on neuronal activity during a behavior focused on specific regions, such as the hippocampus, that are involved in a behavior, instead of the whole brain. The majority of these studies used immunohistochemistry methods for cfos detection, which has low time resolution. Whole-brain activity mapping allows us to examine an unbiased sample of active areas during a behavioral task, such as novel environment exploration. Additionally, in this study, neuronal activity during exposure to the novel environment was compared to the activity caused by repeat exposure to the same environment (now familiar), instead of homecage conditions, as in previous studies. Furthermore, this study utilizes “TRAP” mice (Denardo et al., 2015; Guenthner et al., 2013) to monitor neuronal activity during a distinct time frame, which has a higher time resolution compared to immunohistochemical detection of cfos expressing cells. TRAP is based on a recombination event between two transgenes, one expressing tamoxifen-dependent recombinase CreERT2 from an activity-dependent IEG promoter, Fos, and the other expressing an effector gene, which is tdTomato, in a Cre-dependent manner. In the presence of tamoxifen, active cells expressing cfos are permanently tagged with a fluorescent protein.
In studies using cfos immunostaining, it has been observed that mice exposed to novel stimuli expressed cfos in numerous brain areas, namely, the hippocampus and parietal cortex. On the contrary, repeated exposure to the same stimuli resulted in a reduction of cfos production in those areas (Radulovic et al., 1998). Expression of cfos was higher in regions of the hippocampus, particularly dentate gyrus, and piriform cortex in rodents introduced to a novel environment than in homecage controls (Guenthner et al., 2013; Jaeger et al., 2018; VanElzakker et al., 2008).
The goal of this study was to perform whole-brain mapping of active neurons, expressing cfos, in transgenic TRAP mice during exploration of a novel environment using tissue clearing and advanced microscopy. Using these techniques, we have mapped neurons active during exposure to novel and familiar environments in exploration and memory circuits. We hypothesize that animals will explore the novel environment more than the familiar and that this will be associated with increased neuronal activity in circuits involved in exploration and memory.
2. Results
2.1. Animals
To analyze behavior in novel and familiar environments TRAP2 mice (n=24) were used for behavioral tests (Fig. 1A). After exposure to those environments, mice were randomly selected (n=5 novel environment, n=5 familiar environment) for quantification of active neurons (Fig. 1B). One animal was excluded from the familiar group, as the images had a large amount of background artifact, likely secondary to a poor perfusion, making quantification impossible. In addition, four brains were excluded from analysis due to suboptimal tissue processing or slicing.
Figure 1: Activity in novel vs. familiar environment.
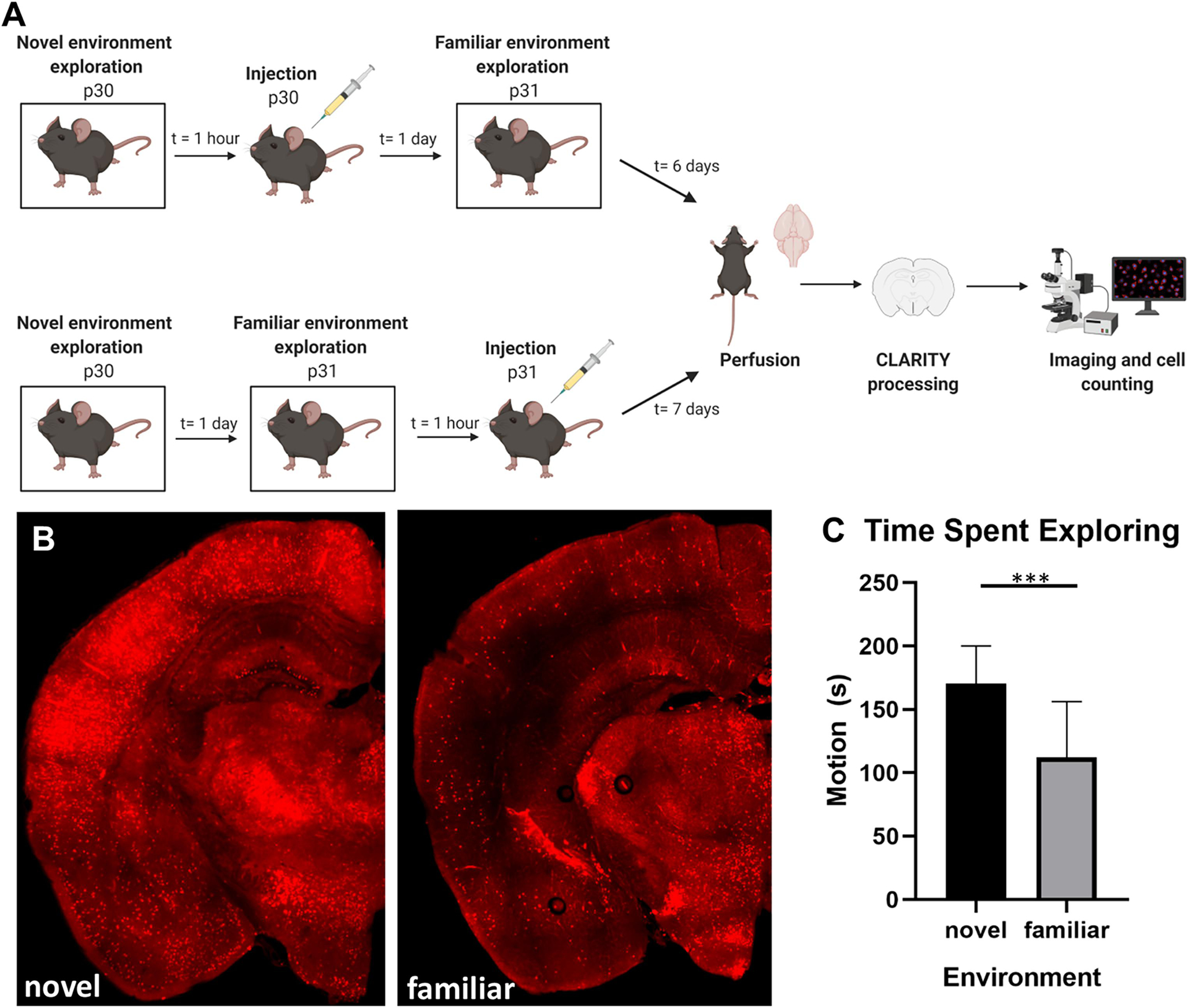
A) Experimental timeline. B) Representative image of clarified brain slice in novel environment condition. TdTomato expression in red, inset image is high power view of the dentate gyrus. 10x, 200μm thick clarified slice. Bregma −2.255. C) Time spent exploring the environment is significantly higher during novel environment exposure compared to familiar, p<0.0001 (plotted as mean with standard deviation) (n=24; 8 males, 13 females).
2.2. Exploration of environment is greater in a novel environment
Behavior in the novel and familiar environments was examined by quantifying the amount of time the animals spent actively moving during the five-minute exposure to the environment. Mice spent significantly more time exploring the environment on the first day of exposure (novel environment condition) compared to repeat exposure to the environment 24 hours later (familiar environment condition) (170.5s±29.64 vs. 112.5s±43.74, p=0.0001) (Fig. 1C).
2.3. Neuronal activity is increased in the memory circuit during novel environment exploration
In order to examine neuronal activity in circuits involved in exploration and memory, we quantified cfos expressing cells. Regions involved in the exploration of the environment, including the primary and secondary motor cortex, somatosensory cortex, prelimbic cortex (Sharpe and Killcross, 2015), olfactory bulb, visual cortex, anterior cingulate cortex (Qadir et al., 2018), and piriform cortex, were examined. In the primary and secondary motor cortex, the number of active neurons during novel environment exploration was increased by 26% and 48%, respectively. These differences in primary motor cortex (345.68±176.94 vs 273.69±164.56, p=0.055) and secondary motor cortex (277.64528 ± 136.4136 vs. 187.66 ± 76.62, p=0.28) were not statistically significant (Fig. 2A&B). The primary and secondary somatosensory cortex did not show significant differences in neuronal activity during novel vs. familiar exploration (p=0.98 and p=0.78, respectively). The olfactory bulb (p=0.72), piriform cortex, a region that receives olfactory projections (p=0.9), and the anterior cingulate cortex (ACC), a region that is involved in response to environmental stimuli (p=0.89) did not show significant differences between the two conditions. The visual cortex had a 40% increase in active neurons in the novel condition compared to familiar (1082.68 ± 421.05 vs. 773.88 ± 534.55, p=0.36) (Fig. 2A&B). The prelimbic cortex (PLC), which uses cues in the environment to modulate a response (Sharpe and Killcross, 2015), had a 46% increase in active neurons during novel environment exposure (217.26 ± 32.33 vs. 49.13 ± 48.66, p=0.052) (Fig. 2A&B).
Figure 2: Neuronal activity in select regions of interest involved in exploration.
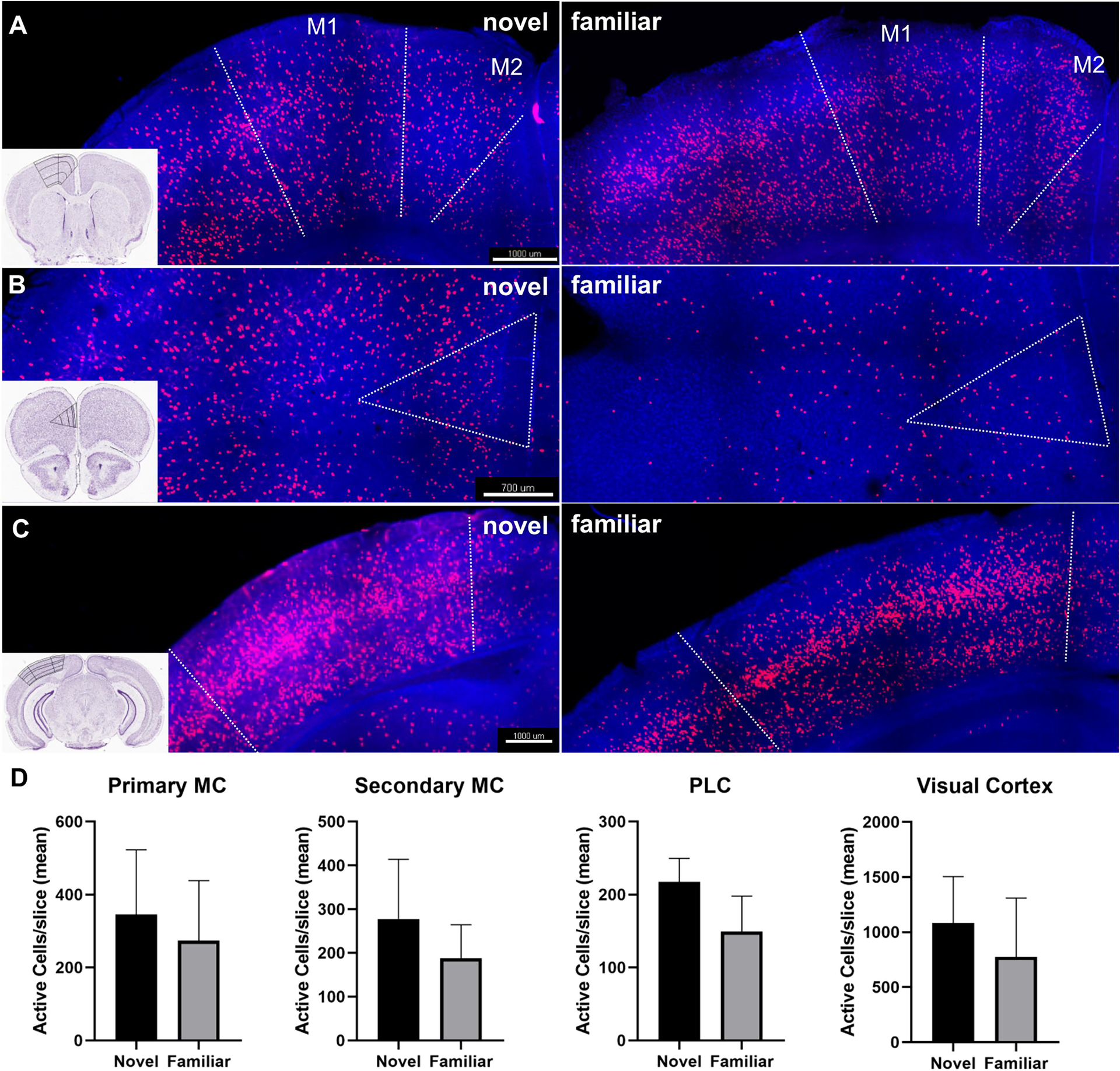
A) Representative image of cfos (tdTomato) expression in primary (M1) and secondary (M2) motor cortex during the novel and familiar exploration. Inset image with an outline of anatomic regions demarcated. Bregma 0.845. B) Representative image of cfos expression in the prelimbic cortex during the novel and familiar environment exploration. Bregma 2.045. C) Representative image of cfos expression in the visual cortex during the novel and familiar environment exploration. Bregma −3.38. 10x, 200μm thick clarified slice. D) Cell count quantification for each region of interest in images A-C. (“Allen Brain Atlas Mouse Brain Reference Atlas,” 2004) (plotted as mean with standard deviation) (novel n=5 (2 males, 3 females), familiar n=4 (1 male, 3 females)).
Examination of regions involved in memory included, the pyramidal cell layer of the dentate gyrus, the CA1 and CA3 regions of the hippocampus, retrosplenial cortex (RSC) (Milczarek et al., 2018; Wirtshafter, 2005), and areas of the medial prefrontal cortex (mPFC) (infralimbic cortex (ILC)(Chao et al., 2020; Wirtshafter, 2005), ACC (Wirtshafter, 2005), and PLC (Riga et al., 2014). The pyramidal layer of the dentate gyrus (both the infra- and suprapyramidal blades) exhibited significantly more active neurons during exploration of the novel environment compared to familiar (115.56 ± 53.84 vs. 32.24 ± 12.32, p=0.02), a 258% increase (Fig. 3A&B). CA1 and CA3 regions both exhibited modest increases in neuronal activity of 24% and 57%, respectively, that were not statistically significant (53.80 ± 8.66 vs. 43.26 ± 14.88, p=0.22 and 50.56 ± 20.22 vs. 32.22± 6.25, p=0.13) (Fig. 3A&B). Portions of the mPFC showed increases in active neurons during novel exploration; the infralimbic cortex exhibited an increase of 97% (168.70 ± 55.10 vs. 85.68 ± 53.90, p=0.058) (Fig. 3A&B) and as mentioned above, the prelimbic cortex exhibited a 46% increase (217.26 ± 32.33 vs. 49.13 ± 48.66, p=0.052) (Fig. 2A&B). Another region of the mPFC, the ACC, did not show a significant difference in neuronal activity in the two conditions (p=0.89). The retrosplenial cortex did not exhibit any difference in neuronal activity between the two conditions (p=0.91).
Figure 3: Neuronal activity in select regions of interest involved in memory.
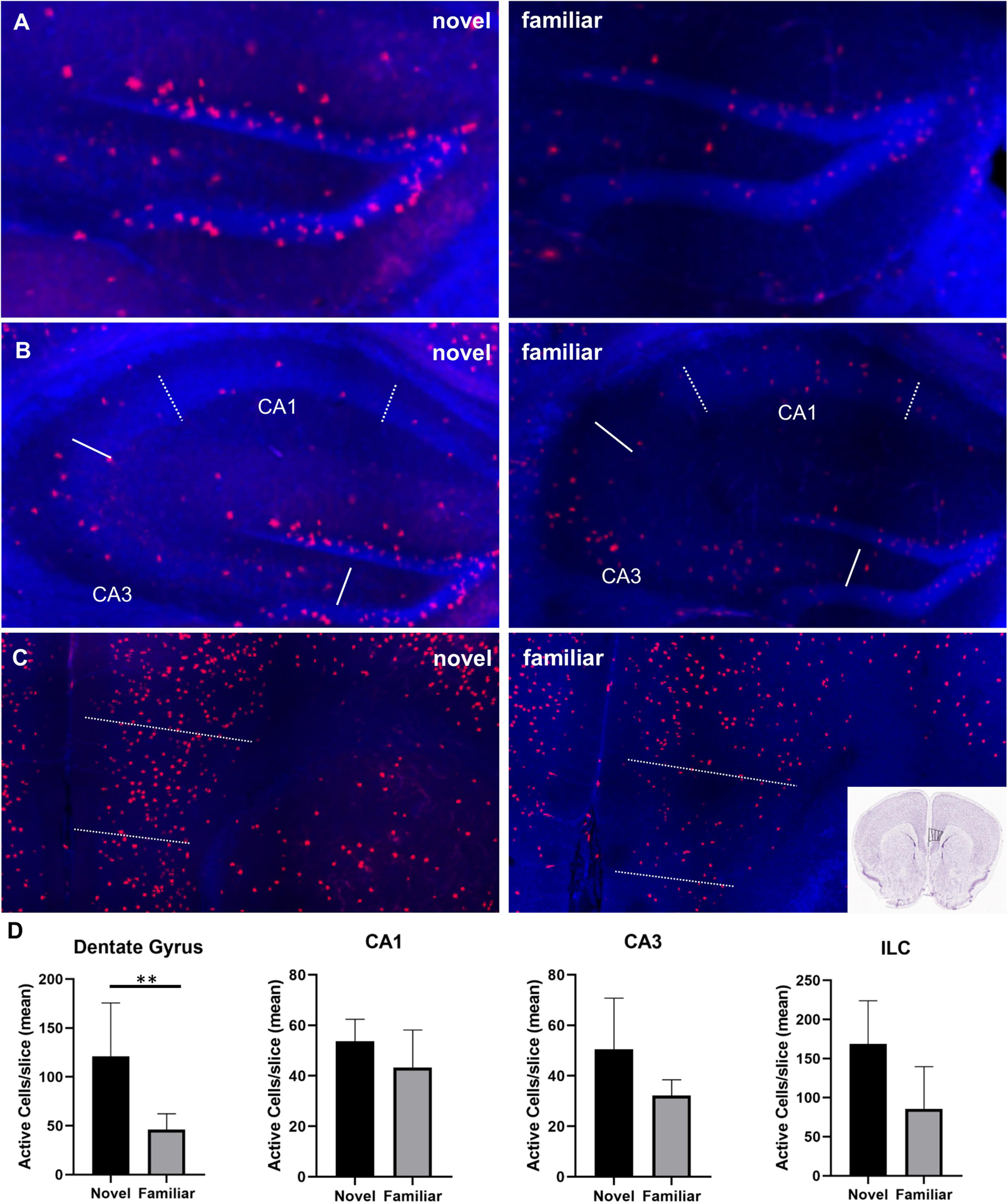
A) Increased cfos expression (tdTomato) in the dentate gyrus during exploration of novel vs. familiar environment. Bregma −1.655. B) cfos expression in CA1 (dotted line) and CA3 (solid line). C) cfos expression in the infralimbic cortex. Bregma 1.42. D) Cell count quantification for each region of interest in images A-C. Dentate gyrus novel vs. familiar, p=0.0159. (“Allen Brain Atlas Mouse Brain Reference Atlas,” 2004) (plotted as mean with standard deviation)
3. Discussion
The goal of this study was to map neuronal activity during the exploration of a novel environment compared to a familiar environment using an unbiased, whole-brain approach. Using tissue clarification and cfos mapping in TRAP transgenic mice, we examined neuronal activity during the exploration of novel and familiar environments. We quantified active cells in regions involved in exploration and memory. The dentate gyrus exhibited a significant increase in neuronal activity during exploration of a novel environment when compared to the familiar. Several other regions important to memory, novelty, and exploration demonstrated modest increases in neuronal activation in the novel environment.
Several previous studies have utilized immediate early genes, such as cfos and Arc, expressed during behavior to examine active regions (Guzowski, J. F., McNaughton, B., Barnes, C., Worley, 1999; Guzowski et al., 2005; Méndez-López et al., 2009; Montag-Sallaz et al., 1999; Naik et al., 2021; Radulovic et al., 1998; VanElzakker et al., 2008). The majority of these studies have taken a focused approach, examining specific regions that may be active during a task. Specific studies that examined neuronal activity during novel environment exposure have focused on discrete regions, such as the dentate gyrus, and compared IEG expression during novel environment exploration to homecage conditions. Here, we have utilized a whole-brain approach to examine areas active during novel environment exploration and compared this to activity during exploration of the same environment on a second exposure (the familiar condition), controlling for differences that may be present when compared to the homecage environment. The majority of studies examining immediate early genes in the context of novel environment exposure have used immunohistochemical approaches; these approaches do not afford the discrete-time resolution and high signal-to-noise ratio as found with the TRAP methods used in this study.
A single, brief exposure to a novel environment results in long-term potentiation (Davis et al., 2004) and increased transcription of the immediate early gene, Arc (Chawla et al., 2005; Jaeger et al., 2018) in the dentate gyrus. A single exposure of TRAP mice to a novel environment was shown to increase neuronal activity in dentate gyrus and CA1 compared to homecage (Guenthner et al., 2013). However, as mentioned above these studies focused largely on dentate gyrus and comparison to homecage exposure. Other studies using a novel gustatory or olfactory stimulus have shown increases in cfos expression in the parietal cortex, cingulate cortex, and several regions of the hippocampus circuit, CA1, CA3, and DG (Hess et al., 1995; Montag-Sallaz et al., 1999).
Regions involved in exploratory behavior, such as the motor cortex, visual cortex, and prelimbic cortex (Sharpe and Killcross, 2015) showed modest increases in cfos expressing cells during novel environment exposure, but these differences were not statistically significant. Other regions important to exploratory behavior, such as the somatosensory cortex, piriform cortex (Illig, K., Wilson, 2014), and anterior cingulate cortex (Qadir et al., 2018) did not exhibit differences in cfos expression between the two environments. When regions in the memory circuitry were examined we found that while the dentate gyrus displayed significant increases in cfos expression during novel environment exploration, other portions of the hippocampal circuit, such as CA1 and CA3, showed only modest increases in cfos activation in the novel environment.
These results are consistent with previous studies, that exploration of a novel environment is associated with significantly increased cfos expression in the dentate gyrus. However, other regions examined showed modest increases in cfos expression which were not statistically significant on cell counting analysis. Several limitations should be taken into consideration when interpreting these results. While the TRAP method allows for a discrete time window of cfos activity to be tagged (roughly 90 minutes), it is possible that our stimulus, a five-minute novel environment exposure, may not be enough to elicit robust cfos activity in all regions important to the exploration of a novel and familiar environment. In addition, a five-minute exposure may not be enough to consider the environment familiar the following day; however, previous studies have shown that a five-minute exposure to an environment is adequate to form a stable spatial representation (Frank et al., 2004). In addition, the exploratory behavior was decreased the following day, supporting the idea that the mice were familiarized with the environment. Lastly, sample sizes for whole-brain mapping and cell counting were small.
Using transgenic mice to map the expression of cfos in clarified tissue, we were able to examine whole-brain neuronal activity during the exploration of a novel environment. This study found that a brief exploration of a novel environment resulted in significantly increased cfos expression in the dentate gyrus. These methods offer an attractive way to examine neuronal activity during behavior and disease states during a discrete time window with a high signal-to-noise ratio. Further studies will use a whole-brain neuronal mapping approach to assess neuronal activation in a novel environment in the context of neurologic injury and diseases.
4. Experimental Procedure
4.1. Animals
All animals used for this study were handled according to a University of Virginia Animal Care and Use Committee approved protocol. Housing was in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals (US Department of Health and Human Services 85–23, 2011). TRAP2 (targeted recombination in active populations) mice (Denardo et al., 2015; Guenthner et al., 2013) used in this study were generated by breeding mice expressing Cre-ER under the regulation of the Fos promoter (B6.129[Cg]-Fostm1.1(cre/ERT2)Luo/J, #021882; Jackson Laboratories, Bar Harbor, ME, USA) with mice expressing tdTomato on the Rosa locus (B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J, #007909; Jackson Laboratories), both on the C57/Bl6 background. Genotyping was performed using a tail sample (KAPA Biosystems, Wilmington, MA, USA). Mice of both sexes were used in all experiments.
4.2. Novel and familiar environment exploration
All novel and familiar environment exposures were performed in a closed chamber equipped with a grid floor and camera to record the behavior of animals. Mice were habituated in their home cage placed in the behavior room for one hour before the experiment. On p30 animals were introduced to the chamber and conditioned for five minutes, individually exploring the chamber and then returned to their homecage (novel condition). Recall was performed after 24 hours, on p31, when mice again explored the chamber for five minutes (familiar condition). The chamber was cleaned with 70% ethanol and 1% acetic acid every time before the animal entered it. Video Freeze (Med Associates Inc) was used to measure active time during exploration. One hour following the experiment, mice were injected with 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich, St. Louis, MO, USA) to induce expression of fluorescent protein tdTomato in neurons that were active and expressing cfos during the exploration period. Mice from the novel environment group were injected on p30 and mice from the familiar environment group on p31 (Fig. 1A).
4.3. Neuronal activity mapping
TRAP2 mice utilize the activity-dependent IEG, cfos, to drive the expression of red fluorescent protein tdTomato. The fos locus is linked to a tamoxifen-inducible recombinase, CreER. Visualization of active neurons, permanently tagged by tdTomato, is possible in the presence of the short-acting tamoxifen metabolite, 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich, St. Louis, MO, USA). Subcutaneous injection of 4-OHT suspended in sesame oil allows tagging of neurons that were active in a 1- to 2-hour window before the time of injection. Mice were subcutaneously injected with 4-OHT 1 hour after novel (p30) or familiar (p31) environment exploration. Mice were subjected to transcardial perfusion seven days after injection, at the time of maximal tdTomato expression. (Guenthner et al., 2013).
4.4. Tissue clarification, processing, and imaging
Brain tissue was processed using the passive clarity technique (PACT) method (Yang et al., 2014), which included tissue-hydrogel polymerization, lipid extraction, and tissue mounting. Horizontal sections (200μm thick) were stained with DAPI and then were mounted in imaging media (RIMS, refractory imaging medium solution)(Yang et al., 2014).
Tissue was imaged using a Zeiss 780 confocal/multiphoton microscope system with Zeiss Zen software for image acquisition (Carl Zeiss, Oberkochen, Germany) under 10x magnification. Excitation wavelengths used for tdTomato were 561nm and for DAPI 358nm. Emission filter ranges for tdTomato were 570 to 691nm and for DAPI 463nm. Zen software was utilized to stitch tiled images with a Z-stack interval of 10μm (Fig. 1B).
4.5. Experimental design and statistics
Imaris 9.2.1(Bitplane Scientific, Zurich, Switzerland) was utilized to generate high-resolution snapshots of coronal sections used later to quantify the number of active cells in Fiji ImageJ’s automated cell counting feature. We quantified active cells in DG, CA1, CA3, infralimbic cortex, prelimbic cortex, primary and secondary motor cortex, primary and secondary somatosensory cortex, anterior cingulate cortex, retrosplenial cortex, visual cortex, piriform cortex, and olfactory bulbs. Regions of interest were chosen based on a qualitative examination of slices for regions of increased activity in both conditions, as well as a review of the literature on regions involved in exploration, novelty recognition, and memory (Guenthner et al., 2013; Guzowski, J. F., McNaughton, B., Barnes, C., Worley, 1999; Hess et al., 1995; Milczarek et al., 2018; Montag-Sallaz et al., 1999; VanElzakker et al., 2008; Wirtshafter, 2005). Researchers performing this quantification and analysis were blinded to experimental group. To eliminate background noise, the intensity threshold was set between 90 and 105. Size threshold was set to 8-infinity pixel units for DG and 2-infinity pixel units for the remaining regions to ensure clusters with the surface too small to be cells were not quantified. The total sum and mean number of active cells per slice were calculated for every region of interest. Statistical analysis was performed in Prism GraphPad Version 8. Descriptive statistics and a Mann-Whitney U test (nonparametric distribution of data) were used to compare novel vs. familiar environment conditions for each region. A p-value of <0.05 was considered statistically significant.
Highlights.
Active neuron mapping during behavior using tissue clarification and TRAP mice.
Mice explore a novel environment more than a familiar environment.
Neuronal activity in the dentate gyrus was increased during novel exploration.
Acknowledgments:
The University of Virginia Keck Center for Cellular Imaging and Dr. A. Periasamy for his guidance (Zeiss 780 microscopy funded by NIH OD016446). David Martin for his statistical advice. Funding: This work was supported by the National Institutes of Health, NINDS K08NS101122 (J.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement: The authors declare no competing financial interests
References
- Allen Brain Atlas Mouse Brain Reference Atlas, 2004. https://mouse.brain-map.org/static/atlas. URL https://mouse.brain-map.org/static/atlas (accessed 5.7.21).
- Burnsed J, Skwarzyńska D, Wagley PK, Isbell L, Kapur J, 2019. Neuronal Circuit Activity during Neonatal Hypoxic–Ischemic Seizures in Mice. Ann. Neurol 10.1002/ana.25601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao OY, de Souza Silva MA, Yang YM, Huston JP, 2020. The medial prefrontal cortex - hippocampus circuit that integrates information of object, place and time to construct episodic memory in rodents: Behavioral, anatomical and neurochemical properties. Neurosci. Biobehav. Rev 10.1016/j.neubiorev.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla MK, Guzowski JF, Ramirez-Amaya V, Lipa P, Hoffman KL, Marriott LK, Worley PF, McNaughton BL, Barnes CA, 2005. Sparse, environmentally selective expression of Arc RNA in the upper blade of the rodent fascia dentata by brief spatial experience. Hippocampus 15, 579–586. 10.1002/hipo.20091 [DOI] [PubMed] [Google Scholar]
- Davis CD, Jones FL, Derrick BE, 2004. Novel environments enhance the induction and maintenance of long-term potentiation in the dentate gyrus. J. Neurosci 24, 6497–6506. 10.1523/JNEUROSCI.4970-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denardo LA, Berns DS, Deloach K, Luo L, 2015. Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat. Neurosci 18, 1687–1697. 10.1038/nn.4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank LM, Stanley GB, Brown EN, 2004. Hippocampal plasticity across multiple days of exposure to novel environments. J. Neurosci 24, 7681–7689. 10.1523/JNEUROSCI.1958-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenthner CJ, Miyamichi K, Yang HH, Heller HC, Luo L, 2013. Permanent genetic access to transiently active neurons via TRAP: Targeted recombination in active populations. Neuron 78, 773–784. 10.1016/j.neuron.2013.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton B, Barnes C, Worley P, 1999. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat. Neuro 2, 1120–1124. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA, 2005. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr. Opin. Neurobiol 10.1016/j.conb.2005.08.018 [DOI] [PubMed] [Google Scholar]
- Hess US, Lynch G, Gall CM, 1995. Regional Patterns of c-fos mRNA Expression in Rat Hippocampus Following Exploration of a Novel Environment versus Performance of a Well-Learned Discrimination for the Neurobiology of Learning and Memory, and 3Depattment of Anatomy and, The Journal of Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illig K, Wilson D, 2014. Olfactory Cortex: Comparative Anatomy. Biomed. Sci 10.1016/B978-0-12-801238-3.04706-1 [DOI] [Google Scholar]
- Jaeger BN, Linker SB, Parylak SL, Barron JJ, Gallina IS, Saavedra CD, Fitzpatrick C, Lim CK, Schafer ST, Lacar B, Jessberger S, Gage FH, 2018. A novel environment-evoked transcriptional signature predicts reactivity in single dentate granule neurons. Nat. Commun 9. 10.1038/s41467-018-05418-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-López M, Méndez M, López L, Arias JL, 2009. Sexually dimorphic c-Fos expression following spatial working memory in young and adult rats. Physiol. Behav 98, 307–317. 10.1016/j.physbeh.2009.06.006 [DOI] [PubMed] [Google Scholar]
- Milczarek MM, Vann SD, Sengpiel F, 2018. Spatial Memory Engram in the Mouse Retrosplenial Cortex. Curr. Biol 28, 1975–1980.e6. 10.1016/j.cub.2018.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag-Sallaz M, Welzl H, Kuhl D, Montag D, Schachner M, 1999. Novelty-Induced Increased Expression of Immediate-Early Genes c-fos and arg 3.1 in the Mouse Brain. [PubMed]
- Naik AA, Sun H, Williams CL, Weller DS, Julius Zhu J, Kapur J, 2021. Mechanism of seizure-induced retrograde amnesia. Prog. Neurobiol 200. 10.1016/j.pneurobio.2020.101984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadir H, Krimmel SR, Mu C, Poulopoulos A, Seminowicz DA, Mathur BN, 2018. Structural connectivity of the anterior cingulate cortex, claustrum, and the anterior insula of the mouse. Front. Neuroanat 12. 10.3389/fnana.2018.00100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radulovic J, Kammermeier J, Spiess J, 1998. Relationship between Fos Production and Classical Fear Conditioning: Effects of Novelty, Latent Inhibition, and Unconditioned Stimulus Preexposure. [DOI] [PMC free article] [PubMed]
- Riga D, Matos MR, Glas A, Smit AB, Spijker S, Van den Oever MC, 2014. Optogenetic dissection of medial prefrontal cortex circuitry. Front. Syst. Neurosci 10.3389/fnsys.2014.00230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe MJ, Killcross S, 2015. The prelimbic cortex uses higher-order cues to modulate both the acquisition and expression of conditioned fear. Front. Syst. Neurosci 8. 10.3389/fnsys.2014.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder KA, Blank MP, Marsolek CJ, 2008. What form of memory underlies novelty preferences? Psychon. Bull. Rev 15, 315–321. 10.3758/PBR.15.2.315 [DOI] [PubMed] [Google Scholar]
- VanElzakker M, Fevurly RD, Breindel T, Spencer RL, 2008. Environmental novelty is associated with a selective increase in Fos expression in the output elements of the hippocampal formation and the perirhinal cortex. Learn. Mem 15, 899–908. 10.1101/lm.1196508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtshafter D, 2005. Cholinergic involvement in the cortical and hippocampal Fos expression induced in the rat by placement in a novel environment. Brain Res. 1051, 57–65. 10.1016/j.brainres.2005.05.052 [DOI] [PubMed] [Google Scholar]
- Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, Shah S, Cai L, Gradinaru V, 2014. Single-cell phenotyping within transparent intact tissue through whole-body clearing. Cell 158, 945–958. 10.1016/j.cell.2014.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]


