Abstract
Major histocompatibility complex class II (MHCII) molecules play a pivotal role in the immune system because they direct the development and activation of CD4+ T cells. There are three human MHCII isotypes, HLA-DR, HLA-DQ, and HLA-DP. Key transcription factors controlling MHCII genes have been identified by virtue of the fact that they are mutated in a hereditary immunodeficiency resulting from a lack of MHCII expression. RFXAP—one of the factors affected in this disease—is a subunit of RFX, a DNA-binding complex that recognizes the X box present in all MHCII promoters. To facilitate identification of conserved regions in RFXAP, we isolated the mouse gene. We then delimited conserved domains required to restore endogenous MHCII expression in cell lines lacking a functional RFXAP gene. Surprisingly, we found that 80% of RFXAP is dispensable for the reactivation of DR expression. Only a short C-terminal segment of the protein is essential for this isotype. In contrast, optimal expression of DQ and DP requires a larger C-terminal segment. These results define an RFXAP domain with an MHCII isotype-specific function. Expression of the three MHCII isotypes exhibits a differential requirement for this domain. We show that this is due to a differential dependence on this domain for promoter occupation and recruitment of the coactivator CIITA in vivo.
Major histocompatibility complex class II (MHCII) molecules are heterodimeric (α-chain–β-chain) transmembrane glycoproteins displayed at the surfaces of specialized cells of the immune system. In humans there are three MHCII isotypes—HLA-DR, HLA-DQ, and HLA-DP—encoded by distinct pairs of α- and β-chain genes (40). MHCII molecules present peptides to the T-cell antigen receptor of CD4+ T cells. The recognition of MHCII-peptide complexes by the T-cell antigen receptor of CD4+ T cells represents an interaction that is of pivotal importance for the adaptive immune system because it controls the development, activation, proliferation, and life span of these cells (6, 44). Given these key functions, it is not surprising that MHCII expression is tightly controlled and restricted to specialized cells of the immune system. In general, expression of the three MHCII isotypes is coordinated.
Expression of MHCII genes is controlled primarily at the level of their transcription by a well-defined regulatory module that is conserved in the promoter-proximal regions of all MHCII genes (reviewed in references 3, 20, 31, and 39). This MHCII regulatory module consists of four cis-acting sequences referred to as the S, X, X2, and Y boxes. The molecular mechanisms that control the MHCII regulatory module have been elucidated to a large extent by the study of a severe hereditary immunodeficiency disease called MHCII deficiency or the bare lymphocyte syndrome (22, 31, 45). As its name implies, this disease is due to the absence of MHCII expression. The genetic defects that are responsible for the disease lie in genes encoding trans-acting regulatory factors that are essential and highly specific for the transcription of MHCII genes (22, 31, 45). Four different transcription factors–CIITA, RFXANK, RFX5, and RFXAP—have been identified and isolated by virtue of the fact that they are defective in four genetically distinct groups (complementation groups A, B, C, and D) of MHCII deficiency patients (9, 21, 27, 37, 38). Three of these factors, RFXANK, RFX5, and RFXAP, assemble into a trimeric DNA-binding complex that binds specifically to the conserved X box sequences of MHCII promoters (8, 9, 21, 26, 28, 32, 37, 43). All three subunits are required for binding of the complex. This RFX complex binds to MHCII promoters cooperatively with the X2 box binding factor X2BP (CREB) and the Y box binding factor NF-Y to generate a stable higher-order nucleoprotein complex referred to as the MHCII enhanceosome (4, 19, 23, 25, 30, 33). The fourth factor, CIITA, functions as a non-DNA-binding transcriptional coactivator that is recruited to the MHCII enhanceosome via protein-protein interactions (8, 13, 23, 49).
The RFXAP gene was identified because it is mutated in MHCII deficiency patients belonging to complementation group D (9, 10, 22, 42). RFXAP is also mutated in an in vitro-generated mutant cell line (6.1.6) characterized by a defect in MHCII expression (9, 11). There is thus no doubt that RFXAP is essential for the transcription of MHCII genes. However, very little is known concerning its mode of action. The primary sequence of RFXAP provides essentially no clues as to how it functions because it contains few, if any, well-defined functional motifs. We therefore performed a detailed structure-function analysis of RFXAP. To identify conserved regions within RFXAP, we first isolated the mouse gene. Minimal essential domains were then mapped by studying their ability to reactivate expression of the endogenous MHCII genes in cell lines having a defect in the RFXAP gene. Surprisingly, we found that over 80% of RFXAP is completely dispensable for the activation of HLA-DR expression. For expression of this isotype, only a short 43-amino-acid C-terminal segment of the protein is required. In contrast, optimal expression of HLA-DQ and HLA-DP requires a larger 130-amino-acid C-terminal segment. This segment defines an 87-amino-acid RFXAP domain having an MHCII isotype-specific function. We demonstrate that the isotype-specific dependence on this domain of RFXAP is due to the facts that it is required in vivo for promoter occupation by RFX and recruitment of CIITA at certain MHCII genes (DQB and DPB) but that it is dispensable for these functions at other genes (DRA).
MATERIALS AND METHODS
Isolation of mouse cDNA clones.
Full-length mouse RFXAP cDNA clones were isolated from a BALB/c mouse spleen cDNA library (34) by screening with a probe spanning nucleotides 516 to 928 of the human RFXAP cDNA (9). Nine independent clones were sequenced on both strands. The sequences were confirmed by the isolation of RFXAP clones from a 129Sv genomic DNA library and sequencing of all exons.
RFXAP expression vectors.
The mouse cDNA clones M-ACG and M-ATG were generated by PCR with the following primers: 5UT1, 5′-GTACGGTTGTGTTTCTCAAG-3′; 5UT2, 5′-GGTGACGGTGCTGGTGATG-3′; and 3UT, 5′-CAGTGTGAGGCTTACGGAG-3′. The PCR fragments were cloned into pBluescript, sequenced, and transferred into the EBO76PL expression vector (38). Human RFXAPs with mutations and deletions were made by PCR using the wild-type cDNA as the template. N-terminal deletions (N1 to N9) were amplified using a downstream primer (5′-GTATAGTCGACAGATGTTCTTGGTAAGTTC-3′ [SalI site is italicized]) and the following upstream primers (NdeI sites at the ATG initiation codons are italicized): N1, 5′-GAATCAACCATATGCAACCCTGTGCTG-3′; N2, 5′-GAAGATTTCATATGAGGTACCTGTGCGAAG-3′; N3, 5′-GAAGAAATCATATGGACGAGGAGACTCAC-3′; N4, 5′-GAAGATTACATATGAGCAAGACCTGCAC-3′; N5, 5′-GAAGATTTCATATGTGCAAGAAACACCGCA-3′; N6, 5′-GAAGACAACATATGAACTGCGGTGGGAC-3′; N7, 5′-GAAGACAACATATGGGAAACGTCAAACTCGAG-3′; N8, 5′-GAAGACAACATATGACAGGATCTTTTGGGGATC-3′; and N9, 5′-GAAGACAACATATGCCTACTCTTTTAGAACAAG-3′.
Constructs with C-terminal deletions (C1 to C4) were amplified using an upstream primer (5′-GCCTGGATCCTCGAGAATTCATATGGAGGCGCAGGGTGTAG-3′ [NdeI site at the ATG initiation codon is italicized]) and the following downstream primers (SalI sites are italicized): C1, 5′-GACTTGTCGACCTACTGCTGTTGTCTTTGCTC-3′; C2, 5′-GACTTGTCGACCTACACTACTTCTGGACTTC-3′; C3, 5′-GACTTGTCGACCTACAGTCTTTTTTGATTTAACAC-3′; and C4, 5′-CTCTTGTCGACTTACTCGAGTTTGACGTTTCC-3′.
Constructs with point mutations (M1 and M2) were made using the following primers to introduce substitutions (italicized) into the upper (M1C and M2C) and lower (M1D and M2D) strands: M1C, 5′-CAAGTATAACAACAAGAAGAGCGACCAG-3′; M1D, 5′-CTCTTCTTGTTGTTATACTTGTCCTTG-3′; M2C, 5′-CAAGTATAACAAGAAGAACAGCGACCAGGCCCTG-3′; and M2D, 5′-CTGTTCTTCTTGTTATACTTGTCCTTG-3′.
PCR was performed with the Expand High Fidelity PCR system (Boehringer Mannheim) under conditions specified by the manufacturer, except that efficient amplification required the addition of 5% dimethyl sulfoxide. PCR products were digested with suitable restriction enzymes (SalI, NdeI), cloned into a pBluescript plasmid downstream of a hemagglutinin (HA) tag (MGYPYDVPDYASLGGPHH), and verified by sequencing. Wild-type RFXAP and RFXAPs with the N1, N2, N3, C3, and C4 deletions were transferred into the EBO76PL expression vector. Wild-type RFXAP and RFXAPs with the M1, M2, N4, N5, N6, N7, N8, N9, C1, and C2 mutations and deletions were transferred into the first cistron of the bicistronic pHRImCD8 lentiviral vector (43). This vector encodes a cell surface marker (mouse CD8 [mCD8]) in the second cistron under the control of an internal ribosome entry site. Expression of mCD8 was used to purify the transduced cells by sorting.
Cell culture, transfection, transduction, and FACS analysis.
Cells were grown at 37°C in the presence of 5% CO2 in RPMI 1640 (Raji, TK6, and 6.1.6 B cells) or Dulbecco's modified Eagles' medium (ABI fibroblasts) supplemented with 10% heat-inactivated fetal calf serum, 0.1 mg (each) of penicillin and streptomycin per ml, and 2 mM glutamine. Four micrograms of EBO76PL-based expression vectors was transfected into 107 6.1.6 cells by electroporation with a Bio-Rad electropulser using a 300-V and 960-μF pulse. Transfected cells carrying the vectors were selected with 50 μg of hygromycin per ml as described previously (38). Transduction of cells with bicistronic lentiviral vectors was done essentially as described previously (43). One week after transduction, the expression of HLA-DR and mCD8 was analyzed by fluorescence-activated cell sorting (FACS). Transduced cells expressing mCD8 were then purified by labeling them with biotin-conjugated anti-mouse CD8a (Ly-2; PharMingen) and sorting them with streptavidin-coated dynabeads (M-280; Dynal). Purified cells were analyzed by FACS 3 to 5 days after being sorted. To induce MHCII expression in ABI fibroblasts, the cells were cotransduced with a bicistronic lentiviral vector encoding CIITA. The following antibodies were used for FACS analysis: HLA-DR monoclonal antibody 2.06 (5), HLA-DP monoclonal antibody BRAFB6 (Serotec), HLA-DQ monoclonal antibody SPVL3 (Serotec), HLA-ABC w6/32 monoclonal antibody (Serotec), R-phycoerythrin-conjugated rat anti-mCD8a (Ly-2) monoclonal antibody (PharMingen), and the polyclonal fluorescein isothiocyanate-conjugated F(ab′)2 rabbit anti-mouse immunoglobulin G STAR9B (Serotec). Dead cells were excluded from the analysis by staining with propidium iodide and by their forward and sideways light-scattering properties.
In vitro transcription-translation.
Transcription of plasmids encoding RFXAP and translation in a nuclease-treated rabbit reticulocyte lysate (Promega) were done as described previously (29). Translation products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cell extracts and EMSA.
Whole-cell extracts (17) and nuclear extracts (36) were prepared as described previously. The DRA-X box probe (WX2), binding, and gel electrophoresis procedures used to study binding of RFX by electrophoretic mobility shift assays (EMSA) have been described in detail elsewhere (30, 33, 43). Binding of the multiprotein RFX–NF-Y–X2BP complex and dissociation rate experiments were performed as described previously (19, 25, 30, 33, 43). The identity of the RFX–NF-Y–X2BP complex was confirmed by competition and supershift experiments as described previously (19, 25, 30, 33, 43).
RNase protection assays.
RNase protection assays were performed with cytoplasmic RNAs prepared as described in reference 47. The probes used to detect HLA-DRA, HLA-DRB, HLA-DQA, HLA-DQB, HLA-A, and GAPDH mRNAs have been described previously (2, 12).
Chromatin immunoprecipitation.
Chromatin immunoprecipitations with antibodies directed against RFX5 and CIITA were performed as described previously (23). Immunoprecipitated promoter fragments were amplified by PCR and analyzed by agarose gel electrophoresis or quantitative real-time PCR using the SYBR Green method (Applied Biosystems, Rotkreuz, Switzerland). Real-time PCR results were normalized with respect to a standard dilution curve generated with input chromatin. The primers used for amplification were as follows: DRA, 5′-ATTTTTCTGATTGGCCAAAGAGTAATT-3′ and 5′-AAAAGAAAAGAGAATGTGGGGTGTAA-3′; DQB, 5′-CTGCCCAGAGACAGATGAGGT-3′ and 5′-TGATGTACCTGGCAGAAAGAATAAAAA-3′; DPB, 5′-CTCATACAAAGCTCAGTGTCCATTG-3′ and 5′-CTGTGACCCTGGGATTGGAC-3′; and CD20, 5′-AACCAAAGTAATTGGAGCGAAGC-3′ and 5′-GCCTGAAGATGAAATCGCTGATA-3′.
Nucleotide sequence accession number.
Nucleotide and amino acid sequences of mouse RFXAP have been submitted to GenBank under the accession number AF335512.
RESULTS
Mouse RFXAP is functionally equivalent to human RFXAP.
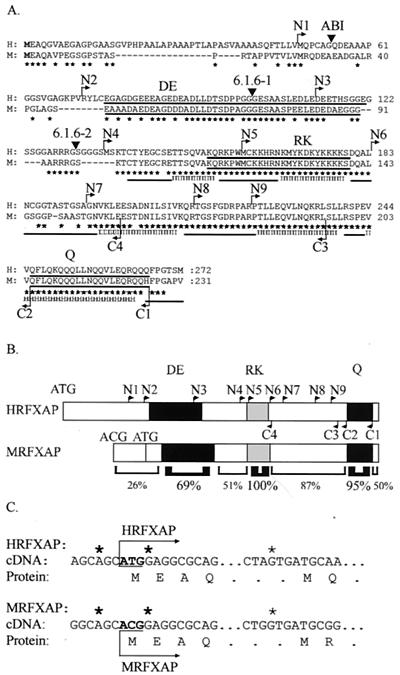
We isolated the mouse RFXAP gene in order to facilitate the identification of conserved regions. Full-length RFXAP cDNA clones were isolated from a mouse spleen cDNA library, and several independent clones were sequenced (Fig. 1A). The sequence was also confirmed by the isolation and sequencing of the genomic RFXAP gene (data not shown). The mouse RFXAP cDNA clones contain a 231-amino-acid open reading frame. The alignment of the human and mouse sequences is quite informative (Fig. 1A). Mouse RFXAP shows an overall homology of 65% amino acid identity to the human protein. Conservation is highest in the C-terminal two-thirds of the protein, particularly in three regions that had been noted previously in the human protein (Fig. 1B). Two of these regions—an acidic segment (69% amino acid identity) and a glutamine-rich segment (95% amino acid identity)—are reminiscent of the transcription activation domains found in certain transcription factors (24). The third region is rich in basic amino acids and encompasses a sequence resembling a bipartite nuclear-localization signal (NLS) (100% amino acid identity) (15). Outside of these regions homology is lower and optimal alignment requires the introduction of gaps. Mutations within the RFXAP gene have been defined in several MHCII deficiency patients and in an in vitro-generated mutant cell line (6.1.6) (Fig. 1A) (9, 10, 22, 42). All of these mutations lead to the synthesis of severely truncated RFXAP proteins lacking the most conserved C-terminal moiety.
FIG. 1.
Sequence homology between human and mouse RFXAP. (A) Alignment of the human (H) and mouse (M) RFXAP amino acid sequences. Stars indicate identical residues. Triangles indicate positions at which the protein is truncated in the ABI cell line and in the two alleles of the 6.1.6 cell line. Arrows indicate ends of the N-terminal (N1 to N9) and C-terminal (C1 to C4) deletions. Regions rich in acidic amino acids (DE), basic amino acids (RK), and glutamine (Q) are underlined. Amino acid coordinates are indicated at the right. A secondary-structure prediction is indicated on the bottom line: H represents α-helices, the line represents coils, and E represents β-strands. (B) Schematic representations of human and mouse RFXAPs are shown. The acidic-amino-acid (DE)-, basic-amino-acid (RK)-, and glutamine (Q)-rich regions are represented as filled boxes. The ends of N- and C-terminal deletions are positioned on the human protein. Potential translation initiation codons are indicated. Homology (percent identity) between mouse and human RFXAPs within different regions is indicated below. (C) Sequences of human and mouse RFXAPs in the vicinity of the translation initiation codon and the next conserved downstream ATG codon. The translation initiation codon of human RFXAP aligns with an ACG codon in mouse RFXAP. Stars indicate nucleotides providing a good Kozak context.
Surprisingly, the AUG translation initiation codon of human RFXAP aligns with an ACG codon rather than an AUG codon in the mouse sequence (Fig. 1C). The first in-frame AUG codon in the mouse RFXAP open reading frame is situated 81 nucleotides downstream of this ACG codon (Fig. 1C). In other systems, it has been shown that certain ACG codons can function as translation initiation codons (7, 14). Moreover, with an A nucleotide at −3 and a G nucleotide at +4, this ACG codon is found in a favorable Kozak context for translation initiation (16). To determine whether translation of mouse RFXAP could be initiated at the ACG codon, we performed in vitro transcription-translation experiments with two RFXAP constructs (Fig. 2). The first construct (M-ACG) contains the entire open reading frame starting at the putative ACG initiation codon, whereas the second (M-ATG) starts at the downstream ATG codon (Fig. 2A). mRNAs transcribed from the two constructs were translated in a rabbit reticulocyte lysate system, and translation products were analyzed by SDS-PAGE (Fig. 2B). The M-ACG construct directs the synthesis of a protein that is clearly larger than the protein encoded by the M-ATG construct, indicating that the unusual ACG codon is indeed used. The shorter protein, initiated from the internal ATG, is not synthesized from the M-ACG construct. This result indicates that the ACG initiation codon is used quite efficiently and that there is thus very little readthrough down to the ATG codon.
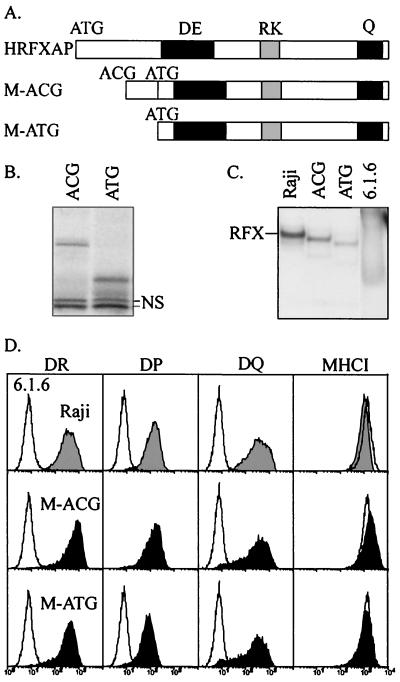
FIG. 2.
Mouse RFXAP is functionally equivalent to the human protein. (A) Schematic maps of human RFXAP and the two mouse RFXAP constructs (M-ACG and M-ATG). The M-ACG construct starts at the nonclassical ACG initiation codon and contains a second ATG codon situated 81 nucleotides further downstream. The M-ATG construct starts at the second ATG codon. (B) In vitro translation products of M-ACG and M-ATG were analyzed by SDS-PAGE. NS corresponds to nonspecific bands labeled in the lysate. (C) Binding of the RFX complex in extracts derived from cells complemented with mouse RFXAP. Binding was analyzed by EMSA with a DRA X box probe. Whole-cell extracts from Raji cells, nontransfected 6.1.6 cells, and 6.1.6 cells transfected with M-ACG or M-ATG were analyzed. All of the lanes are from the same gel at the same length of exposure. (D) Mouse RFXAP restores MHCII expression in 6.1.6 cells. Cell surface expression of HLA-DR, HLA-DP, and HLA-DQ was analyzed by FACS in Raji cells (gray histograms), nontransfected 6.1.6 cells (open histograms), and 6.1.6 cells transfected with M-ACG or M-ATG (black histograms). Expression of MHCI molecules was included as a control.
To test whether mouse RFXAP could substitute functionally for the human protein, the M-ACG and M-ATG versions were cloned into an episomal expression vector and transfected into a mutant human B-cell line (6.1.6) lacking a functional RFXAP gene. The ability of mouse RFXAP to complement the 6.1.6 cells was evaluated by examining cell surface MHCII expression (Fig. 2D). Both the M-ACG and M-ATG constructs restore expression of HLA-DR, HLA-DQ, and HLA-DP to levels that are identical to those found in wild-type B cells (Fig. 2D). Both versions of mouse RFXAP can thus substitute functionally for the human protein. These results also indicate that the first 27 amino acids of mouse RFXAP can be removed without affecting its function. As previously observed for complementation of 6.1.6 with human RFXAP (9), complementation with mouse RFXAP does not affect the level of cell surface MHCI expression (Fig. 2D).
To confirm that mouse RFXAP can associate with the other human RFX subunits to form complexes capable of binding to the X box, we performed EMSA with nuclear extracts from 6.1.6 cells complemented with the M-ACG and M-ATG constructs. As expected from the reactivation of MHCII expression in these complemented cells, both mouse proteins could be incorporated into functional RFX complexes (Fig. 2C). The abundance of these RFX complexes is similar to that of the native RFX complex observed in wild-type human B cells (Raji), indicating that the mouse RFXAP proteins are synthesized at physiological levels. Supershift experiments confirmed that these RFX complexes contain all three subunits (data not shown). Finally, the RFX complexes containing mouse RFXAP could bind cooperatively with X2BP and NF-Y (data not shown).
The relative mobilities of the RFX complexes formed with human RFXAP, M-ACG, and M-ATG, are consistent with their respective sizes (Fig. 2C). Due to the smaller sizes of the mouse proteins, the RFX complexes formed with M-ACG (231 amino acids) and M-ATG (204 amino acids) migrated faster than the native complex containing human RFXAP (272 amino acids). Moreover, the RFX complex formed with M-ACG migrated more slowly than the one formed with M-ATG. The latter observation indicates that, in accordance with the in vitro translation data, the unusual ACG translation initiation codon is used in vivo in the transfected cells.
Mapping of the minimal region of RFXAP required for expression of HLA-DR.
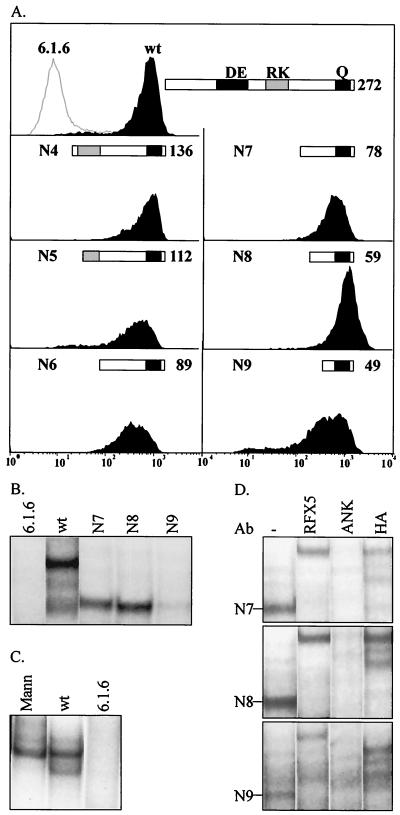
In a previous report we determined that the last 138 C-terminal amino acids of human RFXAP are sufficient for in vitro assembly and DNA binding of the RFX complex (4). Similar results were reported by others (8). To define the minimal essential region of RFXAP more precisely, we engineered a series of N-terminal deletions (N1 to N9) and examined their ability to restore expression of the endogenous MHCII genes in 6.1.6 cells. The endpoints of the deletions were chosen such that they progressively removed either segments exhibiting greater-than-average conservation between the human and mouse proteins or regions containing putative functional motifs (acidic, basic, and glutamine-rich regions) (Fig. 1A and B). The RFXAP deletions were introduced into 6.1.6 cells using a bicistronic lentiviral expression vector (43). In this system, the first cistron encoding RFXAP is followed by a second cistron directing the synthesis of mCD8 via an internal ribosomal entry site. The advantage of this system is that stably transduced cells can be purified by sorting for mCD8 expression. After one round of sorting for mCD8, homogeneous populations of stably transfected cells were obtained. Remarkably, constructs with all deletions restored HLA-DR expression as efficiently as did wild-type RFXAP (Fig. 3A). Over 80% of RFXAP, including the acidic and basic regions, can thus be deleted without affecting its ability to reactivate expression of the endogenous HLA-DR genes. Only the last 49 C-terminal amino acids encompassing the glutamine-rich region are required.
FIG. 3.
A short C-terminal region of RFXAP is sufficient for binding of the RFX complex and activation of HLA-DR expression in RFXAP-deficient 6.1.6 cells. (A) Cell surface HLA-DR expression was analyzed by FACS in nontransfected 6.1.6 cells (open histogram) and in 6.1.6 cells complemented with bicistronic lentiviral vectors encoding wild-type RFXAP (wt) or RFXAPs with the N4, N5, N6, N7, N8, and N9 deletions (black histograms). The various versions of RFXAP are indicated schematically and their sizes in amino acids are provided. (B) Binding of the RFX complex was analyzed by EMSA with nuclear extracts derived from nontransfected 6.1.6 cells, 6.1.6 cells complemented with wild-type RFXAP, and 6.1.6 cells complemented with the versions of RFXAP with the N7, N8, or N9 deletion. All of the lanes are from the same gel at the same length of exposure. (C) Binding of the RFX complex was analyzed by EMSA with nuclear extracts derived from nontransfected 6.1.6 cells (6.1.6), Mann B cells, and 6.1.6 cells complemented with wild-type RFXAP. All lanes come from the same gel at the same length of exposure. (D) The composition of RFX complexes detected in EMSA was analyzed by supershift experiments using antibodies directed against RFX5, RFXANK, and the HA tag carried by the transfected RFXAP subunit. All three antibodies supershift the RFX complexes formed in 6.1.6 cells complemented with the constructs with the N7, N8, and N9 deletions.
The ability of the N-terminal deletions to be incorporated into RFX complexes capable of binding to the X box was analyzed by EMSA using nuclear extracts from the complemented 6.1.6 cells. In agreement with the complementation analysis, the N-terminal deletions were able to generate RFX complexes capable of binding to the X box of the HLA-DRA gene. Results for the last three deletions (N7, N8, and N9) are shown in Fig. 3B. As expected from their size, the N7 (78-amino-acid), N8 (59-amino-acid), and N9 (49-amino-acid) deletions formed RFX complexes that migrated significantly faster than the complex formed with wild-type RFXAP (272 amino acids). Supershift experiments have confirmed that these complexes contain the endogenous RFX5 and RFXANK subunits as well as the transduced RFXAP subunit (Fig. 3D).
The RFX complexes generated in 6.1.6 cells complemented with constructs with the N7 and N8 deletions are similar in abundance to those obtained with wild-type RFXAP, whereas a significantly lower level was obtained with the N9 construct (Fig. 3B). Moreover, the abundance of the RFX complexes restored in complemented 6.1.6 cells is similar to the amount of native RFX present in wild-type B cells (Fig. 3C). The possibility that the severely deleted versions of RFXAP retain their ability to restore HLA-DR expression because they are strongly overexpressed can thus be excluded.
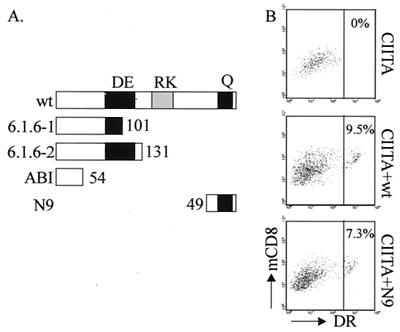
The mutations affecting the two alleles of the RFXAP gene in the 6.1.6 cell line are such that they may in theory lead to the synthesis of two truncated RFXAP proteins containing the first 101 and 131 amino acids (Fig. 4A) (9). There is no evidence that these truncated proteins are indeed produced in 6.1.6 cells. However, these endogenous N-terminal fragments would contain part or all of the conserved acidic region of RFXAP, which raises the possibility that the short N9 fragment can complement only 6.1.6 cells because the acidic region retained in the endogenous N-terminal RFXAP fragments provides a function that is required in conjunction with N9. To exclude the possibility of such an intermolecular complementation, we turned to the ABI cell line, which was derived from an MHCII deficiency patient. In ABI cells, there is a homozygous premature stop codon at amino acid 54 of the RFXAP gene (42). This mutation removes all of the conserved regions of RFXAP (Fig. 4A). The remaining 54 N-terminal amino acids show only poor conservation (30%) between the human and mouse genes. ABI cells were transduced with the bicistronic lentiviral constructs encoding wild-type RFXAP or RFXAP with the N9 deletion (Fig. 4B). Since ABI cells are fibroblasts, which do not express the MHCII transactivator CIITA, the cells were transduced simultaneously with a bicistronic lentiviral vector encoding CIITA. The fraction of cells that were cotransduced by the RFXAP and CIITA expression constructs was relatively small under the conditions used. Nevertheless, the results indicate that the N9 construct restored HLA-DR expression almost as efficiently as wild-type RFXAP (7.3% of the cells for N9 versus 9.5% of the cells for wild-type RFXAP) (Fig. 4B). The HLA-DR-positive cells are those that express the highest level of mCD8. This finding is consistent with the interpretation that they correspond to doubly transduced cells carrying both the CIITA and RFXAP expression vectors because both expression constructs encode mCD8 in the second cistron.
FIG. 4.
The minimal essential C-terminal region of RFXAP is sufficient for activation of HLA-DR expression in RFXAP-deficient ABI fibroblasts. (A) Schematic representation of the truncated RFXAP proteins that could be made in the 6.1.6 cell line and in the ABI fibroblast cell line derived from the MHCII deficiency patient. wt, wild type. (B) FACS analysis of HLA-DR and mCD8 expression on ABI cells transduced with CIITA alone (top) and together with wild-type RFXAP (middle) or RFXAP with the N9 deletion (bottom). Similar numbers of HLA-DR-positive cells are observed when ABI cells are cotransduced with CIITA and wild-type RFXAP (9.5% of mCD8-positive cells) or with CIITA and RFXAP with the N9 deletion (7.3% of mCD8-positive cells). No HLA-DR-positive cells are observed when only CIITA is transduced.
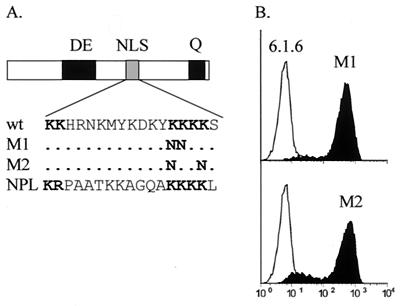
The NLS motif is not essential for activation of MHCII expression.
The basic region of RFXAP contains a 17-amino-acid sequence that resembles a bipartite NLS and is perfectly conserved between the human and mouse proteins. This motif is characterized by two clusters of basic residues separated by a spacer of 10 amino acids (Fig. 5A) and is very similar to the NLSs found in other proteins such as nucleoplasmin (35). Our deletion analysis suggests that this putative NLS is not essential for the function of RFXAP (Fig. 3). However, it remained possible that it is in fact required for nuclear import of the intact protein but that deletions removing it (N6 to N9) generate proteins that are sufficiently small to enter the nucleus by passive diffusion. To exclude this possibility, we introduced point mutations into the NLS-like sequence. Two independent mutant proteins were constructed. Each mutant construct contained lysine-to-asparagine substitutions that are known to destroy the NLS of nucleoplasmin (Fig. 5A). The two NLS mutant constructs were cloned into the bicistronic lentiviral vector and transduced into 6.1.6 cells. After purification of the transduced cells by sorting for mCD8, the expression of HLA-DR was analyzed by FACS. Both mutant constructs retained their ability to restore HLA-DR expression in 6.1.6 cells (Fig. 5B). The expression of HLA-DP and HLA-DQ molecules was also restored (data not shown). Together with the deletion analysis (Fig. 3), the results obtained with the point mutations argue against a crucial role of the NLS-like sequence of RFXAP.
FIG. 5.
The putative NLS present in the basic region is not essential for the function of RFXAP. (A) Sequences of the putative NLS in wild-type RFXAP (wt), of the NLS of nucleoplasmin (NPL), and of the two mutants (M1 and M2) are shown. Bold residues have been shown to be essential for NLS function in nucleoplasmin. (B) FACS analysis of HLA-DR expression on nontransduced 6.1.6 cells (open histograms) and 6.1.6 cells transduced with the M1 and M2 mutants of HRFXAP (black histograms).
The C-terminal glutamine-rich region is essential for the function of RFXAP.
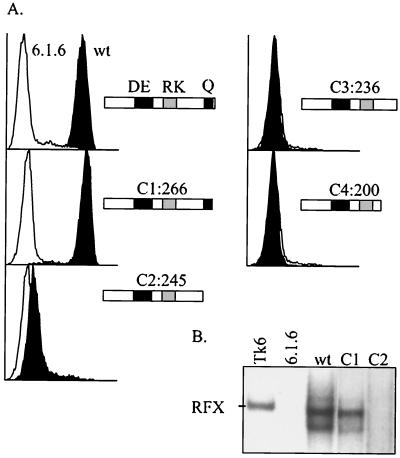
To narrow down further the minimal essential domain of RFXAP, we prepared a series of mutant constructs with C-terminal deletions (C1 to C4) and tested their ability to complement 6.1.6 cells (Fig. 6A). Removal of the last six weakly conserved residues of RFXAP (C1) does not affect the ability to restore HLA-DR expression. In contrast, complementation is completely lost when the glutamine-rich region is deleted (C2, C3, and C4). This result reduces the minimal essential domain of RFXAP to a 43-amino-acid segment encompassing the C-terminal glutamine-rich region. This domain is 100% conserved between the human and mouse proteins (Fig. 1).
FIG. 6.
The glutamine-rich region is essential for the function of RFXAP. (A) FACS analysis of HLA-DR expression on nontransfected 6.1.6 cells (open histograms) and 6.1.6 cells complemented with wild-type RFXAP (wt) or with RFXAPs with deletions C1 to C4 (black histograms). Wild-type RFXAP and RFXAPs with C-terminal deletions are represented schematically, and their sizes in amino acids are provided. (B) Binding of the RFX complex was analyzed by EMSA with nuclear extracts derived from TK6 B cells, nontransfected 6.1.6 cells, 6.1.6 cells complemented with wild-type RFXAP, and 6.1.6 cells complemented with the version of RFXAP with the C1 or C2 deletion. All of the lanes are from the same gel at the same length of exposure.
The ability of the C-terminal deletions to form RFX complexes capable of binding to DNA was analyzed by EMSA using nuclear extracts derived from the complemented 6.1.6 cells (Fig. 6B). As expected from the complementation analysis, C1 was able to form an RFX complex capable of binding to the X box with the same efficiency as that of the wild-type protein. In contrast, no RFX complex was detected in extracts from cells expressing C2 (Fig. 6B) or C3 and C4 (data not shown), suggesting that these mutants cannot assemble into a functional RFX complex.
Differential dependence of MHCII isotypes on RFXAP.
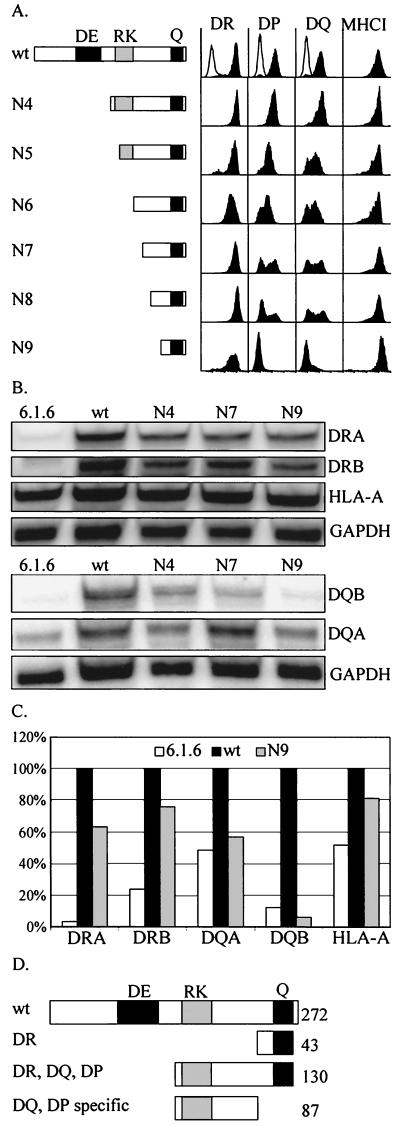
The expression of HLA-DR is typically more robust than that of HLA-DQ and HLA-DP, particularly in complementation experiments of the type described above. In order to reveal more subtle effects of the different versions of RFXAP with deletions, we extended our analysis to HLA-DQ and HLA-DP. The expression of all three MHCII isotypes was analyzed by FACS in 6.1.6 cells stably transduced with RFXAPs with N-terminal deletions (Fig. 7A). All constructs with deletions down to and including N9 were fully capable of restoring HLA-DR expression. In contrast, the result was strikingly different for HLA-DQ and HLA-DP. Whereas constructs with N1 to N4 deleted retained the ability to restore the expression of all three isotypes, further deletions (N5 to N8) led to a progressive reduction in the efficiency with which the expression of HLA-DQ and HLA-DP was reactivated. The N9 construct completely lost the capacity to restore HLA-DQ and HLA-DP expression. Expression of HLA-DQ was lost more quickly than that of HLA-DP (see results for N5 and N6 in Fig. 7A). The progressive loss in the ability to restore HLA-DQ and HLA-DP expression was not simply a reflection of lower expression levels. For instance, the cells transfected with the N7 and N8 constructs contained wild-type amounts of functional nuclear RFX complexes (Fig. 3B).
FIG. 7.
Identification of a segment within RFXAP that has an MHCII isotype-specific function. (A) FACS analysis of the expression of HLA-DR, HLA-DP, HLA-DQ, and MHCI at the surfaces of nontransduced 6.1.6 cells (open histograms) and 6.1.6 cells transduced with wild-type RFXAP (wt) or RFXAPs with the deletions N4 to N9 (black histograms). The transduced RFXAP proteins are represented at the left. (B) RNase protection analysis of the expression of the HLA-DRA, HLA-DRB, HLA-DQA, HLA-DQB, and HLA-A mRNAs in nontransduced 6.1.6 cells and in 6.1.6 cells transduced with wild-type RFXAP or versions of RFXAP with the N4, N7, and N9 deletions. GAPDH mRNA was included as an internal standard. (C) Expression levels measured by RNase protection experiments were quantified by phosphorimager analysis. All values were normalized with respect to GAPDH mRNA expression. For each mRNA, the level attained by complementation with wild-type RFXAP was defined as 100%. (D) Schematic representation of the regions within RFXAP that are required for expression of the three MHCII isotypes. A short 43-amino-acid C-terminal region is sufficient for expression of HLA-DR. In contrast, a 130-amino-acid C-terminal region is required for expression of HLA-DP and HLA-DQ. These two regions define an 87-amino-acid domain having HLA-DQ- and HLA-DP-specific functions.
The isotype-specific reduction in complementation was confirmed for selected deletions (N4, N7, and N9) by RNase protection experiments using probes specific for the mRNAs encoding the α- and β-chains of HLA-DR and HLA-DQ (Fig. 7B). The N4, N7, and N9 constructs were equally efficient at activating expression of the HLA-DRA and HLA-DRB mRNAs. Quantification with respect to the level in the internal GAPDH control indicated that the N9 construct was 65 to 75% as efficient as wild-type RFXAP at activating HLA-DRA and HLA-DRB expression (Fig. 7C). In contrast, sequential deletion from N4 to N9 led to a selective loss in the activation of HLA-DQ mRNA expression. HLA-DQA and HLA-DQB mRNA expression was not activated by the N9 construct above the initial background levels observed in the nontransduced cells (Fig. 7C). Taken together, these results indicate that the region situated between the endpoints of deletions N4 and N9 is required for expression of HLA-DQ and HLA-DP but not for expression of HLA-DR (Fig. 7D).
It is known that RFX contributes to the activity of MHCI promoters (41). Therefore, despite the fact that the level of cell surface MHCI expression was not increased by complementation of 6.1.6 cells (Fig. 2C and 7A), we nevertheless analyzed MHCI (HLA-A) mRNA levels by RNase protection experiments (Fig. 7B to C). A twofold increase in HLA-A expression was observed upon complementation of 6.1.6 cells by intact RFXAP. The N9 construct retained the ability to activate HLA-A expression. The HLA-DQ and -DP-specific region of RFXAP is thus not essential for HLA-A.
Isotype-specific role of RFXAP in promoter occupation in vivo.
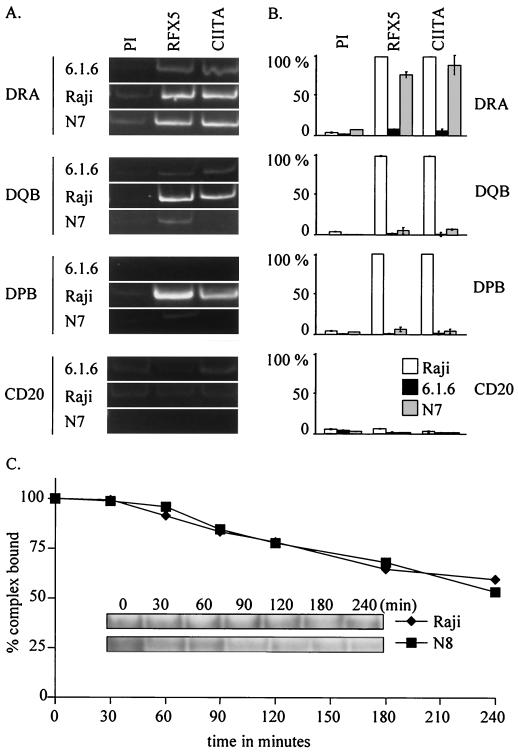
The binding of RFX is essential for the activation of MHCII promoters. We therefore examined whether the isotype-specific dependence on the C-terminal region of RFXAP could be accounted for by differences in levels of promoter occupation by RFX. The binding of RFX in vivo to three endogenous MHCII promoters—DRA, DQB, and DPB—was studied by means of chromatin immunoprecipitation experiments using an antibody directed against the largest subunit of RFX (RFX5). To study the role of the isotype-specific region of RFXAP, we compared wild-type cells and 6.1.6 cells transduced with a deletion mutant RFXAP exhibiting an impaired ability to activate expression of DP and DQ. The N7 transfectant was chosen for the analysis because it exhibits a clear dissociation between the expression of DR and that of the two other MHCII isotypes (Fig. 7) despite the fact that wild-type levels of the mutant RFX complex were restored (Fig. 3). In the wild-type cells, RFX binds to all three MHCII promoters (Fig. 8A). In the N7 transfectant on the other hand, binding of RFX to the DRA promoter was retained at nearly normal levels whereas binding to the DQB and DPB promoters was almost completely lost (Fig. 8A and B). Differential activation of DR versus that of DQ and DP can thus be explained by differences in promoter occupation by the mutant RFX complex.
FIG. 8.
Isotype-specific role of RFXAP in inducing promoter occupation by RFX and CIITA in vivo. (A and B) The in vivo occupation of the DRA, DQB, and DPB promoters by RFX and CIITA was studied by chromatin immunoprecipitation experiments with Raji cells, 6.1.6 cells, and 6.1.6 cells complemented with the N7 construct. The CD20 promoter was chosen as a negative control. Results were visualized by gel electrophoresis of the PCR products (A) or quantified by real-time PCR (B). Results are given in percentages relative to values obtained for RFX and CIITA in Raji cells. PI, control immunoprecipitation performed with preimmune serum. (C) Dissociation rates were determined by EMSA for the higher-order protein-DNA complexes formed by simultaneous binding of RFX, NF-Y, and X2BP to the DRA promoter. The complexes were assembled using extracts from Raji cells containing wild-type RFX or 6.1.6 cells complemented with the N8 construct. The gels were quantified by phosphorimager analysis (graphs). The percentages of protein-DNA complexes remaining are plotted as a function of time. The curves represent the means of results from four independent experiments. Representative gels are shown as an inset.
Binding of RFX is a prerequisite for the occupation of MHCII promoters by a multiprotein enhanceosome complex that serves as a landing pad for the transcriptional coactivator CIITA (23). We therefore examined the effect of the N7 deletion on recruitment of CIITA to the DRA, DQB, and DPB promoters (Fig. 8A and B). As expected, recruitment of CIITA to the DRA promoter could be supported normally by the N7 deletion. In contrast, the recruitment of CIITA to the DQB and DPB promoters was abolished. We conclude that the N7 deletion eliminates binding of RFX to the DQB and DPB promoters in vivo and that promoter occupation and CIITA recruitment can thus not occur at these promoters.
A weak association of RFX and CIITA with the DRA promoter was observed in 6.1.6 cells (Fig. 8A and B). This observation is consistent with results of previous studies that have indicated that this cell line is slightly leaky for DR expression (18).
RFX binds to MHCII promoters cooperatively with two other transcription factors, NF-Y and X2BP (19, 25, 30, 33, 43). An in vitro binding study has suggested that the importance of these cooperative binding interactions varies from one promoter to another (19). It thus seemed plausible that the gene-specific promoter occupation defect of the RFXAP deletions could be due to the loss of cooperative binding with NF-Y and/or X2BP. To examine this possibility, we assembled the RFX–NF-Y–X2BP complex on the DRA promoter using nuclear extracts from cells containing either wild-type RFXAP or RFXAP with deletions. We then compared the levels of stability of these multiprotein-DNA complexes by measuring their dissociation rates. Results for the N8 deletion are shown in Fig. 8C. No difference in dissociation rate was observed between the complexes containing wild-type and mutant RFXAP (Fig. 8C). Conditions for generating the higher-order complexes on MHCII promoters other than DRA are less well established (19). We nevertheless managed to measure the dissociation rates of higher-order complexes formed on the DQB promoter, although the precise contents of these complexes could not be unambiguously defined. As observed for the DRA promoter, the deletions in RFXAP had no effect on the dissociation rate of the higher-order complexes formed on the DQB promoter (data not shown). Taken together, these results indicate that the N8 deletion does not lead to an obvious loss in cooperative binding between RFX, NF-Y, and X2BP in vitro.
DISCUSSION
RFXAP was first identified as a transcription factor for MHCII genes because it was found to be defective in 6.1.6 cells, an in vitro-generated MHCII-negative mutant, and in patients suffering from bare lymphocyte syndrome, a hereditary disease resulting from a deficiency in MHCII expression (9, 10, 11, 42). Here we have used a genetic complementation approach to identify the regions within RFXAP that are necessary for the activation of MHCII genes. In this system, the ability to reactivate expression of the endogenous MHCII genes in RFXAP-deficient cells is evaluated. This has two key advantages. First, activation of the endogenous genes in their native genomic context is evaluated rather than the expression of artificial reporter gene constructs. Second, expression of the genes coding for all three MHCII isotypes can be assayed simultaneously in the same transfected cells. These advantages have permitted us to define two distinct domains within RFXAP. A highly conserved 43-amino-acid segment situated at the C terminus of RFXAP is required for expression of the genes encoding all three MHCII isotypes. Remarkably, this minimal essential domain is sufficient for expression of HLA-DR but not HLA-DQ and HLA-DP. Expression of the last two MHCII isotypes requires an additional region of RFXAP. The three different human MHCII isotypes thus exhibit a differential dependence on RFXAP. This results from the fact that the isotype-specific region of RFXAP is required for promoter occupation by RFX and recruitment of CIITA at certain MHCII genes (DQB, DPB) but not at others (DRA).
To facilitate our structure-function analysis, we isolated the mouse RFXAP gene. Mouse RFXAP can restore expression of all human MHCII isotypes in 6.1.6 cells and is thus functionally equivalent to the human protein. Homology between human and mouse RFXAP is high in the C-terminal moiety of the protein but is very low in its N-terminal half. This pattern of conservation is consistent with the finding that the N-terminal half of RFXAP can be removed without affecting its function. The most conserved segment in the N terminus of RFXAP is a previously noted region rich in acidic amino acids. We have been unable to attribute any function to this acidic domain; it can be removed completely without affecting the ability to complement 6.1.6 and ABI cells.
Surprisingly, the mouse codon corresponding to the translation initiation codon of human RFXAP is ACG rather than AUG. Although only a few examples exist, nonclassical ACG initiation codons have been described for other systems (7, 14). Moreover, this ACG codon of mouse RFXAP is found in an optimal Kozak consensus motif. We therefore determined whether translation of mouse RFXAP was initiated at the ACG codon or at a conserved downstream AUG codon. Our results indicate that the ACG codon is indeed used both in vitro and in transfected cells. The mouse RFXAP gene is thus among the rare examples where translation of a cellular gene is initiated at an ACG codon. However, initiation at this ACG is not essential for the function of RFXAP because initiation at the next downstream AUG generates a truncated protein that is fully capable of complementing 6.1.6 cells.
The basic region of RFXAP contains a sequence resembling an NLS. However, deletion of this sequence (N6 to N9) and point mutations that would inactivate the putative NLS (M1 and M2) do not lead to a reduction in the amount of the RFX complex present in nuclear extracts and do not affect the ability to activate the transcription of the genes encoding HLA-DR. These findings suggest that the basic sequence does not function as an NLS. Alternatively, it is possible that it does function as an NLS but that it is redundant with other NLS sequences present either in RFXAP or in one or both of the other two RFX subunits. The RFX complex assembles in solution by the association of RFXAP, RFX5, and RFXANK (4, 8, 43). RFXAP may thus be carried into the nucleus passively by virtue of its association with RFX5 and/or RFXANK.
Remarkably, over 80% of RFXAP is dispensable for the activation of HLA-DR expression. A short 43-amino-acid C-terminal segment of the protein is sufficient for activating the endogenous HLA-DR genes, both in the B-cell line 6.1.6 and in the fibroblast cell line ABI. It is also sufficient for association with RFXANK and RFX5 and for assembly of an RFX complex that can bind specifically to the X box of the DRA promoter in vivo and in vitro. This has been demonstrated both for transfected cells (Fig. 3 and 8) and with recombinant RFX subunits (data not shown). The minimal essential region of RFXAP must thus mediate at least two functions. First, it must contain protein-protein interaction domains required for assembly of the RFX complex. Second, it must contribute to the DNA binding activity of RFX, either indirectly by inducing a conformational change in one of the other two subunits or directly by contacting the DNA (46). In addition, the minimal domain can sustain stable occupation by RFX and recruitment of CIITA in vivo at the DRA promoter. The essential 43-amino-acid segment contains a glutamine-rich region and is predicted to have a high α-helical content (Fig. 1). It is likely that one or both of these features are characteristic of the domains of RFXAP that contribute to assembly and binding of the RFX complex.
In contrast to the situation observed for HLA-DR, the minimal essential region of RFXAP is not sufficient for the expression of HLA-DQ and HLA-DP. Optimal expression of these two MHCII isotypes requires an additional 87-amino-acid region situated immediately upstream of the minimal domain (Fig. 7). There are several predicted α-helical regions within the isotype-specific domain of RFXAP (Fig. 1). The region rich in basic amino acids is also contained within this domain (Fig. 1). Both the basic region and the potential helices are particularly well conserved between the human and mouse proteins (Fig. 1). It is therefore tempting to speculate that the basic region and/or the putative helices represent structural elements implicated in the function of this domain.
Our results demonstrate that the isotype-specific region of RFXAP is essential in vivo for promoter occupation by RFX and recruitment of CIITA to the DQB and DPB promoters but is dispensable for these functions at the DRA promoter (Fig. 8A). The occupation of MHCII promoters in vivo is known to require cooperative binding between RFX, NF-Y, and X2BP (19, 25, 30, 33, 43, 48). In vitro binding experiments have demonstrated that the importance of these cooperative binding interactions varies for different MHCII promoters (19, 30). It thus seemed likely that the accessory region of RFXAP could be implicated in cooperative binding interactions between RFX, X2BP, and NF-Y. However, our dissociation rate measurements of the multiprotein-DNA complex formed by the simultaneous binding of RFX, X2BP, and NF-Y does not support this model (Fig. 8B). The precise function that is provided by the isotype-specific region of RFXAP thus remains to be determined. Several possibilities come to mind. First, it remains possible that it participates in cooperative binding with X2BP and/or NF-Y but that this contribution is either too subtle to be detected by our in vitro assays or significant only in a native chromatin context. Another possibility is that it mediates interactions with other as yet unidentified MHCII promoter binding factors. These may include either factors binding to all MHCII promoters, such as proteins recognizing the S box, or factors implicated at only a subset of MHCII promoters. Finally, an interesting possibility is that the isotype-specific domain of RFXAP recruits accessory proteins, such as chromatin-remodeling factors, that are required only for permitting occupation of certain MHCII promoters. The differential dependence on this domain may thus reflect differences in levels of promoter accessibility or chromatin structures.
The expression of HLA-DR, HLA-DQ, and HLA-DP is generally coordinated. However, there are also several reports describing a dissociated pattern of expression in certain primary cells, established cell lines, and tumor cells (reviewed in reference 1). In most cases, expression of HLA-DR is higher than that of HLA-DP whereas HLA-DQ is barely detectable or completely absent. This pattern (DR > DP > DQ) is reminiscent of the one observed in our complementation experiments with certain of the RFXAPs with deletions (Fig. 7A, N5 and N6). This suggests that the mechanisms leading to the dissociated pattern of MHCII expression observed in certain cell types may involve the isotype-specific domain we have identified in RFXAP.
ACKNOWLEDGMENTS
We are very grateful to S. Landmann and K. Masternak for their help with the chromatin immunoprecipitation experiments.
This work was supported by a grant from the Swiss National Science Foundation. M. Peretti was supported by a fellowship from the Yamanouchi Research Institute.
REFERENCES
- 1.Alcaide-Loridan C, Lennon A M, Bono M R, Barbouche R, Dellagi K, Fellous M. Differential expression of MHC class II isotype chains. Microbes Infect. 1999;1:929–934. doi: 10.1016/s1286-4579(99)00224-5. [DOI] [PubMed] [Google Scholar]
- 2.Bontron S, Ucla C, Mach B, Steimle V. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol Cell Biol. 1997;17:4249–4258. doi: 10.1128/mcb.17.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 4.Caretti G, Cocchiarella F, Sidoli C, Villard J, Peretti M, Reith W, Mantovani R. Dissection of functional NF-Y–RFX cooperative interactions on the MHC class II E alpha promoter. J Biol Chem. 2000;302:539–552. doi: 10.1006/jmbi.2000.4028. [DOI] [PubMed] [Google Scholar]
- 5.Charron D J, McDevitt H O. Analysis of HLA-D region-associated molecules with monoclonal antibody. Proc Natl Acad Sci USA. 1979;76:6567–6571. doi: 10.1073/pnas.76.12.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 7.Curran J, Kolakofsky D. Ribosomal initiation from an ACG codon in the Sendai virus P/C mRNA. EMBO J. 1988;7:245–251. doi: 10.1002/j.1460-2075.1988.tb02806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeSandro A M, Nagarajan U M, Boss J M. Associations and interactions between bare lymphocyte syndrome factors. Mol Cell Biol. 2000;20:6587–6599. doi: 10.1128/mcb.20.17.6587-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fondaneche M C, Villard J, Wiszniewski W, Jouanguy E, Etzioni A, Le Deist F A, Peijnenburg A, Casanova J L, Reith W, Mach B, Fischer A, Lisowska-Grospierre B. Genetic and molecular definition of complementation group D in MHC class II deficiency. Hum Mol Genet. 1998;7:879–885. doi: 10.1093/hmg/7.5.879. [DOI] [PubMed] [Google Scholar]
- 11.Gladstone P, Pious D. Stable variants affecting B cell alloantigens in human lymphoid cells. Nature. 1978;271:459–461. doi: 10.1038/271459a0. [DOI] [PubMed] [Google Scholar]
- 12.Gonczy P, Reith W, Barras E, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Inherited immunodeficiency with a defect in a major histocompatibility complex class II promoter-binding protein differs in the chromatin structure of the HLA-DRA gene. Mol Cell Biol. 1989;9:296–302. doi: 10.1128/mcb.9.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hake S B, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol. 2000;20:7716–7725. doi: 10.1128/mcb.20.20.7716-7725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y, Armanious M K, Thomas M J, Cress W D. Identification of E2F–3B, an alternative form of E2F-3 lacking a conserved N-terminal region. Oncogene. 2000;19:3422–3433. doi: 10.1038/sj.onc.1203682. [DOI] [PubMed] [Google Scholar]
- 15.Jans D A, Xiao C Y, Lam M H. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 17.Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–156. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- 18.Levine F, Pious D. Revertants from the HLA class II regulatory mutant 6.1.6: implications for the regulation of Ia gene expression. J Immunol. 1984;132:959–962. [PubMed] [Google Scholar]
- 19.Louis-Plence P, Moreno C S, Boss J M. Formation of a regulatory factor X/X2 box-binding protein/nuclear factor-Y multiprotein complex on the conserved regulatory regions of HLA class II genes. J Immunol. 1997;159:3899–3909. [PubMed] [Google Scholar]
- 20.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 21.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 22.Masternak K, Muhlethaler-Mottet A, Villard J, Peretti M, Reith W. Molecular genetics of the bare lymphocyte syndrome. Rev Immunogenet. 2000;2:267–282. [PubMed] [Google Scholar]
- 23.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 24.Mitchell P, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 25.Moreno C S, Emery P, West J E, Durand B, Reith W, Mach B, Boss J M. Purified X2 binding protein (X2BP) cooperatively binds the class II MHC X box region in the presence of purified RFX, the X box factor deficient in the bare lymphocyte syndrome. J Immunol. 1995;155:4313–4321. [PubMed] [Google Scholar]
- 26.Moreno C S, Rogers E M, Brown J A, Boss J M. Regulatory factor X, a bare lymphocyte syndrome transcription factor, is a multimeric phosphoprotein complex. J Immunol. 1997;158:5841–5848. [PubMed] [Google Scholar]
- 27.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 28.Nekrep N, Jabrane-Ferrat N, Peterlin B M. Mutations in the bare lymphocyte syndrome define critical steps in the assembly of the regulatory factor X complex. Mol Cell Biol. 2000;20:4455–4461. doi: 10.1128/mcb.20.12.4455-4461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reith W, Herrero Sanchez C, Kobr M, Silacci P, Berte C, Barras E, Fey S, Mach B. MHC class II regulatory factor RFX has a novel DNA-binding domain and a functionally independent dimerization domain. Genes Dev. 1990;4:1528–1540. doi: 10.1101/gad.4.9.1528. [DOI] [PubMed] [Google Scholar]
- 30.Reith W, Kobr M, Emery P, Durand B, Siegrist C A, Mach B. Cooperative binding between factors RFX and X2bp to the X and X2 boxes of MHC class II promoters. J Biol Chem. 1994;269:20020–20025. [PubMed] [Google Scholar]
- 31.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of mhc expression. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 32.Reith W, Satola S, Herrero Sanchez C, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 33.Reith W, Siegrist C A, Durand B, Barras E, Mach B. Function of major histocompatibility complex class II promoters requires cooperative binding between factors RFX and NF-Y. Proc Natl Acad Sci USA. 1994;91:554–558. doi: 10.1073/pnas.91.2.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reith W, Ucla C, Barras E, Gaud A, Durand B, Herrero Sanchez C, Kobr M, Mach B. RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol. 1994;14:1230–1244. doi: 10.1128/mcb.14.2.1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robbins J, Dilworth S M, Laskey R A, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro D J, Sharp P A, Wahli W W, Keller M J. A high efficiency HELA cell nuclear transcription extract. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 37.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA binding regulatory factor is mutated in primary MHC class II deficiency (Bare Lymphocyte Syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 38.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency. Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 39.Ting J P, Baldwin A S. Regulation of MHC gene expression. Curr Opin Immunol. 1993;5:8–16. doi: 10.1016/0952-7915(93)90074-3. [DOI] [PubMed] [Google Scholar]
- 40.Trowsdale J. Genomic structure and function in the MHC. Trends Genet. 1993;9:117–122. doi: 10.1016/0168-9525(93)90205-v. [DOI] [PubMed] [Google Scholar]
- 41.van den Elsen P J, Peijnenburg A, van Eggermond M C, Gobin S J. Shared regulatory elements in the promoters of MHC class I and class II genes. Immunol Today. 1998;19:308–312. doi: 10.1016/s0167-5699(98)01287-0. [DOI] [PubMed] [Google Scholar]
- 42.Villard J, Lisowska-Grospierre B, Van den Elsen P, Fischer A, Reith W, Mach B. Mutation of RFXAP, a regulator of MHC class II genes, in primary MHC class II deficiency. N Engl J Med. 1997;337:748–753. doi: 10.1056/NEJM199709113371104. [DOI] [PubMed] [Google Scholar]
- 43.Villard J, Peretti M, Masternak K, Barras E, Caretti G, Mantovani R, Reith W. A functionally essential domain of RFX5 mediates activation of major histocompatibility complex class II promoters by promoting cooperative binding between RFX and NF-Y. Mol Cell Biol. 2000;20:3364–3376. doi: 10.1128/mcb.20.10.3364-3376.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Viret C, Janeway C A J. MHC and T cell development. Rev Immunogenet. 1999;1:91–104. [PubMed] [Google Scholar]
- 45.Waldburger J M, Masternak K, Muhlethaler-Mottet A, Villard J, Peretti M, Landmann S, Reith W. Lessons from the bare lymphocyte syndrome: molecular mechanisms regulating MHC class II expression. Immunol Rev. 2000;178:148–165. doi: 10.1034/j.1600-065x.2000.17813.x. [DOI] [PubMed] [Google Scholar]
- 46.Westerheide S D, Boss J M. Orientation and positional mapping of the subunits of the multicomponent transcription factors RFX and X2BP to the major histocompatibility complex class II transcriptional enhancer. Nucleic Acids Res. 1999;27:1635–1641. doi: 10.1093/nar/27.7.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilkinson M. A rapid and convenient method for isolation of nuclear, cytoplasmic and total RNA. Nucleic Acids Res. 1988;16:10934. doi: 10.1093/nar/16.22.10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wright K L, Vilen B J, Itoh-Lindstrom Y, Moore T L, Li G, Criscitiello M, Cogswell P, Clarke J B, Ting J P. CCAAT box binding protein NF-Y facilitates in vivo recruitment of upstream DNA binding transcription factors. EMBO J. 1994;13:4042–4053. doi: 10.1002/j.1460-2075.1994.tb06721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu X S, Linhoff M W, Li G, Chin K C, Maity S N, Ting J P. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]