Abstract
Mutations in the Saccharomyces cerevisiae SNF1 gene affect a number of cellular processes, including the expression of genes involved in carbon source utilization and phospholipid biosynthesis. To identify targets of the Snf1 kinase that modulate expression of INO1, a gene required for an early, rate-limiting step in phospholipid biosynthesis, we performed a genetic selection for suppressors of the inositol auxotrophy of snf1Δ strains. We identified mutations in ACC1 and FAS1, two genes important for fatty acid biosynthesis in yeast; ACC1 encodes acetyl coenzyme A carboxylase (Acc1), and FAS1 encodes the β subunit of fatty acid synthase. Acc1 was shown previously to be phosphorylated and inactivated by Snf1. Here we show that snf1Δ strains with increased Acc1 activity exhibit decreased INO1 transcription. Strains carrying the ACC1 suppressor mutation have reduced Acc1 activity in vitro and in vivo, as revealed by enzymatic assays and increased sensitivity to the Acc1-specific inhibitor soraphen A. Moreover, a reduction in Acc1 activity, caused by addition of soraphen A, provision of exogenous fatty acid, or conditional expression of ACC1, suppresses the inositol auxotrophy of snf1Δ strains. Together, these findings indicate that the inositol auxotrophy of snf1Δ strains arises in part from elevated Acc1 activity and that a reduction in this activity restores INO1 expression in these strains. These results reveal a Snf1-dependent connection between fatty acid production and phospholipid biosynthesis, identify Acc1 as a Snf1 target important for INO1 transcription, and suggest models in which metabolites that are generated or utilized during fatty acid biosynthesis can significantly influence gene expression in yeast.
Cellular responses to environmental signals are often mediated by protein kinases and phosphatases. In Saccharomyces cerevisiae, the Snf1 protein kinase plays a central role in the response to glucose availability. Together with Snf4 and one of three β subunits, Snf1 activates the transcription of glucose-repressed genes under conditions of glucose depletion via the glucose response signal transduction pathway (7, 63). The activity of the Snf1 kinase is itself regulated by the Glc7 phosphatase and its regulatory subunit, Reg1 (36, 48, 61, 77). Snf1 also regulates other events in yeast, including sporulation, glycogen accumulation, peroxisome proliferation, and phospholipid synthesis (8, 67, 69, 75). The mammalian homologue of Snf1, the AMP-activated protein kinase, is activated by environmental conditions that diminish the energy supplies of a cell and raise the AMP-to-ATP ratio (27). Therefore, both the AMP-activated protein kinase and Snf1 have been classified as environmental sensors for eukaryotic cells (26–28). The yeast and mammalian proteins have at least one common substrate, since both can directly phosphorylate and inactivate acetyl coenzyme A (acetyl-CoA) carboxylase (Acc1), the enzyme that catalyzes the rate-limiting step in fatty acid biosynthesis (51, 81).
Snf1 is thought to regulate gene expression by at least two mechanisms. First, Snf1 can directly phosphorylate and alter the activities of gene-specific transcriptional activators and repressors (7). In response to low glucose levels, Snf1 phosphorylates the Mig1 transcriptional repressor, a protein that binds specifically to the promoters of several glucose-repressed genes (70, 76). This event correlates with the translocation of Mig1 from the nucleus to the cytoplasm (16). Second, several observations suggest that Snf1 directly influences the activity of the RNA polymerase II holoenzyme. Genetic selections for extragenic suppressors of a snf1 mutation identified six SSN (suppressors of snf1) genes that encode components of the Srb-mediator complex (9, 43, 72). The Srb-mediator complex is associated with the carboxy-terminal repeat domain (CTD) of RNA polymerase II and is involved in the response to transcriptional activators and repressors (6). More recently, Snf1 has been shown to interact physically with some members of the Srb-mediator complex (42). In addition, mutations in SNF1 and mutations that truncate the CTD cause similar mutant phenotypes, including inositol auxotrophy and defects in galactose-regulated transcription (32, 38, 55, 62).
The inositol auxotrophy of RNA polymerase CTD mutants correlates with a failure to express the INO1 gene (62). Certain mutants defective in the TATA binding protein (TBP), which is encoded by the SPT15 gene, or in histone acetylation are also impaired in INO1 transcription (3, 21, 59). The INO1 gene encodes inositol 1-phosphate synthase, the enzyme that catalyzes the conversion of glucose 6-phosphate to inositol 1-phosphate (17). In yeast, this reaction is rate limiting for the synthesis of inositol-containing phospholipids when inositol is absent from the growth medium. However, when inositol is present, transcription of the INO1 gene is repressed more than 10-fold by a mechanism that requires the negative regulatory protein Opi1 (31). Expression of the INO1 gene requires the Ino2 and Ino4 transcriptional activators (2, 31, 33, 54) that bind to a repeated element, UASINO, found in the promoters of INO1 and other genes subject to regulation by inositol (10, 11, 23, 30).
Previous studies involving the isolation of suppressors of the inositol auxotrophy conferred by specific ino4 and spt15 alleles resulted in the identification of recessive reg1 mutations and a dominant allele of SNF4, establishing a connection between INO1 expression and members of the glucose response pathway (57, 67). To identify potential targets of Snf1 that are important for INO1 transcription, we performed a genetic selection for mutations that suppress the inositol auxotrophy of snf1Δ strains. This work uncovered a connection between genes involved in fatty acid synthesis, notably ACC1 and FAS1, and the regulation of INO1 transcription by Snf1.
MATERIALS AND METHODS
Genetic methods and media.
Media used for the experiment depicted in Fig. 1 were essentially as described by Shirra and Arndt (67). All other experiments were performed in defined synthetic media containing (+I) or lacking (−I) 75 μM inositol, prepared as described previously (24). In some cases 10 μM inositol was used (+I10). Where noted, media also contained 0.5 mM palmitoleic acid (C16:1) dispersed in 1% Brij 58 (final concentrations). Thus, the synthetic media used for these studies contained various combinations of inositol (I) and palmitoleic acid (C16:1) and are abbreviated as follows: (i) −I −C16:1, (ii) +I −C16:1, (iii)−I +C16:1, (iv) +I +C16:1, (v) +I10 −C16:1, and (vi) +I10 +C16:1. Soraphen A, a gift of A. Freund (BASF, Ludwigshafen, Germany), was added to the media from a 10-mg/ml stock solution in methanol. The FY, KY, and PY strains, described in Table 1, are congenic with FY2, a derivative of S288C (80).
FIG. 1.
Suppressor mutations significantly increase transcription of INO1 in strains containing snf1Δ10. Northern analysis of INO1 transcription is shown. Repressed RNA samples (R) were obtained from cells grown in −I media supplemented with 200 μM inositol. Derepressed RNA samples (DR) were obtained from cells that were grown in 200 μM inositol media, washed, resuspended in −I media supplemented with 10 μM inositol, and harvested after incubation at 30°C for an additional 10 h. Strains used were as follows: PY165 (lanes 1 and 2), PY133 (lanes 3 and 4), PY794 (lanes 5 and 6), PY802 (lanes 7 and 8), and PY803 (lanes 9 and 10). The filter from the upper panel was reprobed for SPT15 mRNA as a control. A representative experiment is shown.
TABLE 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Source or reference |
|---|---|---|
| FY454 | MATa snf4Δ1 his4-912δ lys2-128δ leu2Δ1 ura3-52 | F. Winston |
| KY214 | MATα spt15-328 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 | 3 |
| KY231 | MATα spt15-341 his4-917δ lys2-173R2 leu2Δ1 ura3-52 trp1Δ63 | 3 |
| PY129 | MATa snf1Δ10 his3Δ200 leu2Δ1 ura3-52 ade8 | This study |
| PY130 | MATa snf1Δ10 ura3-52 ade8 | This study |
| PY131 | MATα snf1Δ10 his3Δ200 ura3-52 trp1Δ63 | This study |
| PY132 | MATα snf1Δ10 leu2Δ1 ura3-52 | This study |
| PY133 | MATasnf1Δ10 his3Δ200 ura3-52 | This study |
| PY165 | MATahis3Δ200 leu2Δ1 ura3-52 | This study |
| PY188 | MATα ino2Δ::TRP1 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 | This study |
| PY190 | MATα ino4Δ::LEU2 his3Δ200 leu2Δ1 ura3-52 trp1Δ63 | This study |
| PY199 | MATα ACC1-794 leu2Δ1 ura3-52 trp1Δ63 | This study |
| PY731 | MATasnf1Δ10 sup731 ura3-52 ade8 | This study |
| PY794 | MATasnf1Δ10 ACC1-794 his3Δ200 ura3-52 | This study |
| PY802 | MATa snf1Δ10 opi1-802 his3Δ200 ura3-52 | This study |
| PY803 | MATasnf1Δ10 fas1-803 his3Δ200 ura3-52 | This study |
| YUG37 | MATaura3-52 trp1-63 leu2Δ1::tTA-LEU2 | J. Hegemann |
| AUY009 | MATa tetO7-ACC1 ura3-52 trp1-63 leu2Δ1::tTA-LEU2 | This study |
| ENYFB73-4D | MATα cat1(snf1)::HIS3 his3 leu2-3, 112 ura3-52 trp1-289 MAL3 SUC3 MAL2-8 | D. Entian |
| acc1-2150 | MATaacc1-2150 ade | 50 |
| 479-2A | MATα acc1cs his3-11, 15 leu2-3, 112 ura3-1 trp1-1 ade2-1 can1-100 | 64 |
| YAXU008-3a | MATα lys2 leu2 ura3 trp1 | This study |
| YAXU008-3b | MATα acc1-2150 snf1::HIS3 his3 leu2 ura3 ade | This study |
| YAXU008-3d | MATa snf1::HIS3 his3 lys2 leu2 ura3 trp1 ade | This study |
| YAXU009-6a | MATaacc1cs (479-2A) snf1Δ10 ura3 his3 leu2 trp1 ade2 | This study |
| YAXU015-1a | MATa tetO7-ACC1 snf1Δ10 ura3 his3 leu2Δ1::tTA-LEU2 | This study |
Isolation of extragenic suppressors of snf1Δ10.
Five parental strains, PY129 to PY133, of both mating types and with complementing auxotrophies, were used for the selection of snf1Δ10 suppressors. For each strain, 28 individual colonies were patched to yeast extract-peptone-dextrose (YPD) solid medium and replica plated to medium lacking inositol. Patches were mutagenized with UV radiation of 0 to 1,500 μJoules/cm2 in a Stratalinker (Stratagene). No more than one Ino+ colony was purified from each patch to ensure that all suppressor candidates were independently derived. Following purification, 97 lno+ suppressor strains were obtained. These strains were mated to snf1Δ10 parental strains of the opposite mating type to determine if the suppressor mutations were dominant or recessive. Of the 97 suppressor strains, 79 were found to harbor dominant mutations that suppressed the snf1Δ10 inositol auxotrophy. Genetic crosses followed by tetrad analysis showed that the dominant mutations in three of these strains were tightly linked, and tetrad analysis of crosses with snf1Δ10 parental strains showed that the suppressor mutations were in a single gene (data not shown). One of these dominant suppressor strains, PY794, was selected for further study (see below). The remaining 18 suppressor strains contained recessive or partially recessive mutations. For 16 of these strains, the Ino+ phenotype segregated 2:2 in backcrosses with snf1Δ10 parental strains, indicating that the suppressor mutations affected a single gene. Complementation analysis was complicated by the partially recessive Ino+ phenotype of many of these strains; however, three strains were found to contain clearly noncomplementing suppressor mutations: PY731, PY802, and PY803.
Cloning of suppressor genes.
Because previous work had shown that a mutation in OPI1 can suppress the inositol auxotrophy of a snf1Δ10 strain (67), we tested whether a plasmid containing wild-type OPI1, pPS31 (67), would complement the suppressor mutations in PY731, PY802, or PY803. pPS31 fully reversed the Ino+ phenotype of PY802, suggesting that PY802 contains a mutation in OPI1. This assignment was confirmed by linkage analysis of a cross between PY802 and an opi1Δ::HIS3 strain. The Ino+ phenotype of PY731 was partially reversed by pPS31, and this strain was not selected for further study. pPS31 did not alter the Ino+ phenotype of PY803.
To facilitate identification of the suppressor mutation in PY803, we tested this strain for additional mutant phenotypes. We found that PY803 was unable to grow on YPD medium containing 15 mM caffeine, and this caffeine sensitivity cosegregated with suppression of snf1Δ10. Using a plasmid-based yeast genomic library (67), constructed with the pRS316 vector (68), we cloned the PY803 suppressor gene by complementation of the caffeine and inositol phenotypes. Three complementing plasmids, which contained overlapping sequences from chromosome XI, were isolated. The only open reading frame included on all plasmids was the FAS1 gene, which encodes one of two subunits of fatty acid synthase. Linkage of the PY803 suppressor mutation to FAS1 was confirmed by a cross between PY803 and a strain which contained URA3 integrated at the FAS1 locus.
To identify the dominant suppressor in PY794, a plasmid library of PY794 genomic DNA was constructed in pRS316 (68) using a protocol provided by Craig Thompson (74). Upon transformation into a snf1Δ10 strain, PY133, one plasmid was isolated that conferred an Ino+ phenotype to the strain. pPS65 contains yeast genomic sequences from chromosome XIV, 666087 to 654165 (numbering is per the Saccharomyces Genome Database [http://genome-www.stanford.edu/Saccharomyces]), which includes the gene encoding acetyl-CoA carboxylase, ACC1. A plasmid subclone of pPS65, containing ACC1 sequences as the only complete open reading frame, also suppressed the Ino− phenotype of PY133 (data not shown). In addition, the presence of a mutation in ACC1 was supported by results of biochemical assays of Acc1 activity (see Results) and by linkage of the PY794 suppressor mutation to the chromosomal ACC1 locus (data not shown).
Construction of a conditional ACC1 allele.
A conditional allele of ACC1 under the control of the doxycycline-regulatable tetO7 promoter (22) was constructed as follows. A 2,731-bp integration cassette carrying the tetO7-CYC1 promoter and the kanMX4 marker was generated by PCR using a plasmid template, which carries the tetO7-CYC1 hybrid promoter linked to a kanMX4 marker (J. Hegemann et al., unpublished data). The hybrid primers contained 20 nucleotides homologous to the loxP-kanMX4-loxP-tetO7 region of the template. In addition, the hybrid primers contained 50 nucleotide extensions homologous either to the region 50 bp upstream of the start codon or to the first 50 bp of the coding region of the ACC1 gene. The PCR fragment was transformed into strain YUG37 harboring the tetracycline-controlled transactivator gene (tTA) integrated into the LEU2 locus (Hegemann et al., unpublished data). Transformants were selected based on the kanamycin resistance gene, kanMX, on YPD medium containing 200 μg of G418 (Calbiochem)/ml. Correct integration of the tetO7 promoter was verified by colony PCR. ACC1 expression was reduced by addition of 2 to 100 μg of doxycycline/ml.
Northern hybridization analysis.
Cells were grown at 30°C to a density of 1 × 107 to 2 × 107 cells/ml in the appropriate media and harvested or induced as described in the figure legends (see also Results). Isolation of RNA and Northern analyses were performed as described previously (3). Hybridization probes for INO1, TUB2, SPT15, and ACC1 were prepared from pJH310 (31), pYST138 (71), pDE32-1 (18), and YEp352-ACC1 (66), respectively, using a nick translation kit (Roche) or PCR.
Phospholipid analysis.
Wild-type and snf1Δ10 cells grown to mid-logarithmic phase in synthetic medium containing 1% Brij 58 and 75 μM inositol were harvested and washed with sterile water. Each strain was used to inoculate four cultures at an optical density at 600 nm (OD600) of ≈0.5 in the following media: −I −C16:1, +I −C16:1, −I +C16:1, and +I +C16:1. The strains were grown for 1 h at 30°C, at which time 10 μCi of [32P]H3PO4/ml was added to the medium. Following 20 min of labeling, the cells were harvested, suspended in 5% trichloroacetic acid, and placed on ice for 30 min. Lipids were extracted (4), individual phospholipid species were resolved by two-dimensional paper chromatography (73), and phospholipids were quantified by PhosphorImager analysis.
β-galactosidase assays.
Strains were transformed to uracil prototrophy with a plasmid (pJH359) bearing an INO1-CYC1-lacZ fusion (47). Wild-type and snf1Δ10 cells grown to mid-logarithmic phase in synthetic medium containing 1% Brij 58 and 75 μM inositol were harvested and washed with sterile water. Each strain was used to inoculate two cultures at an OD600 of ≈0.2 in −I −C16:1 medium and −I +C16:1 medium. At various times, aliquots of the cultures were removed and assayed for β-galactosidase activity using the Pierce Chemical Company yeast β-galactosidase assay kit. Units of β-galactosidase activity were calculated with the formula A420 × 1,000/(min × ml × OD600).
Acc1 activity determination.
Due to the significant background signal in the standard Acc1 activity assay (44), the enzyme was purified from cytosolic fractions by means of biotin-avidin affinity chromatography as follows. Cells were harvested at 4,000 × g for 10 min, washed with 0.1 M K-PO4 buffer (pH 6.5), mixed with breaking buffer (50 mM Tris-HCl, 100 mM NaF, 1 mM EDTA, 10 mM β-mercaptoethanol, 0.25 M sucrose, 1 mM phenylmethylsulfonyl fluoride [pH 7.5]) and glass beads (0.30-mm diameter) in a ratio of 1:1:1 (wt/vol/wt) and disrupted by four 1-min bursts in a Braun-Melsungen homogenizer under CO2 cooling. Afterwards, the homogenate was centrifuged at 20,000 × g for 20 min. The supernatant (equal amounts of total protein for the various preparations) was loaded onto a preconditioned avidin column (200 μl; Pierce, Inc.), and unbound protein was eluted with 10 ml of phosphate-buffered saline buffer (0.1 M K-PO4 [pH 7.2], 0.15 M NaCl). Acc1 and other biotin enzymes (e.g., pyruvate carboxylase) were eluted with 4 ml of phosphate-buffered saline buffer containing 2 mM biotin (Pierce). All steps were carried out at 4°C. Activity of enriched acetyl-CoA carboxylase was determined using a photometric assay in a coupled enzymatic reaction as described previously (49) immediately after chromatography. All enzyme measurements were carried out at 24°C.
RESULTS
Isolation of suppressors of the inositol auxotrophy of snf1Δ10 strains.
Previously, in two independent studies, we uncovered a role for members of the glucose response pathway and the Snf1-Snf4 kinase complex in the regulation of INO1 transcription (57, 67). However, no substrate was identified for the kinase. The study by Shirra and Arndt (67) suggested that the Opi1 regulatory factor might be a target. However, mutant analyses showed that Opi1 could not be the only target relevant to inositol regulation (67). To further analyze the connection between the Snf1 kinase and INO1 transcription, we performed a genetic selection to identify extragenic suppressors of the inositol auxotrophy of snf1Δ10 mutant strains (see Materials and Methods for details). We reasoned that we might isolate mutations in other negative regulators of INO1 transcription, which might be direct targets of the kinase. Standard cloning procedures, followed by linkage tests, indicated that we isolated recessive suppressor mutations in the OPI1 and FAS1 genes and a dominant mutation in the ACC1gene.
Isolation of an opi1 mutant in a screen for snf1Δ suppressors was expected, as opi1 mutations had previously been shown to suppress the inositol auxotrophy of snf1Δ strains (67). The identification of mutations in FAS1 and ACC1 was more surprising. FAS1 encodes the β subunit of the heteromeric fatty acid synthase enzyme, and ACC1 encodes acetyl-CoA carboxylase (79). Both enzymes are involved in the synthesis of long-chain fatty acids from acetyl-CoA. Importantly, Acc1, in both yeast and mammals, is known to be a direct target of the Snf1 kinase, and phosphorylation by Snf1 inactivates purified Acc1 in vitro (26, 27, 51, 81).
The snf1Δ suppressors restore INO1 transcription.
To determine whether the suppressor mutations act at the level of INO1 transcription, Northern analysis on the parental and double-mutant strains was performed (Fig. 1). Compared to a wild-type strain, the snf1Δ10 strain showed a 3.5-fold-lower level of INO1 transcription under the derepressing conditions used here (Fig. 1, compare lanes 2 and 4). The level of INO1 mRNA in the wild-type strain is probably underestimated, because wild-type strains reach growth saturation during the induction and INO1 is repressed in stationary phase (37). All three suppressor mutations, ACC1-794, opi1-802, and fas1-803, conferred a high level of INO1 transcription in strains containing the snf1Δ mutation (Fig. 1, lanes 6, 8, and 10). As expected from previous studies on OPI1, the opi1-802 strain transcribed INO1 even in the presence of high levels of inositol (Fig. 1, lane 7). Therefore, the suppressor mutations restore the ability of snf1Δ10 strains to grow on medium lacking inositol by increasing INO1 transcription.
Suppression by ACC1-794 is specific to mutations in SNF1 and SNF4.
To determine if the suppression of snf1Δ10 by a mutation in ACC1 is specific to the Snf1 kinase pathway, we investigated whether ACC1-794 could also suppress the inositol auxotrophy caused by other mutations. We chose to examine two mutations in the SPT15 gene, which encodes the general transcription factor TBP. The inositol auxotrophy conferred by these mutations, spt15-328 and spt15-341, was previously shown to be suppressed by a dominant mutation in SNF4, SNF4-204, which enhances the physical interaction between Snf1 and Snf4 (67). We also examined null mutations that remove the transcriptional activators of INO1, Ino2, and Ino4 (2, 31, 33, 54). We transformed the strains with a plasmid containing the dominant ACC1 suppressor mutation. As a control, we also tested suppression by the dominant mutation, SNF4-204. Table 2 shows that ACC1-794 only suppressed mutations in the Snf1-Snf4 pathway, suggesting that the suppression mechanism is specific to Snf1 and requires additional signals supplied by TBP and the Ino2 and Ino4 transcription factors. Furthermore, the strong suppression of the TBP mutants by SNF4-204 suggests an additional role for the Snf1 kinase in INO1 transcription that is independent of its function as an inhibitor of Acc1.
TABLE 2.
Specificity of suppression by ACC1-794
| Relevant genotypea | Growth withb:
|
||
|---|---|---|---|
| Vector | ACC1-794c | SNF4-204d | |
| snf1Δ10 | − | + | − |
| snf4Δ1 | − | + | + |
| spt15-328 | − | −/+ | + |
| spt15-341 | − | −−/+ | + |
| ino2Δ | − | − | − |
| ino4Δ | − | − | − |
The following yeast strains were tested: PY133, FY454, KY214, KY231, PY188, and PY190.
Symbols indicate relative growth on solid media lacking inositol after 3 days at 30°C. Strong, weak, very weak, and no growth are indicated by +, −/+, −−/+, and −, respectively.
pPS65 contains ACC1-794 sequences (see Materials and Methods).
pPS47 contains SNF4-204 sequences (67).
Suppression of the snf1Δ Ino− phenotype by ACC1 mutations is allele specific.
Mutants with defects in Acc1 have been isolated in genetic screens involving diverse phenotypes. While the ACC1-794 allele reported here was isolated as a suppressor of the Ino− phenotype of a snf1Δ mutant, the recessive, cold-sensitive allele, acc1cs, was identified in a screen for mutations that are synthetically lethal with the hyperrecombination mutant, hpr1 (64). In addition, a temperature-sensitive allele of ACC1, mtr7, was isolated as a mutation affecting mRNA transport out of the nucleus (66), and acc1-2150 is a conditional fatty acid auxotroph (50).
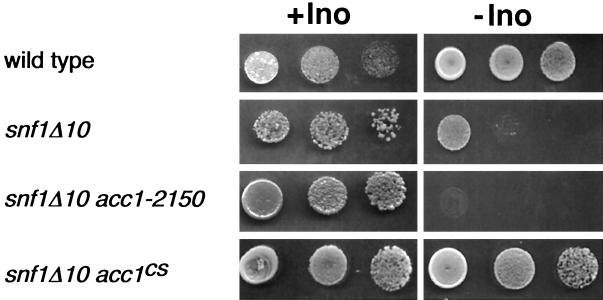
To test the allele specificity of snf1Δ suppression by acc1 alleles, double mutants were constructed by standard genetic crosses. As shown in Fig. 2, the cold-sensitive, recessive acc1cs allele, like the dominant ACC1-794 allele, suppressed the inositol auxotrophy of snf1Δ mutants. However, the acc1-2150 (Fig. 2) and mtr7 alleles (data not shown) did not suppress the snf1Δ phenotype. Therefore, acc1 mutations suppress the inositol auxotrophy of snf1Δ mutants in an allele-specific fashion.
FIG. 2.
Suppression of the snf1Δ10 Ino− phenotype by ACC1 mutations is allele specific. Strains grown overnight in inositol-containing medium were harvested, washed, normalized by OD600, and spotted onto plates in a series of three 10-fold dilutions. All media contained 1% Brij 58 and either contained (+Ino) or lacked (−Ino) 75 μM inositol. Plates were allowed to grow for 3 days at 30°C. Strains used were as follows: YAXU008-3a, YAXU008-3d, YAXU008-3b, and YAXU009-6a.
Suppression of the inositol auxotrophy of the snf1Δ10 mutant results from inactivation of acetyl-CoA carboxylase.
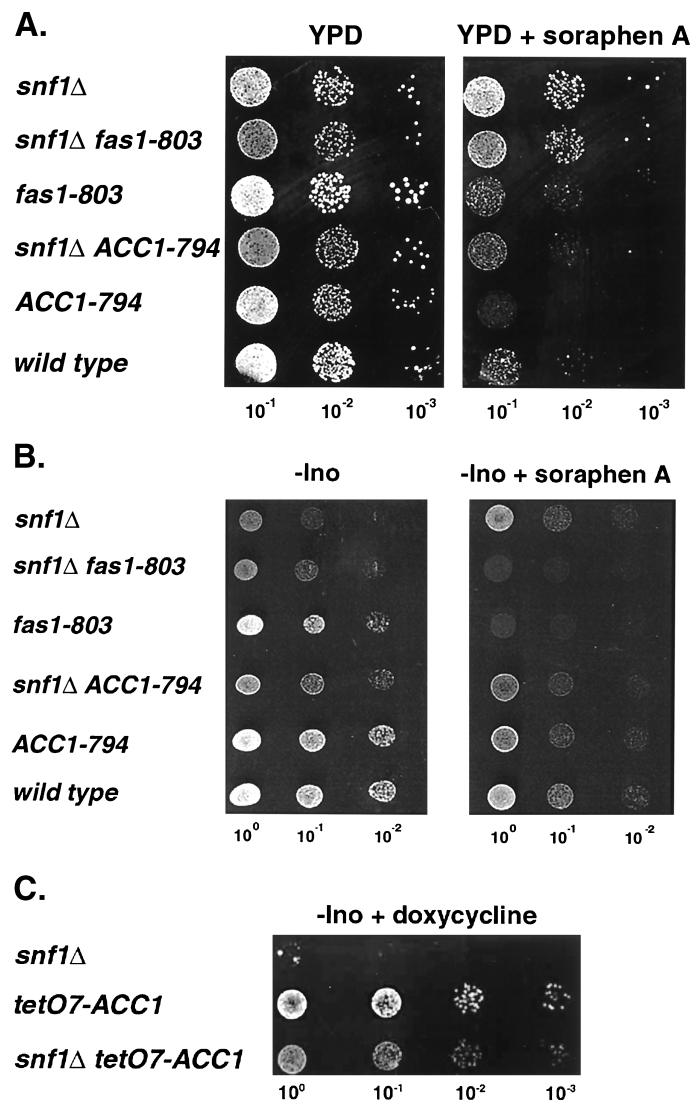
The observation that a recessive, loss-of-function mutation such as acc1cs could suppress the snf1Δ10 mutation suggested that the mechanism of suppression might involve inactivation of acetyl-CoA carboxylase activity, although the ACC1-794 suppression phenotype is dominant. To determine whether inactivation of Acc1 correlates with suppression of the inositol auxotrophy of the snf1Δ mutant, we employed soraphen A, a potent inhibitor of Acc1 activity (78). Cells with elevated Acc1 activity are more resistant to this compound than wild-type cells, while cells with lowered Acc1 activity are more sensitive (64, 65). Relative to a wild-type strain, a snf1Δ strain was significantly more resistant to soraphen A, suggesting an increase in Acc1 activity in the mutant strain (Fig. 3A). The ACC1-794 strain was even more sensitive to soraphen A than the wild type, indicating reduced Acc1 activity, while growth of the fas1-803 mutant was comparable to that of the wild type. The ACC1-794 snf1Δ double-mutant strain exhibited a sensitivity to soraphen A that was intermediate to those of strains containing either single mutation. The fas1-803 snf1Δ10 strain, on the other hand, exhibited a sensitivity to the drug that was comparable to that of the snf1Δ parent.
FIG. 3.
The effect of soraphen A and reduced ACC1 expression on the growth and inositol auxotrophy of snf1Δ10 strains. Yeast cultures, grown overnight in YPD, were diluted in sterile water to the OD600 indicated at the bottom of each lane, and 5-μl samples were spotted onto the following media: YPD and YPD plus 0.25 μg of soraphen A/ml (A); -Ino and -Ino plus 0.25 μg of soraphen A/ml (B); and -Ino + 2 μg of doxycycline/ml (C). The following yeast strains were tested: PY133, PY803, PY170, PY794, PY199, PY165, AUY009, and YAXU015-1a.
Addition of soraphen A to plates lacking inositol partially reversed the Ino− phenotype of snf1Δ strains (Fig. 3B). Since soraphen A is highly specific for Acc1, these findings suggest that a reduction in Acc1 activity suppresses the inositol auxotrophy of snf1Δ strains. On plates lacking inositol, the presence of the fas1 mutation presumably results in reduced production of fatty acids, rendering the cells unable to grow if the flux through the fatty acid synthesis pathway is further reduced by inhibiting Acc1 (Fig. 3B).
To further test the correlation between acetyl-CoA carboxylase activity and snf1Δ suppression, a conditional allele of ACC1 was constructed by replacing the endogenous promoter with the regulatable tetO7 promoter (see Materials and Methods) (22). In this construct, ACC1 expression levels can be modulated by the addition of doxycycline, which interacts with the expression-activation system and results in repression. Acc1 is an essential enzyme, and addition of 50 μg of doxycycline/ml to YPD media completely abolished growth of haploid strains harboring the tetO7-ACC1 allele (data not shown). By addition of limiting amounts of doxycycline (2 μg/ml), the Ino− phenotype conferred by snf1Δ was suppressed on plates lacking inositol (Fig. 3C). Taken together, these in vivo results strongly suggest that the inositol auxotrophy of snf1Δ strains arises, in part, from an elevation in Acc1 enzyme activity and that a reduction in Acc1 activity, caused by mutation, drug inactivation, or conditional expression, suppresses this phenotype.
Supplementation with fatty acid suppresses the inositol auxotrophy and INO1 transcriptional defect of snf1Δ strains.
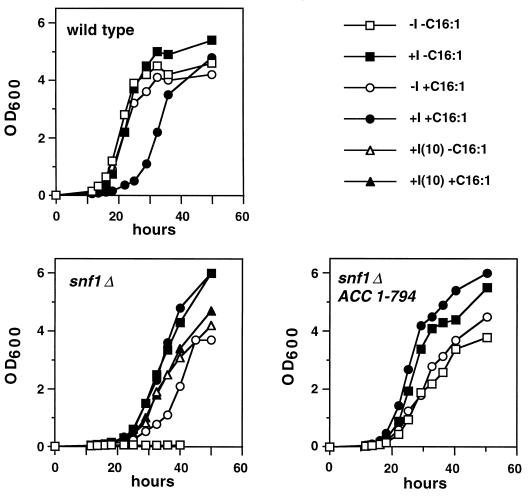
In mammalian cells, fatty acyl-CoAs are known to inhibit Acc1 activity (53, 56), and in S. cerevisiae the addition of exogenous fatty acids to the medium inhibits both Acc1 and fatty acid synthase activity (12) through a mechanism that appears to require acyl-CoA synthase activity (19, 39). Therefore, we asked whether addition of exogenous fatty acids to medium lacking inositol would restore growth of a snf1Δ10 strain. As shown in Fig. 4, supplementation of medium with 0.5 mM palmitoleic acid (C16:1) allowed growth of the snf1Δ mutant strain (lower left panel). In medium that contained C16:1 and lacked inositol, the snf1Δ10 strain grew similarly to the snf1Δ10 ACC1-794 strain in the absence of inositol (Fig. 4, lower panels). However, the snf1Δ strain exhibited a slightly longer lag time and did not grow to as high a density in −I +C16:1 medium as in +I −C16:1 medium (Fig. 4, lower left panel).
FIG. 4.
Palmitoleic acid suppresses the inositol auxotrophy of snf1Δ10 strains. Strains grown overnight in inositol-containing medium (+I −C16:1) were harvested, washed, and used to start liquid cultures at an OD600 of 0.01 for growth at 30°C. All media contained 1% Brij 58 and the indicated combinations of inositol (+I, 75 μM; −I, 0 μM) and palmitoleic acid (+C16:1, 0.5 mM; −C16:1, 0 mM). In the case of the snf1Δ10 mutant, the media contained 10 μM inositol instead of 0 μM inositol. Strains used were as follows: SNF1 (PY165), snf1Δ10 (PY133), fas1-803 (PY170), ACC1-794 (PY199), snf1Δ10 fas1-803 (PY803), and snf1Δ10 ACC1-794 (PY794).
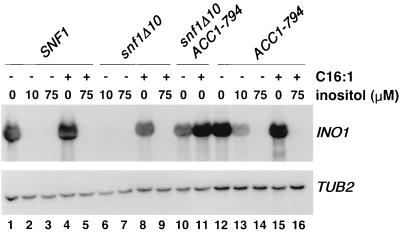
To test whether the growth effect of fatty acid supplementation correlated with an increase in INO1 transcription in snf1Δ strains, Northern analysis was performed. As shown in Fig. 5, addition of palmitoleic acid (C16:1) to medium lacking inositol restored INO1 transcription in snf1Δ10 strains (lane 8). Addition of C16:1 to the growth medium of the snf1Δ10 ACC1-794 strain also increased INO1 transcription (Fig. 5, lanes 10 and 11). However, this higher level of INO1 transcription was no greater than that seen in strains containing ACC1-794 alone, in the presence or absence of exogenous C16:1 (Fig. 5, lanes 12 and 15).
FIG. 5.
Fatty acid supplementation supports a high level of INO1 transcription in snf1Δ10 strains. Northern analysis of INO1 transcription is shown. Cells were grown in media containing 1% Brij 58 detergent in the presence or absence of 0.5 mM palmitoleic acid (C16:1) and the indicated concentrations of inositol. Cells were harvested at a cell density of 1 × 107 to 2 × 107 cells/ml. Strains used were as follows: PY165 (lanes 1 to 5), PY133 (lanes 6 to 9), PY794 (lanes 10 to 11), and PY199 (lanes 12 to 16). The filter from the upper panel was reprobed for TUB2 mRNA as a control. A representative experiment is shown.
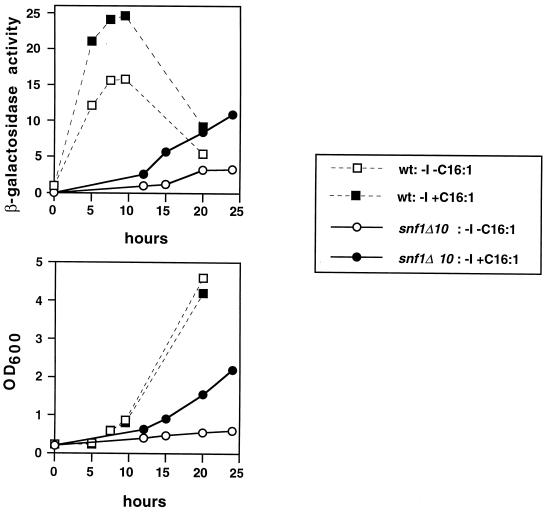
To analyze the kinetics of INO1 derepression in snf1Δ and wild-type strains under conditions of fatty acid supplementation, we employed a strain carrying an INO1-CYC1-lacZ fusion. Cells carrying this fusion were grown in inositol-containing medium (+I −C16:1), shifted to inositol-free medium, and harvested for β-galactosidase assays. As expected, the snf1Δ10 mutant strain was unable to derepress INO1-CYC1-lacZ in inositol-free medium, while the wild-type strain exhibited rapid derepression, as previously reported (58). The snf1Δ10 mutant, however, was able to induce INO1 expression in inositol-free medium when palmitoleic acid was supplied (−I +C16:1). However, the kinetics of induction were not as rapid as those observed for the wild-type strain shifted to −I medium, with or without fatty acid. The increase in β-galactosidase expression driven by the INO1 promoter with the snf1Δ strain grown in −I +C16:1 medium paralleled the increase in optical density of the culture (Fig. 6, lower panel). The wild-type strain also exhibited greater lacZ induction when transferred to −I +C16:1 medium than when transferred to −I −C16:1 medium. Thus, addition of C16:1 fatty acid appears to result in an increase in INO1 transcription in both wild-type and snf1Δ10 cells.
FIG. 6.
Effect of palmitoleic acid on the kinetics of INO1 derepression. Strains bearing the plasmid pJH359 (INO1-CYC1-lacZ) (47) were grown to mid-logarithmic phase in synthetic medium containing 1% Brij 58 and 75 μM inositol. Following harvesting and washing, each strain was used to inoculate two different media (−I −C16:1 and −I +C16:1) at an OD600 of ≈0.2. At various times, aliquots of the cultures were removed and assayed for β-galactosidase activity. Data represent the averages of results of two independent experiments. Strains used were as follows: SNF1 (PY165), snf1Δ10 (PY133), and snf1Δ10 ACC1-794 (PY794). The apparent decrease in β-galactosidase activity [A420 × 1,000/(min × ml × OD600)] in the wild-type culture at the 20-h time point is a reflection of the strain's continued growth, once its β-galactosidase activity has reached a plateau level.
Inhibition of Acc1 enzyme activity correlates with suppression of the snf1Δ inositol auxotrophy.
To confirm data obtained from our in vivo studies, Acc1 activity and protein levels, as well as ACC1 steady state mRNA levels, were determined. Total activity of Acc1 isolated from a snf1Δ10 strain was elevated approximately threefold relative to that of Acc1 isolated from a SNF1+ strain (Fig. 7A), and this activity from snf1Δ10 cells was more resistant to soraphen A in vitro (data not shown). However, ACC1 mRNA and protein levels (Fig. 7C and data not shown) were lower in the snf1Δ mutant strain, suggesting significantly increased specific activity (5- to 7-fold) of acetyl-CoA carboxylase if it remains unphosphorylated by the Snf1 kinase. The level of acetyl-CoA carboxylase activity was lower in the ACC1-794 mutant (Fig. 7A) despite increased ACC1 expression (Fig. 7C) and about fourfold-higher levels of Acc1 protein (data not shown). These data suggest that Acc1 activity controls an autoregulatory loop, leading to reduced expression of ACC1 in a snf1Δ strain where Acc1 activity is stimulated by a lack of phosphorylation. Conversely, increased ACC1 expression is observed in the ACC1-794 strain, which has impaired enzymatic activity.
FIG. 7.
Acc1 enzyme activity and ACC1 expression in the absence and presence of exogenous palmitoleic acid. (A and B) Acc1 enzyme activity was determined as described in Materials and Methods. The activity was determined three to four times and normalized to the protein concentration in the homogenate, and it is depicted as specific activity relative to activity of a wild-type strain grown in the absence of exogenous C16:1 (set at 100%). For the experiment depicted in panel B, C16:1 was added to the growth media to a final concentration of 100 μM without detergent (detergent was found to interfere with the enzyme preparation and resulted in a loss of Acc1 activity). (C) Northern analysis of ACC1 expression. Total RNA was prepared 0 and 4 h after addition of C16:1 (100 μM, where indicated), separated on denaturing agarose gels, blotted, and hybridized with digoxigenin-labeled ACC1 and PMA1 probes. Strains used were as follows: PY133, PY803, PY170, PY794, PY199, and PY165.
Supplementation of the growth medium of the snf1Δ strain with fatty acid (C16:1; 100 μM without detergent) resulted in a reduced level of Acc1 activity, from a level threefold higher than that of the wild type to a level 1.4-fold higher than that of the wild type (Fig. 7B). This reduction in Acc1 activity by more than 50% in the snf1Δ and wild-type strains grown in the presence of fatty acid was comparable to the relative drop in ACC1 expression under these conditions (Fig. 7C).
The phospholipid composition of snf1Δ cells does not reflect the state of INO1 expression.
Changes in the pattern of phospholipid synthesis have been implicated in the mechanism of INO1 derepression (30). Wild-type cells grown in media lacking inositol exhibit increased synthesis of phosphatidic acid (PtdOH) and CDP diacylglycerol (CDP-DAG) and decreased synthesis of phosphatidylinositol (PtdIns) compared to the same cells grown in media containing inositol (5, 40). By pulse labeling phospholipids with 32P, we found that both wild-type and snf1Δ10 strains displayed elevated synthesis of PtdOH and CDP-DAG following transfer to −I media, whether or not C16:1 was present (data not shown). Thus, neither the inositol auxotrophy of snf1Δ10 strains nor the suppression of this phenotype by C16:1 appears to be correlated to alterations in phospholipid synthesis. These data suggest that Snf1 may act downstream of or independently of the signal produced through phospholipid metabolism to affect INO1 transcription.
DISCUSSION
Previous selections for extragenic suppressors of snf1 mutations relied on the inability of snf1 mutants to derepress glucose-repressible genes such as SUC2 (9, 43, 72). These studies resulted in the isolation of mutations in components of the Srb-mediator complex associated with RNA polymerase II. In contrast, we selected for suppressors of the inositol auxotrophy conferred by a snf1Δ mutation and identified components of the fatty acid biosynthetic pathway. We have shown that the inositol auxotrophy of snf1Δ cells, which correlates with decreased expression of the INO1 gene, is suppressed by mutations in genes encoding acetyl-CoA carboxylase (ACC1) and a subunit of fatty acid synthase (FAS1) as well as by provision of exogenous fatty acid. The mutants isolated in the present study define a role in yeast for fatty acid biosynthesis in metabolic signaling and a role for the Snf1 kinase in controlling lipid metabolism. Moreover, we have identified acetyl-CoA carboxylase as a target of the Snf1 kinase that is relevant to transcriptional regulation of phospholipid biosynthesis.
Snf1 is necessary for expression but not regulation of INO1.
Analysis of the pattern of INO1 expression in diverse genetic backgrounds including mutants with defects in phospholipid metabolism supports the hypothesis that PtdOH, or a closely related lipid, generates a signal that results in derepression of UASINO-containing genes such as INO1 (10, 30). Since fatty acids are immediate precursors of PtdOH (Fig. 8) and we have identified a role for fatty acid metabolism as a target for Snf1 signaling, we considered the possibility that Snf1 might transmit the inositol-sensitive signal controlling INO1 expression. However, two lines of evidence, presented here, suggest that this is not the case. First, the response to inositol deprivation is believed to be initiated by a shift in the pattern of phospholipid metabolism that includes increased accumulation of PtdOH and CDP-DAG and decreased synthesis of PtdIns (40). However, the snf1Δ mutant exhibited a pattern of phospholipid synthesis comparable to that of wild-type cells when shifted to inositol-free medium, and this pattern was unaffected by the addition of fatty acid. Second, when INO1 expression is restored in snf1Δ cells by provision of fatty acid (16:1) or by introduction of the ACC1-794 or fas1-803 mutations, regulation in response to inositol is also restored. Thus, an active Snf1 kinase is not needed to transmit the inositol-sensitive signal, and, furthermore, the presence or absence of an active SNF1 gene product does not seem to influence the pattern of phospholipid synthesis that is believed to be involved in the signaling. Thus, the Snf1 kinase appears to affect the overall level of INO1 expression rather than its regulation in response to inositol or phospholipid metabolism.
FIG. 8.
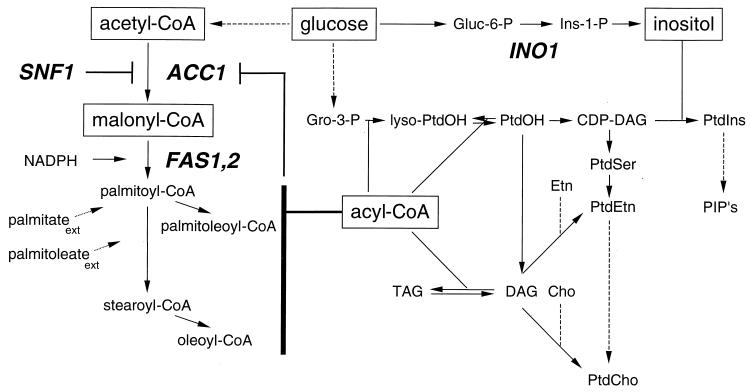
Schematic diagram of phospholipid biosynthesis in S. cerevisiae. Solid arrows indicate direct enzymatic conversions. Dashed arrows indicate conversions that require more than one enzymatic step. Gene designations are in bold italics. Phosphorylation of Acc1 by the SNF1 gene product inhibits Acc1 activity. Acyl-CoAs, including malonyl-CoA, palmitoyl-CoA, palmitoleoyl-CoA, stearoyl-CoA, and oleoyl-CoA, inhibit Acc1 activity. Externally added palmitate (palmitateext) and palmitoleate (palmitoleateext) are converted to their respective CoA derivatives in the cell. Lyso-PtdOH, lysophosphatidic acid; Gro-3-P, glycerol-3-phosphate; Gluc-6-P, glucose-6-phosphate; Ins-1-P, inositol-1-phosphate; PtdOH, phosphatidic acid; DAG, diacylglycerol; TAG, triacylglycerol; Etn, ethanolamine; Cho, choline; PtdIns, phosphatidylinositol; PtdSer, phosphatidylserine; PtdEtn, phosphatidylethanolamine; PtdCho, phosphatidylcholine; PIP's, polyphosphoinositides.
Since enzymes involved in fatty acid biosynthesis clearly play a role in Snf1-dependent transcription of INO1, we considered whether INO1 expression might correlate with fatty acid composition. The snf1Δ mutant had a slightly lower proportion of C16:0 fatty acids than the wild type, but so did the fas1 mutant, whether or not the snf1 mutation was present (unpublished observations). Cells carrying the ACC1-794 mutation, like previously described acc1 mutants (65, 66), exhibited an increased proportion of 16 carbon fatty acids, whether or not the snf1 mutation was also present. Thus, there was no clear correlation between the cellular fatty acid composition and the ability to express INO1 in the snf1 genetic background, which, however, does not exclude potential effects of specifically localized altered lipid species.
Identification of Acc1 as a Snf1 substrate important for INO1 expression.
Initially, it seemed paradoxical that mutations that presumably reduce the rate of fatty acid biosynthesis and provision of exogenous fatty acid had similar effects: namely, suppression of the inositol auxotrophy of snf1Δ cells. However, this apparent paradox was resolved with the recognition of a correlation between INO1 expression and total cellular activity of acetyl-CoA carboxylase. Our first indication of such a relationship came from the growth properties of various yeast strains in the presence of soraphen A, a drug known to inhibit Acc1 specifically (78). As expected, snf1Δ cells, previously reported to have high levels of Acc1 activity (81), were more resistant to soraphen A than wild-type cells. The ACC1-794 mutation increased the soraphen A sensitivity of both SNF1+ and snf1Δ cells. Acc1 activity levels predicted by these phenotypic results were confirmed by enzyme assays.
In yeast, Acc1 is known to be inactivated by the Snf1 kinase (51, 81), and a similar regulatory relationship exists in mammalian cells (15, 52). In the snf1Δ ACC1-794 double mutant strain, the absence of Snf1-dependent phosphorylation of Acc1 increases the activity of the Acc1-794 mutant enzyme to a level comparable to that of the wild type. The ACC1-794 allele clearly reduces Acc1 function despite its apparent dominance as a suppressor of the snf1Δ Ino− phenotype. We propose, therefore, that suppression of the snf1Δ inositol auxotrophy is due to a partial loss of Acc1 function. In support of this conclusion, we have shown that the acc1cs allele, another partial loss-of-function mutation, and reduced expression of ACC1 from a doxycycline-repressible promoter also suppress the snf1Δ inositol auxotrophy.
The provision of exogenous fatty acids also lowers Acc1 activity in wild-type and snf1Δ cells, which is at least in part due to repression of ACC1 expression. Acyl-CoA, the end product of fatty acid synthesis, is known to inhibit mammalian acetyl-CoA carboxylase activity in vitro (53, 56). Kamiryo et al. reported that exogenous fatty acid caused a reduction of Acc1 activity in yeast and, furthermore, that activation of exogenous fatty acid to acyl-CoA was necessary for the reduction in Acc1 activity (39). We found that the level of Acc1 activity in snf1Δ cells grown in the presence of exogenous 16:1 fatty acid is reduced to a level almost comparable to that for the wild-type strain grown in the absence of added fatty acid. Under these conditions, INO1 is expressed. Indeed, in each case (the presence of the ACC1-794 mutation or the provision of fatty acid), the ability of cells to express INO1 is correlated with a reduction of Acc1 activity to a level comparable to or lower than that found in wild-type cells grown in the absence of exogenous fatty acid.
The nature of the Snf1-dependent signal controlling INO1 expression.
Our analysis of the fas1-803 suppressor mutation presents a potential contradiction to the hypothesis that the ability to express INO1 is correlated with Acc1 activity. The fas1-803 strain exhibited Acc1 activity levels comparable to wild-type levels in vitro, despite the ability of the fas1-803 mutation to suppress the Ino− phenotype conferred by snf1Δ10. Consistent with this observation, the fas1 mutant does not appear to have increased soraphen A sensitivity. The fas1-803 snf1Δ10 mutant exhibits resistance to the drug that is, at best, only slightly reduced compared to that of the snf1Δ10 strain.
These observations raise the possibility that the actual basis of suppression may not be reduction of Acc1 enzyme activity, per se, but rather may be related to the overall flux of metabolites through the fatty acid biosynthetic pathway. The step catalyzed by Acc1 is rate limiting for fatty acid biosynthesis in wild-type yeast cells (50, 60) and in mammalian cells (25). In snf1Δ cells which exhibit elevated Acc1 activity, a mutated Fas1 subunit might cause the step catalyzed by fatty acid synthase to become rate limiting. Overall, our results support the hypothesis that Acc1 either is directly involved in the mechanism by which Snf1 controls INO1 expression or exerts its influence by affecting the flux of metabolites through the fatty acid biosynthetic pathway. Interestingly, ACC1 expression itself is repressed by inositol in the growth medium through a regulatory circuit that involves the Ino2 and Ino4 transcription factors as well as Opi1, all of which also control INO1 regulation (13, 29).
We favor the idea that the level of a metabolite(s) produced or utilized in fatty acid biosynthesis, an energy-demanding process, is responsible for generating a signal which affects INO1 transcription. High levels of energy-rich metabolites may favor INO1 transcription, while low levels may tend to repress INO1 expression. Recent reports demonstrating that certain histone deacetylases, including those encoded by SIR2 and its homologues (35, 45), require NAD+ suggest the possibility that metabolic factors affecting the NAD+ levels may globally affect patterns of gene expression through influencing chromatin structure. Malonyl-CoA levels might also serve as a metabolic sensor, as they have been postulated to do in mammalian cells, possibly through inhibition of Acc1 or by triggering other metabolic signals that may influence cellular energy levels and affect chromatin modification. Interestingly, a link between Acc1 activity and expression of another gene, PHO5, has been reported (46). In this case, constitutive PHO5 expression was observed in several acc1 mutant strains, and the authors also concluded that a metabolite(s) of fatty acid biosynthesis might serve as a signaling molecule for transcriptional regulation of this gene (46).
Acetyl-CoA, another metabolite that could potentially affect INO1 expression, serves as a substrate for both fatty acid biosynthesis and histone acetylation. Since acetyl-CoA carboxylase uses acetyl-CoA directly as a substrate, high levels of Acc1 activity might deplete the pools of acetyl-CoA normally reserved for histone acetylation. Transcription of the INO1 gene is known to be sensitive to mutations that affect histone acetyltransferase and histone deacetylase complexes. Mutations in SIN3, which encodes a component of the Sin3-Rpd3 histone deacetylase complex, lead to high levels of INO1 expression (34). In contrast, mutations that remove certain components of the SAGA histone acetyltransferase cause inositol auxotrophy and a severe defect in INO1 activation (21, 59). Because histone acetylation is required for the recruitment of certain transcriptional activators (14, 41) and ultimately for the recruitment of TBP (1) to promoters, metabolic changes that influence this process could lead to dramatic effects on gene regulation. Recently, acetyl-CoA has been shown to stimulate promoter binding by TFIID in vitro, suggesting another mechanism by which Acc1 may regulate transcription (20). Significantly, we initially uncovered a role for the Snf1 kinase pathway in INO1 transcription by searching for suppressors of a mutant Ino4 activator protein and a DNA binding-defective TBP (57, 67). Continued genetic and biochemical studies will help elucidate how the Snf1 kinase pathway and additional signal transduction cascades control chromatin modification or other events that culminate in activation of INO1 transcription.
ACKNOWLEDGMENTS
The first three authors contributed equally to this work.
We thank A. Tartakoff, H. Klein, E. Schweizer, J. Hegemann, F. Winston, and D. Entian for strains and plasmids; A. Jandrositz and G. Gogg for constructing the tetO-ACC1 strain and G. Gogg for fatty acid analyses and support with enzyme preparations; A. Freund (BASF) for the gift of soraphen A; and K. Roinick for technical assistance.
This work was supported by grants from the National Institutes of Health to K.M.A. (GM52593 and AI01816) and S.A.H. (GM19629) and the Austrian Science Fund, FWF (F706), to S.D.K.
REFERENCES
- 1.Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. doi: 10.1016/s0092-8674(00)00169-0. [DOI] [PubMed] [Google Scholar]
- 2.Ambroziak J, Henry S A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 3.Arndt K M, Ricupero-Hovasse S, Winston F. TBP mutants defective for activated transcription in vivo. EMBO J. 1995;14:1490–1497. doi: 10.1002/j.1460-2075.1995.tb07135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atkinson K D, Ramirez R M. Secretion can proceed uncoupled from net plasma membrane expansion in inositol-starved Saccharomyces cerevisiae. J Bacteriol. 1984;160:80–86. doi: 10.1128/jb.160.1.80-86.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Becker G W, Lester R L. Changes in phospholipids of Saccharomyces cerevisiae associated with inositol-less death. J Biol Chem. 1977;252:8684–8691. [PubMed] [Google Scholar]
- 6.Carlson M. Genetics of transcriptional regulation in yeast: connections to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 7.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 8.Carlson M, Osmond B C, Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson M, Osmond B C, Neigeborn L, Botstein D. A suppressor of snf1 mutations causes constitutive high-level invertase synthesis in yeast. Genetics. 1984;107:19–32. doi: 10.1093/genetics/107.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carman G M, Henry S A. Phospholipid biosynthesis in the yeast Saccharomyces cerevisiae and interrelationship with other metabolic processes. Prog Lipid Res. 1999;38:361–399. doi: 10.1016/s0163-7827(99)00010-7. [DOI] [PubMed] [Google Scholar]
- 11.Carman G M, Henry S A. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–669. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 12.Chirala S S. Coordinated regulation and inositol-mediated and fatty acid-mediated repression of fatty acid synthase genes in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1992;89:10232–10236. doi: 10.1073/pnas.89.21.10232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chirala S S, Zhong Q, Huang W, Al-Feel W. Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression. Nucleic Acids Res. 1994;22:412–418. doi: 10.1093/nar/22.3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 15.Davies S P, Carling D, Munday M R, Hardie D G. Diurnal rhythm of phosphorylation of rat liver acetyl-CoA carboxylase by the AMP-activated protein kinase, demonstrated using freeze-clamping. Effects of high fat diets. Eur J Biochem. 1992;203:615–623. doi: 10.1111/j.1432-1033.1992.tb16591.x. [DOI] [PubMed] [Google Scholar]
- 16.De Vit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donahue T F, Henry S A. Myo-inositol-1-phosphate synthase. Characteristics of the enzyme and identification of its structural gene in yeast. J Biol Chem. 1981;256:7077–7085. [PubMed] [Google Scholar]
- 18.Eisenmann D M, Dollard C, Winston F. SPT15, the gene encoding the yeast TATA binding factor TFIID, is required for normal transcription initiation in vivo. Cell. 1989;58:1183–1191. doi: 10.1016/0092-8674(89)90516-3. [DOI] [PubMed] [Google Scholar]
- 19.Faergeman N J, Knudsen J. Role of long-chain fatty acyl-CoA esters in the regulation of metabolism and in cell signalling. Biochem J. 1997;323:1–12. doi: 10.1042/bj3230001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galasinski S K, Lively T N, Grebe De Barron A, Goodrich J A. Acetyl coenzyme A stimulates RNA polymerase II transcription and promoter binding by transcription factor IID in the absence of histones. Mol Cell Biol. 2000;20:1923–1930. doi: 10.1128/mcb.20.6.1923-1930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gansheroff L J, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 23.Greenberg M L, Lopes J M. Genetic regulation of phospholipid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1996;60:1–20. doi: 10.1128/mr.60.1.1-20.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griac P, Swede M J, Henry S A. The role of phosphatidylcholine biosynthesis in the regulation of the INO1 gene of yeast. J Biol Chem. 1996;271:25692–25698. doi: 10.1074/jbc.271.41.25692. [DOI] [PubMed] [Google Scholar]
- 25.Ha J, Kim K H. Inhibition of fatty acid synthesis by expression of an acetyl-CoA carboxylase-specific ribozyme gene. Proc Natl Acad Sci USA. 1994;91:9951–9955. doi: 10.1073/pnas.91.21.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardie D G, Carling D. The AMP-activated protein kinase—fuel gauge of the mammalian cell? Eur J Biochem. 1997;246:259–273. doi: 10.1111/j.1432-1033.1997.00259.x. [DOI] [PubMed] [Google Scholar]
- 27.Hardie D G, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 28.Hardie D G, Carling D, Halford N. Roles of the Snf1/Rkin1/AMP-activated protein kinase family in the response to environmental and nutritional stress. Semin Cell Biol. 1994;5:409–416. doi: 10.1006/scel.1994.1048. [DOI] [PubMed] [Google Scholar]
- 29.Hasslacher M, Ivessa A S, Paltauf F, Kohlwein S D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- 30.Henry S A, Patton-Vogt J L. Genetic regulation of phospholipid metabolism: yeast as a model eukaryote. Prog Nucleic Acid Res Mol Biol. 1998;61:133–179. doi: 10.1016/s0079-6603(08)60826-0. [DOI] [PubMed] [Google Scholar]
- 31.Hirsch J P, Henry S A. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol Cell Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirschhorn J N, Brown S A, Clark C D, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 33.Hoshizaki D K, Hill J E, Henry S A. The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990;265:4736–4745. [PubMed] [Google Scholar]
- 34.Hudak K A, Lopes J M, Henry S A. A pleiotropic phospholipid biosynthetic regulatory mutation in Saccharomyces cerevisiae is allelic to sin3 (sdi1, ume4, rpd1) Genetics. 1994;136:475–483. doi: 10.1093/genetics/136.2.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Imai S-I, Armstrong C M, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 36.Jiang R, Carlson M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 1996;10:3105–3115. doi: 10.1101/gad.10.24.3105. [DOI] [PubMed] [Google Scholar]
- 37.Jiranek V, Graves J A, Henry S A. Pleiotropic effects of the opi1 regulatory mutation of yeast: its effects on growth and on phospholipid and inositol metabolism. Microbiology. 1998;144:2739–2748. doi: 10.1099/00221287-144-10-2739. [DOI] [PubMed] [Google Scholar]
- 38.Johnston M, Flick J S, Pexton T. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3834–3841. doi: 10.1128/mcb.14.6.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kamiryo T, Parthasarathy S, Numa S. Evidence that acyl coenzyme A synthetase activity is required for repression of yeast acetyl coenzyme A carboxylase by exogenous fatty acids. Proc Natl Acad Sci USA. 1976;73:386–390. doi: 10.1073/pnas.73.2.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kelley M J, Bailis A M, Henry S A, Carman G M. Regulation of phospholipid biosynthesis in Saccharomyces cerevisiae by inositol. Inositol is an inhibitor of phosphatidylserine synthase activity. J Biol Chem. 1988;263:18078–18085. [PubMed] [Google Scholar]
- 41.Krebs J E, Kuo M H, Allis C D, Peterson C L. Cell cycle-regulated histone acetylation required for expression of the yeast HO gene. Genes Dev. 1999;13:1412–1421. doi: 10.1101/gad.13.11.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuchin S, Treich I, Carlson M. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 2000;97:7916–7920. doi: 10.1073/pnas.140109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lampl M. Function of acetyl CoA carboxylase in the yeast Saccharomyces cerevisiae. Graz, Austria: Technische Universitat Graz; 1998. [Google Scholar]
- 45.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lau W W, Schneider K R, O'Shea E K. A genetic study of signaling processes for repression of PHO5 transcription in Saccharomyces cerevisiae. Genetics. 1998;150:1349–1359. doi: 10.1093/genetics/150.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopes J M, Hirsch J P, Chorgo P A, Schulze K L, Henry S A. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 1991;19:1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matsuhashi M. Acetyl-CoA carboxylase from yeast. Methods Enzymol. 1969;14:3–8. [Google Scholar]
- 50.Mishina M, Roggenkamp R, Schweizer E. Yeast mutants defective in acetyl-coenzyme A carboxylase and biotin: apocarboxylase ligase. Eur J Biochem. 1980;111:79–87. doi: 10.1111/j.1432-1033.1980.tb06077.x. [DOI] [PubMed] [Google Scholar]
- 51.Mitchelhill K I, Stapleton D, Gao G, House C, Michell B, Katsis F, Witters L A, Kemp B E. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J Biol Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- 52.Munday M R, Campbell D G, Carling D, Hardie D G. Identification by amino acid sequencing of three major regulatory phosphorylation sites on rat acetyl-CoA carboxylase. Eur J Biochem. 1988;175:331–338. doi: 10.1111/j.1432-1033.1988.tb14201.x. [DOI] [PubMed] [Google Scholar]
- 53.Nikawa J, Tanabe T, Ogiwara H, Shiba T, Numa S. Inhibitory effects of long-chain acyl coenzyme A analogues on rat liver acetyl coenzyme A carboxylase. FEBS Lett. 1979;102:223–226. doi: 10.1016/0014-5793(79)80005-8. [DOI] [PubMed] [Google Scholar]
- 54.Nikoloff D M, Henry S A. Genetic analysis of yeast phospholipid biosynthesis. Annu Rev Genet. 1991;25:559–583. doi: 10.1146/annurev.ge.25.120191.003015. [DOI] [PubMed] [Google Scholar]
- 55.Nonet M L, Young R A. Intragenic and extragenic suppressors of mutations in the heptapeptide repeat domain of Saccharomyces cerevisiae RNA polymerase II. Genetics. 1989;123:715–724. doi: 10.1093/genetics/123.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogiwara H, Tanabe T, Nikawa J, Numa S. Inhibition of rat-liver acetyl-coenzyme-A carboxylase by palmitoyl-coenzyme A. Formation of equimolar enzyme-inhibitor complex. Eur J Biochem. 1978;89:33–41. doi: 10.1111/j.1432-1033.1978.tb20893.x. [DOI] [PubMed] [Google Scholar]
- 57.Ouyang Q, Ruiz-Noriega M, Henry S A. The REG1 gene product is required for repression of INO1 and other UASINO containing genes of yeast. Genetics. 1999;152:89–100. doi: 10.1093/genetics/152.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Patton-Vogt J L, Griac P, Sreenivas A, Bruno V, Dowd S, Swede M J, Henry S A. Role of the yeast phosphatidylinositol/phosphatidylcholine transfer protein (Sec14p) in phosphatidylcholine turnover and INO1 regulation. J Biol Chem. 1997;272:20873–20883. doi: 10.1074/jbc.272.33.20873. [DOI] [PubMed] [Google Scholar]
- 59.Roberts S M, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roggenkamp R, Numa S, Schweizer E. Fatty acid-requiring mutant of Saccharomyces cerevisiae defective in acetyl-CoA carboxylase. Proc Natl Acad Sci USA. 1980;77:1814–1817. doi: 10.1073/pnas.77.4.1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sanz P, Alms G R, Haystead T A, Carlson M. Regulatory interactions between the Reg1-Glc7 protein phosphatase and the Snf1 protein kinase. Mol Cell Biol. 2000;20:1321–1328. doi: 10.1128/mcb.20.4.1321-1328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scafe C, Chao D, Lopes J, Hirsch J P, Henry S, Young R A. RNA polymerase II C-terminal repeat influences response to transcriptional enhancer signals. Nature. 1990;347:491–494. doi: 10.1038/347491a0. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt M C, McCartney R R. β-subunits of Snf1 kinase are required for kinase function and substrate definition. EMBO J. 2000;19:4936–4943. doi: 10.1093/emboj/19.18.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schneiter R, Guerra C E, Lampl M, Gogg G, Kohlwein S D, Klein H L. The Saccharomyces cerevisiae hyperrecombination mutant hpr1Δ is synthetically lethal with two conditional alleles of the acetyl coenzyme A carboxylase gene and causes a defect in nuclear export of polyadenylated RNA. Mol Cell Biol. 1999;19:3415–3422. doi: 10.1128/mcb.19.5.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneiter R, Guerra C E, Lampl M, Tatzer V, Zellnig G, Klein H L, Kohlwein S D. A novel cold-sensitive allele of the rate-limiting enzyme of fatty acid synthesis, acetyl coenzyme A carboxylase, affects the morphology of the yeast vacuole through acylation of Vac8p. Mol Cell Biol. 2000;20:2984–2995. doi: 10.1128/mcb.20.9.2984-2995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schneiter R, Hitomi M, Ivessa A S, Fasch E V, Kohlwein S D, Tartakoff A M. A yeast acetyl coenzyme A carboxylase mutant links very-long-chain fatty acid synthesis to the structure and function of the nuclear membrane-pore complex. Mol Cell Biol. 1996;16:7161–7172. doi: 10.1128/mcb.16.12.7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shirra M K, Arndt K M. Evidence for the involvement of the Glc7-Reg1 phosphatase and the Snf1-Snf4 kinase in the regulation of INO1 transcription in Saccharomyces cerevisiae. Genetics. 1999;152:73–87. doi: 10.1093/genetics/152.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Simon M, Binder M, Adam G, Hartig A, Ruis H. Control of peroxisome proliferation in Saccharomyces cerevisiae by ADR1, SNF1 (CAT1, CCR1) and SNF4 (CAT3) Yeast. 1992;8:303–309. doi: 10.1002/yea.320080407. [DOI] [PubMed] [Google Scholar]
- 70.Smith F C, Davies S P, Wilson W A, Carling D, Hardie D G. The SNF1 kinase complex from Saccharomyces cerevisiae phosphorylates the transcriptional repressor protein Mig1p in vitro at four sites within or near regulatory domain 1. FEBS Lett. 1999;453:219–223. doi: 10.1016/s0014-5793(99)00725-5. [DOI] [PubMed] [Google Scholar]
- 71.Som T, Armstrong K A, Volkert F C, Broach J R. Autoregulation of 2μm circle gene expression provides a model for maintenance of stable plasmid copy levels. Cell. 1988;52:27–37. doi: 10.1016/0092-8674(88)90528-4. [DOI] [PubMed] [Google Scholar]
- 72.Song W, Treich I, Qian N, Kuchin S, Carlson M. SSN genes that affect transcriptional repression in Saccharomyces cerevisiae encode SIN4, ROX3, and SRB proteins associated with RNA polymerase II. Mol Cell Biol. 1996;16:115–120. doi: 10.1128/mcb.16.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Steiner M R, Lester R L. In vitro studies of phospholipid biosynthesis in Saccharomyces cerevisiae. Biochim Biophys Acta. 1972;260:222–243. doi: 10.1016/0005-2760(72)90035-5. [DOI] [PubMed] [Google Scholar]
- 74.Thompson C M, Koleske A J, Chao D M, Young R A. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 75.Thompson-Jaeger S, Francois J, Gaughran J P, Tatchell K. Deletion of SNF1 affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Treitel M A, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tu J, Carlson M. REG1 binds to protein phosphatase type 1 and regulates glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vahlensieck H F, Pridzun L, Reichenbach H, Hinnen A. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr Genet. 1994;25:95–100. doi: 10.1007/BF00309532. [DOI] [PubMed] [Google Scholar]
- 79.Wakil S J, Stoops J K, Joshi V C. Fatty acid synthesis and its regulation. Annu Rev Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- 80.Winston F, Dollard C, Ricupero-Hovasse S. Construction of a set of convenient S. cerevisiae strains that are isogenic to S288C. Yeast. 1995;11:53–55. doi: 10.1002/yea.320110107. [DOI] [PubMed] [Google Scholar]
- 81.Woods A, Munday M R, Scott J, Yang X, Carlson M, Carling D. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J Biol Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]