Abstract
We present a genome assembly from an individual female Caprimulgus europaeus (the European nightjar; Chordata; Aves; Caprimulgiformes; Caprimulgidae). The genome sequence is 1,178 megabases in span. The majority of the assembly (99.33%) is scaffolded into 37 chromosomal pseudomolecules, including the W and Z sex chromosomes.
Keywords: Caprimulgus europaeus, European nightjar, Eurasian nightjar, genome sequence, chromosomal
Species taxonomy
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi; Archelosauria; Archosauria; Dinosauria; Saurischia; Theropoda; Coelurosauria; Aves; Neognathae; Caprimulgimorphae; Caprimulgiformes; Caprimulgidae; Caprimulginae; Caprimulgus; Caprimulgus europaeus Linnaeus 1758 (NCBI:txid85660).
Background
The European nightjar ( Caprimulgus europaeus; also known as the Eurasian nightjar and common goatsucker) is an insectivorous, crepuscular, ground-nesting bird distributed throughout the Western Palearctic ( Hagemeijer & Blair, 1997). It breeds in semi-natural dry and open habitats with scattered trees ( Cramp & Brooks, 1985). Little is known about the ecology of the European nightjar ( Cramp & Brooks, 1985; Polakowski et al., 2020), and in general that of the Caprimulgidae family. The family comprises peculiar species such as the only bird known to hibernate, the Common Poorwill ( Phalaenoptilus nuttallii) ( Carey, 2019; French, 2019; Woods et al., 2019), and one of the few birds that uses echo-localization, the South American Oilbird ( Steatornis caripensis) ( Brinkløv et al., 2013). The European nightjar has been found to be more resistant to pathogens than other bird species ( Jiang et al., 2021). Although categorized as ‘least concern’ by the IUCN ( IUCN, 2016), the European nightjar has experienced a steady population decline in the past decades, and is of conservation concern in Europe ( Eaton et al., 2015; Evens et al., 2017; Keller et al., 2010). The availability of a high-quality, chromosome-level reference genome will help to deepen the knowledge on the biology and evolution of this species, boosting studies on the genomics of the peculiar family of Caprimulgidae. Moreover, as genomic resources gain preheminence in conservation efforts ( Allendorf, 2017; Fuentes-Pardo & Ruzzante, 2017; Supple & Shapiro, 2018), we expect that the reference genome presented here will help aid planning conservation actions for the European nightjar.
Genome sequence report
The genome was sequenced from a blood sample taken from a single female C. europaeus collected from a bird ringing station in Ventotene, Italy (latitude 40.79404, longitude 13.42777). A total of 87-fold coverage in Pacific Biosciences single-molecule long reads and 62-fold coverage in 10X Genomics read clouds were generated. Primary assembly contigs were scaffolded with chromosome conformation Hi-C data. Manual assembly curation corrected 144 missing/misjoins and removed 31 haplotypic duplications, reducing the assembly length by 0.15% and the scaffold number by 21.94%, and increasing the scaffold N50 by 26.46%.
The final assembly has a total length of 1,178 Mb in 121 sequence scaffolds with a scaffold N50 of 83 Mb ( Table 1). Of the assembly sequence, 99.3% was assigned to 37 chromosomal-level scaffolds, representing 35 autosomes (numbered by sequence length) and the W and Z sex chromosomes ( Figure 1– Figure 4; Table 2). The assembly has a BUSCO ( Simão et al., 2015) completeness of 97.4% (single 96.9%, duplicated 0.6%) using the aves_odb10 reference set. While not fully phased, the assembly deposited is of one pseudo-haplotype. Contigs corresponding to the alternate haplotype have also been deposited.
Table 1. Genome data for Caprimulgus europaeus, bCapEur3.1.
| Project accession data | |
|---|---|
| Assembly identifier | bCapEur3.1 |
| Species | Caprimulgus europaeus |
| Specimen | bCapEur3 |
| NCBI taxonomy ID | NCBI:txid111811 |
| BioProject | PRJEB44540 |
| BioSample ID | SAMEA7524394 |
| Isolate information | Female, blood |
| Raw data accessions | |
| PacificBiosciences SEQUEL II | ERR6445211 |
| 10X Genomics Illumina | ERR6054683-ERR6054686 |
| Hi-C Illumina | ERR6054687, ERR6054688 |
| Genome assembly | |
| Assembly accession | GCA_907165065.1 |
| Accession of alternate haplotype | GCA_907165095.1 |
| Span (Mb) | 1,178 |
| Number of contigs | 274 |
| Contig N50 length (Mb) | 31 |
| Number of scaffolds | 121 |
| Scaffold N50 length (Mb) | 83 |
| Longest scaffold (Mb) | 126 |
| BUSCO * genome score | C:97.4%[S:96.9%, D:0.6%],F:0.5%,M:2.1%,n:8338 |
*BUSCO scores based on the aves_odb10 BUSCO set using v5.1.2. C= complete [S= single copy, D=duplicated], F=fragmented, M=missing, n=number of orthologues in comparison. A full set of BUSCO scores is available at https://blobtoolkit.genomehubs.org/view/bCapEur3.1/dataset/CAJRAV01/busco.
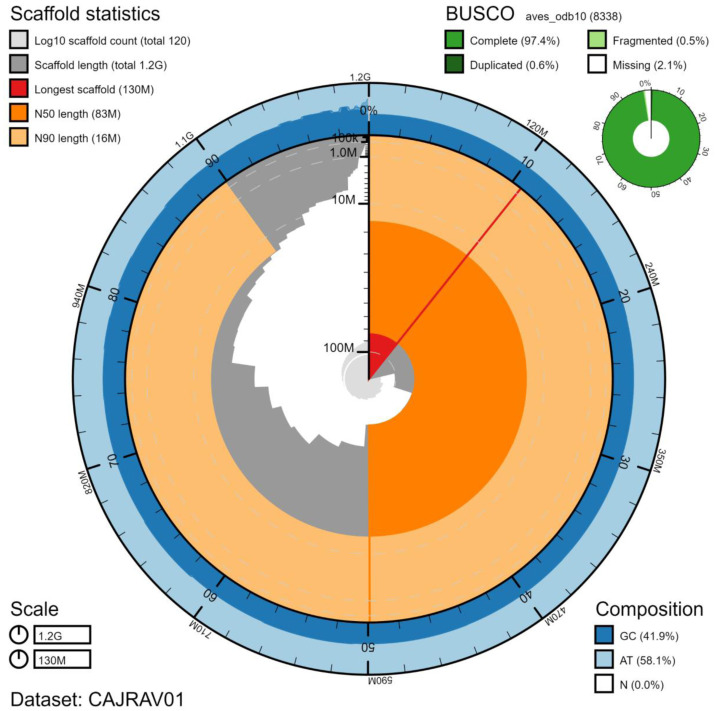
Figure 1. Genome assembly of Caprimulgus europaeus, bCapEur3.1: metrics.
The BlobToolKit Snailplot shows N50 metrics and BUSCO gene completeness. The main plot is divided into 1,000 size-ordered bins around the circumference with each bin representing 0.1% of the 1,177,791,212 bp assembly. The distribution of chromosome lengths is shown in dark grey with the plot radius scaled to the longest chromosome present in the assembly (126,318,510 bp, shown in red). Orange and pale-orange arcs show the N50 and N90 chromosome lengths (82,614,289 and 15,699,869 bp), respectively. The pale grey spiral shows the cumulative chromosome count on a log scale with white scale lines showing successive orders of magnitude. The blue and pale-blue area around the outside of the plot shows the distribution of GC, AT and N percentages in the same bins as the inner plot. A summary of complete, fragmented, duplicated and missing BUSCO genes in the aves_odb10 set is shown in the top right. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/bCapEur3.1/dataset/CAJRAV01/snail.
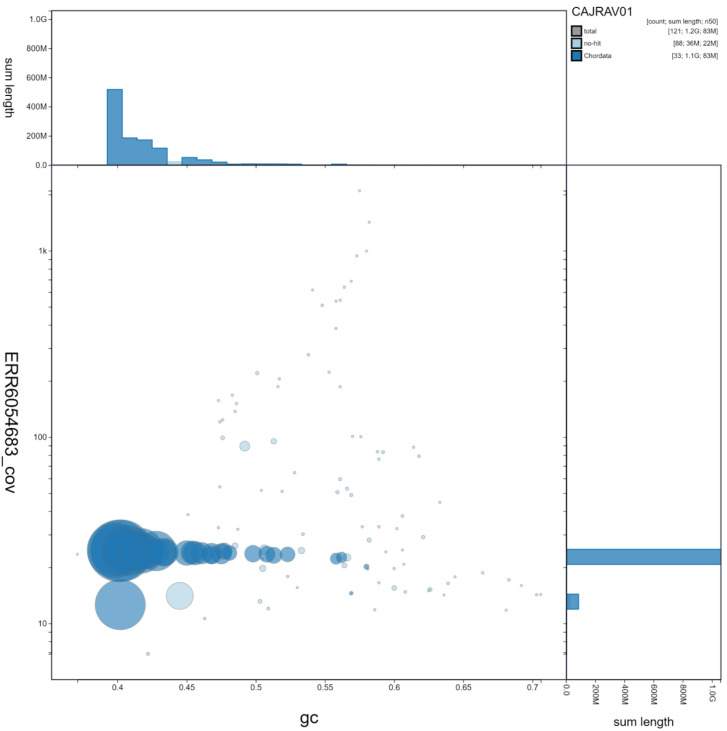
Figure 2. Genome assembly of Caprimulgus europaeus, bCapEur3.1: GC coverage.
BlobToolKit GC-coverage plot. Scaffolds are coloured by phylum. Circles are sized in proportion to scaffold length. Histograms show the distribution of scaffold length sum along each axis. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/bCapEur3.1/dataset/CAJRAV01/blob.
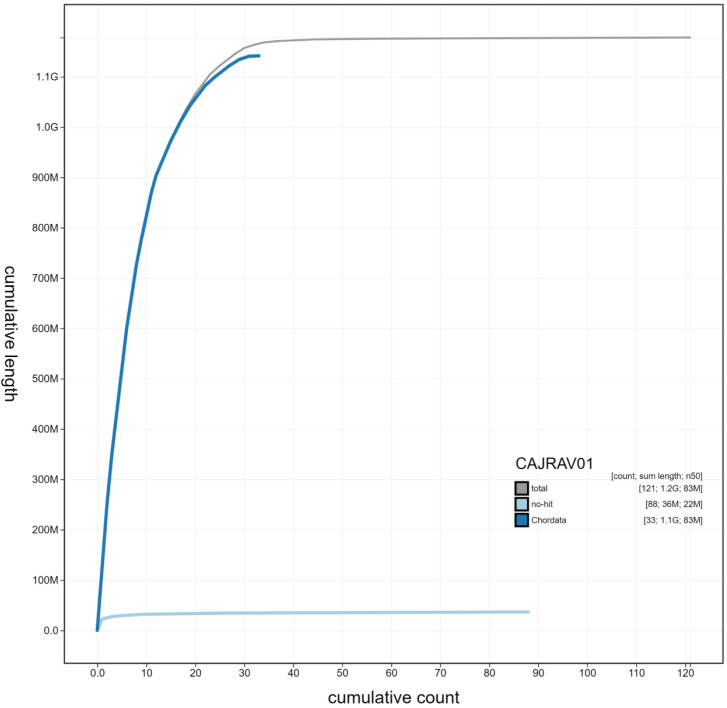
Figure 3. Genome assembly of Caprimulgus europaeus, bCapEur3.1: cumulative sequence.
BlobToolKit cumulative sequence plot. The grey line shows cumulative length for all scaffolds. Coloured lines show cumulative lengths of scaffolds assigned to each phylum using the buscogenes taxrule. An interactive version of this figure is available at https://blobtoolkit.genomehubs.org/view/bCapEur3.1/dataset/CAJRAV01/cumulative.
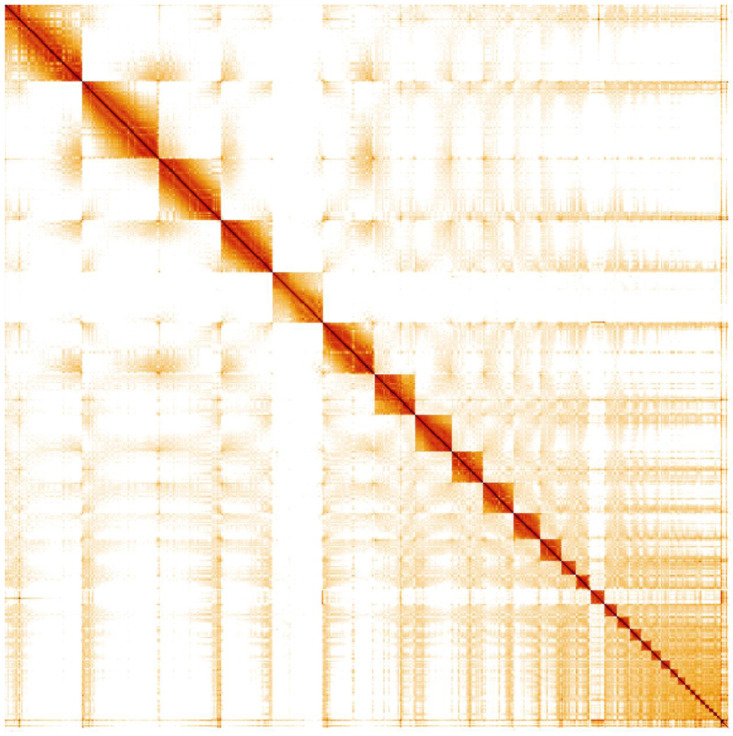
Figure 4. Genome assembly of Caprimulgus europaeus, bCapEur3.1: Hi-C contact map.
Hi-C contact map of the bCapEur3 assembly, visualised in HiGlass. Chromosomes are shown in order of size from left to right and top to bottom.
Table 2. Chromosomal pseudomolecules in the genome assembly of Caprimulgus europaeus, bCapEur3.1.
| INSDC accession | Chromosome | Size (Mb) | GC% |
|---|---|---|---|
| OU015523.1 | 1 | 126.32 | 40.1 |
| OU015524.1 | 2 | 125.37 | 40.3 |
| OU015525.1 | 3 | 100.16 | 39.8 |
| OU015526.1 | 4 | 83.32 | 39.9 |
| OU015528.1 | 5 | 82.61 | 40.7 |
| OU015529.1 | 6 | 65.35 | 41.7 |
| OU015530.1 | 7 | 60.47 | 40.6 |
| OU015531.1 | 8 | 50.91 | 42.8 |
| OU015532.1 | 9 | 48.66 | 41.6 |
| OU015533.1 | 10 | 43.00 | 41.3 |
| OU015534.1 | 11 | 35.23 | 42.1 |
| OU015535.1 | 12 | 23.52 | 43.4 |
| OU015536.1 | 13 | 22.81 | 42.3 |
| OU015538.1 | 14 | 22.35 | 43.3 |
| OU015539.1 | 15 | 19.40 | 42.8 |
| OU015540.1 | 16 | 18.74 | 45 |
| OU015541.1 | 17 | 16.93 | 45.6 |
| OU015542.1 | 18 | 15.70 | 45.4 |
| OU015543.1 | 19 | 13.78 | 46.1 |
| OU015544.1 | 20 | 12.52 | 46.8 |
| OU015545.1 | 21 | 12.35 | 47.5 |
| OU015546.1 | 22 | 9.16 | 46.8 |
| OU015547.1 | 23 | 8.19 | 49.8 |
| OU015548.1 | 24 | 7.57 | 47.7 |
| OU015549.1 | 25 | 7.54 | 51.3 |
| OU015550.1 | 26 | 7.50 | 50.8 |
| OU015551.1 | 27 | 6.26 | 52.3 |
| OU015552.1 | 28 | 6.04 | 48.1 |
| OU015553.1 | 29 | 3.39 | 55.8 |
| OU015554.1 | 30 | 2.94 | 56.1 |
| OU015555.1 | 31 | 2.47 | 49.2 |
| OU015556.1 | 32 | 2.22 | 50.6 |
| OU015557.1 | 33 | 1.26 | 56.6 |
| OU015558.1 | 34 | 0.56 | 51.3 |
| OU015559.1 | 35 | 0.20 | 47.7 |
| OU015537.1 | W | 22.49 | 44.5 |
| OU015527.1 | Z | 82.63 | 40.2 |
| - | Unplaced | 7.86 | 54.9 |
Methods
Sample acquisition
Sampling was performed during the routine activity of the scientific ringing station located in Ventotene island, Latina, Italy (latitude 40.7926°, longitude 13.4241°) during spring migration. Samples have been collected by ISPRA researchers within their institutional activities as from Italian national Law n. 157/92. Bird capture was performed in the evening according to standardized protocols using mist-nets ( Saino et al., 2010; Spina et al., 1993). The sample was collected with a heparinized capillary tube after puncturing the ulnar vein with an intra-epidermal needle. The blood was immediately transferred into 99% ethanol, initially kept at room temperature and then frozen.
DNA extraction and sequencing
High molecular weight DNA was extracted from the blood sample at the Scientific Operations core of the Wellcome Sanger Institute using the Bionano Prep Blood DNA Isolation Kit according to the Bionano Prep Frozen Blood protocol. Pacific Biosciences CLR long read and 10X Genomics read cloud sequencing libraries were constructed according to the manufacturers’ instructions. Sequencing was performed by the Scientific Operations core at the Wellcome Sanger Institute on Pacific Biosciences SEQUEL II and Illumina HiSeq X instruments. Hi-C data were generated from the same blood sample using the Arima Hi-C+ kit and sequenced on HiSeq X.
Genome assembly
Assembly was carried out following the Vertebrate Genome Project pipeline v1.6 ( Rhie et al., 2020) with Falcon-unzip ( Chin et al., 2016), haplotypic duplication was identified and removed with purge_dups ( Guan et al., 2020) and a first round of scaffolding carried out with 10X Genomics read clouds using scaff10x. Scaffolding with Hi-C data ( Rao et al., 2014) was carried out with SALSA2 ( Ghurye et al., 2019). The Hi-C scaffolded assembly was polished with arrow using the PacBio data, with merfin ( Formenti et al., 2021b) applied to avoid a drop in QV, then polished with the 10X Genomics Illumina data by aligning to the assembly with longranger align, calling variants with freebayes ( Garrison & Marth, 2012) and applying homozygous non-reference edits using bcftools consensus. A complete mitochondrion was not found using mitoVGP ( Formenti et al., 2021a), likely due to the sample being sourced from blood tissue, so mitochondrial sequence NC_025773.1 ( Caprimulgus indicus) was used during polishing. The assembly was checked for contamination and corrected using the gEVAL system ( Chow et al., 2016) as described previously ( Howe et al., 2021). Manual curation ( Howe et al., 2021) was performed using gEVAL, HiGlass ( Kerpedjiev et al., 2018) and Pretext. The genome was analysed, and BUSCO scores generated, within the BlobToolKit environment ( Challis et al., 2020). Table 3 gives version numbers of the software tools used in this work.
Table 3. Software tools used.
| Software tool | Version | Source |
|---|---|---|
| Falcon-unzip | 1.8.0 | Chin et al., 2016 |
| purge_dups | 1.2.3 | Guan et al., 2020 |
| SALSA2 | 2.2 | Ghurye et al., 2019 |
| Arrow | GCpp-1.9.0 | https://github.com/PacificBiosciences/GenomicConsensus |
| Merfin | 1.7 | Formenti et al., 2021b |
| longranger align | 2.2.2 | https://support.10xgenomics.com/genome-exome/software/pipelines/latest/advanced/other-pipelines |
| freebayes | 1.3.1-17-gaa2ace8 | Garrison & Marth, 2012 |
| gEVAL | N/A | Chow et al., 2016 |
| HiGlass | 1.11.6 | Kerpedjiev et al., 2018 |
| PretextView | 0.1.x | https://github.com/wtsi-hpag/PretextView |
| BlobToolKit | 2.6.2 | Challis et al., 2020 |
Data availability
European Nucleotide Archive: Caprimulgus europaeus (Eurasian nightjar). Accession number PRJEB44830; https://identifiers.org/ena.embl:PRJEB44830.
The genome sequence is released openly for reuse. The C. europaeus genome sequencing initiative is part of the Darwin Tree of Life (DToL) project and the Vertebrate Genomes Project. All raw sequence data and the assembly have been deposited in INSDC databases. The genome will be annotated and presented through the Ensembl pipeline at the European Bioinformatics Institute. Raw data and assembly accession identifiers are reported in Table 1.
Funding Statement
This work was supported by Wellcome through core funding to the Wellcome Sanger Institute (206194) and the Darwin Tree of Life Discretionary Award (218328).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 2 approved]
Author information
Members of the Darwin Tree of Life Consortium are listed here: https://doi.org/10.5281/zenodo.4783559.
Members of the Darwin Tree of Life Barcoding collective are listed here: https://doi.org/10.5281/zenodo.4893704.
Members of the Wellcome Sanger Institute Tree of Life collective are listed here: https://doi.org/10.5281/zenodo.4783586.
Members of the Sanger Scientific Operations: DNA Pipelines collective are listed here: https://doi.org/10.5281/zenodo.4790456.
Members of the Tree of Life Core Informatics collective are listed here: https://doi.org/10.5281/zenodo.5013542.
References
- Allendorf FW: Genetics and the Conservation of Natural Populations: Allozymes to Genomes. Mol Ecol. 2017;26(2):420–30. 10.1111/mec.13948 [DOI] [PubMed] [Google Scholar]
- Brinkløv S, Fenton MB, Ratcliffe JM: Echolocation in Oilbirds and Swiftlets. Front Physiol. 2013;4:123. 10.3389/fphys.2013.00123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey C: Life In The Cold: Ecological, Physiological, and Molecular Mechanisms. CRC Press,2019. 10.1201/9780429040931 [DOI] [Google Scholar]
- Challis R, Richards E, Rajan J, et al. : BlobToolKit - Interactive Quality Assessment of Genome Assemblies. G3 (Bethesda). 2020;10(4):1361–74. 10.1534/g3.119.400908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin CS, Peluso P, Sedlazeck FJ, et al. : Phased Diploid Genome Assembly with Single-Molecule Real-Time Sequencing. Nat Methods. 2016;13(12):1050–54. 10.1038/nmeth.4035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow W, Brugger K, Caccamo M, et al. : gEVAL — a Web-Based Browser for Evaluating Genome Assemblies. Bioinformatics. 2016;32(16):2508–10. 10.1093/bioinformatics/btw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramp S, Brooks DJ: Vol. IV: Terns to Woodpeckers.1985. Reference Source [Google Scholar]
- Eaton M, Aebischer N, Brown A, et al. : Birds of Conservation Concern 4: The Population Status of Birds in the UK, Channel Islands and Isle of Man. British Birds; an Illustrated Magazine Devoted to the Birds on the British List. 2015;108(12):708–46. Reference Source [Google Scholar]
- Evens R, Beenaerts N, Witters N, et al. : Study on the Foraging Behaviour of the European Nightjar Caprimulgus Europaeus Reveals the Need for a Change in Conservation Strategy in Belgium. J Avian Biol. 2017;48(9):1238–45. 10.1111/jav.00996 [DOI] [Google Scholar]
- Formenti G, Rhie A, Balacco J, et al. : Complete Vertebrate Mitogenomes Reveal Widespread Repeats and Gene Duplications. Genome Biol. 2021a;22(1):120. 10.1186/s13059-021-02336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formenti G, Rhie A, Walenz BP, et al. : Merfin: Improved Variant Filtering and Polishing via K-Mer Validation. bioRxiv. 2021b. 10.1101/2021.07.16.452324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French AR: Hibernation in Birds: Comparisons with Mammals. In: Life in the Cold. CRC Press,2019;43–53. 10.1201/9780429040931-5 [DOI] [Google Scholar]
- Fuentes-Pardo AP, Ruzzante DE: Whole-Genome Sequencing Approaches for Conservation Biology: Advantages, Limitations and Practical Recommendations. Mol Ecol. 2017;26(20):5369–5406. 10.1111/mec.14264 [DOI] [PubMed] [Google Scholar]
- Garrison E, Marth G: Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv: 1207.3907.2012. Reference Source [Google Scholar]
- Ghurye J, Rhie A, Walenz BP, et al. : Integrating Hi-C Links with Assembly Graphs for Chromosome-Scale Assembly. PLoS Comput Biol. 2019;15(8):e1007273. 10.1371/journal.pcbi.1007273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, McCarthy SA, Wood J, et al. : Identifying and Removing Haplotypic Duplication in Primary Genome Assemblies. Bioinformatics. 2020;36(9):2896–98. 10.1093/bioinformatics/btaa025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemeijer WJM, Blair MJ: The EBCC Atlas of European Breeding Birds.Poyser, London,1997;479. Reference Source [Google Scholar]
- Howe K, Chow W, Collins J, et al. : Significantly Improving the Quality of Genome Assemblies through Curation. GigaScience. 2021;10(1):giaa153. 10.1093/gigascience/giaa153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- IUCN: Caprimulgus Europaeus: BirdLife International. IUCN Red List of Threatened Species. IUCN,2016. 10.2305/iucn.uk.2016-3.rlts.t22689887a86103675.en [DOI] [Google Scholar]
- Jiang B, Zhenhua Z, Xu J, et al. : Cloning and Structural Analysis of Complement Component 3d in Wild Birds Provides Insight into Its Functional Evolution. Dev Comp Immunol. 2021;117:103979. 10.1016/j.dci.2020.103979 [DOI] [PubMed] [Google Scholar]
- Keller V, Gerber A, Schmid H, et al. : Rote Liste Brutvögel. Gefährdete Arten Der Schweiz, Stand 2010. Umwelt-Vollzug Nr. 1019.Bundesamt Für Umwelt, Bern, Und Schweizerische Vogelwarte, Sempach.2010. Reference Source [Google Scholar]
- Kerpedjiev P, Abdennur N, Lekschas F, et al. : HiGlass: web-based visual exploration and analysis of genome interaction maps. Genome Biol. 2018;19(1):125. 10.1186/s13059-018-1486-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polakowski M, Broniszewska M, Kirczuk L, et al. : Habitat Selection by the European Nightjar Caprimulgus Europaeus in North-Eastern Poland: Implications for Forest Management. Forests, Trees and Livelihoods. 2020;11(3):291. 10.3390/f11030291 [DOI] [Google Scholar]
- Rao SSP, Huntley MH, Durand NC, et al. : A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159(7):1665–80. 10.1016/j.cell.2014.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, et al. : Towards Complete and Error-Free Genome Assemblies of All Vertebrate Species. bioRxiv. 2020; 2020.05.22.110833. 10.1101/2020.05.22.110833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saino N, Rubolini D, Serra L, et al. : Sex-Related Variation in Migration Phenology in Relation to Sexual Dimorphism: A Test of Competing Hypotheses for the Evolution of Protandry. J Evol Biol. 2010;23(10):2054–65. 10.1111/j.1420-9101.2010.02068.x [DOI] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, et al. : BUSCO: Assessing Genome Assembly and Annotation Completeness with Single-Copy Orthologs. Bioinformatics. 2015;31(19):3210–12. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Spina F, Massi A, Montmaggiori A: Spring Migration across Central Mediterranean: General Results from the" Progetto Piccole Isole. 1993. Reference Source [Google Scholar]
- Supple MA, Shapiro B: Conservation of Biodiversity in the Genomics Era. Genome Biol. 2018;19(1):131. 10.1186/s13059-018-1520-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods CP, Czenze ZJ, Brigham RM: The avian "hibernation" enigma: thermoregulatory patterns and roost choice of the common poorwill. Oecologia. 2019;189(1):47–53. 10.1007/s00442-018-4306-0 [DOI] [PubMed] [Google Scholar]