Abstract
Release of promoter-proximal paused RNA polymerase II (Pol II) during early elongation is a critical step in transcriptional regulation in metazoan cells. Paused Pol II release is thought to require the kinase activity of cyclin-dependent kinase 9 (CDK9) for the phosphorylation of 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) sensitivity-inducing factor (DSIF), negative elongation factor (NELF), and C-terminal domain (CTD) serine-2 of Pol II. Here we demonstrate that Pol II-associated factor 1 (PAF1) is a critical regulator of paused Pol II release, that positive transcription elongation factor b (P-TEFb) directly regulates the initial recruitment of PAF1 complex (PAF1C) to genes, and that the subsequent recruitment of CDK12 is dependent on PAF1C. These findings reveal cooperativity between P-TEFb, PAF1C, and CDK12 in pausing release and Pol II CTD phosphorylation.
Thousands of developmentally regulated genes in metazoans harbor promoter-proximal paused Pol II 30–50 nt downstream of their transcription start sites (TSS) (1–3). Paused Pol IIs are usually phosphorylated on CTD serine-5 and are associated with DSIF and NELF. Release of paused Pol II into productive elongation is believed to require phosphorylation of CTD serine-2, conversion of DSIF into a positive elongation factor by phosphorylation of its SPT5 subunit, and disassociation of NELF (1). Although it was long believed that CTD serine-2 phosphorylation was catalyzed predominantly by CDK9, the mammalian ortholog of yeast Bur1, recent studies have identified CDK12 as the metazoan ortholog of Ctk1, the major CTD serine-2 kinase in yeast, and suggested that CDK9 is a CTD serine-5 kinase (4, 5). The yeast Paf1 complex and the human PAF1 complex, of interest here, have been implicated in transcription elongation on DNA and chromatin templates, recruitment and activation of histone modifiers, mRNA 3’ formation, etc. (6, 7). However, PAF1C has not been considered a critical elongation factor because depletions of PAF1C subunits in yeast and fly, while reducing the level of CTD serine 2-phosphorylated elongating Pol II (8, 10), did not affect the distribution of total Pol II on active genes (8, 9).
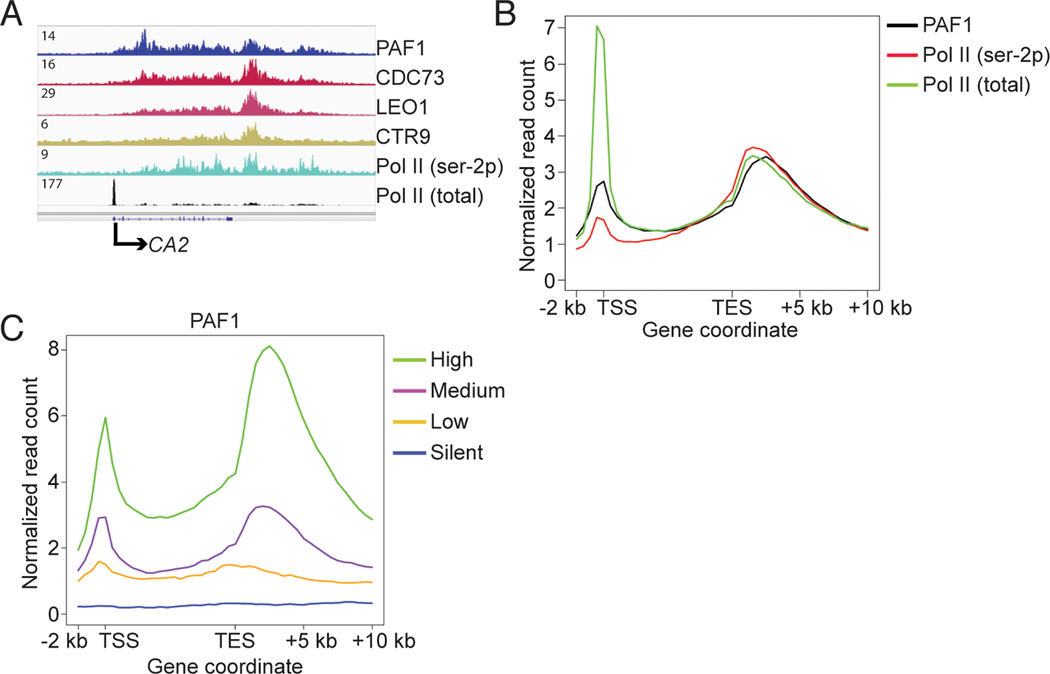
To study the function of human PAF1C, we performed ChIP-seq experiments for PAF1C subunits PAF1, CDC73, LEO1, and CTR9, as well as total Pol II and CTD serine 2-phosphorylated (ser-2p) Pol II, in human acute myeloid leukemia THP1 cells. Similar to Pol II (ser-2p), the four PAF1C subunits occupy transcribed regions of most active genes, and exhibit maximum occupancy downstream of transcription end sites (TESs) (Fig. 1, A to C, and fig. S1, A to F). LEO1 (fig. S1B) and CTR9 (fig. S1C) occupancies do not generally overlap with the promoter-proximal Pol II peaks, as reported previously (2). However, PAF1 and CDC73, the major scaffolding components within human PAF1C (11), do overlap with the promoter-proximal Pol II peaks (Fig. 1, B and C, and fig. S1, A, D, E, and F). Complementary strand-specific mRNA-seq analyses using RNA from THP1 cells identified 19,482 transcripts (RPKM > 1), corresponding to 10,664 genes, of which 9,823 were bound by PAF1. Notably, the PAF1 binding signals on these genes positively correlated with corresponding mRNA levels (Fig. 1C, and fig. S1, A to C), suggesting an involvement of PAF1C in Pol II transcription or transcription-coupled events.
Fig. 1. PAF1C occupancy positively correlates with gene expression level.
(A) An IGV browser snapshot comparing occupancy of PAF1, CDC73, LEO1, CTR9, Pol II (ser-2p), and total Pol II within the CA2 locus in THP1 cells. (B) Occupancy of PAF1, total Pol II, and Pol II (ser-2p) on an average gene. (C) Metagene analyses showing positive correlation between PAF1 occupancy and the mRNA level of genes.
In further functional analyses, two lentiviral shRNAs were employed to reduce the level of the key PAF1 subunit (8, 11) in THP1 cells (Fig. 2A) and global gene expression changes were assessed by RNA-seq. With p < 0.001 and fold-expression change > 1.5, of the 9,823 genes bound by PAF1, only 1,351 showed changes in expression (table S1). The knockdown of PAF1 also resulted in an increased level of promoter-proximal paused Pol II that was not limited to genes whose mRNA levels were affected by PAF1 knockdown (Fig. 2, B and C). Based on a promoter read count change of > 2, of the 9,823 genes bound by PAF1, 5,851 exhibited increased Pol II pausing and only 344 exhibited decreased Pol II pausing (table S2). The increased Pol II pausing, which led to an average 2-fold increase in Pol II occupancy on promoters (Fig 2C), was confirmed by comparison of the traveling ratio of total Pol II in control and knockdown cells (Fig. 2D).
Fig. 2. PAF1C is a critical regulator of promoter-proximal pausing release of Pol II.
(A) Comparison of PAF1 level in control and PAF1 knockdown THP1 cells by Western blot. (B) and (C) Comparison of the occupancy of PAF1 (B) and Pol II (C) on an average gene. (D) Comparison of the traveling ratios of genes bound by Pol II. (E) Comparison of the normalized occupancy of PAF1 on an average gene. (F) Comparison of the occupancy of Pol II (ser-5p) on an average gene. (G) and (H) Comparison of the occupancy of Pol II (G) and LEO1 (H) on an average gene in DMSO- and flavopiridol-treated THP1 cells.
The apparent and seemingly paradoxical increase of PAF1 occupancy near promoters in PAF1 knockdown cells relative to control cells (Fig 2B) raised the possibility that the increased Pol II pausing might not be a direct effect of the decreased PAF1 chromatin association. However, normalization of the PAF1 levels to Pol II levels in control and knockdown cells revealed a relative decrease in PAF1 chromatin association in knockdown cells, suggesting that the increased Pol II pausing is indeed a direct effect of reduced PAF1 association at the promoter proximal region (Fig. 2E). In a further validation of the ChIP-seq results for total Pol II, ChIP-seq for CTD serine 5-phosphorylated Pol II revealed, as expected, a corresponding increase in Pol II (ser-5p) in PAF1 knockdown cells relative to control cells (Fig. 2F, and fig. S2, A and B). Thus, PAF1C is a critical elongation factor that regulates elongation as early as the Pol II pausing release stage.
To determine whether the facilitation of Pol II pausing release by PAF1C reflects a general mechanism that is not cell type-specific, we knocked down PAF1 (by shRNA #2) in human acute lymphoblastic leukemia CCRF-CEM cells and compared PAF1 and Pol II occupancies in control and knockdown cells. Surprisingly, and in contrast to the results in THP1 cells, PAF1 knockdown in these cells resulted in an increase in Pol II pausing on only 142 genes, and a decrease in Pol II pausing on 1,244 genes (fig. S3, A to E and table S3). To rule out any off-target effects, the results were validated by PAF1 knockdown using shRNA #1 (fig. S3, F and G). With respect to the apparent cell/context-dependent variability in effects of PAF1C functions, we note that whereas PAF1C is generally considered a positive effector (6, 7), there are earlier (12) and more recent (13) reports of PAF1C function as a negative regulator of Pol II pausing release. Therefore, the differential effects of PAF1 knockdown on Pol II pausing in THP1 cells (carrying the MLL-AF9 fusion gene) and CCRF-CEM cells (bearing TP53 mutations) likely reflect the distinct genetic backgrounds and physiological states of the two cell types. Thus, the diverse results in current and published studies (12, 13) indicate variable context-dependent effects of PAF1C components as either positive or negative regulators, as further exemplified by a switch in the CDC73 subunit from a positive regulator (oncoprotein) to a negative regulator (tumor suppressor) by tyrosine phosphorylation (14).
The effect of PAF1 knockdown on Pol II pausing resembles that of pan-CDK inhibition by flavopiridol (2, 15). To determine whether flavopiridol treatment affects Pol II pausing in part through PAF1C, we compared the genomic occupancy of Pol II and the LEO1 subunit of PAF1C in DMSO- and flavopiridol-treated THP1 cells. Flavopiridol significantly increased global Pol II pausing (Fig. 2G), and markedly reduced the chromatin occupancy of LEO1 (Fig. 2H, and fig. S4, A to B). These results implicate PAF1C as a key factor for the release of promoter-proximal paused Pol II. The reduced occupancy of PAF1C as a result of flavopiridol treatment suggests that pan-CDK inhibition increases Pol II pausing in part through compromising the recruitment of PAF1C.
In yeast, serine-2 and serine-5 phosphorylation of the Pol II CTD, as well as phosphorylation of the Spt5 subunit of DSIF by Bur1 (ortholog of metazoan CDK9), are critical for the recruitment of PAF1C to target genes (16, 17). To determine whether, as in yeast, CDK9 or SPT5 is required for the recruitment of PAF1C in THP1 cells, we performed independent SPT5 and CDK9 knockdown analyses and evaluated effects on Pol II and PAF1 occupancy. As anticipated (2), SPT5 knockdown reduced the occupancy of Pol II and PAF1 (fig. S5, A to D). Like flavopiridol treatment, CDK9 knockdown (~80%, Fig. 3, A and B) reduced PAF1 occupancy (Fig. 3C) but, unlike flavopiridol, had little effect on Pol II occupancy (Fig. 3D). In addition, and also unexpectedly, CDK9 knockdown only moderately increased global Pol II pausing (fig. S6A). However, a comparison of the levels of BRD4-associated CDK9 (CDK9-active complex) and LARP7-associated CDK9 (CDK9-inactive 7SK snRNP complex) (18) in control and CDK9 knockdown THP1 cells revealed a preferential reduction of the CDK9 fraction in the 7SK snRNP complex relative to the CDK9 fraction in the BRD4 complex (fig. S6B). Thus, the small effect of the CDK9 knockdown on Pol II pausing may be due to the minimal effect on the kinase-active CDK9 fraction, and the regulation of PAF1C recruitment by CDK9 is likely independent of the CDK9 kinase activity.
Fig. 3. P-TEFb contributes to the recruitment of PAF1C.
(A) Comparison of CDK9 level in control and CDK9 knockdown THP1 cells. (B) to (D) ChIP-qPCR data comparing the occupancy of CDK9 (B), PAF1 (C), and Pol II (D). (E) Co-IP of P-TEFb and AF9 with CDC73. Error bars indicate mean ± SD (N=3). (F) Co-IP of PAF1, CDC73, total Pol II, and Pol II (ser-2p) with CDK9. (G) Pull-down assay using immobilized P-TEFb as bait and PAF1C as prey. (H) CDK9 peak distribution in control and PAF1 knockdown cells. Total peak numbers were labeled on top of each column. (I) An IGV browser snapshot comparing CDK9 occupancy within the c-MYC locus. (J) and (K) Comparison of CDK9 occupancy (J) and normalized CDK9 occupancy (K) on an average gene, respectively.
In a further analysis of the role of CDK9 in PAF1C recruitment, a co-immunoprecipitation assay revealed strong association of endogenous PAF1 and P-TEFb (a complex of CDK9 and Cyclin T1) in THP1 cells (Fig. 3E). The much stronger association between PAF1C and P-TEFb relative to the reported (19) PAF1-AF9 interaction (Fig. 3E) raised the possibility of an AF9-independent interaction between these two complexes. The PAF1 and CDC73 subunits of PAF1C, as well as Pol II, were reciprocally co-immumoprecipitated with CDK9 by a CDK9 antibody (Fig. 3F). More importantly, binding assays using purified P-TEFb and PAF1C complexes established a direct AF9-independent interaction between these two complexes (Fig. 3G), indicating that P-TEFb contributes to the recruitment of PAF1C through direct interaction.
These results raised the possibility that PAF1C may regulate promoter-proximal pausing release of Pol II in part by facilitation of P-TEFb extraction from 7SK snRNP, but PAF1C was unable to release P-TEFb from 7SK snRNP in a release assay (fig. S7). Toward a further analysis of the functional consequences of the PAF1C-CDK9 interaction, ChIP-seq experiments for CDK9 in control and PAF1 knockdown cells revealed, first, that CDK9 is mainly associated with both enhancers and promoters (Fig. 3, H and I) (20, 21) and, second, consistent with a previous report (19), that PAF1 depletion reduces normalized CDK9 occupancy on promoters (Fig. 3, J and K). The enhancer- and promoter-association of CDK9, along with Pol II imaging data (21), makes it less likely, as proposed in another study (13), that P-TEFb generally travels with Pol II during elongation. Therefore, we propose (i) that the interaction between PAF1C and P-TEFb is required mainly for the initial recruitment of PAF1C, but may also stabilize the P-TEFb promoter association, and (ii) that CDK9 and other kinases subsequently phosphorylate the CTD of Pol II (16) and the CTR of SPT5 (17), thus creating PAF1C binding sites on Pol II and the associated DSIF that enable PAF1C to facilitate release of paused Pol II into productive elongation.
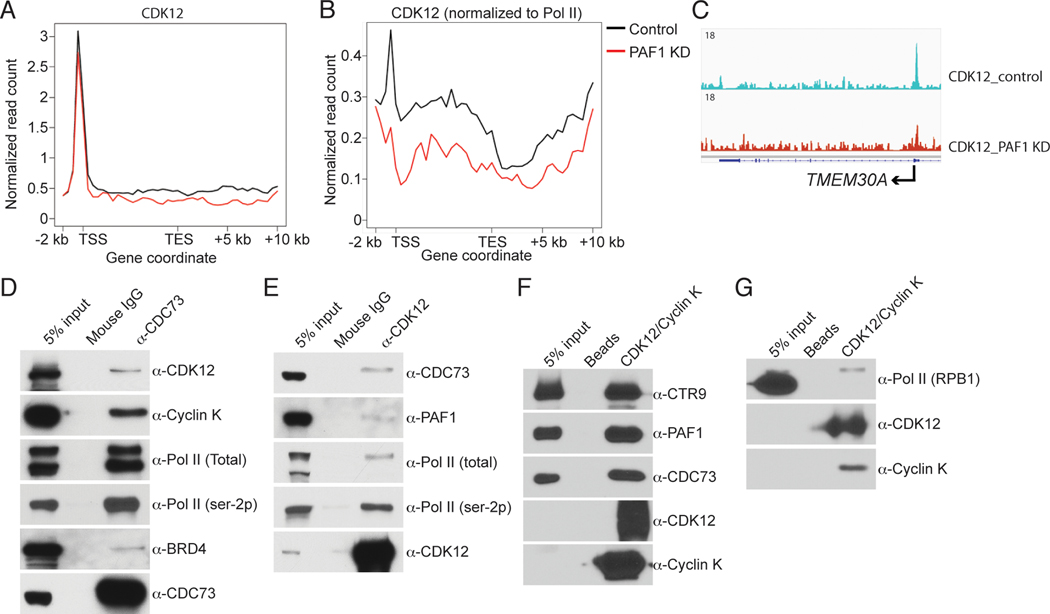
In agreement with previous studies (8, 10), we observed decreased Pol II CTD serine-2 phosphorylation, but increased CTD serine-5 phosphorylation, following PAF1 knockdown (8, 10) (fig. S8, A to C). Thus, PAF1C may be responsible either for the recruitment of CTD serine-2 kinases or for the regulation of their activity. Recent studies have suggested that CDK12 is the predominant serine-2 kinase (4) and that bromodomain-containing protein 4 (BRD4) is an atypical CTD serine-2 kinase (22). A comparison of CDK12 and BRD4 occupancies on the c-MYC gene by ChIP-qPCR revealed reduced occupancy of CDK12, but not BRD4, in PAF1 knockdown cells (fig. S9, A and B). Genomic analysis of the effect of PAF1C on CDK12 recruitment revealed a global decrease in CDK12 occupancy, especially when normalized to Pol II occupancy, in PAF1 knockdown cells (Fig. 4, A to C). These results are consistent with the reduced level of Pol II (ser-2p) and suggest a role for PAF1C-dependent recruitment of CDK12 in Pol II CTD ser-2 phosphorylation.
Fig. 4. PAF1C is responsible for the recruitment of CDK12.
(A) and (B) Comparison of CDK12 occupancy (A) and normalized CDK12 occupancy (B) on an average gene in control and PAF1 knockdown THP1 cells, respectively. (C) An IGV browser snapshot comparing CDK12 occupancy within the TMEM30A locus. (D) Co-IP of CDK12, Cyclin K, total Pol II, Pol II (ser-2p), and BRD4 with CDC73. (E) Co-IP of PAF1, CDC73, total Pol II, and Pol II (ser-2p) with CDK12. (F) and (G) Pull-down assays using immobilized CDK12/Cyclin K complex as a bait, and PAF1C (F) and purified Pol II (G) as preys, respectively.
Experiments to determine whether the PAF1C-dependent recruitment of CDK12 is through a direct interaction showed that CDK12 and Cyclin K, as well as Pol II (ser-2p) and BRD4, were co-immunoprecipitated with an antibody against the CDC73 subunit of PAF1C (Fig. 4D) and, reciprocally, that the PAF1 and CDC73 subunits of PAF1C, as well as Pol II, were coimmunoprecipitated with CDK12/Cyclin K by an antibody against CDK12 (Fig. 4E). The PAF1C association with BRD4 was significantly weaker than its association with CDK12 (Fig. 4D), partially explaining why the recruitment of BRD4 is less dependent on PAF1C (fig. S9B). Analyses with purified recombinant CDK12/Cyclin K and PAF1C complexes revealed a robust direct binding of PAF1C to CDK12/Cyclin K under stringent conditions (Fig. 4F). A parallel binding assay with purified proteins under similar conditions revealed a very weak interaction of CDK12/Cyclin K with Pol II (Fig. 4G) relative to the strong interaction with PAF1C (Fig. 4F). These results strongly suggest (i) that PAF1C, in addition to regulating the release of paused Pol II, is directly involved in the recruitment of CDK12 and (ii) along with the known interaction of human PAF1C with Pol II (11), that the association of CDK12/Cyclin K with Pol II is likely mediated by Pol II-bound PAF1C.
We next asked whether CDK12/Cyclin K affects the recruitment of PAF1C by comparing PAF1 and Pol II occupancy in control and Cyclin K knockdown cells. Despite effecting a globally decreased Pol II (ser-2p) (fig. S8A) and reduced CDK12 occupancy, Cyclin K knockdown had little effect on PAF1 or Pol II occupancy (fig. S10, A to D). These results are in agreement with previously published data showing that a knockout of yeast Ctk1 (homologue of CDK12) does not affect Paf1 occupancy (23), and support a model in which Pol II-bound PAF1C recruits CDK12.
In summary, we report a critical role for PAF1C in Pol II pausing release, a direct role for P-TEFb in PAF1C recruitment, and a PAF1C-CDK12/Cyclin K interaction that is important for CTD serine-2 phosphorylation. These findings complement and extend previous results demonstrating functions for P-TEFb, DSIF/NELF and Pol II CTD serine-2 phosphorylationin Pol II pausing release and are summarized in an updated model (fig. S11) that will guide further mechanistic studies of both positive and negative functions of PAF1C in transcriptional control.
Supplementary Material
Acknowledgments:
The authors thank G. Morin for providing the Cyclin K cDNA and S. Malik, S. Murphy, and M. Guermah for critical reading of the manuscript. This work is supported by a Leukemia and Lymphoma Society SCOR grant to R. G. R. W.Y., T. Ni, and J.Z. are supported by the intramural research program of the National Heart Lung Blood Institute, National Institutes of Health. ChIP-seq and RNA-seq data have been submitted to GEO under accession number GSE62171.
References:
- 1.Adelman K, Lis JT, Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet 13, 720–731 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rahl PB et al. , c-Myc regulates transcriptional pause release. Cell 141, 432–445 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Core LJ, Waterfall JJ, Lis JT, Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartkowiak B. et al. , CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev 24, 2303–2316 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Czudnochowski N, Bosken CA, Geyer M, Serine-7 but not serine-5 phosphorylation primes RNA polymerase II CTD for P-TEFb recognition. Nat Commun 3, 842 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Tomson BN, Arndt KM, The many roles of the conserved eukaryotic Paf1 complex in regulating transcription, histone modifications, and disease states. Biochim Biophys Acta 1829, 116–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jaehning JA, The Paf1 complex: platform or player in RNA polymerase II transcription? Biochim Biophys Acta 1799, 379–388 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mueller CL, Porter SE, Hoffman MG, Jaehning JA, The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14, 447–456 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Adelman K. et al. , Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol Cell Biol 26, 250–260 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nordick K, Hoffman MG, Betz JL, Jaehning JA, Direct interactions between the Paf1 complex and a cleavage and polyadenylation factor are revealed by dissociation of Paf1 from RNA polymerase II. Eukaryotic cell 7, 1158–1167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim J, Guermah M, Roeder RG, The human PAF1 complex acts in chromatin transcription elongation both independently and cooperatively with SII/TFIIS. Cell 140, 491–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bai X. et al. , TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell 142, 133–143 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen FX et al. , PAF1, a Molecular Regulator of Promoter-Proximal Pausing by RNA Polymerase II. Cell 162, 1003–1015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi A. et al. , SHP2 tyrosine phosphatase converts parafibromin/Cdc73 from a tumor suppressor to an oncogenic driver. Mol Cell 43, 45–56 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonkers I, Kwak H, Lis JT, Genome-wide dynamics of Pol II elongation and its interplay with promoter proximal pausing, chromatin, and exons. eLife 3, e02407 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qiu H, Hu C, Gaur NA, Hinnebusch AG, Pol II CTD kinases Bur1 and Kin28 promote Spt5 CTR-independent recruitment of Paf1 complex. The EMBO journal 31, 3494–3505 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y. et al. , Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol Cell Biol 29, 4852–4863 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peterlin BM, Price DH, Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23, 297–305 (2006). [DOI] [PubMed] [Google Scholar]
- 19.He N. et al. , Human Polymerase-Associated Factor complex (PAFc) connects the Super Elongation Complex (SEC) to RNA polymerase II on chromatin. Proc Natl Acad Sci U S A 108, E636–645 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loven J. et al. , Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153, 320–334 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghamari A. et al. , In vivo live imaging of RNA polymerase II transcription factories in primary cells. Genes Dev 27, 767–777 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Devaiah BN et al. , BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 109, 6927–6932 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ahn SH, Kim M, Buratowski S, Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3’ end processing. Mol Cell 13, 67–76 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.