Abstract
The neurogenic gene brainiac was first isolated in Drosophila melanogaster, where it interacts genetically with members of the Notch signaling cascade. We have isolated a murine homologue of the Drosophila brainiac gene and delineated its highly specific expression pattern during development and adult life. We find particularly strong expression in the developing central nervous system, in the developing retina, and in the adult hippocampus. Targeted deletion of mouse Brainiac 1 expression leads to embryonic lethality prior to implantation. Null embryos can be recovered as blastocysts but do not appear to implant, indicating that mouse Brainiac 1, likely a glycosyltransferase, is crucial for very early development of the mouse embryo.
The Notch signaling pathway has been implicated in cell fate decisions in a variety of developmental contexts in Drosophila melanogaster, Caenorhabditis elegans, and vertebrates (4, 22, 52). Notch-dependent signaling events include the induction of cell fates by cell-cell interaction (a process termed “lateral specification”), as well as the formation and maintenance of epithelial cell layers. Notch encodes a transmembrane receptor that binds to the membrane-bound ligands Delta and Serrate. Upon ligand binding, Notch is cleaved by an unknown mechanism and an intracellular portion of the molecule translocates together with the Suppressor of Hairless [Su(H)] gene product to the nucleus, where the protein complex acts as a transcriptional activator (34, 50). Genetic evidence from Drosophila indicates that the genes of the Enhancer of split [E(spl)] complex, which encode basic helix-loop-helix proteins, are targets of the Notch signaling cascade (6, 13, 31, 37). Homologues of Notch signaling molecules have been isolated in Xenopus laevis, mice, and humans and demonstrate a high degree of conservation of the Notch signaling cascade (23, 35, 45). As in Drosophila, Notch signaling in vertebrates is involved in cell fate specifications in a variety of developmental processes.
The neurogenic gene brainiac has been isolated in Drosophila in screens for female sterile or larval lethal mutations and has been shown to be involved in lateral specification and epithelial morphogenesis (21). brainiac mutant flies produce offspring with a neural hyperplasia and epidermal hypoplasia reminiscent of hypomorphic Notch, Delta, or E(spl) alleles, and genetic evidence indicates that brainiac acts in the same genetic pathway as Notch (21). However, in contrast to Notch loss-of-function mutations, brainiac mutant flies do not exhibit altered cell fate specifications during oogenesis or development of the peripheral nervous system, suggesting that the brainiac gene product is only involved in a subset of Notch signaling events in Drosophila. Loss-of-function brainiac mutations in Drosophila cause follicular epithelial cells surrounding the oocyte to lose their epithelial morphology and to acquire a mesenchyme-like shape (18, 19), a phenotype also associated with hypomorphic Notch alleles (53). The follicular epithelium shows loss of epithelial polarity and aberrant cytoskeletal architecture in brainiac mutant flies, and cells accumulate in multiple layers at the posterior end of the oocyte. brainiac mutant flies frequently have discontinuities in the follicular epithelium, presumably caused by a failure of follicle cells to migrate over the top of or adhere to germ cells during early follicular development. brainiac has been shown to act cell nonautonomously in Drosophila, suggesting that brainiac acts on proteins on their transport through the secretory pathway or that brainiac itself is secreted.
In addition to its relationship with Notch signal transduction, brainiac has also been shown to cooperate with the epidermal growth factor (EGF) receptor pathway during morphogenesis of the follicular epithelium in the Drosophila ovary (19, 21). brainiac mutant flies show aberrant dorsal-ventral patterning during oogenesis and show genetic interaction with gurken (the Drosophila homologue of transforming growth factor α [TGF-α]), and the EGF receptor (19, 21).
Genetic evidence indicates that brainiac shares features with fringe, a gene involved in defining dorsal-ventral boundaries during formation of the Drosophila wing and the vertebrate limb (20, 29, 36, 46). Both fringe and brainiac are involved in regulating a subset of Notch-dependent signaling events and also possess significant sequence homology to glycosyltransferases, suggesting that glycosylation of either Notch or its ligands modulates their interactions (11, 25, 41, 56). Recent evidence shows convincingly that Drosophila and vertebrate Fringe proteins are indeed glycosyltranferases modifying Notch itself and that this glycosylation event modulates Notch-Delta interactions (9, 40).
Evidence from Drosophila shows that brainiac is an important modulator of signaling and/or cell adhesion events during embryonic development of the fly. In this report, we describe the functional characterization of a putative murine homologue of the Drosophila brainiac gene. Mouse Brainiac 1 is expressed in the central nervous system (CNS) during development and adult life. We eliminated Brainiac 1 expression by targeted deletion in embryonic stem (ES) cells and show that Brainiac 1 is essential for preimplantation development of the mouse embryo.
MATERIALS AND METHODS
Isolation of murine brainiac cDNA and genomic clones and construction of a targeting vector.
To isolate a murine homologue of the Drosophila brainiac gene, we screened the EST database for vertebrate homologues using the BLAST algorithm. EST clone yr97d07.r1 (Genome Systems), derived from a human spleen and kidney library, possessed the highest sequence homology (at the protein level) to Drosophila brainiac of any vertebrate sequence found in the database at that time. The full-length cDNA insert of this clone was used to screen a mouse 17.5-day-postcoitus (d.p.c.) embryonic library (Clontech) using standard procedures.
An 836-bp cDNA fragment encoding amino acids 31 to 310 of the murine Brainiac 1 gene was used as a probe to screen a mouse 129/SvEv genomic BAC library (Genome Systems). Three positive clones were isolated, and restriction mapped using Southern blot hybridization and the 836-bp mouse Brainiac 1 cDNA probe. All three BAC clones contained an equivalent mouse Brainiac genomic fragment. One BAC clone was used to subclone EcoRI and HindIII genomic fragments of approximately 10 to 12 kbp into pBluescript (Stratagene). Both genomic subclones were restriction mapped using Southern blot hybridization with oligonucleotides corresponding to various regions of the mouse Brainiac 1 coding sequence.
Targeted disruption of mouse Brainiac in ES cells and generation of Brainiac chimeric mice.
A targeting vector (pmbrn11) was constructed in pOSdupdel (a gift kindly provided by O. Smithies), which contains an MCl-neo cassette (flanked by LoxP sites) for positive selection and an outside PGK-TK cassette for negative selection with FIAU. Adjacent genomic HincII fragments of 6 and 2 kb were ligated into the blunted XhoI site and the HpaI site, respectively, of the vector. This construct inserts the PGK-neo cassette into the Brainiac 1 coding sequence 140 bp 3′ of the start codon, with transcription of neo and Brainiac 1 occurring in the same direction.
TC1 ES cells derived from 129/SvEv mice were electroporated with NotI-linearized pmbrn11 and selected with G418 and FIAU as described previously (14). G418- and FIAU-resistant clones were isolated and propagated in 24-well tissue culture dishes. Genomic DNA was isolated from these clones as described elsewhere (15) and sceened for targeting by digestion with EcoRI and AscI, followed by Southern blot analysis using standard conditions. Blots were hybridized with a 1-kbp BanI-AccI fragment (isolated from a BAC subclone) located 3′ of the genomic region used to construct the targeting vector. After screening 230 G418- and FIAU-resistant colonies, we isolated one ES cell clone showing proper genomic rearrangement at the mouse Brainiac 1 locus. This positive ES cell clone was microinjected into C57BL/6J blastocysts, which were subsequently transferred into pseudopregnant Swiss Webster foster mothers (Taconic). We obtained two male and one female high-grade chimeric mice, as judged by agouti coat color. Chimeras were mated to 129/SvEv or NIH Bl/SW mice (Taconic), and germ line transmission was confirmed by Southern blot analysis. As expected, the two male chimeric mice showed germ line transmission of the introduced Brainiac 1 mutation, while the female chimera did not. Heterozygote offspring from this F1 cross were intercrossed to derive the mouse colony.
PCR genotyping was performed using the following primers: mbrn1koF (5′-GGT GAT ATG GTA CCT CAG CCT CCC CCA CTA C-3′), mbrn1koR (5′-GTG AGG TCA CCA GGA TGA CCA GGA ATG GG-3′), and neoR (5′-AAT GAC AAG ACG CTG GGC GGG GTT TGC TCG-3′). Reactions were performed with Taq polymerase (Boehringer Mannheim) in 50-μl total volume, with all three primers simultaneously present at 200 nM each. Primary denaturation was performed at 94°C for 5 min, followed by 40 cycles of 94°C for 1 min, 65°C for 1 min, and 72°C for 1 min. Reaction products were analyzed on 2.2% agarose gels. The wild-type allele (product mbrn1koF-mbrn1koR) was expected to give a PCR product of 166 bp, while the mutant allele (product mbrn1koF-neoR) was expected to give a PCR product of 280 bp.
Isolation and genotyping of bastocysts.
To isolate blastocysts, female Brainiac 1 heterozygote animals were superovulated with pregnant mare serum (PMS) and human chorionic gonatotrophin (HGG) and mated to Brainiac 1 heterozygote males. Blastocysts were isolated at 3.5 d.p.c. by flushing the uterus with CMRL-1066 medium (Gibco) containing 10% heat-inactivated fetal calf serum. Blastocysts were then cultured in 24-well tissue culture dishes at 37°C in CMRL-1066–10% heat-inactivated fetal calf serum–10x MEM Nonessential Amino Acids (Gibco), 10 mM l-glutamine (PenStrep; Gibco).
For genotyping, blastocysts were transferred to PCR tubes and lysed in 10 ml of 100 mM Tris (pH 8.0)–5 mM EDTA–0.2% sodium dodecyl sulfate (SDS)–200 mM NaCl–220 mg of proteinase K (Boehringer Mannheim) per ml overnight at 50°C. The proteinase K was heat-inactivated at 94°C for 159 min. Then, 2 μl of lysate was subjected to 50 cycles of PCR using the primers and conditions described above. PCR products were analyzed on 2.2% agarose gels. For blastocysts at stage 3.5 d.p.c., two separate reactions were performed using primer pairs mbrn1koF-mbrn1koR and mbrn1koF-neoR, respectively. PCR products were visualized by Southern blot hybridization using the following probes: the 836-bp mouse Brainiac 1 cDNA and a 280-bp mutant PCR product amplified from the targeting vector with mbrn1koR and neoR.
Northern blot analysis.
Total cytoplasmic RNA was isolated from cell lines or tissues using RNA STAT (Tel-Test, Inc.), and mRNA was prepared using magnetic poly(T) beads according to the manufacturer's protocols (Boehringer Mannheim). Next, 2 μg of mRNA was loaded on to a 1% agarose gel containing formaldehyde. After electrophoresis, the mRNA was blotted onto GeneScreen Plus membranes (NEN) and hybridized using standard procedures (5). A murine glyceraldehyde-3-phosphate dehydrogenase (GAPDH) full-length cDNA was used as a loading control.
In situ hybridization.
For in situ hybridizations on tissue sections, tissues or embryos were fixed overnight in 4% formaldehyde. For P10 and adult brains, animals were perfused first with ice-cold phosphate-buffered saline (PBS) and then with 4% formaldehyde prior to fixation. After fixation, tissues were dehydrated in ethanol and mounted in paraffin. Samples were sectioned on a microtome (Reichert-Jung; 8-μm sections) and layered on glass microscope slides. Sections were dewaxed with xylene, treated for 7.5 min with proteinase K (20 mg/ml), and postfixed with 4% formaldehyde. Hybridizations were performed in 50% formamide–20 mM Tris (pH 7.4)–10% dextran sulfate–1× Denhardt's solution–10 mM dithiothreitol (DTT)–0.5 mg of yeast tRNA per ml. 35S-labeled riboprobes were added to the hybridization solution at 5 × 104 cpm/μl, and hybridizations were performed overnight at 52°C. After hybridization, slides were washed first with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)– 10 mM DTT at 50°C and then with 2× SSC–50% formamide–10 mM DTT at 65°C and were finally treated with 20 μg of RNase A per ml for 30 min at 37°C. Slides were then dehydrated with ethanol and finally developed in NTP-2 photo emulsion (Kodak). Times of exposure were estimated from signal intensities on Cornex films after exposure overnight. Slides were developed using Kodak developer and counterstained with either hematoxylin and eosin (ovaries and embryos) or toluidine blue (brain sections) using standard procedures.
Control hybridizations were performed using a sense probe to Brainiac 1. In no case could a signal above background be detected (data not shown).
For whole-mount in situ hybridizations, embryos were dissected from pregnant wild-type animals (FVB; Taconic) at various time points of pregnancy (8.5, 9.5, 10.5, and 12.5 d.p.c.) and fixed overnight in 4% paraformaldehyde at 4°C. After fixation, embryos were washed with PBS–0.1% Tween 20 (PBT) several times at 4°C and then dehydrated in a series of methanol-PBT. After incubation overnight in methanol, the embryos were rehydrated in a series of methanol-PBT, bleached with 6% hydrogen peroxide, treated with 10 μg of proteinase K (Boehringer Mannheim) per ml for 15 min at room temperature, and washed with 2 mg of glycine per ml in PBT for 10 min at room temperature. Embryos were then postfixed with 4% paraformaldehyde and 0.2% glutaraldehyde in PBT for 10 min, washed with PBT, and prehybridized in 50% formamide–5× SSC (pH 4.5)–1% SDS–50 μg of yeast RNA (Boehringer Mannheim) per ml–50 μg of heparin per ml for at least 1 h at 70°C.
Mouse Brainiac 1 riboprobes were digoxigenin (DIG)-labeled using T7 and T3 RNA polymerases (DIG RNA Labeling Kit; Boehringer Mannheim) and purified using ethanol precipitation. Embryos were then hybridized in 50% formamide, 5× SSC (pH 4.5), 1% SDS, 50 μg of yeast RNA (Boehringer Mannheim) per ml, and 50 μg of heparin per ml overnight at 70°C.
After hybridization, embryos were washed in 50% formamide–5× SSC (pH 4.5)–1% SDS and blocked in 10% sheep serum-TBST, and transcript was detected using an anti-DIG antibody (Boehringer Mannheim) and nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) staining.
RESULTS
Isolation of a mouse homologue of the Drosophila brainiac gene.
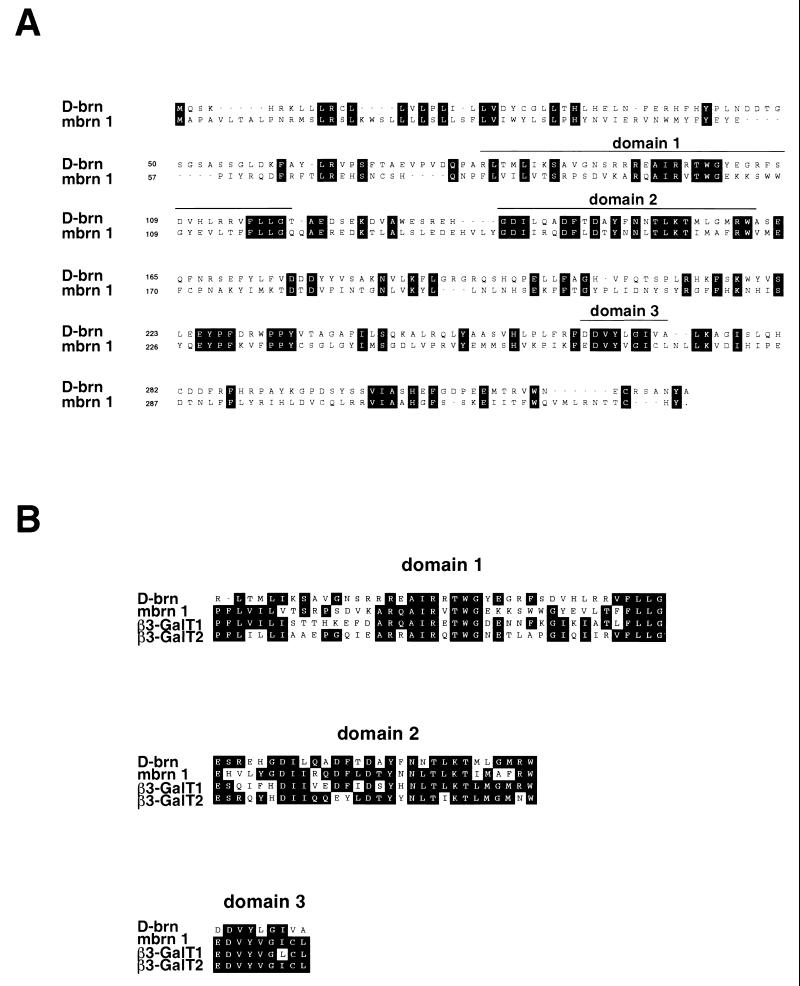
We identified a putative murine homologue of the Drosophila brainiac gene, which we termed mouse Brainiac 1 (Mbrn 1), encoding a protein of 332 amino acids, with a predicted size of 39.4 kDa. The sequence isolated here proved to be identical to the murine β3-GalT-III gene identified recently in an EST database search using human β-galactosyltransferase as the reference sequence (24). Drosophila brainiac and the mouse homologue Brainiac 1 share several regions of high sequence similarity, although the overall sequence identity between the two proteins is only ca. 34% (Fig. 1A). Interestingly, these domains are also highly conserved among different murine β1,3-galactosyltransferase family members but are not found in β1,4- or α1,3-galactosyltransferases (Fig. 1B). Biochemical analysis performed by others showed that mouse Brainiac 1 does indeed specifically catalyze β1,3 links between galactose and N-acetylglucosamine (24). One of the conserved domains (domain 3, Fig. 1B) contains an (E/D)DVXXG motif that has also been found in bacterial galactosyltransferases and may be associated with a catalytic domain (56). These data suggest that Drosophila brainiac and its homologues in higher organisms belong to the same family of β1,3-galactosyltransferases.
FIG. 1.
(A) Alignment of Drosophila brainiac (upper sequence) and mouse Brainiac 1 (lower sequence). Identical amino acids are boxed. Regions showing a high degree of homology between mouse Brainiac 1 and members of the β3-GalT gene family are marked by a line above the sequence. (B) Alignment of domains conserved in murine β3-GalT genes and Drosophila brainiac. The regions showing high sequence similarity include an (E/D)DVXXG motif that has also been found in bacterial galactosyltransferases.
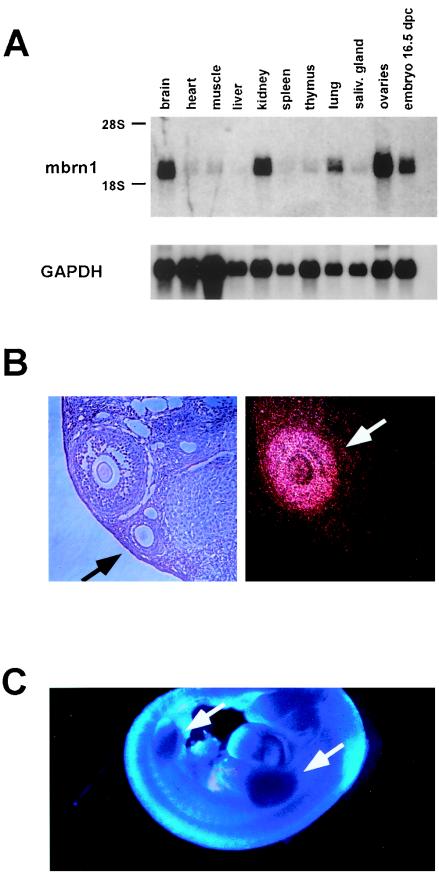
In an expression study of adult mouse tissues by Northern blot analysis, we detected a specific transcript of approximately 2 kb in brain, kidney, lung, and ovaries, as well as in whole 16.5-d.p.c. embryos (Fig. 2A), while all other tissues examined expressed extremely low amounts of the transcript. The size of the transcript is consistent with the cDNA sequence described above. Because of the established role of the Drosophila brainiac gene in oogenesis and neurogenesis, we focused our expression analysis in the mouse on the ovaries and the CNS.
FIG. 2.
Expression of mouse Brainiac 1 in adult mouse tissues and the developing embryo. (A) Northern blot analysis of murine tissues reveals specific expression of a 2-kb transcript. (B) Expression of mouse Brainiac 1 in adult mouse ovaries analyzed by in situ hybridization. The dark-field micrograph shows the expression of mouse Brainiac 1 in follicular granulosa cells in a stage-specific fashion: later-stage follicles with multiple layers of granulosa cells show strong expression of mouse Brainiac 1 (white arrow), while the earlier-stage follicles with single layers of granulosa cells show no expression (black arrow). Control experiments with sense probes were negative (data not shown). (C) Whole-mount in situ hybridization at 12.5 d.p.c. of gestation shows strong expression of mouse Brainiac 1 in the limb buds (arrows).
Mouse Brainiac 1 is expressed in ovarian granulosa cells in a stage-specific manner.
Using in situ hybridization on formalin-fixed sections of murine ovaries, we found mouse Brainiac 1 to be specifically expressed in follicular granulosa cells (Fig. 2B). Interestingly, expression of mouse Brainiac 1 appeared to be highly dependent on the stage of development of the follicle. While follicles containing only a single layer of tightly adhered granulosa cells did not express Brainiac 1, very high expression was exhibited in follicles at later developmental stages in which granulosa cells adhere less tightly to each other (Fig. 2B). Since the Drosophila brainiac gene has been shown to be important in establishing cell adhesion between follicular epithelial cells and germ cells during oocyte development, it is possible that mouse Brainiac 1 has a similar function during murine follicular development and modulates the adhesion properties of granulosa cells.
Expression of mouse Brainiac 1 during development.
To establish the temporal and spatial distribution of mouse Brainiac 1 transcript during embryogenesis and adult life, we performed in situ hybridization on paraffin sections of mouse embryos and tissues.
In 11.5-d.p.c. embryos, we failed to detect a specific signal above background (data not shown). At day 12.5 of gestation, Brainiac 1 transcript could be detected in all four ventricles of the developing brain. At this stage of development, Brainiac 1 transcript was present in the ventricular zone, as well as in the mantle zone, although the signal appeared to be higher in the outer layers of the developing ventricles (Fig. 3A and B). An equivalent expression pattern in the developing CNS was detected at 14.5 d.p.c. (data not shown). This observation suggests that mouse Brainiac 1 is primarily associated with postmitotic cells during the development of the CNS. Thus, mouse Brainiac 1 expression appears to be upregulated when neuroepithelial cells exit the cell cycle and migrate toward the outer layers of the developing brain. This pattern is consistent with expression data from the P19 cell line, which has been used as an in vitro model for neurogenesis (33, 42). P19 cells express Brainiac 1 as undifferentiated precursor cells and at higher levels after retinoic acid-induced differentiation into neurons and glia (data not shown). In addition to this expression pattern in the CNS, we detected strong Brainiac 1 expression in the limb buds of developing embryos at 12.5 d.p.c. (Fig. 2C).
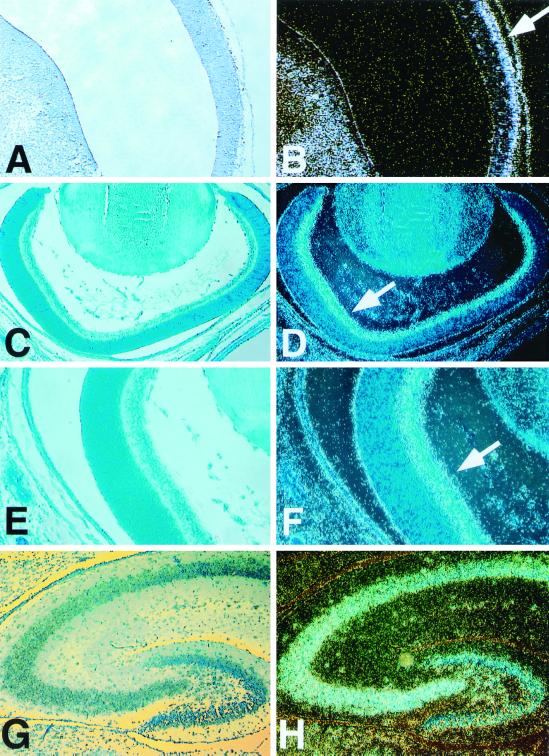
FIG. 3.
Analysis of mouse Brainiac 1 expression in the CNS by in situ hybridization. Throughout the panel, standard phase-contrast micrographs are on the left; dark-field micrographs show the in situ hybridization signal on the right. (A and B) Expression of mouse Brainiac 1 in the ventricles of the developing CNS at day 12.5 of gestation. Expression in the mantle zone appears to be higher than expression in the ventricular zone (B, arrow). (C to F) Mouse Brainiac 1 is expressed in the ganglion cell layer (F, arrow) of the developing retina at postnatal day 1. (G to H) Mouse Brainiac 1 is expressed in the dentate gyrus and the CA regions of the hippocampus at postnatal day 10. An identical expression pattern was observed in the hippocampus at postnatal day 1. Control experiments with sense probes were negative (data not shown).
At postnatal day 1, we detected mouse Brainiac 1 transcript throughout the brain. Particularly high expression of mouse Brainiac 1 was observed in the developing retina (Fig. 3C to F) and the hippocampus (Fig. 3G and H). In the retina, the transcript appeared to be specifically localized to the ganglion cell layer (Fig. 3C to F). Lower amounts of transcript were detected in the outer layers of the retina. At this stage of development, the outer layers of the retina are proliferative, while the ganglion cell layer and the inner plexiform layer consist of postmitotic, differentiated neuronal cells. At postnatal day 1, we also detected particularly strong expression of Brainiac 1 in the hippocampus and the cerebral cortex (data not shown). The expression pattern at this stage is equivalent to that found at postnatal day 10 (see below).
At postnatal day 10, mouse Brainiac 1 transcript was again detected throughout the brain. A particularly strong signal was observed in the hippocampus (Fig. 3G and H), where mouse Brainiac 1 transcript was specifically detected in all CA subfields, as well as the dentate gyrus. Overall, the amount of mouse Brainiac 1 transcript appeared to be lower in the adult brain sections than in the P1 or P10 sections. In adult brain, Brainiac 1 was predominantly expressed in the hippocampus, in a pattern equivalent to that observed in the hippocampus at stage P10 (data not shown).
Targeted disruption of mouse Brainiac 1 leads to embryonic lethality prior to implantation.
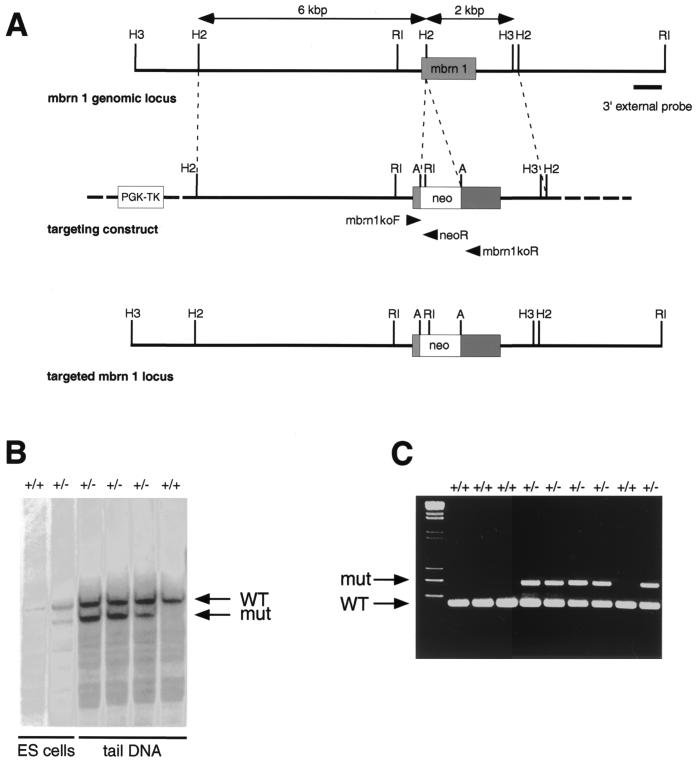
Analysis of the murine Brainiac 1 locus revealed that the entire coding region of the gene is contained in a single exon of 996 bp. We disrupted the murine Brainiac 1 gene by inserting a PGK-neo cassette 140 bp downstream of the start codon using homologous recombination in ES cells (Fig. 4). Since no splicing is anticipated in the stretch of RNA derived from this exon, this insertion of the targeting cassette is expected to abolish expression of the entire carboxy-terminal portion of the protein, leaving only 47 amino acids under the control of the endogenous promoter. This lesion is expected to result in a null allele. ES cells transfected with the targeting vector were selected with G418 for the presence of the neo cassette and with FIAU for the loss of the thymidine kinase gene present in the vector backbone. An ES clone with targeted disruption of the mouse Brainiac 1 gene, as judged by Southern blot analysis, was microinjected into blastocysts, and chimeric mice were generated after transfer into pseudopregnant Swiss Webster foster mothers. Offspring from the F1 generation obtained from matings of male chimeras to NIH BL/SW outbred mice were used to generate the mouse colony.
FIG. 4.
(A) Genomic organization of the mouse Brainiac 1 gene and strategy for targeted deletion of mouse Brainiac 1. The coding region of mouse Brainiac 1 is contained in a single exon. A PGK-neo cassette flanked by LoxP sites was inserted 140 bp downstream from the start codon. Arrows indicate the location of the primers used for PCR genotyping. Thymidine kinase (PGK-TK) was used for negative selection by FIAU. Restriction sites: A, AscI; H2, HincII; H3, HindIII; RI, EcoRI. (B) Southern blot of ES cell DNA digested with EcoRI and AscI. The 3′ external probe was used to identify the targeted allele. (C) PCR assay used to genotype embryos and adult mice.
Mouse Brainiac 1 heterozygote animals are fully viable and fertile and do not appear to have any gross abnormalities. The oldest animals in our colony have reached the age of 1 year now without our being able to detect any signs of illness.
After analyzing over 300 adult mice obtained from F1 or later-generation heterozygote intercrosses, we failed to obtain a single live null animal, indicating that Brainiac 1 expression is essential for development and survival (Table 1). In addition, we never detected perinatal lethality in litters from heterozygote intercrosses, indicating that targeted disruption of the mouse Brainiac 1 gene leads to embryonic lethality.
TABLE 1.
Genotype results from mouse Brainiac 1 heterozygote intercrosses
| Stage | No. of mice with genotype:
|
No. of resorptionsa | ||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| Adult | 75 | 201 | 0 | NA |
| 9.5–11.5 d.p.c | 14 | 67 | 0 | 0 |
| 3.5 d.p.c. | 11 | 18 | 5 | NA |
NA, not applicable.
Analysis of embryos at between 9.5 and 12.5 d.p.c also failed to detect Brainiac 1 null embryos, while heterozygote and wild-type animals were detected using Southern blot hybridization and PCR genotyping (Table 1). In addition, no resorbed embryos were detected, suggesting that death of Brainiac 1 null embryos occurs very early in development, potentially prior to implantation. We therefore focused our attention on the earliest stage accessible to analysis by isolating blastocysts at 3.5 d.p.c. Brainiac 1 heterozygote females were superovulated using PMS and HCG and mated to Brainiac 1 heterozyote males. Blastocysts were isolated at 3.5 d.p.c by flushing the uterus. Blastocysts were either genotyped directly or cultivated in vitro for up to 5 days prior to genotyping.
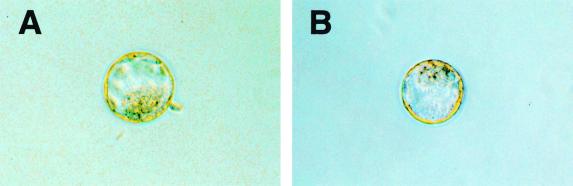
At 3.5 d.p.c., blastocysts of all three genotypes could be detected at the expected frequency (Table 1 and Fig. 5). Brainiac 1 null blastocysts had an easily detectable inner cell mass and cavity and were indistinguishable from blastocysts of wild-type or heterozygote littermates (Fig. 5). The cells of the inner cell mass were clearly adhered in Brainiac 1 null embryos, suggesting intact cell-cell adhesion at this stage of development. In addition, most Brainiac 1 null embryos identified (four of five embryos) at this stage of development had hatched from the zona pellucida, indicating that Brainiac 1 null embryos are viable at 3.5 d.p.c.
FIG. 5.
Targeted deletion of mouse Brainiac 1 results in preimplantation lethality. Blastocysts from Brainiac 1 heterozygote intercrosses isolated at 3.5 d.p.c. appear normal, with a clearly developed inner cell mass and an inner cavity. (A) Blastocyst genotyped as wild type. (B) Blastocyst genotyped as Brainiac 1 null.
We failed to detect any Brainiac null embryos after cultivating blastocysts for 48 h in vitro. At this time point, most embryos have attached to the tissue culture dish, and the trophoblast layer begins to be develop. Embryos heterozygous for the Brainiac 1 mutation or wild-type embryos could be detected in addition to a significant number of embryos that could not be genotyped at all. In most cases where we failed to obtain a clear genotyping result, the embryo had deteriorated significantly and had not attached to the tissue culture dish. This data suggests that Brainiac 1 null embryos die in vivo by between 3.5 and 4.5 d.p.c.
DISCUSSION
In this study, we report the functional characterization of a putative murine homologue of the Drosophila brainiac gene. The mouse gene described here encodes a protein with significant overall amino acid sequence similarity to the Drosophila brainiac protein. Interestingly, regions that show the highest protein sequence conservation between the fly and mouse proteins are also highly conserved in β1,3-galactosyltransferases but not in β1,4- or α1,3-galactosyltransferases, suggesting that brainiac in Drosophila and Brainiac-like molecules in higher organisms belong to a family of β1,3-galactosyltransferase enzymes (12). This suggestion is further supported by the identity of the gene isolated in our screen (for vertebrate Brainiac homologues) with a gene isolated in a database screen (for galactosyltransferases) which encodes in vitro β1,3-galactosyltransferase activity (24). Biochemical analysis revealed that mouse Brainiac 1 does indeed have glycosyltransferase activity in vitro and catalyzes the formation of galactose β1,3-N-actetylglucosamine structures (24). No β1,4 linkage was detected in these in vitro assays, suggesting that the enzymatic activity is highly specific for β1,3 links. A homologous gene has also been isolated in humans but, surprisingly, does not show any glycosyltransferase activity in vitro with the substrates tested (2). Since brainiac acts cell nonautonomously in Drosophila, the putative glycosyltransferase activity suggests that Brainiac might regulate signaling events and/or cell adhesion processes through specific glycosylation of cell-surface proteins.
Protein glycosylation has been shown to be an important modulator of cell surface events in a variety of contexts, and mice carrying targeted deletions in several genes for glycosyltransferases have been described (16). While the loss of some glycosyltransferases appears to lead to embryonic or perinatal lethality, other enzymes of this family are not essential for development and survival (10, 27, 39, 44). Malignant transformation and metastatic potential have also been associated with alterations in protein glycosylation. Ectopic expression of a β1,4-N-acetylglucosaminyltransferase in a B16 mouse melanoma cell line suppresses the metastatic potential of these cells in syngeneic mice (55); This malignant property has been shown to depend on aberrant E-cadherin glycosylation (54), indicating a link between glycosylation events and cell adhesion. In addition, ectopic expression of a β1,4-N-acetylglucosaminyltransferase in PC12 cells has been shown to disrupt NGF/Trk signaling by inhibiting receptor dimerization but not receptor phosphorylation, causing inhibition of neuronal differentiation in these cells. This observation indicates that specific glycosylation events of receptors can modulate signaling events (26). Interestingly, ectopic expression of mouse Brainiac 1 does not interfere with NGF-induced differentiation in PC12 cells, suggesting that glycosyltransferases have very specific targets and modify specific biological events (B. Vollrath and K. Fitzgerald, unpublished observations).
As demonstrated in this report, mouse Brainiac 1 shows strong expression in the developing CNS and retina, two tissues where Notch signaling has been shown to regulate cell fate specifications during development. Although we find Brainiac 1 expression in undifferentiated neuroepithelial cells, Brainiac 1 expression appears to be upregulated in regions containing postmitotic, differentiated cells such as the ganglion cell layer in the retina or the outer layers of the developing ventricles. Notch receptors and its ligands are generally thought to be primarily expressed in undifferentiated neuroepithelial cells in the developing CNS and retina (7, 38). However, several genes involved in Notch signal transduction, such as Manic and Radical fringe are expressed in a pattern very similar to Brainiac 1 in the developing CNS (11). In addition, there is some evidence that Notch 1 is expressed in postmitotic neuronal cell populations in the ganglion cell layer of the developing retina and in the CNS and Notch 1 has been shown to regulate neurite outgrowth in differentiated, postmitotic neurons in vitro (1, 8, 47). In addition to the expression during retinal and CNS development, mouse Brainiac 1 shows strong expression in the limb buds during embryogenesis; in this tissue Notch receptors and their ligands are known to be expressed and function in pattern formation (48, 49).
Mouse Brainiac 1 also shows strong expression in the follicular granulosa cells of the ovary in a stage-dependent fashion. Granulosa cells that adhere tightly in an organized monolayer of epithelial cells show undetectable expression of mouse Brainiac 1, while cells that have lost their cell-cell adherence form less-organized multilayers in later-stage follicles and have a high level of Brainiac 1 expression. Since Drosophila brainiac has been implicated in regulating cell-cell adhesion during oogenesis in the fly, it can be hypothesized that mouse Brainiac 1-dependent glycosylation regulates the adhesive properties of follicular granulosa cells in the mouse.
Our results suggest that Brainiac-dependent glycosylation events are essential for murine development. Loss of Brainiac-dependent glycosylation through targeted deletion of the gene leads either to implantation failure or to embryonic death prior to implantation. This observation suggests that glycosylation events by members of the Brainiac protein family are highly specific, since other members of this protein family are not able to compensate for the loss of Brainiac 1 expression. The phenotype of Brainiac 1 null mice is also strikingly different from phenotypes of mice with targeted deletions of Notch or other components of the Notch signaling cascade. These animals die at midgestation or later during development (30, 32, 51). However, mice with targeted deletions of all murine Notch genes have not been derived. The possibility that Brainiac-dependent glycosylation is important for the function of all Notch receptors, thus leading to the more severe phenotype in Brainiac 1 null mice, cannot be ruled out. It is also possible that Brainiac-dependent glycoslation is important for the function of several different receptor types, thus leading to this very severe phenotype of Brainiac 1 null mice. Since brainiac shows genetic interaction with the EGF receptor and the TGF-α homologue gurken in Drosophila, this pathway is an obvious candidate to be regulated by brainiac or Brainiac-like molecules in Drosophila and higher organisms.
Genetic evidence in Drosophila indicates that brainiac shares features with fringe, an essential gene involved in pattern formation in the fly eye, wing, and leg. Both genes encode molecules which appear to be involved in regulating a subset of Notch signaling activities (20, 28). Fringe has been shown to modify the interaction of Notch, with its ligands Delta and Serrate, by potentiating the activating effects of Delta but repressing those of Serrate and thereby giving specificity to the signal (17, 43). Sequence similarities between brainiac and fringe proteins, which are very weak, can be detected with sensitive motif and profile searches but not with standard alignment algorithms. Alignment of Drosophila brainiac and fringe proteins suggests the existence of a structural motif which is characteristic for procaryotic and eucaryotic glycosyltransferases (41, 56). Since Notch and its ligands Delta, Serrate, and Jagged are all glycoproteins, specific glycosylation events mediated by Brainiac and Fringe could conceivably modulate Notch-ligand interactions and thereby regulate signaling events. Recent evidence in Drosophila and mammalian systems shows that this is true for Fringe: Fringe-dependent glycosylation of Notch modifies Notch signaling and modulates receptor-ligand interactions (9, 25, 40). It remains to be established whether Brainiac can act in similar ways to modify signaling in this pathway. Although Fringe and Brainiac appear to catalyze similar β1,3 glycosylation events, the available in vitro data indicate that their substrate specificity is distinct suggesting distinct biological functions of these two families of proteins (3, 24, 40).
ACKNOWLEDGMENTS
We thank Oliver Smithies for the targeting vector pOSdupdel. In addition, we thank Ann Harrington for performing the blastocyst injections and for expert technical assistance with all mouse procedures, Monteserrat Michelmann for help with targeting ES cells and tissue culture procedures, Jan Pinkas and Nick Chester for helpful comments on the manuscript, and Katy Mclntyre for editing. We also thank all members of the Leder lab and the Harvard Medical School Department of Genetics for helpful comments and suggestions throughout the project.
REFERENCES
- 1.Ahmad I, Zaqouras P, Artavanis-Tsakonas S. Involvement of Notch-1 in mammalian retinal neurogenesis: association of Notch-1 activity with both immature and terminally differentiated cells. Mech Dev. 1995;53:73–85. doi: 10.1016/0925-4773(95)00425-4. [DOI] [PubMed] [Google Scholar]
- 2.Amado M, Almeida R, Carneiro F, Levery S B, Holmes E H, Nomoto M, Hollingsworth M A, Hassan H, Schwientek T, Nielsen P A, Bennett E P, Clausen H. A family of human beta3-galactosyltransferases. Characterization of four members of a UDP-galactose:beta-N-acetyl-glucosamine/beta-nacetyl-galactosamine beta-1,3-galactosyltransferase family. J Biol Chem. 1998;273:12770–12778. doi: 10.1074/jbc.273.21.12770. [DOI] [PubMed] [Google Scholar]
- 3.Amado M, Almeida R, Schwientek T, Clausen H. Identification and characterization of large galactosyltransferase gene families: galactosyltransferases for all functions. Biochim Biophys Acta. 1999;1473:35–53. doi: 10.1016/s0304-4165(99)00168-3. [DOI] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas S, Matsuno K, Fortini M E. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1997. [Google Scholar]
- 6.Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 7.Bao Z Z, Cepko C L. The expression and function of Notch pathway genes in the developing rat eye. J Neurosci. 1997;17:1425–1434. doi: 10.1523/JNEUROSCI.17-04-01425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berezovska O, McLean P, Knowles R, Frosh M, Lu F M, Lux S E, Hyman B T. Notch1 inhibits neurite outgrowth in postmitotic primary neurons. Neuroscience. 1999;93:433–439. doi: 10.1016/s0306-4522(99)00157-8. [DOI] [PubMed] [Google Scholar]
- 9.Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi: 10.1038/35019075. [DOI] [PubMed] [Google Scholar]
- 10.Campbell R M, Metzler M, Granovsky M, Dennis J W, Marth J D. Complex asparagine-linked oligosaccharides in Mgat1-null embryos. Glycobiology. 1995;5:535–543. doi: 10.1093/glycob/5.5.535. [DOI] [PubMed] [Google Scholar]
- 11.Cohen B, Bashirullah A, Dagnino L, Campbell C, Fisher W W, Leow C C, Whiting E, Ryan D, Zinyk D, Boulianne G, Hui C C, Gallie B, Phillips R A, Lipshitz H D, Egan S E. Fringe boundaries coincide with Notch-dependent patterning centres in mammals and alter Notch-dependent development in Drosophila. Nat Genet. 1997;16:283–288. doi: 10.1038/ng0797-283. [DOI] [PubMed] [Google Scholar]
- 12.Cole S E, Mao M S, Johnston S H, Vogt T F. Identification, expression analysis, and mapping of B3galt6, a putative galactosyltransferase gene with similarity to Drosophila brainiac. Mamm Genome. 2001;12:177–179. doi: 10.1007/s003350010241. [DOI] [PubMed] [Google Scholar]
- 13.de Celis J F, de Celis J, Ligoxygakis P, Preiss A, Delidakis C, Bray S. Functional relationships between Notch, Su(H) and the bHLH genes of the E(spl) complex: the E(spl) genes mediate only a subset of Notch activities during imaginal development. Development. 1996;122:2719–2728. doi: 10.1242/dev.122.9.2719. [DOI] [PubMed] [Google Scholar]
- 14.Deng C, Wynshaw-Boris A, Zhou F, Kuo A, Leder P. Fibroblast growth factor receptor 3 is a negative regulator of bone growth. Cell. 1996;84:911–921. doi: 10.1016/s0092-8674(00)81069-7. [DOI] [PubMed] [Google Scholar]
- 15.Deng C X, Wynshaw-Boris A, Shen M M, Daugherty C, Ornitz D M, Leder P. Murine FGFR-1 is required for early postimplantation growth and axial organization. Genes Dev. 1994;8:3045–3057. doi: 10.1101/gad.8.24.3045. [DOI] [PubMed] [Google Scholar]
- 16.Dennis J W, Granovsky M, Warren C E. Protein glycosylation in development and disease. Bioessays. 1999;21:412–421. doi: 10.1002/(SICI)1521-1878(199905)21:5<412::AID-BIES8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 17.Fleming R J, Gu Y, Hukriede N A. Serrate-mediated activation of Notch is specifically blocked by the product of the gene fringe in the dorsal compartment of the Drosophila wing imaginal disc. Development. 1997;124:2973–2981. doi: 10.1242/dev.124.15.2973. [DOI] [PubMed] [Google Scholar]
- 18.Goode S, Meinick M, Chou T B, Perrimon N. The neurogenic genes egghead and brainiac define a novel signaling pathway essential for epithelial morphogenesis during Drosophila oogenesis. Development. 1996;122:3863–3879. doi: 10.1242/dev.122.12.3863. [DOI] [PubMed] [Google Scholar]
- 19.Goode S, Morgan M, Liang Y P, Mahowald A P. Brainiac encodes a novel, putative secreted protein that cooperates with Grk TGF alpha in the genesis of the follicular epithelium. Dev Biol. 1996;178:35–50. doi: 10.1006/dbio.1996.0196. [DOI] [PubMed] [Google Scholar]
- 20.Goode S, Perrimon N. Brainiac and fringe are similar pioneer proteins that impart specificity to notch signaling during Drosophila development. Cold Spring Harbor Symp Quant Biol. 1997;62:177–184. [PubMed] [Google Scholar]
- 21.Goode S, Wright D, Mahowald A P. The neurogenic locus brainiac cooperates with the Drosophila EGF receptor to establish the ovarian follicle and to determine its dorsal-ventral polarity. Development. 1992;116:177–192. doi: 10.1242/dev.116.1.177. [DOI] [PubMed] [Google Scholar]
- 22.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 23.Gridley T. Notch signaling in vertebrate development and disease. Mol Cell Neurosci. 1997;9:103–108. doi: 10.1006/mcne.1997.0610. [DOI] [PubMed] [Google Scholar]
- 24.Hennet T, Dinter A, Kuhnert P, Mattu T S, Rudd P M, Berger E G. Genomic cloning and expression of three murine UDP-galactose: beta-N-acetylglucosamine beta-1,3-galactosyltransferase genes. J Biol Chem. 1998;273:58–65. doi: 10.1074/jbc.273.1.58. [DOI] [PubMed] [Google Scholar]
- 25.Hicks C, Johnston S H, diSibio G, Collazo A, Vogt T F, Weinmaster G. Fringe differentially modulates Jagged1 and Delta1 signalling through Notch1 and Notch2. Nat Cell Biol. 2000;2:515–520. doi: 10.1038/35019553. [DOI] [PubMed] [Google Scholar]
- 26.Ihara Y, Sakamoto Y, Mihara M, Shimizu K, Taniguchi N. Overexpression of N-acetylglucosaminyltransferase III disrupts the tyrosine phosphorylation of Trk with resultant signaling dysfunction in PC12 cells treated with nerve growth factor. J Biol Chem. 1997;272:9629–9634. doi: 10.1074/jbc.272.15.9629. [DOI] [PubMed] [Google Scholar]
- 27.Ioffe E, Stanley P. Mice lacking N-acetylglucosaminyltransferase I activity die at mid-gestation, revealing an essential role for complex or hybrid N-linked carbohydrates. Proc Natl Acad Sci USA. 1994;91:728–732. doi: 10.1073/pnas.91.2.728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irvine K D. Fringe, Notch, and making developmental boundaries. Curr Opin Genet Dev. 1999;9:434–441. doi: 10.1016/S0959-437X(99)80066-5. [DOI] [PubMed] [Google Scholar]
- 29.Irvine K D, Wieschaus E. fringe, a Boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- 30.Ishibashi M, Ang S L, Shiota K, Nakanishi S, Kageyama R, Guillemot F. Targeted disruption of mammalian hairy and Enhancer of split homolog-1 (HES-1) leads to up-regulation of neural helix-loop-helix factors, premature neurogenesis, and severe neural tube defects. Genes Dev. 1995;9:3136–3148. doi: 10.1101/gad.9.24.3136. [DOI] [PubMed] [Google Scholar]
- 31.Jennings B, Preiss A, Delidakis C, Bray S. The Notch signalling pathway is required for Enhancer of split bHLH protein expression during neurogenesis in the Drosophila embryo. Development. 1994;120:3537–3548. doi: 10.1242/dev.120.12.3537. [DOI] [PubMed] [Google Scholar]
- 32.Jiang R, Lan Y, Chapman H D, Shawber C, Norton C R, Serreze D V, Weinmaster G, Gridley T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev. 1998;12:1046–1057. doi: 10.1101/gad.12.7.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnson J E, Zimmerman K, Saito T, Anderson D J. Induction and repression of mammalian achaete-scute homologue (MASH) gene expression during neuronal differentiation of P19 embryonal carcinoma cells. Development. 1992;114:75–87. doi: 10.1242/dev.114.1.75. [DOI] [PubMed] [Google Scholar]
- 34.Kidd S, Lieber T, Young M W. Ligand-induced cleavage and regulation of nuclear entry of Notch in Drosophila melanogaster embryos. Genes Dev. 1998;12:3728–3740. doi: 10.1101/gad.12.23.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lardelli M, Williams R, Lendahl U. Notch-related genes in animal development. Int J Dev Biol. 1995;39:769–780. [PubMed] [Google Scholar]
- 36.Laufer E, Dahn R, Orozco O E, Yeo C Y, Pisenti J, Henrique D, Abbott U K, Fallon J F, Tabin C. Expression of Radical fringe in limb-bud ectoderm regulates apical ectodermal ridge formation. Nature. 1997;386:366–373. doi: 10.1038/386366a0. [DOI] [PubMed] [Google Scholar]
- 37.Lecourtois M, Schweisguth F. The neurogenic suppressor of hairless DNA-binding protein mediates the transcriptional activation of the enhancer of split complex genes triggered by Notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 38.Lindsell C E, Boulter J, diSibio G, Gossler A, Weinmaster G. Expression patterns of Jagged, Delta1, Notch1, Notch2, and Notch3 genes identify ligand-receptor pairs that may function in neural development. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 39.Metzler M, Gertz A, Sarkar M, Schachter H, Schrader J W, Marth J D. Complex asparagine-linked oligosaccharides are required for morphogenic events during post-implantation development. EMBO J. 1994;13:2056–2065. doi: 10.1002/j.1460-2075.1994.tb06480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moloney D J, Panin V M, Johnston S H, Chen J, Shao L, Wilson R, Wang Y, Stanley P, Irvine K D, Haltiwanger R S, Vogt T F. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- 41.Munro S, Freeman M. The notch signalling regulator fringe acts in the Golgi apparatus and requires the glycosyltransferase signature motif DXD. Curr Biol. 2000;10:813–820. doi: 10.1016/s0960-9822(00)00578-9. [DOI] [PubMed] [Google Scholar]
- 42.Nye J S, Kopan R, Axel R. An activated Notch suppresses neurogenesis and myogenesis but not gliogenesis in mammalian cells. Development. 1994;120:2421–2430. doi: 10.1242/dev.120.9.2421. [DOI] [PubMed] [Google Scholar]
- 43.Panin V M, Papayannopoulos V, Wilson R, Irvine K D. Fringe modulates Notch-ligand interactions. Nature. 1997;387:908–912. doi: 10.1038/43191. [DOI] [PubMed] [Google Scholar]
- 44.Priatel J J, Sarkar M, Schachter H, Marth J D. Isolation, characterization and inactivation of the mouse Mgat3 gene: the bisecting N-acetylglucosamine in asparagine-linked oligosaccharides appears dispensable for viability and reproduction. Glycobiology. 1997;7:45–56. doi: 10.1093/glycob/7.1.45. [DOI] [PubMed] [Google Scholar]
- 45.Robey E. Notch in vertebrates. Curr Opin Genet Dev. 1997;7:551–557. doi: 10.1016/s0959-437x(97)80085-8. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Esteban C, Schwabe J W, De La Pena J, Foys B, Eshelman B, Belmonte J C. Radical fringe positions the apical ectodermal ridge at the dorsoventral boundary of the vertebrate limb. Nature. 1997;386:360–366. doi: 10.1038/386360a0. [DOI] [PubMed] [Google Scholar]
- 47.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 48.Shawber C, Boulter J, Lindsell C E, Weinmaster G. Jagged2: a serrate-like gene expressed during rat embryogenesis. Dev Biol. 1996;180:370–376. doi: 10.1006/dbio.1996.0310. [DOI] [PubMed] [Google Scholar]
- 49.Sidow A, Bulotsky M S, Kerrebrock A W, Bronson R T, Daly M J, Reeve M P, Hawkins T L, Birren B W, Jaenisch R, Lander E S. Serrate2 is disrupted in the mouse limb-development mutant syndactylism. Nature. 1997;389:722–725. doi: 10.1038/39587. [DOI] [PubMed] [Google Scholar]
- 50.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 51.Swiatek P J, Lindsell C E, del Amo F F, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707–719. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 52.Weinmaster G. The ins and outs of notch signaling. Mol Cell Neurosci. 1997;9:91–102. doi: 10.1006/mcne.1997.0612. [DOI] [PubMed] [Google Scholar]
- 53.Xu T, Caron L A, Fehon R G, Artavanis-Tsakonas S. The involvement of the Notch locus in Drosophila oogenesis. Development. 1992;115:913–922. doi: 10.1242/dev.115.4.913. [DOI] [PubMed] [Google Scholar]
- 54.Yoshimura M, Ihara Y, Matsuzawa Y, Taniguchi N. Aberrant glycosylation of E-cadherin enhances cell-cell binding to suppress metastasis. J Biol Chem. 1996;271:13811–13815. doi: 10.1074/jbc.271.23.13811. [DOI] [PubMed] [Google Scholar]
- 55.Yoshimura M, Nishikawa A, Ihara Y, Taniguchi S, Taniguchi N. Suppression of lung metastasis of B16 mouse melanoma by N-acetylglucosaminyltransferase III gene transfection. Proc Natl Acad Sci USA. 1995;92:8754–8758. doi: 10.1073/pnas.92.19.8754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Y P, Schultz J, Mlodzik M, Bork P. Secreted fringe-like signaling molecules may be glycosyltransferases. Cell. 1997;88:9–11. doi: 10.1016/s0092-8674(00)81852-8. [DOI] [PubMed] [Google Scholar]