Abstract
In most animal species, newly formed primordial germ cells (PGCs) acquire the special characteristics that distinguish them from the surrounding somatic cells. Proper fate specification of the PGCs is coupled with transcriptional quiescence, whether they are segregated by determinative or inductive mechanisms. Inappropriate differentiation of PGCs into somatic cells is thought to be prevented due to repression of RNA polymerase (Pol) II-dependent transcription. In the case of a determinative mode of PGC formation (Drosophila, Caenorhabditis elegans, etc.), there is a broad downregulation of Pol II activity. By contrast, PGCs display only gene-specific repression in organisms that rely on inductive signaling-based mechanism (e.g., mice). In addition to the global block of Pol II activity in PGCs, gene expression can be suppressed in other ways, such as chromatin remodeling and Piwi-mediated RNAi. Here, we discuss the mechanisms responsible for the transcriptionally silent state of PGCs in common experimental animals, such as Drosophila, C. elegans, Danio rerio, Xenopus, and mouse. While a PGC-specific downregulation of transcription is a common feature among these organisms, the diverse nature of underlying mechanisms suggests that this functional trait likely evolved independently on several instances. We discuss the possible biological relevance of these silencing mechanisms vis a vis fate determination of PGCs.
Keywords: embryo development, primordial germ cells, transcriptional quiescence, chromatin remodeling, Piwi-mediated RNAi
Introduction
Transmission of genetic information is entirely reliant upon proper specification and functioning of germline stem cells (GSCs), which serve as totipotent precursors for all cell types in the next generation. GSCs have two hallmark traits: a) limited self-renewal and, b) ability to divide asymmetrically to produce cell types capable of differentiating into one or several terminal cell types.
GSCs arise from a special group of cells referred to as primordial germ cells (PGCs). PGCs are segregated from the surrounding soma during early embryonic development. As PGCs serve as the GSC precursors, the unique functional traits of the GSCs can be traced back to the formation and specification of the PGCs. Proper specification of the PGCs is of utmost significance because it allows them to navigate a unique developmental course divergent from the surrounding somatic cells. This early distinction sets the PGCs apart and endows them with the capability of eventual transition into GSCs. The mechanisms underlying early segregation and specification of PGCs are being analysed systematically as they are likely to shed light on how their special identity is established and maintained. Moreover, the acquisition of special traits is crucial for their association with specialized somatic niche cells of the gonad and their gradual transition into GSCs. In the following we will discuss how different modes of PGC specification confer unique properties on the PGCs. Specifically, we will focus on the establishment and/or maintenance of transcriptional quiescence, which distinguishes germ cells from the surrounding soma. Interestingly, although transcriptional silencing of early PGCs is observed in a variety of organisms, distinct molecular mechanisms seem to be employed. Thus, the establishment and/or maintenance of transcriptional quiescence in PGCs may have evolved independently across the evolutionary scale.
There are two modes of PGCs specification, inductive and determinative. The inductive mode, or epigenesis, is widespread among animals. In this mode, PGCs arise de novo from a subset of seemingly equipotent embryonic cells, which come under the influence of inductive signals emanating from the surrounding cells. Supposedly, this is the ancestral way of germline formation that is observed in axolotls and mammals. The determinative mode, or preformation, is characteristic of widespread model organisms, such as the fruit fly Drosophila melanogaster (Drosophila), nematode Caenorhabditis elegans (C. elegans), zebrafish Danio rerio (D. rerio), and clawed frogs Xenopus laevis and Xenopus tropicalis (Xenopus). In these organisms, germ cell-specific mRNAs and proteins are synthesized during oogenesis and deposited in oocytes in the form of ribonucleoprotein complexes, called the germ plasm. The blastomeres that receive the germ plasm give rise to PGCs (Extavour and Akam 2003; Extavour 2007).
One of the unique features of PGCs that is important for their survival and identity is their ability to repress the somatic program. In organisms with the preformation mode of development, global Pol II-dependent transcriptional repression takes place, while transcriptional repression in mammals is not comprehensive, and some germline-specific genes are still transcribed. Transcriptional quiescence of PGCs is thought to prevent their differentiation into soma and, thus, is maintained until the germline program is initiated in late PGCs presumably due to transcriptional activation of germline-specific genes. Moreover, the available data suggest that transcriptional repression in various animals may also prevent premature differentiation of PGCs into meiotic cells and protect the genome from promiscuous transposon activity. In this review, we will focus on the mechanisms that underlie the silencing of transcription that specifies and/or preserves the identity of PGCs during embryogenesis.
PGC formation and specification
The determinative mode of PGC formation is accomplished through the maternally deposited germ plasm, which is characterized by an elevated levels of mitochondria and contains ribonucleoprotein complexes with a number of germline-specific components (Beams and Kessel 1974). Individual constituents of these complexes are synthesized during the middle to late stages of oogenesis and aggregate into the germ plasm, which assembles at the posterior of the oocytes (Little et al. 2015).
In Drosophila, PGCs are the first cells to form in a fertilized embryo. Fertilization is followed by a series of 13 rapid, synchronous nuclear division cycles, with the embryo developing as a syncytium. By nuclear division cycle 9/10, a few nuclei from the center of the embryo migrate into the posteriorly localized germ plasm to give rise to pole buds, which subsequently cellularize to form pole cells (PGCs) at the end of cycle 10. The pole cells divide mitotically one or two more times, and then their divisions cease (Su et al. 1998), while the remaining nuclei continue to divide, reach the periphery, and eventually cellularize by nuclear division cycle 14. While a few genes are transcribed between nuclear cycles 6 and 10 (e.g., sex determination genes including sisterless-a, scute, etc.) (Cline and Meyer 1996), zygotic genome activation (ZGA) is initiated at nuclear division cycle 11 and genome-wide activation of transcription ensues by nuclear cycle 14. By contrast, Pol II-dependent mRNA transcription is switched off in the nuclei that enter the germ plasm (Erickson and Cline 1993; Seydoux and Dunn 1997). For instance, segmentation genes responsible for patterning are transcribed in the somatic nuclei by nuclear cycle 10/11, whereas their transcription is blocked in newly formed pole buds and pole cells. Transcription is not initiated in PGCs until they migrate through the midgut primordium at stage 9 of embryonic development.
Unlike in the case of D. melanogaster, zygotic nuclear divisions are accompanied by cytokinesis in C. elegans, D. rerio, and Xenopus. However, the germ plasm components are selectively partitioned into a few daughter cells /blastomeres whose descendants acquire the PGC fate. In C. elegans, the germline is segregated from the soma after four asymmetric cleavages. The P4 blastomere inherits the germ plasm and gives rise to PGCs. After each asymmetric division, embryonically transcribed mRNAs are detected in the somatic blastomeres, but not in the germline (Seydoux et al. 1996). At the 8-cell stage, the P4 blastomere divides once to produce two PGCs, Z2 and Z3. Global transcriptional silencing is maintained in PGCs, although a few germline-specific genes (e.g., pgl-1 and nos-1) begin to be transcribed in the germline lineage at this stage (Kawasaki et al. 2004). Global ZGA in Z2 and Z3 starts at the first larval stage (L1), only after the larva begins feeding (reviewed in (Wang and Seydoux 2013)).
In Xenopus, the germ plasm is accumulated in the vegetal sub-cortex of oocytes and retains its localization during the four first cleavages. The germ plasm is then translocated from the vegetal subcortical region into the endodermal yolky mass. At the blastula stage, four to five PGCs containing the germ plasm are segregated from the endodermal lineage. PGCs subsequently divide a few times and migrate out of the endoderm (reviewed in (Yang et al. 2015; Butler et al. 2017)).
As in Drosophila, the 12 first zygotic divisions in Xenopus occur very quickly, without ZGA occurring either in somatic cells or in PGCs. Transcription in somatic cells starts at the mid-blastula transition (MBT) stage, when PGCs are still transcriptionally repressed. The zygotic transcription program in PGCs is initiated at the late gastrula stage.
In D. rerio, germ plasm components in oocytes are localized at the vegetal pole and move into the blastodisc after fertilization. At the four-cell stage, the germ plasm concentrates in four cleavage furrows, with this localization leading to the appearance of four PGCs. Prior to gastrulation (after zygotic division 9), each PGC divides mitotically to produce four clusters of PGCs. At the gastrula stage, these four PGCs clusters begin to migrate (reviewed in (Dosch 2015). ZGA in the soma begins at the MBT stage (zygotic division 10), as in Xenopus, whereas ZGA in PGCs does not occur until zygotic division 13 (Knaut et al. 2000).
Organisms that depend on inductive mechanisms of PGC formation, by contrast, do not rely upon maternally accumulated determinants in the zygote. Rather PGCs are induced de novo from undifferentiated, equipotent embryonic cells and the PGC specification is orchestrated by inductive signals. Among the organisms wherein the PGC specification is not determinative but depends on inductive signaling based mechanism, mouse is the best studied model. In mice, after fertilization, the zygote produces the blastocyst, which subsequently differentiates into the epiblast, trophectoderm, and primitive endoderm. The germ cell lineage arises during gastrulation from cells in the proximal epiblast. However, at this stage, the germ cells are not irreversibly determined and can give rise to other embryonic and extra-embryonic tissues. Moreover, cells transplanted into the proximal epiblast from elsewhere can also give rise to PGCs. Interestingly, the extraembryonic ectoderm adjacent to the proximal epiblasts expresses two members of the TGF-Beta superfamily, BMP4 and BMP8, and inactivation of either gene leads to embryos lacking PGCs. Similarly, isolated epiblasts can be induced to form PGCs by adding recombinant BMP4 to the culture medium. Conversely, inactivation of the downstream targets for TGFb signaling, the SMAD transcription factors SMAD1 and SMAD5, leads to a loss of PGCs. Analysis of the pattern of SMAD expression indicates that SMAD1 expression increases in proximal epiblasts during the time when PGC fate is being specified while Smad5 expression is induced. By contrast, SMAD expression in the distal epiblasts decreases during this same period. The BMP-dependent specification process is thought to remodel the transcriptome, generating a unique transcriptional program that differs from that of the surrounding somatic cells. For instance, while the Hox genes are turned on in the nearby somatic cells, they are kept off in the PGCs in part through the action of Blimp1 histone methyltransferase. By contrast, three genes implicated in establishment of pluripotency—Oct4, Nanog, and Sox2—are kept on in the PGCs (reviewed in (Gunesdogan et al. 2014; Gunesdogan and Surani 2016)).
Importantly, a group of genes that directs differentiation of the PGCs into the germline is specifically activated in mouse PGCs immediately after the formation of these cells, whereas repression of somatic and meiotic genes is sustained both in PGC precursors and in PGCs themselves (Nakaki and Saitou 2014; Suzuki et al. 2015; Tu et al. 2016; Endoh et al. 2017).
Thus, PGCs develop almost synchronously with somatic cells in C. elegans, Drosophila, Xenopus, and D. rerio, but activation of the zygotic genome (ZGA) is asynchronous and is considerably delayed in PGCs. It is likely that germ plasm components, which are maternally deposited in the PGCs, play a major role in this process. The germ plasm in Drosophila, C. elegans, D. rerio, and Xenopus embryos determines the germline fate of blastomeres and maintains the identity and number of early PGCs until zygotic transcription of germline genes is activated. Functions of the germ plasm, which is similar in composition to the perinuclear nuage in germ cells, are related mainly to RNA metabolism. Most of the known germ plasm proteins belong to conserved RNA-binding families of transcription and translation factors and components of RNAi machinery.
However, because the germ plasm is capable of repressing transcription on a chromatin template (Lamb and Laird 1976), it may also contain specific factors that attenuate transcription. The exact composition of germinal RNP complexes is unknown. They contain not only proteins and noncoding RNAs but also various mRNAs that are translated at an appropriate time during development (Voronina et al. 2011). RNP complexes in Drosophila may contain up to 200 different transcripts (Frise et al. 2010; Trcek et al. 2015), and several, but not all, of these RNAs are translated immediately post-fertilization, i.e., stage 2 of embryonic development, even prior to the formation of pole cells (Amikura et al. 2001). This early translational activity generates components that are responsible for the establishment and/or maintenance of transcriptional quiescence, including proteins, such as Nanos, Polar granule component, and Germ-cell-less and a pool of chromatin-binding proteins likely responsible for the maintenance of genome-wide silencing in PGCs. Taken together, several regulators involved in maintaining germ cell fate and behavior are generated via this translational control mechanism.
Transcriptional repression
Transcriptional repression at the transcription machinery level
Eukaryotic protein-coding genes are transcribed by Pol II to yield protein-coding mRNAs. Therefore, transcriptional repression applies in this case to Pol II-dependent transcription, which is a multistep process and involves a number of factors. In general, activation of Pol II-dependent transcription proceeds through four steps: preinitiation complex (PIC) assembly on the promoter, transcription initiation and promoter clearance, 5’ mRNA capping and Pol II pausing, and onset of productive elongation (Fig. 1) (Kwak and Lis 2013; Bentley 2014; Gupta et al. 2016; Griesenbeck et al. 2017).
Figure 1.
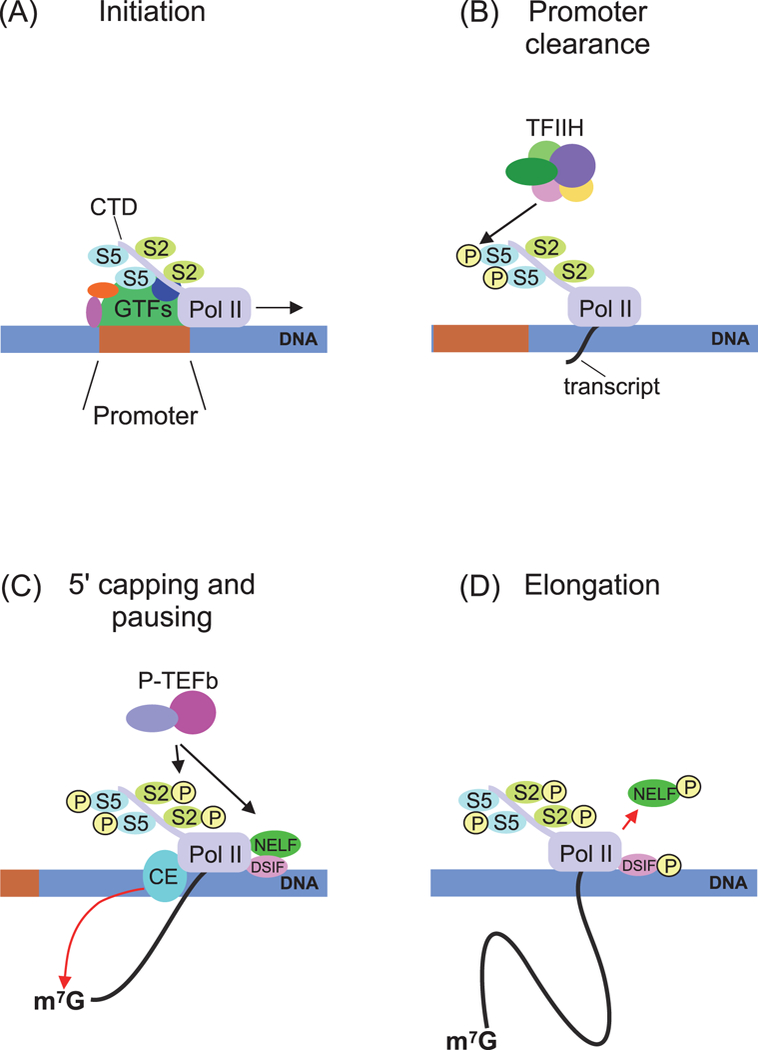
Scheme of Pol II-dependent transcription activation in Drosophila. (A) PIC assembly on a promoter. The complex consists of Pol II and general transcription factors (GTFs). The C-terminal domain (CTD) of Pol II is not phosphorylated at Ser2 and Ser5. (B) Initiation and promoter clearance step. TFIIH phosphorylates Ser5 in the CTD repeats, and then Pol II starts transcript synthesis. (C) 5’ capping and pausing step. Pol II is paused under the action of the negative factors NELF and DSIF. Pausing is resolved by P-TEFb kinase, which phosphorylates CTD Ser2, DSIF, and NELF. Capping enzymes modify the nascent transcript. (D) Elongation step. Phosphorylated NELF moves away from the transcription complex, phosphorylated DSIF acts as a positive factor, and Pol II starts productive transcription elongation.
Global transcriptional repression may occur by blocking PIC assembly. The general transcription factor TFIID is one of the important players belonging to this class. TFIID is composed of the TATA-binding protein (TBP) and a number of TBP-associated factors (TAFs). The TAF4 subunit of TFIID is inactivated in the C. elegans germ plasm-containing blastomeres P0 and P1, which form after the first and second asymmetric zygotic divisions, respectively. Two germ plasm proteins, OMA1 and OMA2, retain TAF4 in the cytoplasm (Wright et al. 2006; Guven-Ozkan et al. 2008) (Fig. 2). In daughter somatic blastomeres, these proteins are phosphorylated and subsequently degraded, TAF4 moves into the nucleus, and ZGA is initiated. In the next generation, the OMA1/2 proteins degrade quickly in the germ plasm-containing blastomere P2, and transcriptional repression is maintained possibly via other mechanism(s).
Figure 2.
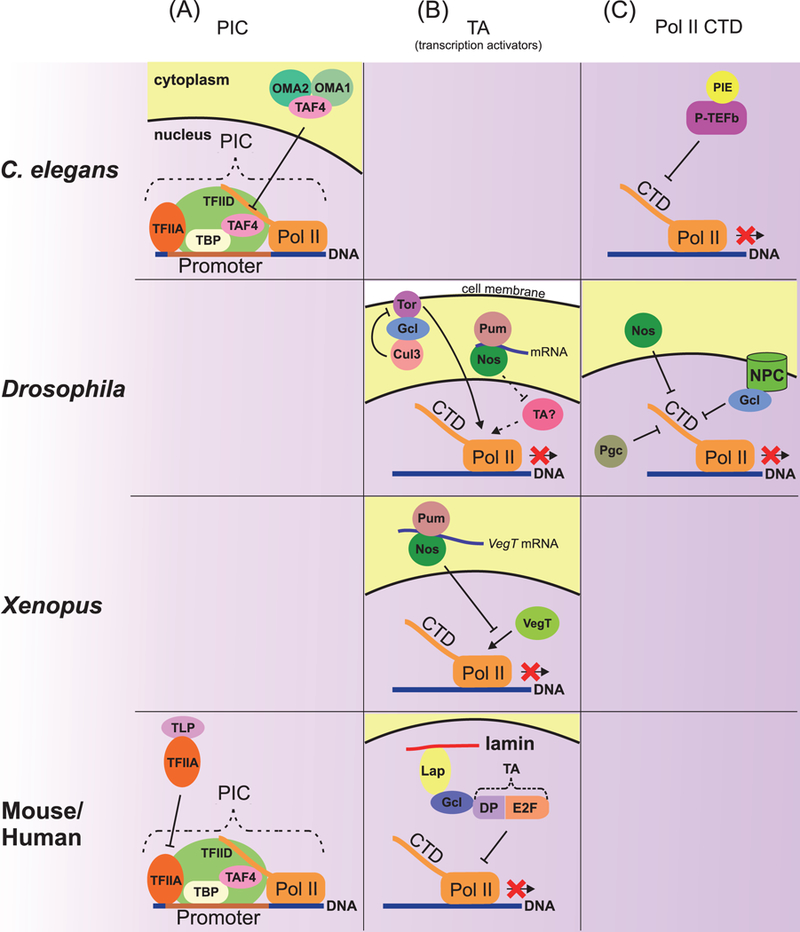
Mechanisms of repression of Pol II-dependent transcription. (A) Repression of transcription at the PIC assembly level. In C. elegans, germ plasm proteins OMA1 and 2 bind and retain the PIC component TAF4 in the cytoplasm, thus affecting PIC assembly in the nucleus. In mouse and human, the ТВР-related factor TLP binds with the unprocessed form of the general transcription factor TFIIA, thereby preventing completion of PIC assembly. (B) Repression of transcriptional activators. In Drosophila, Nos and Pum probably repress translation of an unknown transcriptional activator. In the Drosophila pole plasm, Gcl inactivates Torso signaling, which mediates somatic differentiation in terminal regions of the embryo. Gcl binds both Torso and the Cul3 ubiquitin ligase complex, which degrades Torso in the cell membrane. In Xenopus, Nos and Pum repress translation of the VegT transcriptional activator. In mouse and human, nuclear Gcl anchored to lamin through Lap physically interacts with the transcriptional factor DP-E2F and then inhibits its activity in the nuclear periphery. (C) Repression by a regulation of Pol II CTD phosphorylation. To start mRNA elongation, Pol II has to be modified by phosphorylation of Ser2 in CTD (shown in Fig. 1). In C. elegant, the germ plasm protein PIE inhibits P-TEFb kinase activity, which is required for phosphorylation of CTD Ser2. In Drosophila, the germ plasm proteins Nos, Gcl, and Pgc are involved in regulating CTD phosphorylation. Nuclear Gcl associated with NPC and cytoplasmic Nos inhibit phosphorylation of CTD by unknown mechanisms. Pgc affects CTD phosphorylation via inhibition of P-TEFb kinase activity (shown in detail in Fig. 4). Dashed lines indicate presumptive regulations.
Several Tbp-related factors (Trf2 in Drosophila, Tlp in mammals, and Tlf in C. elegans and D. rerio) may be involved in transcriptional silencing at the transcription initiation step. These proteins perform the functions of Tbp on TATA-less promoters (Muller et al. 2001) and are known to act as transcriptional activators (Kaltenbach et al. 2000; Kopytova et al. 2006). However, human Tlp has been shown to bind an unprocessed form of the general transcription factor TFIIA. Only the processed TFIIA activates transcription by stabilizing the TATA-TBP association. Tlp interferes with TFIIA processing and thereby suppresses transcription of TATA box-containing genes (Suzuki et al. 2015) (Fig. 2). In C. elegans, maternal Tlf may also act as a general transcriptional repressor, but the mechanism of its action is unknown In D. rerio embryos, overexpression of Tlf results in inhibition of ZGA in early embryos presumably because Tlf is capable of suppressing transcription (Muller et al. 2001). In Drosophila, Trf2 is is preferentially enriched on condensed chromatin in late primary spermatocytes (Kopytova et al. 2006), and its enrichment correlates with their transcriptionally silent status and suggests a potential role for Trf2 in transcriptional repression at this stage. Interestingly, a high level of the trf2 mRNA is also detected in the Drosophila germ plasm and PGCs (Lecuyer et al. 2007), but it is still unclear what the functional significance of this trf2 mRNA enrichment is, i.e., whether Trf2 functions as a general transcriptional repressor in Drosophila embryonic PGCs.
It is proposed that transcriptional output is modulated by phosphorylation of the C-terminal domain (CTD) of the largest Pol II subunit. CTD contains multiple copies of the heptapeptide motif YSPTSPS, which are phosphorylated at Ser5 (pSer5 Pol II) during transcription initiation and subsequently at Ser2 (pSer2 Pol II) during transcription elongation (Fig. 1) (for a review, see (Heidemann et al. 2013)). The pSer5 Pol II levels are considerably reduced in Drosophila PGCs and C. elegans P-line blastomeres, whereas pSer2 Pol II is undetectable in these cells. Taken together, these data suggest that Pol II-depended transcription is nearly absent in the worm and fly PGCs (Seydoux and Dunn 1997). Several components of the germ plasm are known to affect the level of Pol II phosphorylation (Fig. 2).
A crucial distinction that sets apart the Drosophila and C. elegans PGCs from the somatic cells concerns the pattern of gene activity. In the period spanning between fertilization and nuclear cycle 9-10, there is little RNA Pol II transcription in the embryo and only a few special genes are expressed (Erickson and Cline 1993). However, once the somatic nuclei arrive at the surface, upregulation of Pol II commences and transcription is fully activated by the cellular blastoderm. Significantly, the mechanisms that turn on somatic transcription are blocked in PGCs, and the PGCs remain transcriptionally quiescent until later in development. Consequently, Pol II genes that are actively transcribed in somatic blastoderm nuclei are not expressed in newly formed PGCs.
Studies from different laboratories over the past couple of decades have uncovered individual regulators that control different characteristic traits, which collectively define the PGC identity. For instance, the Gcl protein appears to be necessary for pole bud formation and proper cellularization of pole cells (Jongens et al. 1992). In addition, the actin binding protein Anillin and two microRNA pathway components, namely, dFMR and Ago2, also contribute to the same process (Deshpande et al. 2005; Deshpande et al. 2006). While Nos/Pum do not influence the precocious cellularization of pole buds, both these genes are needed to restrict the mitotic potential of PGCs by inhibiting the translation of mitotic cyclin, i.e., cyclin B (Kadyrova et al. 2007). On the other hand, the establishment of transcriptional quiescence in fly PGCs is mediated by all the three maternal determinants Gcl, Pgc, and Nos/Pum. These three maternal factors have a few unique targets in addition to some overlapping ones. They function during early nuclear division cycles albeit at different time points. More importantly, they employ distinct mechanisms to influence transcription. Gcl functions when PGCs form, and its target genes are activated prior to the mid-blastula transition. Pgc and Nos are required somewhat later, and while they both prevent transcription of somatic mid-blastula transition genes, there are differences in their targets. Pgc blocks zen and tailless, whereas Nos is required to prevent pair rule genes, like even-skipped, from being activated. Blocking Pol II activity appears to be important for PGC development in Drosophila. Evidently, while PGCs from gcl and pgc mothers can go on to form functional germline stem cells (GSCs), the total number of PGCs in the mutant gonads at stage 15 is considerably reduced. Even more drastic effects are evident in nos embryos. Nos PGCs fail to maintain PGC identity, and unlike either gcl or pgc PGCs, they never develop into functional GSCs.
Though cells in the C. elegans P germline lineage differ from those of flies in that they continue dividing (asymmetrically), transcriptional activity is still downregulated. Like in Drosophila, the elongation CTD marker, pSer2, is absent in the P lineage, while the initiation marker, pSer5, is present at only low levels (Seydoux and Dunn 1997). (Seydoux and Dunn, 1997). Transcriptional quiescence in the germ-line, i.e., P lineage depends upon Pie-1 (Ghosh and Seydoux 2008). When the Z1/Z2 PGCs form at the 100-cell stage, two worm Nos proteins are responsible for maintaining quiescence and re-programming the genome (Wang and Seydoux 2013). In mammals, there is evidence that newly formed PGCs are also subject to transcriptional repression. Initially there is a selective downregulation and reprogramming of gene expression including activation of one of the mammalian nos genes (Yamaguchi et al. 2005). However, unlike Drosophila and many other species, which rely on maternally derived cell autonomous factors, the first steps in PGC determination are controlled by signals from the BMP pathway (for review (Surani et al. 2007)). Once PGC identity is established and the cells begin migrating to the gonad, there is a general inhibition of transcription and a G2 arrest.
Thus, it seems clear that different strategies are employed to establish and maintain the soma-germline distinction. Flies and worms depend upon maternal deposition of specialized germ plasm and subsequent segregation of the germ plasm within the PGCs to establish and maintain germline identity. The use of non-autonomous somatic signals during PGC specification in higher organisms suggests that there are vast differences between mammals and other animals. Since PGCs (and GSCs) perform identical function across the evolutionary scale, it will be interesting to explore possible similarities in the regulatory strategies and underlying mechanisms than anticipated. PGCs specification and maintenance of PGC identity require a proper programming of the transcriptome, presumably, to inactivate the genes/pathways that promote differentiation, while at the same time selectively turning on genes/pathways needed to confer and protect “totipotent” potential. The cross-species comparison between processes leading up to PGC specification raises a number of important questions pertinent to the underlying mechanisms as well as fidelity and biological relevance of this process. While transcriptionally quiescent nature of the PGCs seems to be a conserved trait among model organisms, it is unclear if it is an essential feature, especially in higher organisms. Besides deploying similar strategies to program a totipotent state, the process of PGC specification in different species depends upon many of the orthologous genes (e.g., vasa, nos, and gcl) and conserved signalling pathways. As together the three Drosophila proteins, i.e., Gcl, Nos and Pgc, constitute a group of proteins involved in the establishment and/or maintenance of transcriptional quiescence, in the following section we will summarize salient functional features of these proteins.
Germ cell less (Gcl):
Among the different components involved in PGC determination, Germ cell-less (Gcl) (Jongens et al. 1992) is particularly interesting because it is the only protein known to be involved in the actual formation of PGCs (Jongens et al. 1992; Jongens et al. 1994; Robertson et al. 1999; Cinalli and Lehmann 2013; Lerit et al. 2017)). Interestingly, Gcl encodes a nuclear envelope protein that is well conserved and specifically associates with the nuclei of future PGCs. Nuclear envelope association of Gcl (Jongens et al. 1992; Jongens et al. 1994) dovetailed nicely with its possible influence on transcriptional silencing in light of the transcriptional repression mechanisms that operate at the nuclear periphery (for a review, see (Fedorova and Zink 2008)) (Fig. 2). Moreover, the physical proximity to the nuclear pore complexes (NPCs) (Jongens et al. 1994; Nili et al. 2001) is also conserved in higher organisms; however, direct functional relevance of this association has not been tested.
NPCs influence transcriptional landscape by contributing to the positioning of active euchromatin at the nuclear periphery (Brown and Silver 2007; Ptak et al. 2014). It was recently shown that pre-assembled NPCs from annulate lammelae (AL) are inserted into the nuclear envelope (NE) in Drosophila blastoderm embryos (Hampoelz et al. 2016). Moreover, the NE localization, as well as germline specific function of Gcl, seems to be conserved across evolution (Li et al. 2006). It remains to be determined if the Gcl protein contributes to transcriptional quiescence in newly formed pole buds/PGCs by modulating NPC function, however.
Subsequent analysis performed to uncover the Gcl function in early embryos from Drosophila revealed that it is required for the establishment of transcriptional quiescence correlated with attenuated Pol II activity in early pole buds. Specifically, two sex determination pathway numerator elements, Sisterless A and Sisterless B, are ectopically upregulated in the gcl pole cells (Leatherman et al. 2002). These two X-linked numerator elements have been shown to regulate the activity of the master switch gene Sex-lethal, which controls female identity (Barbash and Cline 1995). Lastly, supporting the conclusion that its role in the establishment/maintenance of transcriptional quiescence is likely a critical function of Gcl, the failure to establish quiescence is tightly correlated with ultimate failure to form the pole cells.
Recent studies showed that Gcl mediates proper germ cell formation by regulating the spatial organization of centrosomes to promote efficient germ plasm segregation and PB formation (Lerit et al. 2017). The ability of the Gcl protein to influence centrosome dynamics is compatible with its subsequent role in cytokinesis and proper execution of the cleavage (Cinalli and Lehmann 2013). Curiously, however, the latter function of Gcl was also thought to be independent of its involvement in the establishment/maintenance of transcriptional quiescence, raising a possibility that Gcl performs two distinct functions that can be mechanistically uncoupled. This possibility gathered further steam when the Gcl protein was shown to mediate degradation of the Torso receptor via the Cul3 ubiquitin ligase complex (Pae et al. 2017). It was also suggested that inactivation of the terminal patterning pathway is at the heart of germline soma distinction engineered by Gcl activity. Consistent with this claim, simultaneous removal of gcl and the torso receptor ligand torso-like (tsl) rescued the loss of germ cells, resulting in elevated numbers of PGCs in stage 5 embryos. It, however, remained unresolved if loss of pole buds seen at stage 3 in gcl mutants was in fact rescued by compromising torso-like levels or the rescue was a result of a ‘bypass’, i.e., enhanced mitosis of the existing ‘unrescued’ buds. Future studies will be necessary to resolve this pertinent question as the answer will shed light on the primary function of Gcl protein during pole bud/PGC formation and specification.
Nanos:
A possible germline-specific function of Nanos during the establishment and/or maintenance of transcriptional quiescence in PGCs was first appreciated by Satoru Kobyashi and colleagues They reported precocious activation of gene expression in the PGCs maternally compromised for nos during mid-embryogenesis. The activity of Nos during silencing of Pol II-dependent transcription was subsequently traced back to syncytial blastoderm-stage embryos (Deshpande et al. 1999). Since these early observations, Nos has proven to be the most conserved factor involved in different aspects of germline development and nos homologues have been reported in most, if not all, model organisms, including humans (Asaoka et al. 1998; Tsuda et al. 2003; Kawasaki et al. 2004; Lai et al. 2012; Bhandari et al. 2014; Campbell et al. 2015). The mechanism underlying the ability of Nos to repress transcription is still unclear although Nos likely influences different stages of the transcription cycle, including the initiation as well as elongation phase. Consistently, Drosophila embryos maternally compromised for the nos gene have an elevated level of Pol II phosphorylated not only at Ser 2 (elongation form) but also at Ser5 (initiation of transcription) (Deshpande et al. 2005). It probably acts by repressing translation of mRNA(s) encoding an unknown transcription factor that regulates Pol II binding to promoter DNA, as shown for Xenopus Nanos1. In Xenopus, transcription of endoderm-specific genes is regulated by the maternal transcription factor VegT. The VegT mRNA is present in PGCs, but its translation is repressed by Nanos-1 and its protein partner, Pumilio (Lai et al. 2012). Recent studies from Seydoux lab on worm homologs of nos, namely, nos1 and nos 2, have provided very interesting clues in this regard (Lee et al. 2017). These authors analyzed the transcriptome profiles of primordial germ cells (PGCs) lacking the nanos homologs nos-1 and nos-2 and demonstrated that Nos homologs are responsible for silencing oocyte specific due to impaired RNA degradation and inappropriate transcriptional activation. They further showed that defective transcriptional downregulation in the germline is due to delayed turnover of the maternally inherited transcription factor LIN-15B, which, in turn, inhibits PRC2 activity (see below). PRC2-dependent reprogramming of the genome downstream of nos1 and nos2 is essential for a proper transcriptional regulation and, hence, the functioning of the germline.
Polar granule component (Pgc):
Pgc was initially discovered as a non-coding RNA whose function is essential during Drosophila germline development (Nakamura et al. 1996; Martinho et al. 2004). It was subsequently shown that Pgc activity was necessary to establish and/or maintain transcriptional quiescence in early embryonic PGCs. Consistently, loss of pgc resulted in ectopic activation of transcription in PGCs. A number of somatic transcripts (zen, tailless, etc.) were observed in germ cells compromised for pgc (Deshpande et al. 2004; Martinho et al. 2004). Subsequent careful molecular analysis demonstrated that the pgc transcript, in fact, encodes a short peptide (Hanyu-Nakamura et al. 2008). The molecular mechanism of transcriptional repression by Pgc is perhaps the best documented. The CDK9-cyclin T complex or transcription elongation factor b (P-TEFb) phosphorylates the Ser2 residue in Pol II CTD (Heidemann et al. 2013) to activate transcription elongation (Fig. 1). Pgc encodes a peptide that resembles Pol II CTD and thus can function as a competitive inhibitor of CDK9 kinase. As a result, in the presence of Pgc, Pol II CTD phosphorylation is attenuated and transcription elongation is repressed (Hanyu-Nakamura et al. 2008) (Fig. 3). The transcriptional regulator PIE-1 from C. elegans also behaves in a similar manner (Ghosh and Seydoux 2008), although it is not a structural homolog of Pgc. PIE-1 repressor recognizes and binds P-TEFb, inhibiting phosphorylation of Pol II CTD. Interestingly, three structurally distinct proteins from different organisms, i.e., Pgc in Drosophila, PIE1 in C. elegans, and PEM in ascidians, perform an analogous function (Ghosh and Seydoux 2008; Hanyu-Nakamura et al. 2008; Kumano et al. 2011). Altogether these observations underscore the fact that, although transcriptional quiescence appears to be a conserved trait, it likely arose independently on many occasions.
Figure 3.
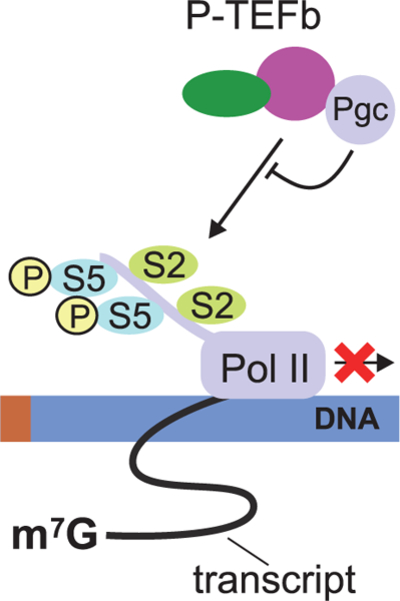
Transcription repression in Drosophila germ cells: Pgc binds to P-TEFb, thereby preventing P-TEFb recruitment onto chromatin and consequently blocking Ser2 phosphorylation.
Global transcriptional repression is not an essential feature of organisms where PGCs form and are specified under the influence of non-autonomous inductive signaling. In mammals, only a subset of genes is repressed in the PGCs that are competent to activate the somatic developmental program. For instance, hox, myc, and evx1 are repressed in mice (Surani et al. 2007), whereas otx2, atct1, and gata4 are repressed in human PGCs (Fang et al. 2018). On the other hand, genes necessary for maintaining the stem cell state and acquisition of a germ cell status (oct3/4, nanog, sox2/17 (sox2 and sox17 are expressed in mice and human respectively)) are activated (for a review, see (Surani et al. 2007)). In mice, the transcription repressor Blimp1, also known as Prdm1, is a key regulator of PGC specification (for a review, see (Wang and Cao 2016)). Prdm1 is thought to control the genes for transcriptional regulators, such as Myc, Hox, and Evx1, and other cell proliferation genes. In addition to the key regulator Blimp1, there are other molecular components responsible for germline specific transcriptional regulation. As shown in an in vitro system, three proteins, namely, Blimp1, AP2 gamma, and Prdm14 (with its corepressor CBFA2T2 (Burton and Torres-Padilla 2016)), together engineer PGC specification (Magnusdottir et al. 2013) by modulating the activity of different cis-regulatory elements involved in transcriptional regulation: Blimp1 binds to the promoter regions; Prdm14, to the enhancers; and AP2 gamma, to distal regulatory elements as well as to the promoters (Ma et al. 2011; Magnusdottir et al. 2013). In human PGCs, BLIMP1 (PRDM1) also represses somatic genes, but its activity is regulated by other transcription factors, including SOX17 and PAX5 (Irie et al. 2015; Fang et al. 2018). The precise mode of action of these factors and how they cooperate to ultimately achieve PGC specification is currently being investigated.
Transcriptional repression at the epigenetic level
Another level of Pol II-depended transcriptional repression in early germ cells is chromatin-based repression through posttranslational modifications of histones and increased chromatin compaction. PGCs carry a number of markers of inactive chromatin and proteins involved in chromatin compaction (Fig. 4). A prominent member of this class of proteins is heterochromatin protein 1 (HP1), which is represented by several variants in Metazoa. Indeed, Drosophila PGCs are enriched with HP1а (Brower-Toland et al. 2007); while mouse PGCs, with HP1alfa, which displays a disperse nuclear localization, which probably reflects the distribution of repression sites (Seki et al. 2005). Mouse HP1gamma, by contrast, is essential for proper germ cells proliferation, and both male and female mutants in the HP1gamma gene are sterile (Abe et al. 2011). Thus, the relative amount of heterochromatin in the nuclei of PGCs is apparently greater than in somatic nuclei.
Figure 4.
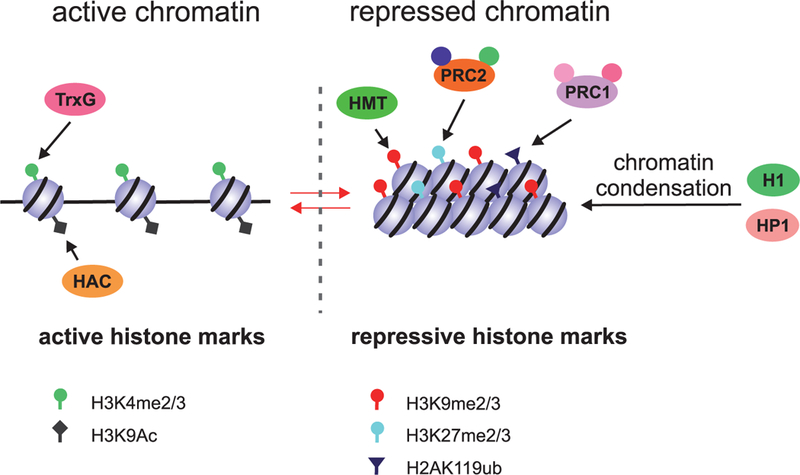
Regulation of transcription at the chromatin level. Transcriptionally active chromatin is characterized by a low level of organization. This state has active histone marks, such as H3K4me2/3 and H3K9ac, which are introduced by the Trithorax group proteins (TrxGs) and histone acetyltransferases (HACs), respectively. Transcriptionally silent chromatin undergoes compaction by the mechanisms that are mediated by HP1 and linker histone H1. Core histones have the inactive chromatin marks H3K9me2/3, H3K27me2/3, and H2AK119ub, which are introduced by lysine histone methyltransferases (HMTs), PRC2, and PRC1, respectively.
An important role in transcriptional repression at the chromatin level is played by diverse variants of the linker histone H1, which is involved in producing compact chromatin. Drosophila has two forms of the protein, dH1 and dBigH1. The dBigH1 form is present in the nuclei of the syncytial blastoderm embryo; upon cellularization and ZGA in the somatic nuclei, it is replaced by dH1 in somatic cells and retained in PGCs. Loss-of-function mutants for dBigH1 show premature ZGA in both somatic cells and PGCs (Perez-Montero et al. 2013). In Xenopus embryos, an embryo-specific H1 variant called B4 is replaced by somatic linker histone H1 during the MBT stage (Dimitrov et al. 1993). However, it is unclear as in the case of Drosophila BigH1, whether B4 is also retained in the Xenopus PGCs. Mouse oocytes contain two specific variants of H1, H1oo and H1°, which are replaced by other variants of H1 in the two-cell stage embryos and, hence, are dispensable for fertility. Mouse H1 histones in different combinations are necessary for embryonic stem cell differentiation. They account for local heterochromatin compaction, e.g., on the promoters of pluripotency genes (for a review, see (Pan and Fan 2016)). Thus, it appears that a unique combination of H1 histone variants contributes to gene repression essential for the proper development of mouse PGCs.
Posttranslational modifications like methylation, acetylation, phosphorylation, and ubiquitynation, of nucleosome core histones at the N-terminal tails mark euchromatin and heterochromatin states (Fig. 4). We will summarize some well-documented modifications that correlate with and potentially regulate transcriptional activation and repression, respectively. Active chromatin is characterized by low compaction and a high level of histone H3 acetylation at Lys9 (H3K9ac) and histone H3 di- and trimethylation at Lys4 (H3K4me2, H3K4me3). Unlike somatic cells, Drosophila PGCs have a very low level of total H3K4me2/3 (Martinho et al. 2004), whereas the levels of H3K9me2 and H3K9me3 are high, indicating an inactive chromatin status (Schaner et al. 2003; Rudolph et al. 2007). In C. elegans, germ plasm-containing P blastomeres retain the markers of active chromatin. Histone modifications indicative of chromatin-based repression appear in PGCs Z2 and Z3 (Schaner et al. 2003; Checchi and Kelly 2006). Chromatin compaction in these cells occurs simultaneously with PIE-1 degradation, but these processes are apparently not interdependent (Checchi and Kelly 2006). It is noteworthy that the translational repressor Nanos seems to play a significant role in the regulation of histone modifications in PGCs of Drosophila and C. elegans. While the precise mechanism of its action is unclear, both in flies and worm PGCs (in nos- mutants), the level of H3K4me3 remains high (Schaner et al. 2003). It also remains to be determined whether this is a consequence of an altered level of transcription.
Many developmental processes and the regulation of gene expression in adult tissues are under the control of Trithorax group (TrxG) and Polycomb group (PcG) of proteins. These two conserved protein complexes act at the epigenetic level in an antagonistic manner. The TrxG factors activate gene expression through H3K4 methylation, while the PcG proteins modify chromatin into an inactive state via other marks in core histones. PcGs form protein complexes, Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) being well known among them. The PcG proteins are recruited to particular DNA sequences called the Polycomb response elements (PREs) in target genes. For gene repression, PRC1 and PRC2 act both interdependently and independently by using diverse enzymatic activities to ubiquitinate and methylate histones, respectively (Schaner et al. 2003; Checchi and Kelly 2006; Rudolph et al. 2007; Tavares et al. 2012; Kassis and Brown 2013; Schwartz and Pirrotta 2014; Kahn et al. 2016; Dorafshan et al. 2017) (Fig. 4). Recent studies on Drosophila embryonic cell cultures have shown that both PRC1 and PRC2 have a global effect on transcriptional repression by extensive chromatin remodeling in intergenic and gene regions, and the functions of these complexes are likely to be independent (Lee et al. 2015). Curiously, transcriptional quiescence and the underlying mechanisms seem to have evolved multiple times in an independent manner. Evidently, transcriptional silencing that operates at the level of transcriptional elongation by inhibiting the function of P-TEFb appears to have originated several times, because this process is regulated in different species by nonhomologous proteins, i.e., Pgc in Drosophila (Hanyu-Nakamura et al. 2008), PIE-1 in C. elegans (Ghosh and Seydoux 2008), and PEM in ascidians (Kumano et al. 2011).
PRC2 controls transcriptional repression in PGCs of C. elegans. The PRC2 proteins MES-2, MES-3, and MES-6 are localized in all nuclei of early embryos, including the germline. Functions of the MES proteins are essential for germline fate to prevent differentiation into somatic cells (Korf et al. 1998; Patel et al. 2012). The Drosophila genome contains at least 16 PcG-coding genes (reviewed in (Pirrotta 1997; Kassis and Brown 2013)). It has been shown that genes encoding the PRC1 proteins are not required for PGCs formation and germline development (Haynie 1983; Breen and Duncan 1986; Soto et al. 1995). PRC2 regulates the development of germ cells in adults (Iovino et al. 2013; Eun et al. 2017), but the role of PRC2 in transcription regulation in Drosophila PGCs is still unknown.
In mouse embryonic stem cells, the promoters of transcriptionally silent developmental regulatory genes are in a bivalent (poised) state. Poised chromatin is characterized by the presence of both activating histone H3 (H3K4me3) and repressive PRC-dependent (H3K27me3) modifications. During embryonic stem cell differentiation, poised promoters may be converted into the active or silent state (Bernstein et al. 2006). Meiotic genes, such as Rec8, Stra8, and Sycp3, have been found to have poised promoters in PGCs (Lesch et al. 2013). PRC1 represses the Stra8 gene and, probably, other meiotic genes in these cells, thereby counteracting the induction of meiosis by retinoic acid (Yokobayashi et al. 2013). As shown recently, a noncanonical PRC1 (containing the ring finger protein PCGF6 and the Myc heterodimerization partner Max) represses late PGC and meiotic genes in mouse embryonic stem cells and germline stem cells by preventing a premature onset of meiosis through mono-ubiquitination of H2AK119 (H2AK119ub) and further recruitment of PRC2, which, in turn, trimethylates H3K27 (Suzuki et al. 2016; Endoh et al. 2017).
Transcriptional repression by RNAi machinery
One of the possible roles for global Pol II inactivation in germ cell survival and maintenance is to repress the activity of transposable elements (TEs), particularly retrotransposons, that are transcribed by this enzyme (Haig 2016). Excessive transcription of TE loci poses a danger of insertional mutagenic events due to generation of new copies of TEs. RNA interference (RNAi) is a conserved mechanism employed in TE silencing. RNAi machinery acts at the transcriptional and posttranscriptional levels as interrelated nuclear and cytoplasmic mechanisms, providing for TE repression through degradation of their transcripts. Nuclear RNAi may be stage- or cell-specific in different species. Some nuclear RNAi proteins are also implicated in transcriptional control of developmental genes, and their functions may be partly or entirely independent of other RNAi components (Castel and Martienssen 2013).
TE silencing in germ cells is achieved mainly via the Piwi-interacting noncoding RNA (piRNA) pathway of RNAi. Piwi is a member of the Argonaute protein family and acts together with other Argonautes to ensure the two-step “ping-pong” cycle of piRNA biogenesis. In general, the piRNA pathway is specific for germ cells, though Piwi has been found in somatic cells in Drosophila (Cox et al. 1998; Brower-Toland et al. 2007; Qi et al. 2011).
Piwi induces nuclear repression of TEs in a complex with piRNAs, which complementarily recognizes elongating mRNAs of TEs (reviewed in (Toth et al. 2016)). Furthermore, the piRNA-Piwi complex induces heterochromatization of several TE loci (Klenov et al. 2007; Shpiz et al. 2009). Notably, Piwi can interact with linker histone H1-associated HP1 and can probably recruit histone methyltransferases (HMTs) to produce H3K9me3 (Brower-Toland et al. 2007; Iwasaki et al. 2016; Toth et al. 2016) (Fig. 5). In Drosophila PGCs, suppression of TE transcription probably results in heterochromatization because Piwi and HP1 (HP1a in Drosophila) have been observed to co-localize in these cells (Brower-Toland et al. 2007). Recently, some other protein players have been found to participate in Piwi-mediated transcriptional repression (Donertas et al. 2013; Sienski et al. 2015). Moreover, Piwi affects TE silencing by inhibiting Pol II-dependent transcription. In ovarian germline cells, the Maelstrom (Mael) protein, a component of nuage, shuttles between the cytoplasm and the nucleus, where it associates with Piwi and blocks Pol II occupancy at TE loci (Sienski et al. 2012).
Figure 5.
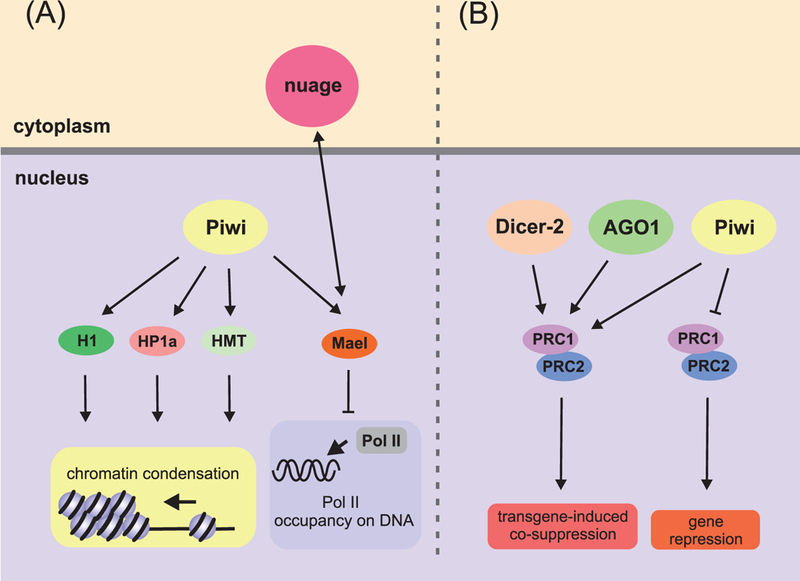
Role of RNAi proteins in repression of Pol II-mediated transcription in Drosophila. (A) Piwi in complex with piRNA interacts with histone H1, heterochromatin protein HP1, and histone methyltransferases (HMTs), thus leading to chromatin condensation in TE-coding regions. The piRNA-Piwi complex can additionally interact with Mael, thereby blocking Pol II association with DNA in TE loci. (B) Regulation of PcG-dependent gene repression by RNAi proteins. Dicer-2, AGO1, and Piwi stabilize contacts between PRC complexes bound to transgenic and endogenous Fab-7 regulatory elements, promoting PcG-dependent repression of the hox gene copies. Piwi can interact with PRC2 proteins independently of RNAi machinery in ovarian stem cells. These interactions lead to a “switching off” of the PgC-dependent silencing of numerous genes.
The mechanism of TE transcriptional silencing in C. elegans differs from that in Drosophila. In adult germ cells of C. elegans, the Piwi homolog PRG-1 in complex with 21U-RNAs (piRNAs) produces 22G-RNAs (siRNAs) (Das et al. 2008) (Fig. 6). 22G-RNAs interact with the germ cell-specific Argonaute proteins WAGO-9 (also referred to as HRGE-1) and WAGO-10, and the 22G-RNA-WAGO-9/10 complexes move into the nuclei, where they recognize elongating target RNAs and initiate methylation of H3K9, which, in turn, is recognized by HPL-2 (HP1 ortholog); thus, stable transgenerational silencing is established (reviewed in (Castel and Martienssen 2013; Kasper et al. 2014)). The nuclear RNAi mechanism is likely to be activated in PGCs because PRG-1 and WAGO-9 are detectable in these cells (Batista et al. 2008; Buckley et al. 2012).
Figure 6.
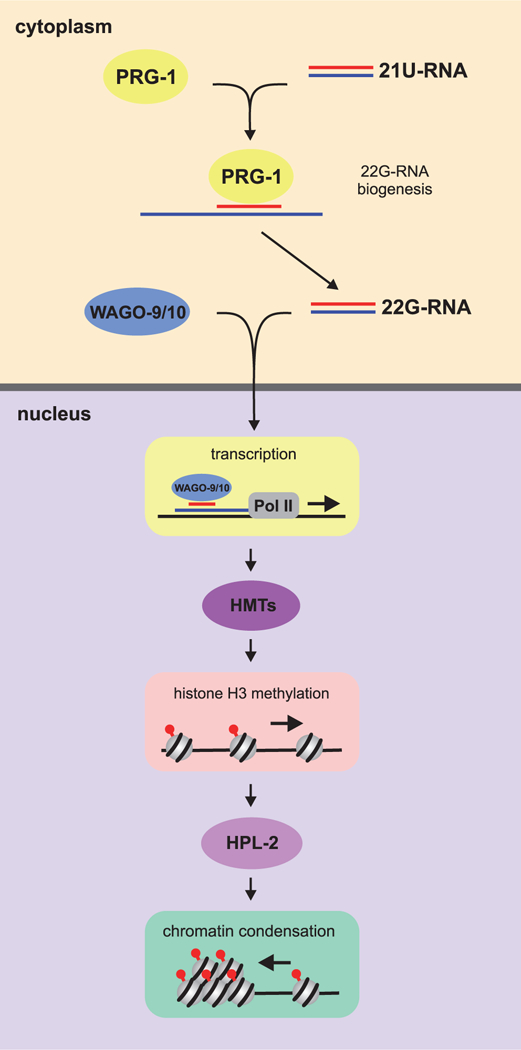
Transcriptional gene silencing through the siRNA-mediated RNAi pathway in the C. elegans germline. This pathway is initiated by 21U-RNAs (piRNAs), whose precursors are transcribed from the 21U-RNA genes of chromosome IV and then processed in the cytoplasm. 21U-RNAs form a complex with the Piwi protein PRG-1, which is loaded on foreign transcripts through imperfect base-pairing, with consequent production of 22G-RNAs (siRNAs). The germ cell-specific Argonaute proteins WAGO-9 and WAGO-10 form complexes with 22G-RNAs, which are then moved into the nucleus and recognize nascent transcripts there. Once loaded onto the target transcripts, these complexes recruit methyltransferases, which trimethylate H3K9. HPL-2 binds to the H3K9me3 mark, contributing to stable transgenerational gene silencing.
The Xenopus Piwi homologs Xiwi and Xili are included in different protein complexes found in eggs and embryos. It is known that Xiwi, which interacts with piRNAs, is a nucleocytoplasmic protein in the oocytes and a component of germ plasm (Lau et al. 2009; Wilczynska et al. 2009; Faunes et al. 2012; Wei et al. 2012). Ziwi, a D. rerio homolog of Piwi, is a protein of maternally deposited germ plasm. Another Piwi homolog, Zili, appears on day 3 after fertilization, has nucleocytoplasmic localization in PGCs during the first week, and then becomes predominantly cytoplasmic. It is suggested that Zili may regulate epigenetic events that affect TE transcription in PGCs (Houwing et al. 2007; Houwing et al. 2008). piRNA biogenesis is activated in eggs and early embryos of D. rerio because their transcriptomes contain abundant piRNAs (Batista et al. 2008).
In mouse embryos, TE activities are repressed at the epigenetic level by DNA methylation in CpG sites inherited from the parental genomes. Somatic and germline cells undergo demethylation, and their DNAs are subsequently methylated de novo during early development, except in female germ cells (reviewed in (Schaefer et al. 2007)). The piRNA pathway is crucial for de novo methylation of TE loci in male germ cells, but seems to be unnecessary in females. The Piwi proteins appear when PGCs reach gonadal rings (reviewed in (Aravin and Hannon 2008)). Beginning from 15.5 dpc to postnatal day 4, the mouse Piwi homologs Mili and Miwi2 guide de novo methylation of several types of TEs in males (Aravin and Hannon 2008; Manakov et al. 2015). In female germ cells, de novo methylation occurs later than in males, at the growing oocyte stage, and key players in this process have not yet been identified (for review, see (Schaefer et al. 2007; Yang and Wang 2016)).
It is noteworthy that some RNAi proteins can regulate the PRC functions, but particular mechanisms of the regulation are context dependent. In Drosophila ovarian germline stem cells and proliferating germ cells, Piwi has been shown to be responsible for a genome-wide decrease in H3K27me3 level and to increase Pol II transcription by downregulating PRC2 independently of other RNAi proteins (Peng et al. 2016). In contrast, the three RNAi proteins Dicer-2, Piwi, and Argonaute1 in Drosophila larvae and adults stabilize long-distance interactions between PRC complexes during transgene-induced co-suppression of multiple copies of PRE-containing Fab-7 elements, which is induced by transgene (Grimaud et al. 2006). On the whole, our current knowledge is that the components of the piRNA pathway are present in germline cells from the time they segregate from the soma, and the transmission of these components to the next generation suggests the involvement of Piwi-dependent processes in TE silencing.
Conclusions
Pol II-dependent transcription is attenuated in the PGCs possibly to maintain their identity during embryogenesis and, in particular, to prevent execution of any somatic program, regardless of determinative or inductive mode of development. Repression of Pol II-dependent transcription in germ cells is a conserved feature among model organisms. As shown recently, global gene repression at the initial stages of gametocyte formation is essential for sexual development in Plasmodium berghei, a protozoan parasite causing malaria in rodents (Yuda et al. 2015).
In PGCs of Drosophila, C. elegans, and Xenopus, global transcriptional repression is due to a combination of genome-wide and gene-specific repressive mechanisms that prevent acquisition of somatic fate and/or apoptotic cell death. On the other hand, the same machinery likely prevents premature activation of the germline-specific transcriptional program. Several overlapping mechanisms, such as disruption of PIC assembly, inhibition of Pol II, epigenetic modifications, and heterochromatin assembly, seem to cooperate to achieve the transcriptional silencing in PGCs.
Gene-specific transcriptional repression in PGCs has been documented in embryonic PGCs from mice, worms and flies. It is accomplished by Polycomb family proteins through promoter inhibition and consequent chromatin remodeling, which leads to inactivation of transcription. The contribution of the Polycomb proteins to specific repression of somatic genes in PGCs of other model organisms has not been analyzed in detail, but their functions appear to be conserved throughout Metazoa, regardless of the mechanisms of germline specification. However, the Polycomb proteins do not appear to exert their repressive influence in a genome-wide manner to silence most, if not all, of the somatic genes, and other as yet unknown players may be involved in this process.
Transcriptional repression of protein-coding genes is global, but apparently not total. For example, the BMP signaling pathway is activated in Drosophila PGCs and is required to maintain their identity (Deshpande et al. 2014). In the PGC, nuclei at stage 5 display substantially elevated levels of phosphorylated Mad (pMad), which is a BMP-regulated transcriptional regulator. Although BMP pathway components are maternally deposited, the thickveins (tkv) gene for the BMP signaling pathway receptor is expressed in the zygote, and its expression is necessary for the accumulation of pMad in PGCs at that stage (Dorfman and Shilo 2001). Moreover, PGCs in Drosophila have elevated levels of at least two components of the Jak/Stat pathway: phosphorylated STAT and its coactivator SAYP (Li et al. 2003; Vorobyeva et al. 2009; Panov et al. 2012), which may contribute to gene-specific activation in PGCs. It appears that expression of a select subset of genes necessary for maintaining the PGC identity may be activated despite global transcriptional repression under conditions where Pol II activity is broadly downregulated.
Transcriptional repression in PGCs can also be viewed as a molecular strategy to prevent excessive amplification of transposable elements by maternal factors (Haig 2016). This alternative view concerning the biological relevance of transcriptional quiescence is consistent with the fact that germ cells need to continually preserve the genome integrity. This is presumably achieved due to the presence the maternally deposited germ plasm until the initiation of zygotic transcription of the germline genes. Taking into account that no transcriptional silencing of TEs takes place in female mouse PGCs, the significance of global transcriptional silencing in PGCs arises probably to repress both somatic genes and TEs, but activities of TEs may be eliminated only at posttranscriptional level in some cases. Moreover, the involvement of RNAi proteins in the regulation of protein-coding genes suggests that global transcriptional silencing is achieved by complex machinery, with certain proteins being necessary to repress the activities of both protein-coding genes and TEs. While global transcriptional repression in PGCs is likely to be ancient, it is clear that it was evolved multiple times, and the underlying mechanisms are still under investigation. Nonetheless, based on the data accumulated from the model organisms, transcriptional silencing may prove to be a widespread approach that PGCs adopt to specify and maintain their fate and eventual successful transition into GSCs.
Acknowledgments
Funding
Y.S. was supported by a Russian Science Foundation grant (16-14-10346). P.S. and G.D were supported by a NIH grant (GM126975).
Footnotes
Disclosure statement
The authors declare no conflict of interest.
References
- 1.Abe K, Naruse C, Kato T, Nishiuchi T, Saitou M, Asano M. 2011. Loss of heterochromatin protein 1 gamma reduces the number of primordial germ cells via impaired cell cycle progression in mice. Biol Reprod. 85:1013–1024. [DOI] [PubMed] [Google Scholar]
- 2.Amikura R, Kashikawa M, Nakamura A, Kobayashi S. 2001. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci USA. 98:9133–9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravin AA, Hannon GJ. 2008. Small RNA silencing pathways in germ and stem cells. Cold Spring Harb Symp Quant Biol. 73:283–290. [DOI] [PubMed] [Google Scholar]
- 4.Asaoka M, Sano H, Obara Y, Kobayashi S. 1998. Maternal Nanos regulates zygotic gene expression in germline progenitors of Drosophila melanogaster. Mech Dev. 78:153–158. [DOI] [PubMed] [Google Scholar]
- 5.Barbash DA, Cline TW. 1995. Genetic and molecular analysis of the autosomal component of the primary sex determination signal of Drosophila melanogaster. Genetics. 141:1451–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, Conte D, Jr., Luo S, Schroth GP, Carrington JC, Bartel DP, Mello CC. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell. 31:67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beams HW, Kessel RG. 1974. The problem of germ cell determinants. Int Rev Cytol. 39:413–479. [DOI] [PubMed] [Google Scholar]
- 8.Bentley DL. 2014. Coupling mRNA processing with transcription in time and space. Nat Rev Genet. 15:163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 125:315–326. [DOI] [PubMed] [Google Scholar]
- 10.Bhandari D, Raisch T, Weichenrieder O, Jonas S, Izaurralde E. 2014. Structural basis for the Nanos-mediated recruitment of the CCR4-NOT complex and translational repression. Genes Dev. 28:888–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breen TR, Duncan IM. 1986. Maternal expression of genes that regulate the bithorax complex of Drosophila melanogaster. Dev Biol. 118:442–456. [DOI] [PubMed] [Google Scholar]
- 12.Brower-Toland B, Findley SD, Jiang L, Liu L, Yin H, Dus M, Zhou P, Elgin SC, Lin H. 2007. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 21:2300–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brown CR, Silver PA. 2007. Transcriptional regulation at the nuclear pore complex. Curr Opin Genet Dev. 17:100–106. [DOI] [PubMed] [Google Scholar]
- 14.Buckley BA, Burkhart KB, Gu SG, Spracklin G, Kershner A, Fritz H, Kimble J, Fire A, Kennedy S. 2012. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 489:447–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton A, Torres-Padilla ME. 2016. A Pluripotency Platform for Prdm14. Dev Cell. 38:3–5. [DOI] [PubMed] [Google Scholar]
- 16.Butler AM, Aguero T, Newman KM, King ML. 2017. Primordial Ggrm cell isolation from Xenopus laevis embryos. Methods Mol Biol. 1463:115–124. [DOI] [PubMed] [Google Scholar]
- 17.Campbell PD, Chao JA, Singer RH, Marlow FL. 2015. Dynamic visualization of transcription and RNA subcellular localization in zebrafish. Development. 142:1368–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castel SE, Martienssen RA. 2013. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 14:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Checchi PM, Kelly WG. 2006. emb-4 is a conserved gene required for efficient germline-specific chromatin remodeling during Caenorhabditis elegans embryogenesis. Genetics. 174:1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cinalli RM, Lehmann R. 2013. A spindle-independent cleavage pathway controls germ cell formation in Drosophila. Nat Cell Biol. 15:839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cline TW, Meyer BJ. 1996. Vive la difference: males vs females in flies vs worms. Annu Rev Genet. 30:637–702. [DOI] [PubMed] [Google Scholar]
- 22.Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H. 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev. 12:3715–3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, Matthews N, Berezikov E, Ketting RF, Tavare S, Miska EA. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell. 31:79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deshpande G, Calhoun G, Schedl P. 2004. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development. 131:1247–1257. [DOI] [PubMed] [Google Scholar]
- 25.Deshpande G, Calhoun G, Schedl P. 2005. Drosophila argonaute-2 is required early in embryogenesis for the assembly of centric/centromeric heterochromatin, nuclear division, nuclear migration, and germ-cell formation. Genes Dev. 19:1680–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deshpande G, Calhoun G, Schedl P. 2006. The drosophila fragile X protein dFMR1 is required during early embryogenesis for pole cell formation and rapid nuclear division cycles. Genetics. 174:1287–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deshpande G, Calhoun G, Yanowitz JL, Schedl PD. 1999. Novel functions of nanos in downregulating mitosis and transcription during the development of the Drosophila germline. Cell. 99:271–281. [DOI] [PubMed] [Google Scholar]
- 28.Deshpande G, Willis E, Chatterjee S, Fernandez R, Dias K, Schedl P. 2014. BMP signaling and the maintenance of primordial germ cell identity in Drosophila embryos. PloS One. 9:e88847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dimitrov S, Almouzni G, Dasso M, Wolffe AP. 1993. Chromatin transitions during early Xenopus embryogenesis: changes in histone H4 acetylation and in linker histone type. Dev Biol. 160:214–227. [DOI] [PubMed] [Google Scholar]
- 30.Donertas D, Sienski G, Brennecke J. 2013. Drosophila Gtsf1 is an essential component of the Piwi-mediated transcriptional silencing complex. Genes Dev. 27:1693–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dorafshan E, Kahn TG, Schwartz YB. 2017. Hierarchical recruitment of Polycomb complexes revisited. Nucleus. 8:496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dorfman R, Shilo BZ. 2001. Biphasic activation of the BMP pathway patterns the Drosophila embryonic dorsal region. Development. 128:965–972. [DOI] [PubMed] [Google Scholar]
- 33.Dosch R 2015. Next generation mothers: Maternal control of germline development in zebrafish. Crit Rev Biochem Mol Biol. 50:54–68. [DOI] [PubMed] [Google Scholar]
- 34.Endoh M, Endo TA, Shinga J, Hayashi K, Farcas A, Ma KW, Ito S, Sharif J, Endoh T, Onaga N, Nakayama M, Ishikura T, Masui O, Kessler BM, Suda T, Ohara O, Okuda A, Klose R, Koseki H. 2017. PCGF6-PRC1 suppresses premature differentiation of mouse embryonic stem cells by regulating germ cell-related genes. eLife. 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erickson JW, Cline TW. 1993. A bZIP protein, sisterless-a, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 7:1688–1702. [DOI] [PubMed] [Google Scholar]
- 36.Eun SH, Feng L, Cedeno-Rosario L, Gan Q, Wei G, Cui K, Zhao K, Chen X. 2017. Polycomb group gene E(z) is required for spermatogonial dedifferentiation in Drosophila adult testis. J Mol Biol. 429:2030–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Extavour CG. 2007. Evolution of the bilaterian germ line: lineage origin and modulation of specification mechanisms. Integr Comp Biol. 47:770–785. [DOI] [PubMed] [Google Scholar]
- 38.Extavour CG, Akam M. 2003. Mechanisms of germ cell specification across the metazoans: epigenesis and preformation. Development. 130:5869–5884. [DOI] [PubMed] [Google Scholar]
- 39.Fang F, Angulo B, Xia N, Sukhwani M, Wang Z, Carey CC, Mazurie A, Cui J, Wilkinson R, Wiedenheft B, Irie N, Surani MA, Orwig KE, Reijo Pera RA. 2018. A PAX5-OCT4-PRDM1 developmental switch specifies human primordial germ cells. Nat Cell Biol. 20:655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Faunes F, Almonacid LI, Melo F, Larrain J. 2012. Characterization of small RNAs in X. tropicalis gastrulae. Genesis. 50:572–583. [DOI] [PubMed] [Google Scholar]
- 41.Fedorova E, Zink D. 2008. Nuclear architecture and gene regulation. Biochim Biophys Acta. 1783:2174–2184. [DOI] [PubMed] [Google Scholar]
- 42.Frise E, Hammonds AS, Celniker SE. 2010. Systematic image-driven analysis of the spatial Drosophila embryonic expression landscape. Mol Syst Biol. 6:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh D, Seydoux G. 2008. Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation. Genetics. 178:235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Griesenbeck J, Tschochner H, Grohmann D. 2017. Structure and Function of RNA Polymerases and the Transcription Machineries. Subcell Biochem. 83:225–270. [DOI] [PubMed] [Google Scholar]
- 45.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. 2006. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 124:957–971. [DOI] [PubMed] [Google Scholar]
- 46.Gunesdogan U, Surani MA. 2016. Developmental competence for primordial germ cell fate. Curr Top Dev Biol. 117:471–496. [DOI] [PubMed] [Google Scholar]
- 47.Gunesdogan U, Magnusdottir E, Surani MA. 2014. Primordial germ cell specification: a context-dependent cellular differentiation event. Philos Trans R Soc Lond B Biol Sci. 369:20130543–20130543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta K, Sari-Ak D, Haffke M, Trowitzsch S, Berger I. 2016. Zooming in on transcription preinitiation. J Mol Biol. 428:2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guven-Ozkan T, Nishi Y, Robertson SM, Lin R. 2008. Global transcriptional repression in C. elegans germline precursors by regulated sequestration of TAF-4. Cell. 135:149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Haig D 2016. Transposable elements: Self-seekers of the germline, team-players of the soma. Bioessays. 38:1158–1166. [DOI] [PubMed] [Google Scholar]
- 51.Hampoelz B, Mackmull MT, Machado P, Ronchi P, Bui KH, Schieber N, Santarella-Mellwig R, Necakov A, Andres-Pons A, Philippe JM, Lecuit T, Schwab Y, Beck M. 2016. Pre-assembled nuclear pores insert into the nuclear envelope during early development. Cell. 166:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hanyu-Nakamura K, Sonobe-Nojima H, Tanigawa A, Lasko P, Nakamura A. 2008. Drosophila Pgc protein inhibits P-TEFb recruitment to chromatin in primordial germ cells. Nature. 451:730–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haynie JL. 1983. The maternal and zygotic roles of the gene Polycomb in embryonic determination in Drosophila melanogaster. Dev Biol. 100:399–411. [DOI] [PubMed] [Google Scholar]
- 54.Heidemann M, Hintermair C, Voss K, Eick D. 2013. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 1829:55–62. [DOI] [PubMed] [Google Scholar]
- 55.Houwing S, Berezikov E, Ketting RF. 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J. 27:2702–2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, Plasterk RH, Hannon GJ, Draper BW, Ketting RF. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell. 129:69–82. [DOI] [PubMed] [Google Scholar]
- 57.Iovino N, Ciabrelli F, Cavalli G. 2013. PRC2 controls Drosophila oocyte cell fate by repressing cell cycle genes. Dev Cell. 26:431–439. [DOI] [PubMed] [Google Scholar]
- 58.Irie N, Weinberger L, Tang WW, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, Surani MA. 2015. SOX17 is a critical specifier of human primordial germ cell fate. Cell. 160:253–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iwasaki YW, Murano K, Ishizu H, Shibuya A, Iyoda Y, Siomi MC, Siomi H, Saito K. 2016. Piwi modulates chromatin accessibility by regulating multiple factors including histone H1 to repress transposons. Mol Cell. 63:408–419. [DOI] [PubMed] [Google Scholar]
- 60.Jongens TA, Hay B, Jan LY, Jan YN. 1992. The germ cell-less gene product: a posteriorly localized component necessary for germ cell development in Drosophila. Cell. 70:569–584. [DOI] [PubMed] [Google Scholar]
- 61.Jongens TA, Ackerman LD, Swedlow JR, Jan LY, Jan YN. 1994. Germ cell-less encodes a cell type-specific nuclear pore-associated protein and functions early in the germ-cell specification pathway of Drosophila. Genes Dev. 8:2123–2136. [DOI] [PubMed] [Google Scholar]
- 62.Kadyrova LY, Habara Y, Lee TH, Wharton RP. 2007. Translational control of maternal Cyclin B mRNA by Nanos in the Drosophila germline. Development. 134:1519–1527. [DOI] [PubMed] [Google Scholar]
- 63.Kahn TG, Dorafshan E, Schultheis D, Zare A, Stenberg P, Reim I, Pirrotta V, Schwartz YB. 2016. Interdependence of PRC1 and PRC2 for recruitment to Polycomb Response Elements. Nucleic Acids Res. 44:10132–10149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaltenbach L, Horner MA, Rothman JH, Mango SE. 2000. The TBP-like factor CeTLF is required to activate RNA polymerase II transcription during C. elegans embryogenesis. Mol Cell. 6:705–713. [DOI] [PubMed] [Google Scholar]
- 65.Kasper DM, Gardner KE, Reinke V. 2014. Homeland security in the C. elegans germ line: insights into the biogenesis and function of piRNAs. Epigenetics. 9:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kassis JA, Brown JL. 2013. Polycomb group response elements in Drosophila and vertebrates. Adv Genet. 81:83–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kawasaki I, Amiri A, Fan Y, Meyer N, Dunkelbarger S, Motohashi T, Karashima T, Bossinger O, Strome S. 2004. The PGL family proteins associate with germ granules and function redundantly in Caenorhabditis elegans germline development. Genetics. 167:645–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, Gvozdev VA. 2007. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 35:5430–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C. 2000. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol. 149:875–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kobayashi S, Yamada M, Asaoka M, Kitamura T. 1996. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature. 380:708–711. [DOI] [PubMed] [Google Scholar]
- 71.Kopytova DV, Krasnov AN, Kopantceva MR, Nabirochkina EN, Nikolenko JV, Maksimenko O, Kurshakova MM, Lebedeva LA, Yerokhin MM, Simonova OB, Korochkin LI, Tora L, Georgiev PG, Georgieva SG. 2006. Two isoforms of Drosophila TRF2 are involved in embryonic development, premeiotic chromatin condensation, and proper differentiation of germ cells of both sexes. Mol Cell Biol. 26:7492–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Korf I, Fan Y, Strome S. 1998. The Polycomb group in Caenorhabditis elegans and maternal control of germline development. Development. 125:2469–2478. [DOI] [PubMed] [Google Scholar]
- 73.Kumano G, Takatori N, Negishi T, Takada T, Nishida H. 2011. A maternal factor unique to ascidians silences the germline via binding to P-TEFb and RNAP II regulation. Curr Biol. 21:1308–1313. [DOI] [PubMed] [Google Scholar]
- 74.Kwak H, Lis JT. 2013. Control of transcriptional elongation. Annu Rev Genet. 47:483–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lai F, Singh A, King ML. 2012. Xenopus Nanos1 is required to prevent endoderm gene expression and apoptosis in primordial germ cells. Development. 139:1476–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lamb MM, Laird CD. 1976. Increase in nuclear poly(A)-containing RNA at syncytial blastoderm in Drosophila melanogaster embryos. Dev Biol. 52:31–42. [DOI] [PubMed] [Google Scholar]
- 77.Lau NC, Ohsumi T, Borowsky M, Kingston RE, Blower MD. 2009. Systematic and single cell analysis of Xenopus Piwi-interacting RNAs and Xiwi. EMBO J. 28:2945–2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leatherman JL, Levin L, Boero J, Jongens TA. 2002. germ cell-less acts to repress transcription during the establishment of the Drosophila germ cell lineage. Curr Biol. 12:1681–1685. [DOI] [PubMed] [Google Scholar]
- 79.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 131:174–187. [DOI] [PubMed] [Google Scholar]
- 80.Lee CS, Lu T, Seydoux G. 2017. Nanos promotes epigenetic reprograming of the germline by down-regulation of the THAP transcription factor LIN-15B. eLife. 6:e30201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lee HG, Kahn TG, Simcox A, Schwartz YB, Pirrotta V. 2015. Genome-wide activities of Polycomb complexes control pervasive transcription. Genome Res. 25:1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lerit DA, Shebelut CW, Lawlor KJ, Rusan NM, Gavis ER, Schedl P, Deshpande G. 2017. Germ cell-less promotes centrosome segregation to induce germ cell formation. Cell Rep. 18:831–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lesch BJ, Dokshin GA, Young RA, McCarrey JR, Page DC. 2013. A set of genes critical to development is epigenetically poised in mouse germ cells from fetal stages through completion of meiosis. Proc Natl Acad Sci USA. 110:16061–16066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J, Xia F, Li WX. 2003. Coactivation of STAT and Ras is required for germ cell proliferation and invasive migration in Drosophila. Dev Cell. 5:787–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li W, Deng F, Wang H, Zhen Y, Xiang F, Sui Y, Li J. 2006. Germ cell-less expression in zebrafish embryos. Dev Growth Differ. 48:333–338. [DOI] [PubMed] [Google Scholar]
- 86.Little SC, Sinsimer KS, Lee JJ, Wieschaus EF, Gavis ER. 2015. Independent and coordinate trafficking of single Drosophila germ plasm mRNAs. Nat Cell Biol. 17:558–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ma Z, Swigut T, Valouev A, Rada-Iglesias A, Wysocka J. 2011. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat Struct Mol Biol. 18:120–127. [DOI] [PubMed] [Google Scholar]
- 88.Magnusdottir E, Dietmann S, Murakami K, Gunesdogan U, Tang F, Bao S, Diamanti E, Lao K, Gottgens B, Azim Surani M. 2013. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol. 15:905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Manakov SA, Pezic D, Marinov GK, Pastor WA, Sachidanandam R, Aravin AA. 2015. MIWI2 and MILI have differential effects on piRNA biogenesis and DNA methylation. Cell Rep. 12:1234–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Martinho RG, Kunwar PS, Casanova J, Lehmann R. 2004. A noncoding RNA is required for the repression of RNApolII-dependent transcription in primordial germ cells. Curr Biol. 14:159–165. [DOI] [PubMed] [Google Scholar]
- 91.Muller F, Lakatos L, Dantonel J, Strahle U, Tora L. 2001. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol. 11:282–287. [DOI] [PubMed] [Google Scholar]
- 92.Nakaki F, Saitou M. 2014. PRDM14: a unique regulator for pluripotency and epigenetic reprogramming. Trends Biochem Sci. 39:289–298. [DOI] [PubMed] [Google Scholar]
- 93.Nakamura A, Amikura R, Mukai M, Kobayashi S, Lasko PF. 1996. Requirement for a noncoding RNA in Drosophila polar granules for germ cell establishment. Science. 274:2075–2079. [DOI] [PubMed] [Google Scholar]
- 94.Nili E, Cojocaru GS, Kalma Y, Ginsberg D, Copeland NG, Gilbert DJ, Jenkins NA, Berger R, Shaklai S, Amariglio N, Brok-Simoni F, Simon AJ, Rechavi G. 2001. Nuclear membrane protein LAP2beta mediates transcriptional repression alone and together with its binding partner GCL (germ-cell-less). J Cell Sci. 114:3297–3307. [DOI] [PubMed] [Google Scholar]
- 95.Pae J, Cinalli RM, Marzio A, Pagano M, Lehmann R. 2017. GCL and CUL3 control the switch between cell lineages by mediating localized degradation of an RTK. Dev Cell. 42:130–142.e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Pan C, Fan Y. 2016. Role of H1 linker histones in mammalian development and stem cell differentiation. Biochim Biophys Acta. 1859:496–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Panov VV, Kuzmina JL, Doronin SA, Kopantseva MR, Nabirochkina EN, Georgieva SG, Vorobyeva NE, Shidlovskii YV. 2012. Transcription co-activator SAYP mediates the action of STAT activator. Nucleic Acids Res. 40:2445–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patel T, Tursun B, Rahe DP, Hobert O. 2012. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep. 2:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peng JC, Valouev A, Liu N, Lin H. 2016. Piwi maintains germline stem cells and oogenesis in Drosophila through negative regulation of Polycomb group proteins. Nat Genet. 48:283–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Perez-Montero S, Carbonell A, Moran T, Vaquero A, Azorin F. 2013. The embryonic linker histone H1 variant of Drosophila, dBigH1, regulates zygotic genome activation. Dev Cell. 26:578–590. [DOI] [PubMed] [Google Scholar]
- 101.Pirrotta V 1997. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 13:314–318. [DOI] [PubMed] [Google Scholar]
- 102.Ptak C, Aitchison JD, Wozniak RW. 2014. The multifunctional nuclear pore complex: a platform for controlling gene expression. Curr Opin Cell Biol. 28:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Qi H, Watanabe T, Ku HY, Liu N, Zhong M, Lin H. 2011. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J Biol Chem. 286:3789–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Robertson SE, Dockendorff TC, Leatherman JL, Faulkner DL, Jongens TA. 1999. germ cell-less is required only during the establishment of the germ cell lineage of Drosophila and has activities which are dependent and independent of its localization to the nuclear envelope. Dev Biol. 215:288–297. [DOI] [PubMed] [Google Scholar]
- 105.Rudolph T, Yonezawa M, Lein S, Heidrich K, Kubicek S, Schafer C, Phalke S, Walther M, Schmidt A, Jenuwein T, Reuter G. 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3-3. Mol Cell. 26:103–115. [DOI] [PubMed] [Google Scholar]
- 106.Schaefer CB, Ooi SK, Bestor TH, Bourc’his D. 2007. Epigenetic decisions in mammalian germ cells. Science. 316:398–399. [DOI] [PubMed] [Google Scholar]
- 107.Schaner CE, Deshpande G, Schedl PD, Kelly WG. 2003. A conserved chromatin architecture marks and maintains the restricted germ cell lineage in worms and flies. Dev Cell. 5:747–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Schwartz YB, Pirrotta V. 2014. Ruled by ubiquitylation: a new order for polycomb recruitment. Cell Rep. 8:321–325. [DOI] [PubMed] [Google Scholar]
- 109.Seki Y, Hayashi K, Itoh K, Mizugaki M, Saitou M, Matsui Y. 2005. Extensive and orderly reprogramming of genome-wide chromatin modifications associated with specification and early development of germ cells in mice. Dev Biol. 278:440–458. [DOI] [PubMed] [Google Scholar]
- 110.Seydoux G, Dunn MA. 1997. Transcriptionally repressed germ cells lack a subpopulation of phosphorylated RNA polymerase II in early embryos of Caenorhabditis elegans and Drosophila melanogaster. Development. 124:2191–2201. [DOI] [PubMed] [Google Scholar]
- 111.Seydoux G, Mello CC, Pettitt J, Wood WB, Priess JR, Fire A. 1996. Repression of gene expression in the embryonic germ lineage of C. elegans. Nature. 382:713–716. [DOI] [PubMed] [Google Scholar]
- 112.Shpiz S, Kwon D, Rozovsky Y, Kalmykova A. 2009. rasiRNA pathway controls antisense expression of Drosophila telomeric retrotransposons in the nucleus. Nucleic Acids Res. 37:268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sienski G, Donertas D, Brennecke J. 2012. Transcriptional silencing of transposons by Piwi and maelstrom and its impact on chromatin state and gene expression. Cell. 151:964–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sienski G, Batki J, Senti KA, Donertas D, Tirian L, Meixner K, Brennecke J. 2015. Silencio/CG9754 connects the Piwi-piRNA complex to the cellular heterochromatin machinery. Genes Dev. 29:2258–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Soto MC, Chou TB, Bender W. 1995. Comparison of germline mosaics of genes in the Polycomb group of Drosophila melanogaster. Genetics. 140:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Su TT, Campbell SD, O’Farrell PH. 1998. The cell cycle program in germ cells of the Drosophila embryo. Dev Biol. 196:160–170. [DOI] [PubMed] [Google Scholar]
- 117.Surani MA, Hayashi K, Hajkova P. 2007. Genetic and epigenetic regulators of pluripotency. Cell. 128:747–762. [DOI] [PubMed] [Google Scholar]
- 118.Suzuki A, Hirasaki M, Hishida T, Wu J, Okamura D, Ueda A, Nishimoto M, Nakachi Y, Mizuno Y, Okazaki Y, Matsui Y, Izpisua Belmonte JC, Okuda A. 2016. Loss of MAX results in meiotic entry in mouse embryonic and germline stem cells. Nat Commun. 7:11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Suzuki H, Isogai M, Maeda R, Ura K, Tamura TA. 2015. TBP-like protein (TLP) interferes with Taspase1-mediated processing of TFIIA and represses TATA box gene expression. Nucleic Acids Res. 43:6285–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell. 148:664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Toth KF, Pezic D, Stuwe E, Webster A. 2016. The piRNA pathway guards the germline genome against transposable elements. Adv Exp Med Biol. 886:51–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Trcek T, Grosch M, York A, Shroff H, Lionnet T, Lehmann R. 2015. Drosophila germ granules are structured and contain homotypic mRNA clusters. Nat Commun. 6:7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y. 2003. Conserved role of nanos proteins in germ cell development. Science. 301:1239–1241. [DOI] [PubMed] [Google Scholar]
- 124.Tu S, Narendra V, Yamaji M, Vidal SE, Rojas LA, Wang X, Kim SY, Garcia BA, Tuschl T, Stadtfeld M, Reinberg D. 2016. Co-repressor CBFA2T2 regulates pluripotency and germline development. Nature. 534:387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vorobyeva NE, Soshnikova NV, Kuzmina JL, Kopantseva MR, Nikolenko JV, Nabirochkina EN, Georgieva SG, Shidlovskii YV. 2009. The novel regulator of metazoan development SAYP organizes a nuclear coactivator supercomplex. Cell Cycle. 8:2152–2156. [DOI] [PubMed] [Google Scholar]
- 126.Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. 2011. RNA granules in germ cells. Cold Spring Harb Perspect Biol. 3:a002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang JQ, Cao WG. 2016. Key signaling events for committing mouse pluripotent stem cells to the germline fate. Biol Reprod. 94:24. [DOI] [PubMed] [Google Scholar]
- 128.Wang JT, Seydoux G. 2013. Germ cell specification. Adv Exp Med Biol. 757:17–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wei C, Salichos L, Wittgrove CM, Rokas A, Patton JG. 2012. Transcriptome-wide analysis of small RNA expression in early zebrafish development. RNA. 18:915–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wilczynska A, Minshall N, Armisen J, Miska EA, Standart N. 2009. Two Piwi proteins, Xiwi and Xili, are expressed in the Xenopus female germline. RNA. 15:337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wright KJ, Marr MT, 2nd, Tjian R. 2006. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc Natl Acad Sci USA. 103:12347–12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yamaguchi S, Kimura H, Tada M, Nakatsuji N, Tada T. 2005. Nanog expression in mouse germ cell development. Gene Expr Patterns. 5:639–646. [DOI] [PubMed] [Google Scholar]
- 133.Yang F, Wang PJ. 2016. Multiple LINEs of retrotransposon silencing mechanisms in the mammalian germline. Semin Cell Dev Biol. 59:118–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yang J, Aguero T, King ML. 2015. The Xenopus maternal-to-zygotic transition from the perspective of the germline. Curr Top Dev Biol. 113:271–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yokobayashi S, Liang CY, Kohler H, Nestorov P, Liu Z, Vidal M, van Lohuizen M, Roloff TC, Peters AH. 2013. PRC1 coordinates timing of sexual differentiation of female primordial germ cells. Nature. 495:236–240. [DOI] [PubMed] [Google Scholar]
- 136.Yuda M, Iwanaga S, Kaneko I, Kato T. 2015. Global transcriptional repression: An initial and essential step for Plasmodium sexual development. Proc Natl Acad Sci USA. 112:12824–12829. [DOI] [PMC free article] [PubMed] [Google Scholar]


