Abstract
Triple-negative breast cancer (TNBC) and malignant melanoma are highly aggressive cancers that widely express the cell surface chondroitin sulfate proteoglycan 4 (CSPG4/NG2). CSPG4 plays an important role in tumor cell growth and survival and promotes chemo- and radiotherapy resistance, suggesting that CSPG4 is an attractive target in cancer therapy. In the present work, we applied the drug delivery technology photochemical internalization (PCI) in combination with the novel CSPG4-targeting immunotoxin 225.28-saporin as an efficient and specific strategy to kill aggressive TNBC and amelanotic melanoma cells. Light-activation of the clinically relevant photosensitizer TPCS2a (fimaporfin) and 225.28-saporin was found to act in a synergistic manner, and was superior to both PCI of saporin and PCI-no-drug (TPCS2a + light only) in three TNBC cell lines (MDA-MB-231, MDA-MB-435 and SUM149) and two BRAFV600E mutated malignant melanoma cell lines (Melmet 1 and Melmet 5). The cytotoxic effect was highly dependent on the light dose and expression of CSPG4 since no enhanced cytotoxicity of PCI of 225.28-saporin compared to PCI of saporin was observed in the CSPG4-negative MCF-7 cells. The PCI of a smaller, and clinically relevant CSPG4-targeting toxin (scFvMEL-rGel) validated the CSPG4-targeting concept in vitro and induced a strong inhibition of tumor growth in the amelanotic melanoma xenograft A-375 model. In conclusion, the combination of the drug delivery technology PCI and CSPG4-targeting immunotoxins is an efficient, specific and light-controlled strategy for the elimination of aggressive cells of TNBC and malignant melanoma origin. This study lays the foundation for further preclinical evaluation of PCI in combination with CSPG4-targeting.
Introduction
Triple-negative breast cancer (TNBC) and malignant melanoma that do not respond to immune checkpoint inhibition are highly aggressive cancer types that have an unmet need for better treatment options. TNBC constitutes 10–20% of all breast cancers and is known for its particularly unfavourable clinical prognosis with high rates of metastasis.1-3 TNBC is characterized as oestrogen receptor- (ER−), progesterone receptor-(PR−) and human epidermal growth factor receptor 2-negative (HER2−).3 Due to the absence of ER and HER2, TNBC does not benefit from the otherwise effective anti-breast cancer drugs targeting these receptors, such as tamoxifen and trastuzumab, and surgery in combination with cytotoxic therapy remains the only clinically available treatment option.4
Pathological complete responses following neoadjuvant and adjuvant chemotherapy have been observed in 17 to 58% of patients.3 However, patients who exhibit the chemoresistant subtype of TNBC have an exceptionally poor outcome, with a shorter overall survival compared to non-TNBC.5
Malignant melanoma, the most lethal form of skin cancer, is one of the deadliest forms of human cancers showing exceptional abilities to metastasize and develop drug resistance.6 The conventional chemotherapeutic agent dacarbazine induces a response in very few patients (7–20%).7 Important advances have been achieved with targeted drugs, such as the mutated BRAF inhibitor vemurafenib.8 Although vemurafenib induces a strong response in melanoma patients with the BRAFV600E mutation, acquired resistance will develop leading to relapse over a few months.9 Recently, immunotherapies with the anti-CTLA4 antibody ipilimumab or antibodies against the PD-1/PD-L1 axis have received considerable attention in malignant melanoma. These agents can lead to durable clinical responses, but only in a fraction of melanoma patients. In addition, severe adverse events including immune related toxicities, particularly for anti-CTL4 are often observed.10,11 Thus, achieving prolonged therapeutic responses with limited adverse effects and in a larger fraction of patients remains to be a major challenge for malignant melanoma and especially for TNBC.
Chondroitin sulphate proteoglycan 4 (CSPG4, also known as neuron glia antigen-2 (NG2) or melanoma-associated chondroitin sulfate proteoglycan (MCSP)) is a trans-membrane protein,12 expressed in a majority of melanoma lesions and has been associated with melanoma therapy resistance and aggressiveness.13,14 CSPG4 induces signalling cascades through FAK and MAPK, potentially leading to events promoting survival and chemoresistance, invasion, migration, proliferation and epithelial–mesenchymal transition.14 CSPG4 has also been shown to be a novel marker of normal epidermal stem cells of the skin.15 Recently, CSPG4 was shown to be expressed in triple-negative breast cancer, where the antigen was expressed in 32 of the 44 lesions tested, as well as in several cell lines and tumour cells from pleural effusions.16 Additionally, CSPG4 was strongly associated with the proposed cancer stem cell phenotype CD44high/CD24low.16,17 Restricted distribution of CSPG4 in normal tissues makes CSPG4 a potentially valuable target for tailored TNBC and melanoma treatment.12
Photochemical internalization (PCI) is a drug delivery technology based on the use of photosensitizing compounds and targeted illumination, where the illumination induces the release of drugs from endo–lysosomal compartments into the cell cytosol.18,19 Photosensitizers used with PCI accumulate in endocytic vesicle membranes and subsequent light exposure leads to the generation of reactive oxygen species (ROS), causing membrane disruption and release of the drug of interest into target cells. The PCI technology therefore enhances the endosomal and lysosomal escape of entrapped molecules before they are recycled or degraded following endocytosis. Examples of molecules unable to reach their target within the cell include proteins such as type I ribosome inactivating proteins (RIP), e.g. saporin20 or gelonin, nucleic acids and nanoparticles.19 The delivery of toxins linked to antibodies or growth factors using PCI may enhance the specificity of the PCI treatment and has previously been successfully demonstrated for targeting of EpCAM,21,22 EGFR,23,24 HER2,25 VEGFR-1 and −2,26 CD133,27-29 CD44,30 and CSPG4.31 In addition, the PCI technology has been verified as an efficient strategy to kill therapy-resistant cancer cells.25,32-36 The PCI technology is currently under evaluation in several clinical trials, where PCI of bleomycin for solid localized tumours was recently shown to be safe and encouraging with respect to clinical outcomes in a Phase I study.37
In the present study, we aimed to target and eliminate CSPG4-expressing TNBC and drug-resistant malignant melanoma cells by combining the PCI technology with the novel CSPG4-targeting immunotoxin 225.28-saporin. The efficacy and specificity of the PCI-based targeting of CSPG4 was found promising, and may therefore represent a rational strategy for the elimination of aggressive and therapy-resistant TNBCs and malignant melanoma.
Materials and methods
Cell culture
The mammary cancer cell lines MDA-MB-231 (HTB-26), MDA-MB-435 (HTB-129), MCF-7 (HTB-22) and the non-pigmented malignant melanoma A-375 cells were all from ATCC (Manassas, VA, USA). MDA-MB-231 is described as a TNBC38 and a mesenchymal stem-like cell line1 with expression of putative cancer stem cell markers such as ALDH39,40 and CD133.41 SUM149, a TNBC cell line established from primary inflammatory breast cancer,42 was obtained from the Department of Medical Genetics (Oslo University Hospital). The metastatic melanoma cell lines, Melmet 1 and Melmet 5, were established in-house as previously described.43 All cell lines were routinely tested for Mycoplasma infections and cell ID. MDA-MB-231, MDA-MB-435, Melmet 1 and Melmet 5 were cultured in RPMI-1640 medium (Sigma-Aldrich, St Louis, MO, USA) supplied with L-glutamine, 10% fetal bovine serum (FBS) (PAA Laboratories, Pasching, Austria), 100 IU ml−1 penicillin (Sigma-Aldrich) and 100 μg ml−1 streptomycin (Sigma-Aldrich). MCF-7 was cultured in MEM with Earle’s salts (PAA Laboratories), containing phenol red, supplied with l-glutamine, 10% FBS, 100 IU ml−1 penicillin/100 μg ml−1 streptomycin and 10 μg ml−1 recombinant human insulin (Sigma-Aldrich). SUM149 cells were cultured in a Ham’s F12 nutrient mixture (Sigma-Aldrich) supplied with l-glutamine, 5% FBS, 100 IU ml−1 penicillin/100 μg ml−1 streptomycin, 1 μg ml−1 hydrocortisone (H0888, Sigma-Aldrich) and 5 μg ml−1 insulin. A-375 cells were cultured in DMEM (Sigma-Aldrich) supplied with 10% FBS and 100 IU ml−1 penicillin/100 μg ml−1 streptomycin. All cell lines were cultured as monolayers in Nunclon™ surface treated tissue culture flasks (NUNC A/S, Thermo Fisher Scientific, Roskilde, Denmark) in incubators at 37 °C with 5% (v/v) CO2 in a humidified atmosphere.
We wish to stress that there is a controversy in the literature concerning the origin of the MDA-MB-435 cell line.38,44 However, the focus of this work is the use of the PCI-technology to enhance the efficacy of CSPG4-targeting immunotoxins in both breast (TNBCs) and melanoma cancers and not on the biology of the included cell lines. Hence, independent of the origin of this cell line, the use of the MDA-MB-435 in this study as a CSPG4-expressing cell line is valid.
Reagents and chemicals
A stock solution of hydrocortisone for cell culture was prepared by dissolving 1 mg hydrocortisone powder per 1 ml absolute ethanol and stored as aliquots at −20 °C. TPCS2a (Fimaporfin) was provided by PCI Biotech AS (Lysaker, Norway) and stored protected from light as a stock solution (0.35 mg ml−1 in 3% polysorbate 80, 2.8% mannitol, 50 mM Tris pH 8.5) at 4 °C. TPPS2a (LumiTrans) from PCI Biotech was dissolved in dimethyl sulfoxide to make a 0.35 mg ml−1 stock solution. AlPcS2a (disulfonated aluminum phthalocyanine) provided by Frontier Scientific (Logan UT, USA) was dissolved in 0.1 M NaOH and thereafter diluted in phosphate-buffered saline, pH 7.5, to a final concentration of 5 mg ml−1. TPPS2a and AlPcS2a were stored protected from light as aliquots at −20 °C. LysoTracker Green (DND-26) was obtained from Life Technologies (Eugene, Oregon, USA) and stored as single-use aliquots at −20 °C. Streptavidin-conjugated Alexa Fluor 488 (Life Technologies) was stored at 4 °C according to the manufacturer’s instructions.
Preparation of 225.28-saporin and scFvMEL-rGel
CSPG4-specific mouse mAb 225.28, an IgG2a, was developed and characterized as described.45 mAb was purified from ascitic fluid by affinity chromatography on a Protein G column. mAb was biotinylated as described.46 To obtain the immunotoxin 225.28-saporin, streptavidin-saporin (Advanced Targeting Systems, San Diego, CA, USA), with an average of 2.5 molecules of saporin per molecule of streptavidin, was combined with biotinylated 225.28 mAbs at a biotin : streptavidin ratio of 4 : 1 by mixing and incubating at room temperature for 15–20 minutes. The reaction was assumed to be complete due to the high affinity of streptavidin to biotin. In all calculations and presentations, the molar concentration of the immunotoxin was set equal to the concentration of streptavidin-saporin in the mixture. The scFvMEL-rGel targeted toxin was expressed and purified as previously described.47
Flow cytometry investigation of antigen expression
CSPG4 expression was investigated using live cell flow cytometry. Monolayer cells were harvested using EDTA solution (0.54 mM) to avoid enzymatic degradation of the antigen. Subsequently, cells were incubated for 30 min in 1 μg ml−1 225.28-biotin, followed by incubation in 0.5 μg ml−1 streptavidin-Alexa Fluor® 488 (Life Technologies) for 20 minutes, and were then thoroughly washed in PBS prior to examination. Samples were immediately analyzed on a BD LSR II flow cytometer (BD Biosciences) using a 488 nm laser, 505 nm dichroic filter and a 525/50 band pass excitation filter. Doublet cells, debris and unviable cells were excluded from the analyses based on forward/side scatter.
Intracellular localization studies by confocal and fluorescence microscopy
To validate the flow cytometry data, specific binding of the CSPG4-targeting antibody was compared by 4 h incubation of 225.28-Alexa488 in the CSPG4high MDA-MB-435 cells with the CSPG4low MCF-7 cells by epi-fluorescence microscopy. The cells were washed with cold 1× PBS containing Ca2+ and Mg2+ prior to the fluorescence microscopy using a 63× oil-immersion objective with an Axioimager Z1 Microscope (Carl Zeiss, Oberkochen, Germany). A 450–490 nm band pass excitation filter, 495 nm dichroic mirror and a 500–550 band pass emission filter were used for the measurement of the Alexa-488-labeled 225.28 Ab. The micrographs were processed by use of the Axiovision Software (Carl Zeiss).
The intracellular localization of TPCS2a and the 225.28 mAb was investigated by confocal microscopy of live cells. MDA-MB-435 cells were seeded directly on 0.17 (±0.01) mm thick coverslips (Assistent, Sondheim, Germany) in 4-well dishes (NUNC A/S). To label biotinylated mAbs, 18 μg ml−1 225.28 mAb was mixed with 1.8 μg ml−1 streptavidin-Cy3 (Jackson ImmunoResearch Europe, Newmarket, United Kingdom), followed by 15–20 minutes of incubation at room temperature as described in the Results section. For co-localization studies, cells were co-incubated with the Cy3-labelled 225.28 and 1 μg ml−1 TPCS2a for 18 h, followed by a 4–6 h chase prior to examination. LysoTracker® Green (Life Technologies) was added to a concentration of 0.5–5 μM, 30–120 minutes before examination as described in the Results section. The cover slips were washed briefly in PBS, and placed in 35 mm glass bottom MatTek dishes (MatTek Corp. Ashland, MA, USA) for live cell examination with a Zeiss LSM 710 confocal microscope (Carl Zeiss Microscopy GmbH, Jena, Germany), equipped with an Ar-Laser Multiline (458/488/514 nm), a DPSS-561 10 (561 nm), a Laser diode 405-30 CW (405 nm), and a HeNe-laser (633 nm). The objective used was a Zeiss C-Apochromat 40×/1.20 Water Imm DIC III. Image processing was performed with basic software ZEN 2009 (Carl Zeiss MicroImaging GmbH, Jena, Germany) and Photoshop CS2 (Adobe, Mountain View, CA).
Photochemical internalization (PCI) and cytotoxicity assays
Prior to each experiment the photochemical dose (light dose and concentration of TPCS2a) inducing around 50% reduction of cell viability was found for each cell line. Based on this, TPCS2a was diluted in medium to a final concentration of 0.05 μg ml−1 for SUM149 cells, 0.6 μg ml−1 for MCF-7 cells or 0.2 μg ml−1 for the additional cell lines. Cells were incubated in TPCS2a for 18 h, after which they were washed twice with fresh medium and chased for 4 h to allow the photosensitizer to be internalized and cleared from the plasma membrane. Treatment with TPCS2a and light as described above, in the absence of toxin or immunotoxin, is referred to as PCI-no-drug (photochemical treatment alone = TPCS2a + light only). With PCI treatment, cells were co-incubated with TPCS2a and toxin or immunotoxin for 18 h prior to chase. The concentration of 225.28-saporin and streptavidin-saporin was 0.1 nM in MDA-MB-231, Melmet 1 and Melmet 5 cells and 1 nM in MDA-MB-435, SUM149 and MCF-7 cells. With an average of 2.5 molecules of saporin per complex, this is equivalent to 0.25 nM and 2.5 nM saporin, respectively. With the clonogenic assay, MDA-MB-435 cells were treated with 0.1 nM 225.28-saporin and streptavidin-saporin. The concentration of scFvMEL-rGel and rGelonin was 1 nM in the MDA-MB-231 and SUM149 cell lines, and 5 nM in the MCF-7 cell line. Cells were exposed to light using a LumiSource® light source (PCI Biotech AS), consisting of four light tubes with a peak emission at 435 nm, irradiance 12.6 mW cm−2. 1 J cm−2 = 75 s light exposure.
The cell viability was assessed by the MTT assay as described earlier.32 For MTT-based assays, cells were seeded on 96-well plates (NUNC A/S) at 13 000 cellsper well for MDA-MB-231 cells or 10 000 cellsper well for MDA-MB-435 cells 6 h prior to drug incubation, alternatively 8000 cellsper well for MCF-7, SUM149 and Melmet 5 cells, or 4000 cellsper well for Melmet 1 cells 24 h prior to drug incubation. The optical density at 570 nm was recorded using a Powerwave XS2 microplate spectrophotometer (BioTek, Winooski, VT, USA). Following normalization to blanks on each plate, the cell viability was calculated relative to the absorbance of untreated cells.
For the clonogenic survival assay, a modified version of Franken et al. was used.48 In brief, the cells were seeded in 6-well plates (NUNC) at 1000 cellsper well for MDA-MB-231 and MDA-MB-435 cells, and 2000 cellsper well for SUM149 and Melmet 5 cells. In brief; 9–14 days post treatment, colonies were washed once in saline, fixed by absolute ethanol, stained by a methylene blue solution and counted manually. Colonies consisting of less than 50 cells were excluded.
Immunofluorescence
Melmet 1 and Melmet 5 cells grown on glass coverslips were fixed in 4% paraformaldehyde for 15 min on ice before staining with mouse anti-MITF (Thermo Scientific), or rabbit anti-Axl (Cell Signaling), both diluted 1 : 100 in PBS/0.05% saponin. After overnight incubation at 4 °C, the samples were stained for 1 h with donkey anti-mouse 488 or donkey anti-rabbit 549 (both Jackson ImmunoResearch) diluted 1 : 1500 or 1 : 400, respectively. The coverslips were mounted in Prolong Gold mounting medium (Life Technologies) containing DAPI and examined using a laser-scanning confocal microscope (LSM710; Carl Zeiss), equipped with a Plan-Apochromat ×63/1.4 Oil DICIII objective. Image processing and visualization were performed by using the ZEN Light 2011 software.
Drug response in melanoma cell lines
The growth inhibitory effect of dacarbazine (10, 50 and 100 μg ml−1) and vemurafenib (0.33, 0.66, 3.3 and 6.6 μM) was measured by using CellTiter 96 Aqueous One solution (MTS assay, Promega, Madison, WI, USA). Melmet 1 or Melmet 5 was seeded in 96-well plates and treated with the indicated drugs. Seventy two hours after treatment CellTiter 96 Aqueous One Solution was added to the wells and the absorbance was measured at 490 nm after approximately 2 hours using a micro plate reader (Victor2 1420 Multilabel Counter, PerkinElmer). The viability of the treated cells is reported as the percentage of viable cells relative to untreated control cells. Experiments were performed in four parallels and repeated at least in three independent experiments for each treatment condition.
In vivo PCI of scFv-rGel
Female athymic nude mice of the Hsd: Athymic nude Foxn1nu strain were bred at the Department of Comparative Medicine at Oslo University Hospital, Radiumhospitalet. All procedures involving mice were carried out in agreement with the protocols approved by the animal care unit at the Norwegian Radium Hospital—Oslo University Hospital, under control by the National Ethical Committee’s guidelines on animal welfare. The mice were on average 20–25 g (6–8 weeks old) at the start of the experiment. The mice were kept under specific pathogen-free conditions. Water and food were given ad libitum. Exponentially growing A-375 cells were treated and transplanted s.c. as previously described.31 Mice with 75 mm3 (±25 mm3) A375 tumours were subjected to only one PCI of scFvMEL-rGel treatment and tumour growth was measured twice per week as previously described.31
Statistical analysis
Each experiment evaluating PCI-induced cytotoxicity was performed at least three times (n = 3–9 for each cell line) and the same trend in cytotoxic response was observed. Due to the multiple variables with the PCI protocol, such as photosensitizer concentration, toxin concentration, light source efficiency, the cytotoxicity data presented in Fig. 3, 4E-G and 5A (and Fig. ESI-4, 5 and 7†) stem from a single, representative experiment, while ESI-6† shows the average (3–9 replicated experiments) targeting ratio of PCI of saporin/PCI of 225.28-saporin for each individual experiment: briefly, we calculated the ratio between the time of light exposure needed to kill/reduce viability of 50% of the cells (LD50) after PCI of saporin (toxin) and the time of light exposure needed to kill/reduce viability of 50% of the cells after PCI of 225.28-saporin (immunotoxin) giving a targeting ratio for each cell line with standard error (SE). By using SigmaPlot 12.5 software, the two-sided Student’s t-test was applied on technical replicates within each experiment, where the samples were found normally distributed by the Shapiro–Wilk Normality Test. Where normality test failed, a Mann–Whitney Rank Sum Test was used. The null hypothesis was rejected when P was <0.05. The synergistic toxicity in the 6 CSPG4 + cell lines was evaluated by comparing the theoretical additive effect of combining PCI (no drug) and IT with the observed combination (PCI of IT).49-51 It was assumed that PCI(no drug) and the IT had distinct mechanisms of actions and the calculated additive effect was therefore assessed by the product of the survival fractions (SF) of the two treatments;
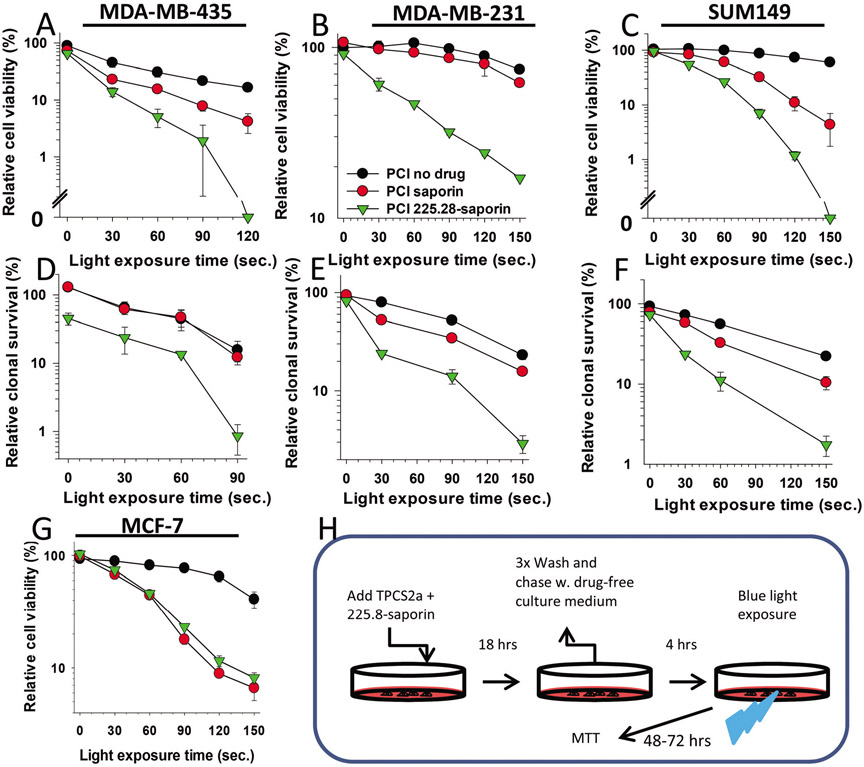
Fig. 3.
Specific and efficient cytotoxic effects induced by PCI of 225.28-saporin in CSPG4-expressing TNBC cell lines. (A), MDA-MB-435, (B) MDA-MB-231 or (C) SUM149 cells were incubated with TPCS2a (PCI no drug), or TPCS2a in combination with streptavidin–saporin or 225.28-saporin (PCI), chased in drug-free medium for 4 h and illuminated as indicated on the X-axis. Relative cell viability was examined using the MTT assay 48 h post illumination. The concentration of TPCS2a was 0.2 μg ml−1 for MDA-MB-231 and MDA-MB-435 and 0.05 μg ml−1 for SUM149. The concentration of toxins was 0.1 nM for MDA-MB-231 and 1 nM for MDA-MB-435 and SUM149. (D–F) Clonogenic survival in (D) MDA-MB-435 (E) MDA-MB-231 or (F) SUM149 cells assessed 10–14 days post light exposure. (G) CSPG-negative MCF-7 control cells after treatments as described in A–C. Each figure shows a single representative experiment out of three independent experiments. Bars = SD of three technical replicates. (H) Experimental protocol of PCI of 225.8-saporin is the same as for PCI of saporin. For PCI no drug (TPCS2a + light only), TPCS2a was incubated alone.
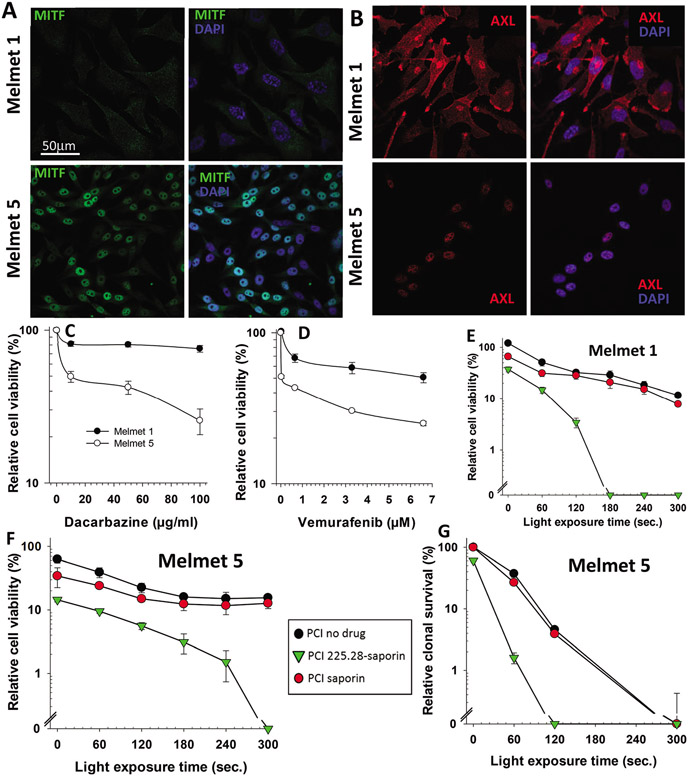
Fig. 4.
PCI of 225.28-saporin is highly cytotoxic in drug-resistant melanoma cells that are BRAFV600E mutated and MITFlow/AXLhigh. (A) MITF (green) or (B) AXL (red) expression was examined by immunofluorescence (left panels). Right panels, merge of MITF or AXL and nuclear staining with DAPI (blue). Melmet q and Melmet 5 cells were treated with either (C) the chemotherapeutic agent dacarbazine or (D) the BRAF inhibitor vemurafenib for 72 h prior to examination of cell viability by the MTS assay. Figures show average of three independent experiments with ±SE bars. (E) Melmet 1 and (F) Melmet 5 cells incubated with 0.2 μg mL−1 TPCS2a (PCI no drug), or TPCS2a in combination with 100 pM streptavidin-saporin (PCI saporin) or the CSPG4-targeting immunotoxin 225.28-saporin (PCI 225.28-saporin) for 18 h, followed by a 4 h chase in drug-free medium prior to light exposure as indicated in the X-axis. Cell metabolic activity/viability was examined using the MTT assay 48 h post illumination. (G) Melmet 5 cells seeded at clonal density (1000 cells per 9.6 cm2) and treated as in F. Cells were assessed for clonogenicity 10–14 days post light exposure. Each figure shows a single representative experiment out of three independent experiments. Bars = SD of three technical replicates.
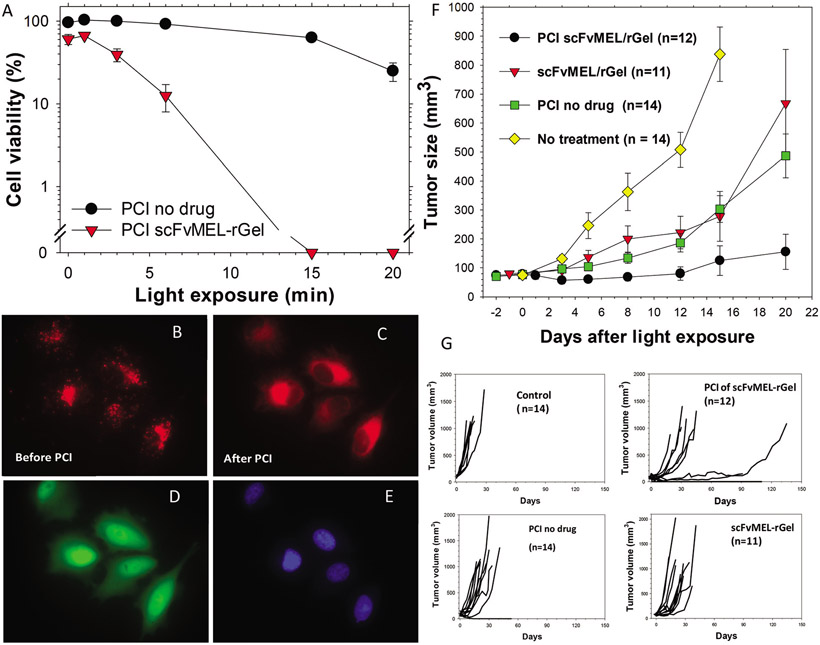
Fig. 5.
Potent in vitro and in vivo effects of the CSPG4-targeting recombinant immunotoxin scFvMEL-rGel. (A) A-375GFP cells were incubated with the PCI-photosensitizer AlPcS2a without (PCI no drug) or with 100 nM scFvMEL-rGel for 18 h and subsequently washed and incubated in drug-free medium for 4 h prior to red broad band light (γmax around 670 nm) exposure. Cytotoxic responses were examined by the MTT assay 72 h post light (data are based on one representative experiment out of five independent experiments (the other 4 experiments are presented in ESI-8†)). Error bars are SD of triplets. (B) A-375 cells with granular (AlPcS2a (red) fluorescence prior to 30 s. microscopy light (650–690 nm) exposure and C) 5 min after PCI-induced cytosolic release of photosensitizer. (D) GFP- and (E) Hoechst 33342 signal. (F) Strong delay of A-375 tumour growth after only one session with PCI of scFvMEL-rGel. These data are extracted from a Kaplan–Meier plot published previously Selbo PK et al. in PLoS One, 4(8), e6691. Athymic nude mice with subcutaneously growing A-375 tumours (approximately 75 mm3) were given AlPcS2a (5 mg kg−1 i.p. 48 h before laser exposure) and scFvMEL-rGel (2 mg kg−1 i.v, 24 h before laser exposure). Laser wavelength: 670 nm, Irradiance: 100 mW cm−2, Light dose: 20 J cm−2. n, number of animals in each group. (G) Individual tumour growth curves over time. The flat line up to day 110 in the PCI of the scFvMEL-rGel group represents 4 mice with complete tumour responses (33%). In the same group, one mouse had a tumour that started to grow slowly at day 100. Another PCI-treated mouse where the tumour size was reduced by 65%, gained a slow tumour growth between day 60 and 90, but had to be sacrificed due to a nose infection. The number of long time survivors (animals with tumours less than 800 mm3 at day 90) was n = 6 (50%) in the PCI group.
The difference in logarithm (DL) of the SFadd and SFPCI IT was calculated;
and the standard error of DL was assessed by;
where
A synergistic combination of the two treatments (PCI of IT) was defined as a significant positive DL as evaluated by a two-sided student t test
The data were considered significant when p ≤ 0.05. All synergy calculations were estabIished at a PCI(no drug)-dose reducing the viability to 20, 30 or 50% (LD50 for the TNBC and LD30 or LD30 for Melmet 1 and Melmet 5, respectively) and extrapolation was used in cases where the LD50/30/20 was not reached. The SF was set to 0.001 (99.9% toxicity) in experiments where no MTT signals or colonies were detected.
Results
CSPG4 expression in breast cancer and melanoma cell lines
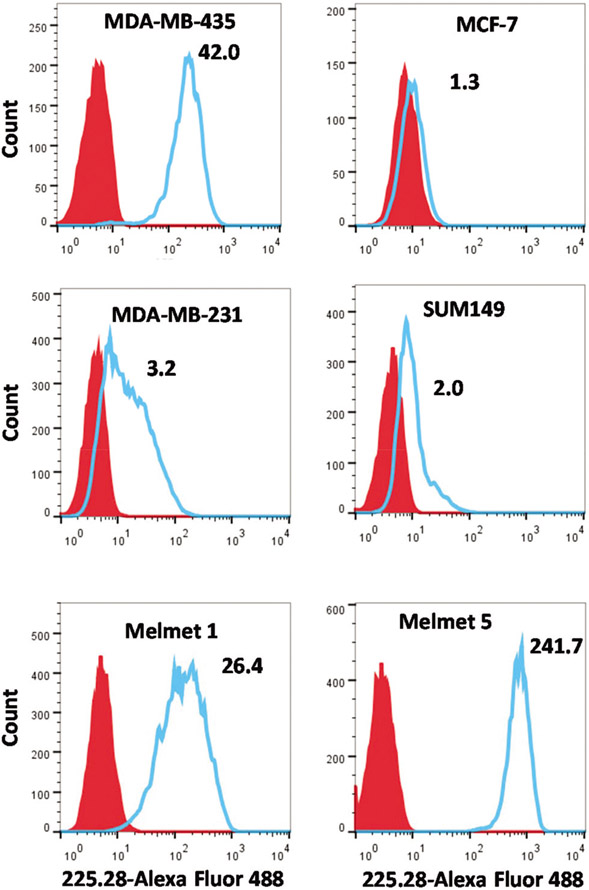
The CSPG4 surface expression levels in four breast cancer and two melanoma cell lines were evaluated by flow cytometry (Fig. 1). The median fluorescence intensity (MFI) was determined in cells stained with Alexa488-225.28-mAbs, compared to the MFI in unstained control cells, and a fluorescence ratio was calculated (presented in Fig. 1). The MDA-MB-435 cells showed the highest expression of CSPG4 of all TNBC cell lines tested, with a fluorescence ratio of 42.0 (Fig. 1), and where 97% of the cells expressed the antigen (Fig. ESI-1†).
Fig. 1.
Cell surface expression of CSPG4 evaluated by flow cytometry. Cells stained with Alexa Fluor 488-linked 225.28 mAb are shown in blue. Red histogram represents unstained cells. The ratio of median fluorescence intensity of stained cells versus unstained cells is provided for each cell line. Figure shows representative histograms out of three independent experiments.
In the TNBC cell lines MDA-MB-231 and SUM149 the MFI ratio was 3.2 and 2.0, respectively. These cells showed a heterogeneous expression of CSPG4, where approximateIy 37% of MDA-MB-231 and 14% of SUM149 cells were strongly positive (Fig. ESI-1†). The luminal breast cancer cell line MCF-7 showed a negligible expression of CSPG4, with a fluorescence ratio of 1.3 and therefore further referred to as CSPG4-negative. The whole cell populations of the malignant melanoma cell lines Melmet 1 and Melmet 5 expressed high levels of CSPG4, with the fluorescence ratios of 26.4 and 241.7, respectively (Fig. 1).
The CSPG4-targeting mAb 225.28 binds to CSPG4-positive cells and is captured in endosomes/lysosomes together with the PCI photosensitizer TPCS2a.
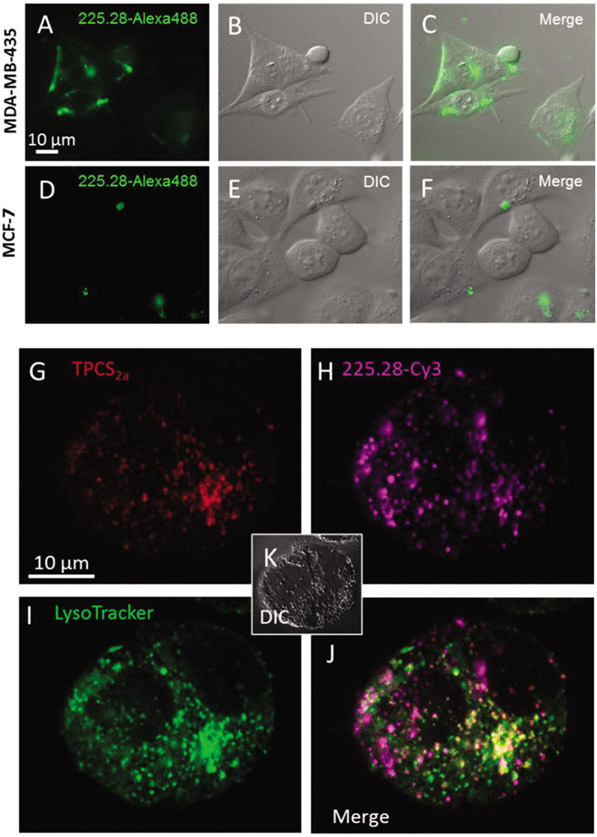
To compare the binding of 225.28mAb to the cell surface of CSPG4-positive MDA-MB-435 cell line versus the CSPG4-negative MCF-7, the cells were incubated for 4 h with Alexa Fluor-labelled 225.28mAb (225.28-Alexa488) and observed by epi-fluorescence microscopy (Fig. 2A-C).
Fig. 2.
Specific binding of mAb 225.28 and co-localization of TPCS2a, mAb 225.28-Cy3 and LysoTracker. (A–F) MDA-MB-435 and MCF-7 cells were incubated with fluorochrome-linked 225.28 for 4 h prior to examination by epi-fluorescence microscopy. (G–K) MDA-MB-435 cells incubated for 18 h in medium containing (G) TPCS2a (1 μg ml−1) and (H) 225.28 mAbs labeled by cyanine 3 (Cy3) (30 nM), washed and chased in fresh medium for 4 h prior to examination by confocal microscopy. (I) LysoTracker Green (1 μM) was added 1 h prior to investigation. (J) Merge of TPCS2a, 225.28-Cy3 and LysoTracker Green. (K) Corresponding DIC micrograph.
While a strong fluorescence signal was detected along the surface of MDA-MB-435 cells, CSPG4-targeting 225.28-Alexa488 could not be detected on the surface of the MCF-7 cell line (Fig. 2D-F). This indicates that the surface binding of 225.28-mAb was specific and depended on the CSPG4 level. The majority of the cell were completely negative, however, in some MCF-7 cells granular fluorescence was observed (Fig. 2D), which may be due to the aggregation and non-specific binding of the complex on the surface of the cells.
Drug delivery by PCI is dependent on co-localization of the photosensitizer and drug of interest within endo–lysosomal compartments. To study the intracellular localization of the 225.28 mAb-delivered moiety, 225.28 mAb was linked to streptavidin-Cyanine 3, referred to as 225.28-Cy3, and used as a surrogate for the immunotoxin. The intracellular localization of 225.28-Cy3 and the photosensitizer TPCS2a was investigated by confocal microscopy in the CSPG4-positive MDA-MB-435 cell line (Fig. 2G-J and ESI-2†). Following 18 h incubation and 4 h chase, TPCS2a and the 225.28-Cy3 complex was detected as intracellular fluorescent puncta. LysoTracker Green was used to stain endosomes and lysosomes (Fig. 2 and ESI-3†).
PCI of 225.28-saporin induces a specific and strong synergistic effect in TNBC cell lines
To generate the CSPG4-targeted immunotoxin, we combined biotinylated 225.28 with streptavidin-saporin, further referred to as 225.28-saporin.
It is important that the binding of the targeting moiety (in this respect: biotinylated 225.28) to streptavidin-saporin do not destroy the enzymatic activity (inhibition of protein synthesis) of saporin. Thus, we evaluated the ribosome-inactivating protein (RIP) activity of 225.28-saporin versus streptavidin-saporin in a cell-free system. No substantial difference or RIP activity between the targeted and non-targeted toxins was observed (Fig. ESI-4†).
To evaluate the potential of PCI to enhance the cytotoxicity of 225.28-saporin in TNBC cell lines, we combined light, TPCS2a and the immunotoxin and measured cell viability by the MTT assay (Fig. 3A).
In the CSPG4-positive MDA-MB-435 cell line, PCI of 225.28-saporin with a light dose of 120 s resulted in a total elimination of the MTT signal (Fig. 3A). Incubation of the 225.28 mAb alone for 18 h (as in the PCI protocol) did not induce any effects on cell viability (Fig. ESI-5A†). Blocking CSPG4 with excess 225.28 mAb abolished the antigen-targeting effect of PCI with 225.28-saporin as the difference of cytotoxicity compared to PCI of saporin was not statistically significant (P = 0.62, Fig. ESI-5B†). In the MDA-MB-231 cell line, the PCI of 225.28-saporin resulted in a strong attenuation of cell viability despite the 10-fold reduction of the concentration of the 225.28-saporin immunotoxin compared with the concentration used for the PCI-based targeting of the MDA-MB-435 cells (Fig. 3B).
To evaluate the long-term effect on cell survival, we performed the clonogenic assay. In accordance with the MTT data, a reduction of colony-forming abilities by close to 99% was achieved in all three CSPG4-positive TNBC cell lines treated by PCI of 225.28-saporin (Fig. 3D-F). PCI of 225.28-saporin induced a synergistic cytotoxic effect in all three cell lines (Table ESI-1†).
In all three TNBC cell lines, there was a substantial and statistically significant difference when comparing PCI of the immunotoxin 225.28-saporin and PCI of the non-targeted toxin streptavidin-saporin (Fig. ESI-6†) or PCI-no-drug. The effect of the single treatments in the TNBC cell lines was minor compared to the effect of PCI of 225.28-saporin. PCI-no-drug induced a modest cytotoxic effect across the different cell lines (Fig. 3). There was no marked cytotoxic effect of the immunotoxin combined with TPCS2a in the absence of light (shown as 0 s. light exposure in Fig. 3), except for the MDA-MB-435 cell line, where the clonogenic assay revealed a 50% reduction in clonal cell survival (Fig. 3E). In the CSPG4-negative MCF-7 cell line, the PCI efficacy of 225.28-saporin versus streptavidin-saporin was indistinguishable (Fig. 3G). The observed difference between PCI of immunotoxin and non-targeted toxin in CSPG4-positive cell lines, but not the CSPG4-negative cell line, indicates that the PCI-enhanced toxicity of the immunotoxin depends on CSPG4 expression.
PCI of 225.28-saporin is highly cytotoxic in BRAFV600E mutated MITFlow/AXLhigh melanoma cells
To evaluate the PCI efficacy in another aggressive cancer type, we used two highly aggressive BRAFV600E mutated malignant melanoma cell lines, Melmet 1 and Melmet 5, originally derived from subcutaneous and lymph node metastases, respectively.43,52 Melmet 5 exhibits a strong nuclear staining of Microphthalmia-associated transcription factor (MITF), the main regulator of melanocytic differentiation, while the MITF level in Melmet 1 was very low (Fig. 4A). The CSPG4 surface experiments including incubation with only secondary antibody-Alexa488 (resulting in no signal, data not shown) we ruled out that the low MITF signal of the Melmet 1 cells are neither unspecific staining nor autofluorescence or background staining. In contrast, the level of AXL receptor tyrosine kinase was high in Melmet 1 and negligible in Melmet 5 (Fig. 4B). This indicates that Melmet 1 and Melmet 5 represent two opposite phenotypes, known as MITFlow/AXLhigh and MITFhigh/AXLlow, respectively.53 It was reported previously that the MITFlow/AXLhigh phenotype is associated with more aggressive, stem-like melanoma cells54,55 showing higher resistance to therapy.53,56 In accordance with these reports, we observed that Melmet 1 cells were more resistant to clinically used drugs, the chemotherapeutic agents dacarbazine, and the mutated BRAF inhibitor vemurafenib than Melmet 5 (Fig. 4C and D). Both Melmet 1 and Melmet 5 melanoma cell lines were sensitive to 225.28-saporin + TPSC2a in the absence of light. However, the cytotoxic efficacy was strongly enhanced by PCI in a light-dose dependent manner (Fig. 4E-G). The MTT assay revealed that the cell viability was below detectable levels following a 180 s and 300 s light exposure in Melmet 1 and Melmet 5, respectively (Fig. 4E and F). This suggests that the drug resistant Melmet 1 was more sensitive to PCI of 225.28-saporin than Melmet 5, even though the sensitivity to TPCS2a + light (PCI-no-drug) and PCI of streptavidin–saporin was comparable in both cell lines, and despite the fact that Melmet 1 expressed approximately 1/10 of the CSPG4 antigen compared to the Melmet 5 cells (Fig. 1). Furthermore, the efficacy of PCI of 225.28-saporin to inhibit colony formation was found superior to PCI-no-drug and PCI of streptavidin–saporin in Melmet 5 cells (Fig. 4G). Of notice, treatment with the immunotoxin alone resulted in relatively high cytotoxic responses in the Melmet cell lines (cells with very CSPG4 expression), which may explain that synergistic responses were not obtained when assessing synergy at LD50 doses. However, at higher light-doses (LD70–80) the PCI of 225.28-saporin resulted in strong synergistic responses (Table ESI-1†). The clonogenic assay could not be performed in Melmet 1, which shows a highly motile mesenchymal phenotype and is therefore not compatible with this assay. Collectively, these data indicate that 225.28-saporin combined with PCI is an effective strategy for targeting malignant melanoma cells that show resistance or only partial response to clinically-used drugs.
PCI-based targeting of CSPG4 by the recombinant fusion toxin scFvMEL-rGel in TNBC cells and a malignant melanoma xenograft
To validate this PCI-based CSPG4 targeting concept and to ensure that the treatment response to 225.28-saporin was not influenced by the streptavidin-biotin linkage or the type of toxin we used; we tested the PCI of the fusion toxin scFvMEL-rGel. scFvMEL-rGel consists of the scFv fragment of the 225.28 mAb, recombinantly fused to the ribosome-inhibiting protein gelonin,47 which belongs to the same RIP type I family as saporin. A similar cytotoxic response was observed with PCI of scFvMEL-rGel as with PCI of 225.28-saporin in the TNBC cell lines MDA-MB-231 and SUM149 and in the CSPG4-negative control cell line MCF-7 (Fig. ESI-7†).
The efficacy of PCI of scFvMEL-rGel was also validated in another malignant melanoma cell line, A-375 (Fig. 5A + ESI-8†), which represent the MITFlow/AXLhigh phenotype as Melmet 1.56 In these experiments we used the PCI photosensitizer AlPcS2a, which also localize to lysosomes.57 Here we demonstrate that AlPcS2a, indeed is released to the cytosol of A375-GFP cells after light exposure (Fig. 5B and C).
In addition, the ability of PCI of scFvMEL-rGel (CSPG4-targeting toxin) to delay tumour growth was demonstrated in the A-375 xenograft model (Fig. 5F and G + Fig. ESI-9† for experimental design overview). While non-treated A-375 tumours (n = 14) had a >12-fold increase in tumour size at day 15, only one treatment with PCI of scFvMEL-rGel (n = 12) resulted in a substantial tumour growth delay where there was no statistical significant (P = 0.39) increase in the tumour size at day 15 compared to day 0. Altogether, the PCI of the scFvMEL-rGel group (n = 12) had a response rate of 83% of which 1/3 of the mice achieved complete responses (Fig. 5G), one mouse had a tumour that did not display aggressive growth (followed until day 91) and another long surviving mouse which reached the study endpoint at day 135. At day 20, the anti-tumour effect of PCI of scFvMEL-rGel was significantly (P < 0.001) better than PCI-no-drug (~7-fold increase at day 20, n = 14) or scFvMEL-rGel (~9-fold increase at day 20, n = 11) treatment groups.
Discussion
CSPG4 is recognized as an attractive target in cancer therapy due to its very low or no expression in normal tissues and overexpression in several cancer types,12 including triple-negative breast cancer (TNBC),16 malignant melanoma,13 malignant mesothelioma,58 sarcomas12 and squamous cell carcinoma of the head and neck,16 and known to correlate with radio- and chemoresistance and poor clinical outcome.59,60 In this study, we developed the CSPG4-targeting model immunotoxin, 225.28-saporin and evaluated its use in combination with the clinically relevant photosensitizer TPCS2a (fimaporfin) for PCI-based targeting of TNBC and malignant melanoma. In addition, we studied the anti-cancer effect of CSPG4-targeting toxin, scFvMEL-rGel delivered by PCI both in vitro and in vivo.
PCI of 225.28-saporin was a highly cytotoxic treatment in all five tested CSPG4 expressing cancer cell lines, with no signs of resistance to the treatment. Interestingly, in malignant melanoma we observed a stronger response to PCI of 225.28-saporin in Melmet 1 cells than in the Melmet 5 cells, despite the lower sensitivity of Melmet 1 to conventional drugs used in the clinic and the fact that these cells have a 10-fold reduced expression of CSPG4 compared to Melmet 5. As shown in this study and reported previously,52 these two malignant melanoma cell lines display opposite phenotypes: mesenchymal-like/invasive de-differentiated (Melmet 1 as well as A-375) and non-invasive differentiated (Melmet 5).52 The mesenchymal-like phenotype has been linked to drug resistance, particularly to targeted therapies.53,56 Hence, the efficient eradication of Melmet 1 and A-375 cells by PCI of CSPG4-targeting immunotoxins suggests that resistance mechanisms characteristic for this phenotype might not interfere with PCI-based therapies. The PCI approach might therefore be effective in targeting cancer cells in a mesenchymal-like de-differentiated state. This observation is also highly relevant in breast cancer, where malignant cells can acquire mesenchymal properties due to epithelial–mesenchymal transition (EMT), which is also linked with drug resistance.61 The TNBC cell lines MDA-MB-231 and SUM149 have been shown to consist of stem cell-like subpopulations capable of self-renewal, increased tumourigenicity and resistance to radio- and chemotherapy.62-64 Furthermore, MDA-MB-231 and MDA-MB-435 have been sub-classified as claudin-low, while the SUM149 cell line has been shown to have a claudin-low subpopulation.65 Claudin-low TNBC cells are low in differentiation markers, high in EMT markers and show stem-cell like properties.65 This is especially interesting, given that PCI of the CSPG4-targeting immunotoxin was effective even in MDA-MB-231 and SUM149, where only a fraction of cells were expressing CSPG4. This may indicate purging of a subpopuIation important for cell-renewing and long-term survival. Taken together, these data underscores the high potential of targeting CSPG4 by PCI in aggressive cancers as triple-negative breast cancer and malignant melanoma, and implies a novel approach for targeting tumour cells with stem cell/-mesenchymal properties.
The PCI of 225.28-saporin induced a stronger cytotoxic effect compared to PCI of streptavidin–saporin. This was especially apparent in cell lines with a high CSPG4 expression, i.e. MDA-MB-435 and the Melmet cell lines. Also in the absence of light, 225.28-saporin exhibited a moderate to high cytotoxic effect in cell lines with high CSPG4 expression levels. The difference in cytotoxic responses to streptavidin–saporin versus 225.28-saporin is most likely explained by different cellular uptakes, as in the cell-free system no difference in the ribosome inhibiting activity was observed (Fig. ESI-4†). The immunotoxin 225.28-saporin is most probably taken up into cells by receptor-mediated endocytosis after binding to CSPG4. This conclusion is supported by the cellular localization studies, showing that the 225.28-targeting moiety binds to the cell surface dependent on CSPG4-levels, and eventually accumulates in the endo–lysosomal compartment. In contrast, streptavidin–saporin, which is not known to bind to any cell surface receptor, is most likely taken up through the slower and less efficient process of pinocytosis. There might be also other reasons for differences in cytotoxicity between 225.28-saporin and streptavidin–saporin, e.g. a different route of intracellular trafficking following endocytosis and/or inhibition of FAK and ERK pathways by the 225.28 mAb itself.16 The latter is, however, less likely as treatment with the 225.28 mAb did not influence the photochemical treatment alone (PCI-no-drug) induced cytotoxic response (Fig. ESI-5†). An elimination of the PCI based targeting of CSPG4 was obtained by blocking target cells with excess 225.28 mAb. In addition, no enhanced targeting effects of PCI of 225.28-saporin was achieved in the CSPG4-neg MCF-7 cells. Altogether, the survival data demonstrates the CSPG4-dependent specificity of 225.28-saporin and implies PCI of 225.28-saporin as a promising targeted treatment option.
The concept of TPCS2a-PCI-enhanced delivery of a CSPG4-targeting toxin should be highly relevant for clinical applications since it offers; (1) the advantage of a high tumour selectivity of the therapeutics due to preferential accumulation of the photosensitizer in tumour tissues, primarily by the enhanced permeability and retention phenomenon;66,67 (2) a mAb or scFv-fragment, which is linked to the ribosome-inactivating toxin, ensures tumour cell targeting; (3) a spatiotemporal controlled laser-activation of the PCI-photosensitizer followed by endosomal escape and activation of the entrapped toxin. Furthermore, pre-clinical data indicates that a single injection of photosensitizer and toxin is sufficient for the anti-tumor effect with PCI,19 reducing adverse effects and the formation of neutralizing antibodies, which is often a challenge with in vivo applications of immunotoxins.26 Of relevance, also clinical data confirm that only one session with PCI-drug combination is sufficient to obtain strong anti-tumour responses.37
The molecular size of 225.28-saporin (>700 kDa) is most probably not optimal for targeting cancer cells in solid tumours, due to the limited transport across vascular endothelium and high interstitial fluid pressure within the tumours, as well as accumulation in organs presenting with a fenestrated vasculature. To overcome this obstacle, a smaller immunotoxin, e.g. scFvMEL-rGel (a 56 kDa) should be used in a preclinical/clinical setting. The in vivo studies with scFvMEL-rGel demonstrated the feasibility of the PCI-based targeting of CSPG4-expressing melanoma tumours. In immunocompromised mice lacking mature T-cells, PCI of scFvMEL-rGel delayed the tumour growth and was superior to all control treatments, including scFvMEL-rGel alone, PCI-no-drug or the combination of PCI-photosensitizer + scFvMEL-rGel without light. The superior anti-cancer effect of PCI was also reflected by enhanced animal survival in the PCI group compared to all other treatment groups as demonstrated previously.31 Hence, the PCI- and CSPG4-based targeting approach may have great potential against CSPG4-expressing tumours.
Late stage TNBC and melanoma are often metastasized at presentation. This represents a problem for focal therapies. However, it has been demonstrated that by photodynamic therapy (PDT, photosensitizer + light) it is possible to stimulate naïve and memory T-cells for the attack on tumour cells distant from the original site of treatment.68 As a proof-of-concept we recently showed that PCI is able to potentiate ovalbumin-based model cancer vaccines,69,70 and to enhance HPV peptide-based vaccines resulting in strong activation of tumour-suppressing CD8 killer cells, both in therapeutic and prophylactic settings.71 The PCI method has therefore potential to be used in more advanced and metastasized cancers. Another challenge for light-based treatment strategies in melanotic melanoma is limited penetration of light into pigmented tissue.72 However, as seen in this work, cytotoxic effects induced by PCI are substantially larger compared to PCI without drug-combination (PCI-no-drug), and requires a lower light dose to achieve anti-tumour effects in amelanotic melanoma. This indicates that the limited light-penetration should be less of a problem with PCI compared to photochemical treatment alone. In either case, localized non-pigmented melanomas should be a clinically relevant indication for PCI.
Conclusions
PCI of CSPG4-based immunotoxins is a highly efficient and selective targeting approach for site-directed killing of drug-resistant cells of TNBC and malignant melanoma origin, as shown in vitro and in vivo. These results lay the foundation for further preclinical evaluation of this targeting approach for the treatment of aggressive melanomas and TNBCs.
Supplementary Material
Acknowledgements
We thank Nina Iversen at the Department of Medical Genetics (Division of Diagnostics and Intervention, Oslo University Hospital) for providing the SUM149 cell line. We also thank the Flow Cytometry Core Facility (Institute for Cancer Research) by Idun D. Rein and Kirsti S. Landsverk for the assistance with flow cytometry analyses, Kotryna Seip for the technical assistance with the Melmet cell lines and Solveig Pettersen for help with the immunofluorescence. We thank the Norwegian Radium Hospital Research Foundation (FU 0803), the Norwegian Research Council (SFI-CAST) for funding this project.
Footnotes
Conflicts of interest
Dr Anders Høgset is the Chief Scientific Officer of PCI Biotech AS of which he also is a minority shareholder. The other authors have no conflicts to declare.
Electronic supplementary information (ESI) available. See DOI: 10.1039/c7pp00358g
References
- 1.Lehmann BD, Bauer JA, Chen X, Sanders ME, Chakravarthy AB, Shyr Y and Pietenpol JA, J. Clin. Invest, 2011, 121, 2750–2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA, Clin. Cancer Res, 2007, 13, 4429–4434. [DOI] [PubMed] [Google Scholar]
- 3.Irshad S, Ellis P and Tutt A, Curr. Opin. Oncol, 2011, 23, 566–577. [DOI] [PubMed] [Google Scholar]
- 4.Reddy KB, Curr. Oncol, 2011, 18, e173–e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gluz O, Liedtke C, Gottschalk N, Pusztai L, Nitz U and Harbeck N, Ann. Oncol, 2009, 20, 1913–1927. [DOI] [PubMed] [Google Scholar]
- 6.Gray-Schopfer V, Wellbrock C and Marais R, Nature, 2007, 445, 851–857. [DOI] [PubMed] [Google Scholar]
- 7.Avril MF, Aamdal S, Grob JJ, Hauschild A, Mohr P, Bonerandi JJ, Weichenthal M, Neuber K, Bieber T, Gilde K, Guillem Porta V, Fra J, Bonneterre J, Saiag P, Kamanabrou D, Pehamberger H, Sufliarsky J, Gonzalez Larriba JL, Scherrer A and Menu Y, J. Clin. Oncol, 2004, 22, 1118–1125. [DOI] [PubMed] [Google Scholar]
- 8.Flaherty KT, Hodi FS and Fisher DE, Nat. Rev. Cancer, 2012, 12, 349–361. [DOI] [PubMed] [Google Scholar]
- 9.Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, Chodon T, Nelson SF, McArthur G, Sosman JA, Ribas A and Lo RS, Nature, 2010, 468, 973–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, van den Eertwegh AJ, Lutzky J, Lorigan P, Vaubel JM, Linette GP, Hogg D, Ottensmeier CH, Lebbe C, Peschel C, Quirt I, Clark JI, Wolchok JD, Weber JS, Tian J, Yellin MJ, Nichol GM, Hoos A and Urba WJ, N. Engl. J. Med, 2010, 363, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robert C, Thomas L, Bondarenko I, O’Day S, Weber J, Garbe C, Lebbe C, Baurain JF, Testori A, Grob JJ, Davidson N, Richards J, Maio M, Hauschild A, Miller WH Jr., Gascon P, Lotem M, Harmankaya K, Ibrahim R, Francis S, Chen TT, Humphrey R, Hoos A and Wolchok JD, N. Engl. J. Med, 2011, 364, 2517–2526. [DOI] [PubMed] [Google Scholar]
- 12.Campoli M, Ferrone S and Wang X, Adv. Cancer Res, 2010, 109, 73–121. [DOI] [PubMed] [Google Scholar]
- 13.Campoli MR, Chang CC, Kageshita T, Wang X, McCarthy JB and Ferrone S, Crit. Rev. Immunol, 2004, 24, 267–296. [DOI] [PubMed] [Google Scholar]
- 14.Price MA, Colvin Wanshura LE, Yang J, Carlson J, Xiang B Li G Ferrone S Dudek AZ Turley EA and McCarthy JB, Pigm. Cell Melanoma Res, 2011, 24, 1148–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Legg J, Jensen UB, Broad S, Leigh I and Watt FM, Development, 2003, 130, 6049–6063. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Osada T, Wang Y, Yu L, Sakakura K, Katayama A, McCarthy JB, Brufsky A, Chivukula M, Khoury T, Hsu DS, Barry WT, Lyerly HK, Clay TM and Ferrone S, J. Natl. Cancer Inst, 2010, 102, 1496–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ and Clarke MF, Proc. Natl. Acad. Sci. U. S. A, 2003, 100, 3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berg K, Selbo PK, Prasmickaite L, Tjelle TE, Sandvig K, Moan J, Gaudernack G, Fodstad O, Kjølsrud S, Anholt H, Rodal GH, Rodal SK and Høgset A, Cancer Res, 1999, 59, 1180–1183. [PubMed] [Google Scholar]
- 19.Selbo PK, Weyergang A, Hogset A, Norum OJ, Berstad MB, Vikdal M and Berg K, J. Controlled Release, 2010, 148, 2–12. [DOI] [PubMed] [Google Scholar]
- 20.Polito L, Bortolotti M, Mercatelli D, Battelli MG and Bolognesi A, Toxins, 2013, 5, 1698–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selbo PK, Sivam G, Fodstad O, Sandvig K and Berg K, Int. J. Cancer, 2000, 87, 853–859. [DOI] [PubMed] [Google Scholar]
- 22.Lund K, Bostad M, Skarpen E, Braunagel M, Kiprijanov S, Krauss S, Duncan A, Hogset A and Selbo PK, mAbs, 2014, 6, 1038–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yip WL, Weyergang A, Berg K, Tonnesen HH and Selbo PK, Mol. Pharmaceutics, 2007, 4, 241–251. [DOI] [PubMed] [Google Scholar]
- 24.Weyergang A, Selbo PK and Berg K, J. Controlled Release, 2006, 111, 165–173. [DOI] [PubMed] [Google Scholar]
- 25.Berstad MB, Weyergang A and Berg K, Biochim. Biophys. Acta, 2012, 1820, 1849–1858. [DOI] [PubMed] [Google Scholar]
- 26.Weyergang A, Selbo PK, Berstad ME, Bostad M and Berg K, Lasers Surg. Med, 2011, 43, 721–733. [DOI] [PubMed] [Google Scholar]
- 27.Stratford EW, Bostad M, Castro R, Skarpen E, Berg K, Hogset A, Myklebost O and Selbo PK, Biochim. Biophys. Acta, 2013, 1830, 4235–4243. [DOI] [PubMed] [Google Scholar]
- 28.Bostad M, Berg K, Hogset A, Skarpen E, Stenmark H and Selbo PK, J. Controlled Release, 2013, 168, 317–326. [DOI] [PubMed] [Google Scholar]
- 29.Bostad M, Olsen CE, Peng Q, Berg K, Høgset A and Selbo PK, J. Controlled Release, 2015, 206, 37–48. [DOI] [PubMed] [Google Scholar]
- 30.Bostad M, Kausberg M, Weyergang A, Olsen CE, Berg K, Hogset A and Selbo PK, Mol. Pharm, 2014, 11, 2764–2776. [DOI] [PubMed] [Google Scholar]
- 31.Selbo PK, Rosenblum MG, Cheung LH, Zhang W and Berg K, PLoS One, 2009, 4, e6691, DOI: 10.1371/journal.pone.0006691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Selbo PK, Weyergang A, Bonsted A, Bown SG and Berg K, J. Pharmacol. Exp. Ther, 2006, 319, 604–612. [DOI] [PubMed] [Google Scholar]
- 33.Lou PJ, Lai PS, Shieh MJ, MacRobert AJ, Berg K and Bown SG, Int. J. Cancer, 2006, 119, 2692–2698. [DOI] [PubMed] [Google Scholar]
- 34.Selbo PK, Weyergang A, Eng MS, Bostad M, Mælandsmo GM, Høgset A and Berg K, J. Controlled Release, 2012, 159, 197–203. [DOI] [PubMed] [Google Scholar]
- 35.Olsen CE, Weyergang A, Edwards VT, Berg K, Brech A, Weisheit S, Høgset A and Selbo PK, Biochem. Pharmacol, 2017, 144, 63–77. [DOI] [PubMed] [Google Scholar]
- 36.Selbo PK, Bostad M, Olsen CE, Edwards VT, Hogset A, Weyergang A and Berg K, Photochem. Photobiol. Sci, 2015, 14, 1433–1450. [DOI] [PubMed] [Google Scholar]
- 37.Sultan AA, Jerjes W, Berg K, Hogset A, Mosse CA, Hamoudi R, Hamdoon Z, Simeon C, Carnell D, Forster M and Hopper C, Lancet Oncol, 2016, 1217–1229, DOI: 10.1016/S1470-2045(16)30224-8. [DOI] [PubMed] [Google Scholar]
- 38.Chambers AF, Cancer Res, 2009, 69, 5292–5293. [DOI] [PubMed] [Google Scholar]
- 39.Croker AK and Allan AL, Breast Cancer Res. Treat, 2012, 133, 75–87. [DOI] [PubMed] [Google Scholar]
- 40.Hwang-Verslues WW, Kuo WH, Chang PH, Pan CC, Wang HH, Tsai ST, Jeng YM, Shew JY, Kung JT, Chen CH, Lee EY, Chang KJ and Lee WH, PLoS One, 2009, 4, e8377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohlfest JR, Zellmer DM, Panyam J, Swaminathan SK, Oh S, Waldron NN, Toma S and Vallera DA, Drug Delivery Transl. Res, 2013, 3, 195–204. [DOI] [PubMed] [Google Scholar]
- 42.van Golen KL, Davies S, Wu ZF, Wang Y, Bucana CD, Root H, Chandrasekharappa S, Strawderman M, Ethier SP and Merajver SD, Clin. Cancer Res, 1999, 5, 2511–2519. [PubMed] [Google Scholar]
- 43.Prasmickaite L, Skrbo N, Hoifodt HK, Suo Z, Engebraten O, Gullestad HP, Aamdal S, Fodstad O and Maelandsmo GM, Pigm. Cell Melanoma Res, 2010, 23, 449–451. [DOI] [PubMed] [Google Scholar]
- 44.Rae JM, Creighton CJ, Meck JM, Haddad BR and Johnson MD, Breast Cancer Res. Treat, 2007, 104, 13–19. [DOI] [PubMed] [Google Scholar]
- 45.Wilson BS, Imai K, Natali PG and Ferrone S, Int. J. Cancer, 1981, 28, 293–300. [DOI] [PubMed] [Google Scholar]
- 46.Wang X, Katayama A, Wang Y, Yu L, Favoino E, Sakakura K, Favole A, Tsuchikawa T, Silver S, Watkins SC, Kageshita T and Ferrone S, Cancer Res, 2011, 71, 7410–7422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rosenblum MG, Cheung LH, Liu Y and Marks JW III, Cancer Res, 2003, 63, 3995–4002. [PubMed] [Google Scholar]
- 48.Franken NA, Rodermond HM, Stap J, Haveman J and van Bree C, Nat. Protoc, 2006, 1, 2315–2319. [DOI] [PubMed] [Google Scholar]
- 49.Steel GG and Peckham MJ, Int. J. Radiat. Oncol., Biol., Phys, 1979, 5, 85–91. [DOI] [PubMed] [Google Scholar]
- 50.Selbo PK, Kaalhus O, Sivam G and Berg K, Photochem. Photobiol, 2001, 74, 303–310. [DOI] [PubMed] [Google Scholar]
- 51.Weyergang A, Berstad ME, Bull-Hansen B, Olsen CE, Selbo PK and Berg K, Photochem. Photobiol. Sci, 2015, 14, 1465–1475. [DOI] [PubMed] [Google Scholar]
- 52.Nygaard V, Prasmickaite L, Vasiliauskaite K, Clancy T and Hovig E, Oncoscience, 2014, 1, 82–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Geukes Foppen MH, Kemper K, Goding CR, McDermott U, Blank C, Haanen J, Graeber TG, Ribas A, Lo RS and Peeper DS, Nat. Commun, 2014, 5, 5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoek KS and Goding CR, Pigm. Cell Melanoma Res, 2010, 23, 746–759. [DOI] [PubMed] [Google Scholar]
- 55.Zipser MC, Eichhoff OM, Widmer DS, Schlegel NC, Schoenewolf NL, Stuart D, Liu W, Gardner H, Smith PD, Nuciforo P, Dummer R and Hoek KS, Pigm. Cell Melanoma Res, 2011, 24, 326–333. [DOI] [PubMed] [Google Scholar]
- 56.Konieczkowski DJ, Johannessen CM, Abudayyeh O, Kim JW, Cooper ZA, Piris A, Frederick DT, Barzily-Rokni M, Straussman R, Haq R, Fisher DE, Mesirov JP, Hahn WC, Flaherty KT, Wargo JA, Tamayo P and Garraway LA, Cancer Discovery, 2014, 4, 816–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hung YH, Chen LM, Yang JY and Yang WY, Nat. Commun, 2013, 4, 2111. [DOI] [PubMed] [Google Scholar]
- 58.Rivera Z, Ferrone S, Wang X, Jube S, Yang H, Pass HI, Kanodia S, Gaudino G and Carbone M, Clin. Cancer Res, 2012, 18, 5352–5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Svendsen A, Verhoeff JJ, Immervoll H, Brogger JC, Kmiecik J, Poli A, Netland IA, Prestegarden L, Planaguma J, Torsvik A, Kjersem AB, Sakariassen PO, Heggdal JI, Van Furth WR, Bjerkvig R, Lund-Johansen M, Enger PO, Felsberg J, Brons NH, Tronstad KJ, Waha A and Chekenya M, Acta Neuropathol, 2011, 122, 495–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J, Svendsen A, Kmiecik J, Immervoll H, Skaftnesmo KO, Planaguma J, Reed RK, Bjerkvig R, Miletic H, Enger PO, Rygh CB and Chekenya M, PLoS One, 2011, 6, e23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS and Kalluri R, Nature, 2015, 527, 525–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Phillips TM, McBride WH and Pajonk F, J. Natl. Cancer Inst, 2006, 98, 1777–1785. [DOI] [PubMed] [Google Scholar]
- 63.Woodward WA, Debeb BG, Xu W and Buchholz TA, Cancer, 2010, 116, 2840–2845. [DOI] [PubMed] [Google Scholar]
- 64.Fillmore CM and Kuperwasser C, Breast Cancer Res, 2008, 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X and Perou CM, Breast Cancer Res, 2010, 12, R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kessel D, Thompson P, Saatio K and Nantwi KD, Photochem. Photobiol, 1987, 45, 787–790. [DOI] [PubMed] [Google Scholar]
- 67.Berg K, Nordstrand S, Selbo PK, Tran DT, Angell-Petersen E and Hogset A, Photochem. Photobiol. Sci, 2011, 10, 1637–1651. [DOI] [PubMed] [Google Scholar]
- 68.Castano AP, Mroz P and Hamblin MR, Nat. Rev. Cancer, 2006, 6, 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Håkerud M, Selbo PK, Waeckerle-Men Y, Contassot E, Dziunycz P, Kundig TM, Høgset A and Johansen P, J. Controlled Release, 2015, 198, 10–17. [DOI] [PubMed] [Google Scholar]
- 70.Håkerud M, Waeckerle-Men Y, Selbo PK, Kundig TM, Høgset A and Johansen P, J. Controlled Release, 2014, 174, 143–150. [DOI] [PubMed] [Google Scholar]
- 71.Haug T, Brede G, Håkerud M, Nedberg AG, Gederaas OA, Flo TH, Edwards V, Selbo PK, Høgset A and Halaas Ø, Front. Immunol, DOI: 10.3389/fimmu.2018.00650, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Huang YY, Vecchio D, Avci P, Yin R, Garcia-Diaz M and Hamblin MR, Biol. Chem, 2013, 394, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.