Abstract
In the yeast Saccharomyces cerevisiae, glycogen is accumulated as a carbohydrate reserve when cells are deprived of nutrients. Yeast mutated in SNF1, a gene encoding a protein kinase required for glucose derepression, has diminished glycogen accumulation and concomitant inactivation of glycogen synthase. Restoration of synthesis in an snf1 strain results only in transient glycogen accumulation, implying the existence of other SNF1-dependent controls of glycogen storage. A genetic screen revealed that two genes involved in autophagy, APG1 and APG13, may be regulated by SNF1. Increased autophagic activity was observed in wild-type cells entering the stationary phase, but this induction was impaired in an snf1 strain. Mutants defective for autophagy were able to synthesize glycogen upon approaching the stationary phase, but were unable to maintain their glycogen stores, because subsequent synthesis was impaired and degradation by phosphorylase, Gph1p, was enhanced. Thus, deletion of GPH1 partially reversed the loss of glycogen accumulation in autophagy mutants. Loss of the vacuolar glucosidase, SGA1, also protected glycogen stores, but only very late in the stationary phase. Gph1p and Sga1p may therefore degrade physically distinct pools of glycogen. Pho85p is a cyclin-dependent protein kinase that antagonizes SNF1 control of glycogen synthesis. Induction of autophagy in pho85 mutants entering the stationary phase was exaggerated compared to the level in wild-type cells, but was blocked in apg1 pho85 mutants. We propose that Snf1p and Pho85p are, respectively, positive and negative regulators of autophagy, probably via Apg1 and/or Apg13. Defective glycogen storage in snf1 cells can be attributed to both defective synthesis upon entry into stationary phase and impaired maintenance of glycogen levels caused by the lack of autophagy.
Cells constantly abstract information about their environment and modify their cellular and metabolic programs to cope with the prevailing conditions. For unicellular organisms like the budding yeast, Saccharomyces cerevisiae, much of the information about nutritional status is carried by the nutrients themselves. Depending on the type and availability of carbon, nitrogen, sulfur, and other requirements, the appropriate metabolic and cellular programs are elicited. Exhaustion of a preferred carbon source, like glucose, signals the induction of numerous genes needed to change to other metabolic regimes, in part by derepression of glucose-repressed genes and in part by cyclic AMP (cAMP) pathway control of gene expression (18, 30). Another decision linked to nutritional deprivation is the synthesis of storage compounds like glycogen and trehalose, which are the primary carbohydrate reserves of the yeast (15). Glycogen is a branched polymer of glucose units that acts as a reserve of glucose and energy (47). In yeast growing on glucose, glycogen is synthesized late in the logarithmic phase and begins to be utilized when cells enter the stationary phase (7, 8, 37). However, glycogen stores can be preserved very late into starvation and are thought to be utilized during sporulation (10). In dividing cells, there is also some evidence for a relationship between accumulation of carbohydrate reserves and the cell cycle (52). Glycogen and trehalose are synthesized in G1 and depleted at the stage of bud emergence.
The pathway of glycogen biosynthesis, conserved between yeast and mammals, starts with glycogenin, a self-glucosylating initiator protein that forms an oligosaccharide primer that is the substrate for elongation and branching by glycogen synthase and the branching enzyme, respectively (7, 42). Glycogen synthase is an important site of control in both yeast and mammals. In yeast, glycogen synthase is encoded by two genes, GSY1 and GSY2, of which Gsy2p accounts for 80% of the glycogen synthase activity in the stationary phase (13). GSY2 transcription is induced by stresses, such as nutrient limitation, high salt, or heat shock (13, 43, 46), and the Gsy2p enzyme is also negatively regulated by covalent phosphorylation (25). A third control is by allosteric activation, the most important ligand being Glc-6-P, the binding of which overcomes inactivation due to phosphorylation (25, 49). This property leads to the use of the ratio of the activity in the absence of Glc-6-P divided by that in its presence as a kinetic index of the phosphorylation state of the enzyme (−/+ Glc-6-P activity ratio).
Snf1p is, by sequence similarity, the closest yeast homologue of the mammalian AMP-activated protein kinase (23) and has been studied mostly in relation to its role in glucose repression (5, 18, 23). In cells lacking SNF1, glucose-repressed genes cannot be derepressed in the absence of glucose (5, 6). SNF1 also has a role in glycogen metabolism and one of the original set of glycogen-deficient mutant strains (GLC mutants), glc2, carried a mutant allele of SNF1 (4). Hardy et al. (24) showed that the impaired glycogen accumulation in snf1 cells correlated with hyperphosphorylation of glycogen synthase, as evidenced by a low −/+ Glc-6-P activity ratio, rather than control of glycogen synthase expression. In an effort to identify protein kinases that phosphorylate glycogen synthase, an snf1 strain was thus used in a screen for second site suppressors of the glycogen defect (26). We identified PHO85, and the snf1 pho85 double mutants selected in the screen had both a wild-type glycogen synthase activity ratio and normal glycogen levels. PHO85 encodes a cyclin-dependent protein kinase (CDK) catalytic subunit that, like other CDKs (1), acts in concert with cyclin partners, called Pcls, of which 10 are known (40). PHO85 was first identified through its involvement in phosphate metabolism, for which it pairs with the cyclin Pho80p to phosphorylate the Pho4p transcription factor (30a, 56). Subsequent work showed that Pcl8p and Pcl10p link Pho85p to inactivation of glycogen synthase (27). Thus, deletion of PCL8 and PCL10 in a wild-type strain results in a high glycogen synthase activity ratio and hyperaccumulation of glycogen (27). Similarly, loss of PCL8 and PCL10 in an snf1 strain restores glycogen synthase activity, but, in an unexpected result that represents the starting point for this study, did not restore glycogen accumulation (27). Therefore, we proposed that there must be some other factor or process, controlled by Snf1p, that is not corrected by mutation of PCL8 and PCL10 (27).
Another response to nutrient starvation by both yeast and mammalian cells is the induction of autophagy (11, 12, 53). Autophagy is a process whereby cells randomly engulf cytosol and organelles to form autophagosomes that are delivered to the vacuole (in yeast) or lysozome (in mammals). There the autophagosomes are degraded to recycle some components, such as amino acids, and to generate energy during starvation. A number of genes have been implicated in this process through different genetic screens (34). One of the first yeast autophagy genes identified was APG1 which encodes a Ser/Thr protein kinase (57), whose kinase activity is required for its function (39). By epistasis, APG1 has been placed downstream of another autophagy gene, APG13 (17), whose sequence matches nothing in the databases. In the course of our efforts to identify novel factors controlled by Snf1p that affected glycogen metabolism, a high-copy suppressor screen with an snf1 pc18 pcl10 host identified APG1 and APG13. We described here how the process of autophagy is related to the ability of cells to maintain glycogen stores, and we propose that autophagy is controlled by Snf1p and Pho85p.
MATERIALS AND METHODS
Strains, media, and genetic methods.
The S. cerevisiae strains used in this study are listed in Table 1. Rich medium, YPD, contains 1% (wt/vol) yeast extract, 2% (wt/vol) Bacto Peptone, and 2% (wt/vol) glucose. Synthetic complete medium, SD, contains 0.67% (wt/vol) yeast nitrogen base, 2% (wt/vol) glucose, and complete supplement mix (Bio 101, Inc). Synthetic selective medium consists of 0.67% (wt/vol) yeast nitrogen base, 2% (wt/vol) of the indicated carbon source (glucose in SD) and the indicated complete supplement mix lacking appropriate amino acids or uracil in SD-Ura. For analysis of glycogen accumulation on plates, aliquots (5 to 10 μl) were spotted onto plates, and cells were grown for the indicated time before detection of glycogen by exposing plates to iodine vapor. SD(−N) medium contains 2% (wt/vol) glucose and 0.17% (wt/vol) yeast nitrogen base without amino acids and ammonia sulfate. Plasmids were maintained in Escherichia coli DH5α. Standard methods of yeast genetic analysis and transformation were used.
TABLE 1.
Yeast trains used in this study
| Straina | Genotype | Source |
|---|---|---|
| EG328-1A | MATα trp1 leu2 ura3-52 | K. Tatchell |
| EG353-1C | MATα trp1 leu2 ura3-52 snf1::Ura3 | K. Tatchell |
| TN125 | MATaUra3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pho8::PHO8Δ60 | Y. Ohsumi |
| ZW102-1 | MATaUra3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pho8::PHO8Δ60 pho85::Ura3 | This study |
| ZW115-1 | MATaUra3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pho8::PHO8Δ60 pho85::Trp apg::Ura3 | This study |
| ZW101-3 | MATaUra3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pho8::PHO8Δ60 snf1::TRP1 | This study |
| ZW105-3 | MATaUra3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pho8::PHO8Δ60 apg7::URA3 | This study |
| ZW107-9 | MATaUra3-52 lys2-801 ade2-101 trp1-Δ1 his3-Δ200 leu2-Δ1 pho8::PHO8Δ60 apg1::URA3 | This study |
| ZW34-263 | MATatrp1 leu2 ura3-52 snf1::TRP1 pcl8::URA3 pcl10::URA3 gph1::TRP1 | This study |
| ZW41-6 | MATα trp1 leu2 ura3-52 sga1::URA3 | This study |
| ZW45-7 | MATα trp1 leu2 ura3-52 gph1::TRP1 | This study |
| ZW48-3 | MATα trp1 leu2 ura3-52 sga1::URA3 apg1::URA3 | This study |
| ZW49-161 | MATα trp1 leu2 ura3-52 gph1::TRP1 apg1::URA3 | This study |
| ZW52-14 | MATα trp1 leu2 ura3-52 gph1::TRP1 sga1::URA3 | This study |
| ZW74-2 | MATα trp1 leu2 ura3-52 apg7::URA3 | This study |
| ZW75-3 | MATα trp1 leu2 ura3-52 aut2::URA3 | This study |
| ZW76-2 | MATα trp1 leu2 ura3-52 prb1::URA3 | This study |
| ZW77-4 | MATα trp1 leu2 ura3-52 pra1::URA3 | This study |
| ZW83-1 | MATα trp1 leu2 ura3-52 apg17::URA3 | This study |
| ZW84-4 | MATα trp1 leu2 ura3-52 cvt9::URA3 | This study |
| ZWP1-5 | MATα trp1 leu2 ura3-52 apg1::URA3 | This study |
| ZWP13-2 | MATα trp1 leu2 ura3-52 apg13::URA3 | This study |
| ZWS2 | MATα trp1 leu2 ura3-52 snf1::TRP1 pcl8::URA3 pcl10::URA3 | This study |
TN125, ZW101-3, ZW102-1, ZW105-3, ZW107-9, and ZW115-1 are isogenic. All other strains are isogenic to EG328-1A.
Gene disruptions.
For disruption of genes, a PCR method (60) was used to generate a DNA fragment from primers that contain 45 nucleotides of flanking sequence from the gene of interest, followed by 21 nucleotides that match pBluescript sequences straddling the chosen marker gene in an appropriate pRS plasmid (51). The URA3 gene in vector pRS306, TRP1 gene in vector pRS304, and LEU2 gene in pRS305 were used as templates for PCR. The resulting PCR products contain the 5′ and 3′ sequence of the genes of interest with the chosen marker gene in the middle. The DNA fragment was then used to transform yeast cells to generate strains with the desired gene disrupted. For each disruption, at least two, and usually more, independent mutants were analyzed.
Screen for multicopy suppressors of the glycogen-deficient phenotype of snf1 pcl8 pcl10 cells.
The snf1 pc18 pcl10 cells (ZWS2) were transformed with a yeast genomic library constructed in the 2μm vector pYEP13 obtained from the American Type Culture Collection. After 3 days of growth on SD-Leu plates at 30°C, approximately 105 transformants were analyzed for glycogen accumulation by staining with iodine vapor. Of these, 104 candidate transformants were selected based on darker staining with iodine. Plasmids were isolated from the candidate transformants and transformed back into ZWS2 to confirm that altered glycogen accumulation was plasmid dependent. A total of 46 positive candidates were finally confirmed. The insert sequences in the candidate plasmids were sequenced from both ends to identify the region of the genome present. For those plasmids with more than one gene in the DNA insert, restriction fragments were subcloned where necessary to identify the gene responsible. DNA corresponding to the genes of interest was cloned into pYEP13 or pRS425 and then transformed into snf1 pcl8 pcl10 cells to confirm that the gene conferred increased glycogen accumulation.
Enzyme and other assays.
For the assay of glycogen synthase and glycogen phosphorylase activity in yeast cell extracts, cultures were grown in 150 ml of the indicated medium in 500-ml flasks at 30°C with full access to oxygen. Aliquots of 7 ml were removed at the indicated times, and cells were harvested by centrifugation for 2 min at room temperature at 1,500 × g in a clinical benchtop centrifuge. The cell pellet was frozen on dry ice and stored at −80°C prior to analysis. The frozen cells were thawed on ice and then resuspended in 400 μl of homogenization buffer (50 mM Tris-HCl, 1 mM EDTA, 5 mM dithiothreitol, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 0.1 mM Nα-p-tosyl-l-lysine chloromethyl ketone, 5 mM benzamidine, 0.25 μg of leupeptin per ml, and 0.5 μg of aprotinin per ml [pH 7.4]). The cells were broken with glass beads as described previously (24).
Glycogen synthase was assayed by the method of Thomas et al. (55), as described by Hardy et al. (24). A unit of activity is defined as the amount of enzyme that catalyzes the transfer of 1 μmol of glucose from UDP-glucose to glycogen per min under the conditions of the standard assay. The total activity of glycogen synthase is measured in the presence of 7.2 mM Glc-6-P. The −/+ Glc-6-P activity ratio is defined as the activity measured in the absence of Gl-6-P divided by the activity measured in its presence. Each measurement was the average of duplicate assays.
Glycogen phosphorylase was assayed in the direction of glycogen synthesis by published methods (22, 29) with minor modification. A unit of activity is defined as the amount of enzyme that catalyzes the transfer of 1 μmol of glucose from Glc-1-P to glycogen per min under the conditions of the standard assay. Each measurement was the average of duplicate assays.
Measurement of alkaline phosphatase activity (change in optical density of 420 nm [ΔOD420] per minute per milligram of protein) was carried out as described by Noda et al. (44). For measurement of activity in extracts from cells grown in synthetic complete medium, cells were grown, collected, and stored as described for glycogen measurement. To measure stimulation of alkaline phosphatase activity upon starvation, 5-ml cultures were grown in YPD medium to log phase (∼1 × 107 to 2 × 107 cells/ml of culture), collected by centrifugation, and washed twice with sterilized water. Cells were resuspended in 5 ml of SD(−N) medium, and incubation continued for 4 to 5 h prior to determination of alkaline phosphatase activity.
Invertase activity was measured in glucose-repressed and derepressed cells as described by Huang et al. (26). Protein concentration was measured by the method of Bradford (3), with bovine serum albumin used as the standard.
Measurement of glycogen, ATP, and Glc-6-P.
Quantitative determination of the glycogen content of yeast cells was determined by a modification of published methods (45). Aliquots (∼1 × 107 cells) of cells grown in YPD medium to the stationary phase were used to inoculate 40-ml liquid cultures of synthetic complete medium (unless noted otherwise). The cultures were incubated at 30°C, and at the indicated times, 1 ml of culture was harvested by centrifugation. The cell pellet was frozen on dry ice and stored at −80°C. For measurement of glycogen, the frozen samples were resuspended with 200 μl of 20% (wt/vol) KOH and boiled for 1 h in a water bath with occasional shaking. The samples were cooled on ice for 2 min, and 200 μl of 4 M HCl was added. Glycogen was precipitated by addition of 1 ml of ice-cold 95% (vol/vol) ethanol. The pellet was collected by centrifugation at 17,500 × g at room temperature for 15 min. After two washes with 66% (vol/vol) ethanol, the pellet was dried, resuspended in 400 μl of 50 mM sodium acetate–5 mM CaCl2 (pH 5.0), and digested with 30 μg of amyloglucosidase and 2 μg of amylase at 56.5°C for 12 h. The glucose released was then determined as described previously (24). The glycogen concentration was calculated and normalized to cell number.
For measurement of ATP and Glc-6-P, yeast cells were grown to the indicated phase in synthetic complete medium and collected by rapid filtration. Cells were then rapidly frozen in liquid nitrogen. The assay of Glc-6-P and ATP was carried out as described previously (61).
RESULTS
Screen for multicopy suppressors of the glycogen-deficient phenotype of snf1 pcl8 pc110 cells.
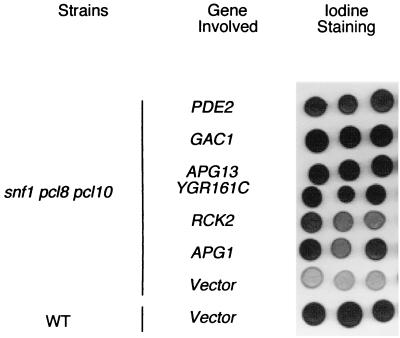
Deletion of PHO85 restored both normal glycogen synthase activity and the ability to accumulate glycogen to snf1 mutants (27). Elimination of the cyclins Pc18p and Pcl10p, which are thought to direct Pho85p to the control of glycogen synthase, resulted in activation of glycogen synthase in either wild-type or snf1 cells, but still glycogen accumulation in snf1 pcl8 pcl10 cells was defective. These results suggest that other pathways or processes, which are positively regulated by SNF1, contribute to glycogen accumulation in S. cerevisiae (27). To identify these potential downstream effectors of Snf1p, we sought multicopy suppressors of the glycogen-deficient phenotype of snf1 pcl8 pcl10 cells. A yeast genomic library constructed in the 2μm vector pYEP13 was used to transform snf1 pcl8 pcl10 cells, and candidates with increased glycogen storage, as judged by iodine staining of colonies, were selected as described in Materials and Methods. Partial sequencing of the plasmids allowed identification of the relevant genomic region, and in most instances, the overlap between independently selected plasmids identified the gene responsible. In addition to SNF1, an expected positive in the screen that was identified 12 times, six other genes were found that restored glycogen accumulation in snf1 pcl8 pcl10 cells (Fig. 1). These multicopy suppressors also increased glycogen accumulation in snf1 mutants, although to a lesser extent (data not shown). The gene most frequently recovered was APG1, a protein kinase that has been implicated in the process of autophagy (39). Interestingly, the screen also yielded APG13, another autophagy gene that has been linked genetically with APG1 (17). Of the other positives from the screen, Gac1p is a targeting subunit of the type 1 protein phosphatase Glc7p responsible for dephosphorylation of glycogen synthase, and its link to glycogen synthesis is well established (16). Similarly, PDE2 encodes a phosphodiesterase that degrades cAMP, and reduced cAMP is known to elevate glycogen stores (35, 55a). RCK2 encodes a calmodulin-dependent kinase-like protein (41) whose connection with glycogen stores has not been reported, but which is implicated in the Hog1p pathway (2). A final positive was a gene, YGR161C, of unknown function. The link between glycogen storage and autophagy is novel and connects two processes that are closely regulated by nutritional status. The present study represents our efforts to understand more about the possible role of autophagy in determining glycogen stores.
FIG. 1.
Glycogen accumulation in snf1 pcl8 pcl10 cells with multicopy suppressor plasmids. Strains are EG328-1A (wild type [WT]) or ZWS2 (snf1 pcl8 pcl10). The plasmids are pRS425 (vector) or pYEP13 with the indicated gene inserted. In the screen, APG1 was found 17 times, APG13 was found 1 time, PDE2 was found 8 times, RCK2 was found 3 times, GAC1 was found 2 times, and YGR161C was found 2 times. The cells were grown on SD-Leu plates for 2 days at 30°C and then exposed to iodine vapor for 2 min to assess glycogen accumulation, with the darker the staining, the greater the glycogen accumulation.
Apg1p is a Ser/Thr kinase that, from sequence alignment, is remotely related to Snf1p, and so formally the two kinases could have some overlapping functions, thus explaining the identification of Apg1p in the screen. Therefore, we tested whether APG1 affected other cellular properties characteristic of snf1 mutants, namely the inability to grow on nonfermentable carbon sources like glycerol and constitutive expression of a glucose-repressed gene like that coding for invertase. First, the presence of a multicopy plasmid bearing the APG1 gene did not restore glycerol growth to snf1 pcl8 pcl10 cells (data not shown). Second, the constitutive expression of invertase was unaffected by overexpression of APG1 from a multicopy plasmid (data not shown). Furthermore, apg1 cells have normally repressible invertase expression and can grow on glycerol. Therefore, we consider it unlikely that APG1 was identified because it substituted for SNF1 function.
Effects of APG1 and APG13 mutation on glycogen accumulation.
To confirm roles for APG1 and APG13 in glycogen accumulation, we deleted APG1 or APG13 in EG328-1A, the strain used for much of our work. The apg1 and apg13 null mutants had no defect in vegetative growth in YPD or synthetic complete medium, but had almost zero viability after being transferred to starvation medium, SD(−N), for 3 days (data not shown). Likewise, diploid homozygous null strains could not sporulate (data not shown). No autophagosomes were visible in the vacuoles of apg1 and apg13 strains after transfer of cells into starvation medium SD(−N) in the presence of the protease inhibitor phenylmethyl sulfonyl fluoride (data not shown). These findings are all consistent with previous characterization of apg1 and apg13 mutants (17, 39, 57).
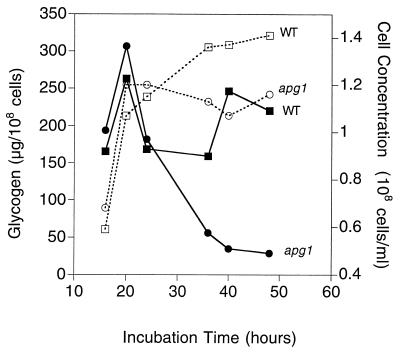
The effect of disruption of APG1 on glycogen accumulation was analyzed by measuring the glycogen content of cells grown in liquid culture (Fig. 2). Cells were grown in synthetic complete medium, and samples were collected at the indicated times starting from the late exponential phase. In both wild-type and apg1 cells, glycogen began to accumulate and increase when the cells reached the late exponential phase and reached similar maximum levels. In wild-type cells, there was a reproducible decrease in glycogen level in the early stationary phase, after which the level was increased slightly until its utilization late in the stationary phase (Fig. 3). We also analyzed another strain, TN125 (44), which was used for assays of autophagy (see below). TN125, which was originally derived from YPH499, had a qualitatively similar pattern of glycogen accumulation, although the time to reach the maximum glycogen level was longer (see below). In apg1 cells (Fig. 2), glycogen was accumulated normally at first, but was rapidly depleted in the early stationary phase and never resynthesized. Measurement of glycogen in apg13 cells gave similar results (data not shown). Therefore, the defect in the apg1 mutant was not its ability to synthesize glycogen, but rather its ability to maintain the stores late into the stationary phase. To check that the apg1 cells were not simply depleted of ATP during the period of glycogen breakdown, we measured ATP and Glc-6-P concentrations after 24 and 48 h of growth and found the values in the apg1 mutant to be indistinguishable from those of wild-type cells (data not shown). Since some of the incubations were for prolonged periods, we also checked viability to ensure that measurements were not influenced simply by cell death. After 5 days in experiments such as those shown in Fig. 3 and 4, both wild-type and apg1 cells had similar viabilities of 40 to 50% (data not shown). These results clearly demonstrate that mutation of APG1 or APG13 affects glycogen storage in S. cerevisiae and that these genes are required for the maintenance of glycogen during prolonged incubations.
FIG. 2.
Glycogen accumulation in apg1 cells after prolonged incubation in synthetic complete medium. Wild-type (EG328-1A) and apg1 (ZWP1-5) cells were grown in liquid SD medium. At the indicated times, cell concentrations were measured by counting cell numbers (□, wild-type cells; ○, apg1 cells). Aliquots were taken for determination of glycogen content (■, wild-type cells; ●, apg1 cells), as described as in Materials and Methods. Representative data from one of three independent experiments are shown.
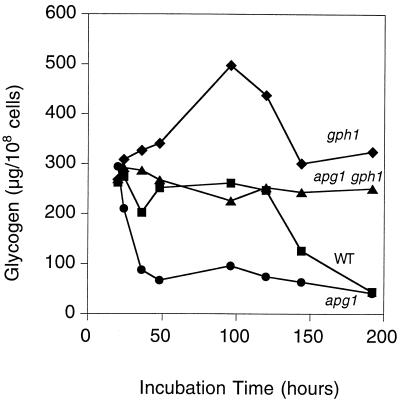
FIG. 3.
Loss of GPH1 reverses glycogen depletion in apg1 cells. Cells were grown in liquid SD medium, and aliquots were taken at the indicated times for determination of glycogen content as described in Materials and Methods. The following strains were used: EG328-1A (wild type [■]), ZWP1-2 (apg1 [●]) ZW49-161 (apg1 gph1 [▴]), and ZW45-7 (gph1 [⧫]). Representative data from one of three independent experiments are shown.
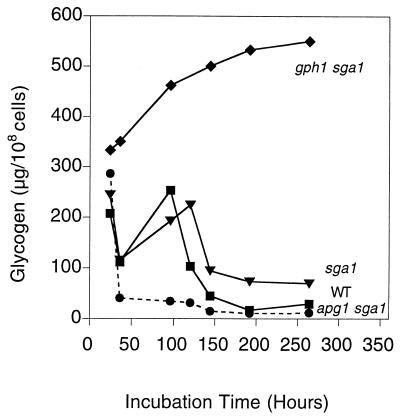
FIG. 4.
Both GPH1 and SGA1 affect glycogen degradation. Cells were grown in liquid SD medium, and aliquots were taken at the indicated times for glycogen measurement. The following strains were used: EG328-1A (wild type [■]), ZW41-6 (sga1 [▾]), ZW48-3 (apg1 sga1 [●]), and ZW48-3 (gph1 sga1 [⧫]). Representative data from one of three independent experiments are shown.
It is worth noting that the kinetics of glycogen accumulation are different between cells grown in liquid culture and those grown on solid plates. On plates, the decreased glycogen accumulation associated with APG1 deletion is not observed until 4 to 5 days on synthetic complete medium. However, in liquid synthetic complete medium, the decreased glycogen accumulation occurs much earlier and depletion is quicker. Several hours after reaching the stationary phase, almost no glycogen can be detected.
Deletion of the glycogen phosphorylase gene, GPH1, reverses the rapid depletion of glycogen in apg1 cells.
As for any metabolite, the amount of glycogen is determined not only by its synthesis, but also its rate of degradation. Cleavage of the α-1,4-glycosidic linkages of glycogen can be achieved by phosphorylysis, catalyzed by glycogen phosphorylase Gph1p (29), or by hydrolysis. S. cerevisiae has three genes known to encode hydrolytic glucoamylases, SGA1, STA1, and STA2. The STA genes are exoenzymes thought to be involved in the utilization of starch as a nutrient (59), whereas SGA1 encodes a very similar enzyme reported to be sporulation-specific and associated with the vacuole (9, 48). Lack of GPH1 eliminated the transient drop in glycogen in the early stationary phase and resulted in an exaggerated resynthesis phase (Fig. 3). Maximal glycogen accumulation was about twice that of wild-type cells, and although a decrease after 5 to 6 days paralleled the late phase depletion seen in wild-type cells, the gph1 strain still retained significantly higher glycogen levels. We infer first that phosphorylase is operating to decrease glycogen levels during the early stationary phase and further that there is active synthesis occurring at this time. Later in the stationary phase, when glycogen levels fall, some other degradative process must be operating.
When GPH1 was deleted in an apg1 mutant strain, the ability to maintain glycogen in the stationary phase was restored (Fig. 3). The level of glycogen accumulation in apg1 gph1 double mutants was nonetheless significantly lower than in the gph1 strain, arguing that Apg1p also exerts a positive effect on the glycogen synthesis occurring at this stage. We measured glycogen synthase and glycogen phosphorylase activities in wild-type and apg1 cells grown in synthetic complete medium (data not shown). There was no difference in enzyme activities after 24 or 48 h of culture. We also measured glycogen synthase and phosphorylase activities in snf1 pcl8 pcl10 cells carrying APG1 or APG13 expressed from multicopy plasmids. Again, there was no difference between the vector control and the cells expressing APG1 or APG13 (data not shown). We infer that Apg1p does not affect glycogen levels by direct effects on either glycogen synthase or phosphorylase, even though the presence of phosphorylase is required for the glycogen maintenance defect of apg1 cells.
Both GPH1 and SGA1 are involved in glycogen degradation.
Elimination of the other glycogen degradative enzyme, encoded by SGA1, had a more complex effect (Fig. 4). The loss of glycogen in sga1 cells during early stationary phase was essentially the same as in wild-type cells. Very late in the stationary phase (over 8 days), there was protection of the glycogen stores in sga1 mutants. This effect was much clearer in a gph1 sga1 double mutant, where there was both the early hyperaccumulation of glycogen seen in gph1 strains (Fig. 3) and continued glycogen accumulation late into the stationary phase. These results suggest that the actions of the degradative enzymes Gph1p and Sga1p are temporally separated and, if Sga1p is vacuolar, linked to physically separate glycogen pools. Deletion of SGA1 in an apg1 strain resulted in a double mutant whose glycogen accumulation was essentially the same as that of an apg1 mutant, arguing that SGA1 is not needed for the phenotype of apg1 strains.
Autophagy affects glycogen storage.
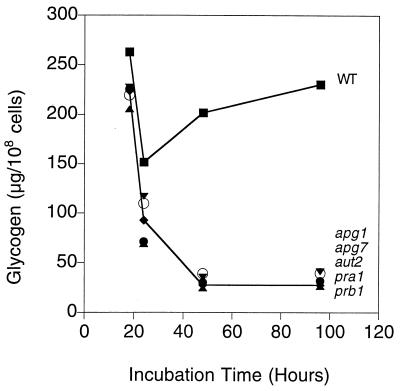
Apg1p is a Ser/Thr protein kinase that has been reported to interact with Apg13p directly by the yeast two-hybrid assay (30b, 58). Thus, these genes could function in some regulatory role, governing both glycogen metabolism and autophagy. Alternatively, it could be that the process of autophagy itself was the factor affecting glycogen storage. Therefore, we checked whether deletion of other autophagy genes could cause a similar defect in glycogen accumulation as seen in apg1 or apg13 cells. Of the autophagy genes, we selected APG7 and AUT2 (32, 36, 54). Apg7p has been proposed to act as a conjugating enzyme in the conjugation of Apg12p and Apg5p, both of which are also required for autophagy. Aut2p has been shown to interact with microtubules and probably is involved in the delivery of autophagic vesicles to the vacuole. Although their exact roles are still not clear, Apg7p and Aut2p are two proteins likely involved in the initiation and delivery of autophagesomes to the vacuole. We also selected the two proteases, proteinase A (PRA1) and proteinase B (PRB1), which are thought to be the master proteinases in the vacuole responsible for the activation of several proenzymes (34). We constructed apg7, aut2, pra1, and prb1 cells in our strain background and measured glycogen levels (Fig. 5). The time course of glycogen accumulation in these mutants was indistinguishable from that observed for apg1 cells, and glycogen was rapidly depleted in early stationary phase.
FIG. 5.
Glycogen accumulation in cells disrupted for various autophagy genes. Cells were grown in liquid SD medium, and aliquots were taken at the indicated times for determination of glycogen content as described in Materials and Methods. The following strains were used: EG328-1A (wild type [■]), ZWP1-5 (apg1) and ZW74-2 (apg7) (○), ZW75-3 (aut2 [▾]), ZW76-2 (prb1 [▴]), and ZW77-4 (pra1 [⧫]). Glycogen accumulation of other mutant cells has essentially the same kinetics as that in prb1 cells.
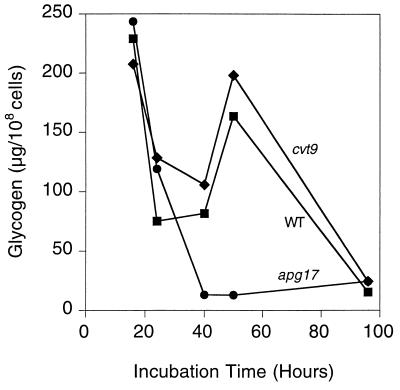
Many of the genes originally implicated in autophagy are also involved in other vesicular trafficking processes (34). Notably, there is considerable overlap with genes involved in cytosol-to-vesicle targeting (Cvt pathway). For example, APG1 and APG13 are implicated in both processes. Recently APG17 and CVT9 (30b) have been identified as being specifically involved in autophagy and the Cvt pathway, respectively. We therefore analyzed glycogen accumulation in corresponding null mutant strains (Fig. 6). The cvt9 cells had normal glycogen accumulation, whereas apg17 cells behaved like the other autophagy-defective mutants that we had examined. This result implicates autophagy, rather than the Cvt pathway, in the maintenance of glycogen stores. We also conclude that it is the process of autophagy itself that influences glycogen storage rather than functions specifically related to APG1 and APG13.
FIG. 6.
Glycogen accumulation in apg17 and cvt9 cells. The cells were grown in liquid SD medium, and aliquots were taken at the indicated times for determination of glycogen content as described in Materials and Methods. The following strains were used: EG328-1A (wild type [■]), ZW83-1 (apg17 [●]), and ZW84-4 (cvt9 [⧫]). Representative data from one of three independent experiments are shown.
Autophagy is induced during the transition from exponential growth to the stationary phase.
We have shown that the ability to perform autophagy is required for glycogen maintenance during the stationary phase. Studies of autophagy have generally used quite extreme experimental conditions, with exponentially growing cells being transferred to a starvation medium SD(−N) completely lacking nitrogen (53, 57). Under conditions such as those used in this investigation, cells grown in synthetic complete medium deplete nutrients more slowly and transition more gradually into a prolonged stationary phase, during which time significant metabolic reprogramming occurs prior to complete loss of viability. It is therefore relevant to ask whether autophagy is induced after exponential growth under our culture conditions. To address this question, we measured the autophagic activity of yeast cells grown in synthetic medium. To monitor autophagy, we used the TN125 strain (44). In this strain, the endogenous PHO8 gene, which encodes a major alkaline phosphatase, has been replaced by a mutated form of the gene, PHO8Δ60, that lacks its vacuolar targeting sequences. In addition, the PHO8 promoter is replaced by the strong TDH3 promoter. The truncated Pho8Δ60p protein can only reach the vacuole via the process of autophagy. Moreover, only in the vacuole can the Pho8p be converted from an inactive zymogen to an active phosphatase by proteolysis. Therefore, measurement of alkaline phosphatase activity, for which Pho8Δ60 is primarily responsible, can be used as an index of autophagy activity.
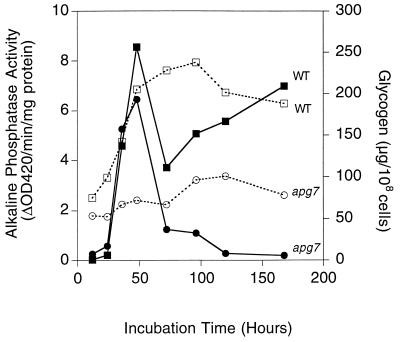
Strains TN125 and TN125 with APG7 deleted were grown in synthetic complete medium, and samples were collected starting from the late exponential growth phase for measurement of glycogen and alkaline phosphatase (Fig. 7). As was true for the EG328-1A strain background, the apg7 mutation in TN125 did not substantially affect glycogen synthesis, but profoundly influenced the ability of the cells to maintain their glycogen reserves during stationary phase. This behavior is qualitatively the same as the EG328-1A strain, except that the time frame is slower, and more time is needed for complete loss of glycogen in the apg7 mutants with the TN125 background. The alkaline phosphatase activity was increased three- to fourfold as the cells exited from the exponential phase and remained elevated throughout the stationary phase, suggesting that autophagy is induced under these conditions. In contrast, a significantly blunted increase of alkaline phosphatase activity was observed in apg7 cells. Similar results were obtained with cells carrying an apg1 allele in this background (data not shown). The increased autophagy is likely due to limitation of nitrogen, since a supplement of nitrogen, but not of carbon, given to cells in the early stationary phase caused rapid cell proliferation (data not shown). The induction of alkaline phosphatase activity in TN125 cells was relatively low compared to the increase elicited by transferring yeast cells grown in rich medium to starvation medium. Indeed, there was a further two- to threefold increase in alkaline phosphatase activity when stationary-phase cells were transferred to starvation medium SD(−N) (data not shown). Nonetheless, these results clearly indicate that there is a significant induction of autophagy during the transition from exponential growth to the early stationary phase in yeast under standard laboratory growth conditions.
FIG. 7.
Induction of autophagy upon entry into the stationary phase. Wild-type (TN125) and apg7 (ZW105-3) cells were grown in liquid SD medium. At the indicated times, cells were harvested. Glycogen content (■, wild-type strain TN125 ●, apg7) and alkaline phosphatase activity (□, wild-type strain TN125; ○, apg7) were measured as described in Methods and Materials. Representative data from one of three independent experiments are shown.
SNF1 and PHO85 regulate autophagy.
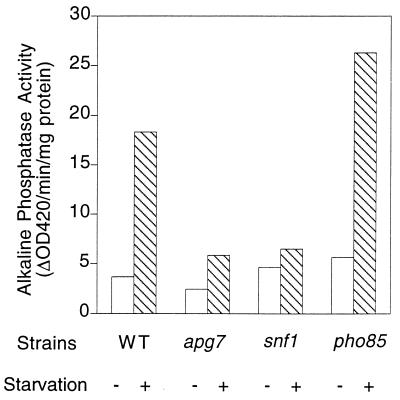
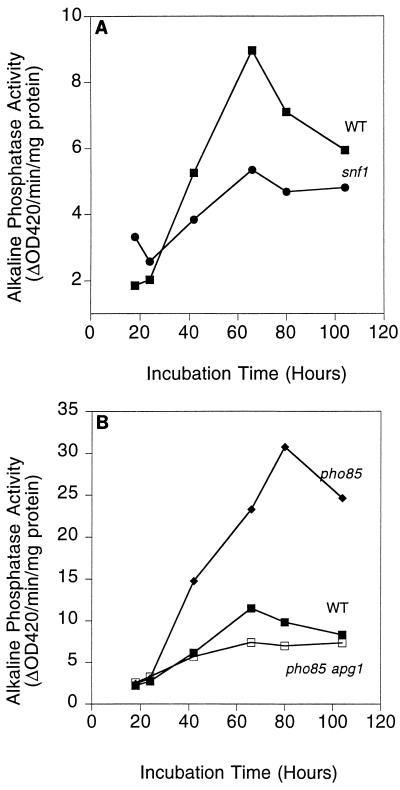
The APG1 and APG13 genes are suppressors of the diminished glycogen accumulation in snf1 and snf1 pcl8 pcl10 cells. An important question, then, is whether SNF1 has any effect on autophagy. Like snf1 cells, autophagy mutants are more sensitive to starvation (57). Therefore, the SNF1 gene was deleted in the TN125 background. TN125 and the corresponding snf1 mutant were grown in YPD medium to the mid-exponential growth phase (∼ 1 × 107 to 2 × 107 cells/ml of culture), collected, and transferred to starvation SD(−N) medium to induce autophagy. Based on measurement of alkaline phosphatase activity, there was a strong induction of autophagy in the TN125 strain, but this was largely blocked in snf1 cells (Fig. 8). An apg7 strain served as a control, also exhibiting reduced induction of alkaline phosphatase upon starvation. These results indicate that SNF1 is a positive regulator of autophagy. In addition, the induction of alkaline phosphatase in snf1 cells upon entry into the stationary phase was blunted compared to that in wild-type cells (Fig. 9). We also tested whether some of the other genes identified as high-copy-number suppressors of snf1 pcl8 pcl10 affected autophagy by making deletions of RCK2 and YGR161C in the TN125 background. Induction of alkaline phosphatase activity in rck2 or ygr161c null mutants was similar to that in wild-type cells upon transfer to starvation medium (data not shown). These results suggest that there are yet other SNF1-mediated mechanisms affecting glycogen storage. The most important conclusion, though, is that the Snf1p protein kinase, implicated in glucose derepression, is involved in controlling autophagy, another key cellular process linked to nutritional regulation.
FIG. 8.
Effect of SNF1 and PHO85 on autophagy. Wild-type (TN125), snf1 (ZW101-3), pho85 (ZW102-1), and apg7 (ZW105-3) cells were grown in YPD to the logarithmic phase before transfer into SD(−N). Incubation was continued for 4 h. Cells were then harvested, and alkaline phosphatase activity was measured as described in Materials and Methods. Representative data from one of three independent experiments are shown.
FIG. 9.
Effects of SNF1 and PHO85 on autophagy upon entry into the stationary phase. (A) Wild-type (TN125) and snf1 (ZW101-3) cells were grown in liquid SD medium. At the indicated times, cells were harvested. Alkaline phosphatase activity (■, wild-type strain TN125 ●, snf1), was measured as described in Materials and Methods. Representative data from one of three independent experiments are shown. (B) Wild-type (TN125), pho85 (ZW102-1), and pho85 apg1 (ZW115-1) cells were grown in liquid SD medium. At the indicated times, cells were harvested. Alkaline phosphatase activity (■, wild-type strain; ⧫, pho85; □, pho85 apg1) was measured as described in Materials and Methods. Representative data from one of three independent experiments are shown.
Since loss of PHO85 suppresses the glycogen defect of snf1 cells, it was natural to test whether PHO85 has any effect on autophagy. By using a TN125 strain with PHO85 deleted, induction of autophagy by starvation was slightly enhanced in the pho85 mutant according to the standard assay (Fig. 8). However, there was an exaggerated induction of alkaline phosphatase activity compared with that in wild-type cells, corresponding to as much as a threefold elevation, as a cell culture entered the stationary phase (Fig. 9). Neither the basal, uninduced level nor the timing of induction was greatly altered—only the maximum activity level and the duration. With a pho85 apg1 double mutant, alkaline phosphatase induction was reduced to the impaired level of an autophagy-defective strain, indicating that mutation of APG1 is epistatic to a pho85 mutant with respect to autophagy. These results suggest that Pho85p exerts a negative control of autophagy, possibly acting through Apg1.
Impairment of glycogen storage in snf1 mutants involves both synthesis and maintenance.
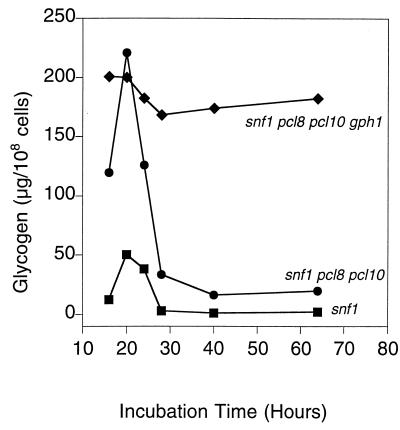
Since impaired glycogen storage in autophagy mutants is linked to the inability to maintain glycogen, we examined in more detail the time course of glycogen accumulation in snf1 mutant strains (Fig. 10). We had previously attributed the inability of snf1 mutants to accumulate glycogen solely to the hyperphosphorylation and inactivation of glycogen synthase. Glycogen synthesis was significantly reduced in snf1 cells, but any glycogen made was rapidly lost and was completely absent after 28 h. Deletion of PCL8 and PCL10 in an snf1 strain restored the initial synthesis of glycogen close to wild-type levels, consistent with our observation that glycogen synthase in this strain was activated, with a normal −/+ Glc-6-P activity ratio (27). However, the strain was unable to retain the glycogen, and this triple mutant behaved exactly like autophagy mutants in this regard. As was true for the autophagy mutants, the decrease in glycogen accumulation was reversed by deletion of GPH1 (Fig. 10). Thus, the inability of snf1 pcl8 pcl10 strains to accumulate glycogen is not due to a defect in synthesis, but to the inability to maintain glycogen, most likely because of defective autophagy.
FIG. 10.
Glycogen accumulation in snf1, snf1 pc18 pcl10, and snf1 pcl8 pcl10 gph1 cells. Cells were grown in YPD overnight and then diluted 1/200 into synthetic complete medium. Samples were collected at the indicated times, and glycogen content was measured as described in Materials and Methods. The following strains were used: EG353-1C (snf1 [■]), ZWS2 (snf1 pcl8 pcl10 [●]), and ZW34-263 (snf1 pcl8 pcl10 gph1 [⧫]). Representative data from one of three independent experiments are shown.
DISCUSSION
The present study has revealed an unexpected link between the process of autophagy and glycogen metabolism, and it has also uncovered novel controls of autophagy. One is a positive regulation mediated by Snf1p, the closest yeast homologue of the mammalian AMP-activated protein kinase. The second is negative regulation by the cyclin-dependent protein kinase Pho85p. Autophagy, which is stimulated by deprivation for nitrogen, carbon or sulfur, provides a mechanism for the cell to recycle its cytosol and organelles (33, 34). Our study shows that autophagic activity is induced during the transition from logarithmic to stationary phase, consistent with the cells adapting to an altered nutritional environment.
SNF1 has long been known for its roles in glucose repression and glycogen accumulation, and so linking it to another response associated with nutritional controls is perhaps not surprising. As regards defective glycogen accumulation, we had originally considered that snf1 mutants were simply defective in glycogen synthesis and that snf1 pcl8 pcl10 mutants had a similar phenotype (27). In other words, we imagined the triple mutant to have some unappreciated impairment of the biosynthetic pathway. From the present study, however, we have recognized that the initial synthesis of glycogen in snf1 pcl8 pcl10 cells is essentially normal, consistent with highly active glycogen synthase. The defect in these cells is their inability to maintain glycogen stores. The snf1 cells are defective in both glycogen synthesis and the ability to preserve what little glycogen they produce. The most likely explanation is the impaired autophagy associated with the snf1 mutation. Since other autophagy genes were not identified by our genetic screen, it is likely that SNF1 controls autophagy via APG1 and APG13, which themselves perform some regulatory function. Mechanistically, SNF1 would be upstream of APG1 and APG13, since multicopy APG1 and APG13 increased stationary-phase glycogen accumulation in snf1 pcl8 pcl10 and snf1 cells in the EG328-1A background (Fig. 11). Note that SNF1 may exert yet other controls over the maintenance of glycogen stores, since deletion of two of the genes identified in the genetic screen, RCK2 and YGR161C, does not affect autophagy. The fact that a pho85 null mutation restores glycogen storage to snf1 cells suggested that PHO85 might negatively regulate the maintenance phase of glycogen metabolism, via cyclins distinct from Pcl8p and Pcl10p. Indeed, deletion of PHO85 generated a strain in which autophagic activity was induced at the normal time, but was exaggerated and persistent. In the more standard autophagy assay, with transfer to starvation medium, induction of alkaline phosphatase was only slightly elevated over the level of the wild-type control, which is consistent with this extreme condition causing a maximum level of autophagic activity. In the more gradual transition to the stationary phase, the pho85 mutants behave as though they are more sensitive to starvation signals. We infer that a protein kinase composed of Pho85p and cyclins other than Pcl8p or Pcl10p normally exerts a negative control over autophagy upon entry into the stationary phase. Furthermore, mutation of APG1 was epistatic to a PHO85 mutation, suggesting that Pho85p might be acting through Apg1p. Interestingly, this would imply a similar antagonism between SNF1 and PHO85 in the control of autophagy, as has been found for the control of glycogen accumulation.
FIG. 11.
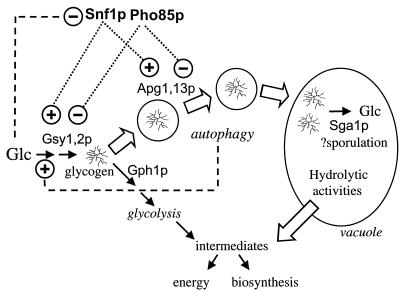
Possible relationships between glycogen metabolism and autophagy. The synthesis and possible fates of glycogen are depicted, with solid arrows representing metabolic interconversion, open arrows indicating physical translocation, and dashed lines indicating regulatory connections. Glycogen is synthesized in the late logarithmic phase through the regulation of glycogen synthase (Gsy1,2p) and other biosynthetic enzymes. At saturation of the culture, there is utilization of glycogen concomitant with activation of phosphorylase (Gph1p) followed by a phase of reaccumulation during which both synthesis and degradation occur simultaneously. Over the same period, starvation signals also lead to increased autophagy, which has at least two effects on glycogen levels. First, it generates, by recycling of cellular components, small molecules such as amino acids that can be returned to the cytosol as a source of energy and/or synthetic intermediates. By an unknown mechanism, autophagy exerts a positive control over glycogen synthesis. A second potential consequence is the sequestration of glycogen in the vacuole, where it is inaccessible to Gph1p, for degradation by the vacuolar glucosidase (Sga1p) very late in the stationary phase. In the absence of autophagy, neither process operates. In the early to mid stationary phase, glycogen is degraded by phosphorylase and is no longer stored in the vacuole, both leading to decreased maintenance of the total glycogen level.
This study also focuses attention on the temporal and spatial aspects of glycogen metabolism during long-term liquid culture. It has long been recognized that glycogen and the other major storage carbohydrate of yeast, trehalose, are accumulated during the late logarithmic phase (15, 37). Upon entry into the stationary phase, there is partial consumption of glycogen, between 24 and 48 h (Fig. 2), presumably in correspondence with the metabolic reprogramming necessitated by depletion of glucose and limitation of other nutrients. Glycogen phosphorylase is active, and it is during this period that mutants defective in autophagy rapidly deplete their glycogen stores. In wild-type cells, there is a phase of resynthesis and maintenance of glycogen stores up to 5 to 6 days. Glycogen phosphorylase is still active during this time, and it is likely that both degradation and synthesis occur simultaneously, since loss of phosphorylase causes hyperaccumulation of glycogen. After 5 to 6 days, the glycogen levels begin to fall, whether in the wild type or in cells with GPH1 deleted, suggesting that phosphorylase is not the only enzyme responsible for glycogen degradation. The prime candidate for this phase of glycogen breakdown is the vacuolar glucosidase Sga1p, since elimination of SGA1, in the wild type and more markedly in a gph1 mutant, protects glycogen levels in the late stationary phase. That the SGA1 gene can have a significant role suggests that it may not be strictly sporulation specific, as has been suggested (9), but simply activated under conditions of relative starvation, as in the late stationary phase. Consistent with this proposal, expression profiling has also indicated that SGA1 transcription is induced in nitrogen starvation (19).
The mechanism by which autophagy is linked to glycogen storage is still not entirely clear. We found no evidence for any direct effect of Apg1p on glycogen synthase or phosphorylase activities. In the absence of autophagy, cells are deprived of a normal mechanism for recycling cytosolic material to provide both monomeric building blocks, such as free amino acids, as well as a source of metabolic energy. A teleological rationale for the depletion of glycogen under these conditions would be to provide missing intermediary metabolites and energy. However, there are also indications that glycogen synthesis is affected in mutants defective for autophagy. For example, apg1 gph1 mutants have lower glycogen accumulation during the early to mid-stationary phase than gph1 cells, suggesting that glycogen synthesis is reduced. In addition, the ability of rapamycin treatment, which induces autophagy, to cause glycogen accumulation in logarithmically growing cells, is blocked in apg1 mutants (Z. Wang and P. J. Roach, unpublished observations), suggesting a link between autophagy and glycogen synthesis. How can autophagy influence glycogen synthesis and degradation? One possibility is through changes in the levels of metabolites associated with different intermediary metabolite fluxes (Fig. 11). A key metabolite for glycogen metabolism is Glc-6-P, a potent activator of glycogen synthase (25, 28, 50) and inhibitor of phosphorylase (14, 38). However, measurements of Glc-6-P levels at the critical time, early stationary phase, indicated no difference between wild-type cells and apg1 mutants. Other metabolites or signals of unappreciated importance, however, could also act to control glycogen metabolism. Our results also lead us to postulate the existence of spatially distinct glycogen pools, which could have an impact on overall glycogen accumulation (Fig. 11). Since autophagy is random, glycogen should be delivered to the vacuole like any other cellular constituent. There is experimental support for this hypothesis, since glycogen has been detected in the vacuole by electron microscopy (53). In mammalian hepatocytes, some 10% of the glycogen is present in lysosomes (20, 21). Cells defective for autophagy would thus be incapable of transporting glycogen to the vacuole. Although the vacuole is an organelle rich in hydrolytic activities and active in degradation, it also serves as a reservoir for some compounds such as amino acids, certain ions, and polyphosphate (31). Perhaps a portion of the glycogen synthesized in the cytosol is actually intended to be stored and protected in the vacuole, until its degradation is signaled very late in starvation, to be used, for example, in sporulation. In the absence of autophagy, none of the glycogen would be protected from phosphorylase in the cytosol and, together with impaired synthesis and increased energetic needs, would be degraded. An attractive feature of this model is that it can explain effects on both synthesis and degradation of glycogen in the absence of altered glycogen synthase and phosphorylase activities. Within the vacuole, degradation of glycogen very late in the stationary phase would be mediated by the vacuolar enzyme Sga1p. Essentially, this model suggests that the temporal separation of glycogen degradation by Gph1p and Sga1p mentioned previously also involves a spatial separation, with the existence of two pools of glycogen, one cytosolic and one vacuolar. This model, although admittedly far from proven, does serve as a framework to guide future work.
ACKNOWLEDGMENTS
We thank Takeshi Noda and Yoshinori Ohsumi for providing the TN125 yeast strain and Yoshiaki Kamada for providing information about the specificities of APG17 and CVT9 prior to publication. We are also grateful to Mark G. Goebl and Ronald C. Wek for many helpful discussions.
This work was supported by NIH grant DK42576 and the Indiana University Diabetes Research and Training Center (DK20542).
REFERENCES
- 1.Andrews B, Measday V. The cyclin family of budding yeast: abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- 2.Bilsland-Marchesan E, Ariño J, Saito H, Sunnerhagen P, Posas F. Rck2 kinase is a substrate for the osmotic stress-activated mitogen-activated protein kinase Hog1. Mol Cell Biol. 2000;20:3887–3895. doi: 10.1128/mcb.20.11.3887-3895.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cannon J F, Pringle J R, Fiechter A, Khalil M. Characterization of glycogen-deficient glc mutants of Saccharomyces cerevisiae. Genetics. 1994;136:485–503. doi: 10.1093/genetics/136.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 6.Carlson M, Osmond B C, Botstein D. Mutants of yeast defective in sucrose utilisation. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng C, Mu J, Farkas I, Huang D, Goebl M G, Roach P J. Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6632–6640. doi: 10.1128/mcb.15.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chester V E. The dissimilation of the carbohydrate reserves of a strain of Saccharomyces cerevisiae. Biochem J. 1963;86:153–160. doi: 10.1042/bj0860153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clancy M J, Smith L M, Magee P T. Developmental regulation of a sporulation-specific enzyme activity in Saccharomyces cerevisiae. Mol Cell Biol. 1982;2:171–178. doi: 10.1128/mcb.2.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Colonna W J, Magee P T. Glycogenolytic enzymes in sporulating yeast. J Bacteriol. 1978;134:844–853. doi: 10.1128/jb.134.3.844-853.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn W A., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn W A., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. J Cell Biol. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farkas I, Hardy T A, Goebl M G, Roach P J. Two glycogen synthase isoforms in Saccharomyces cerevisiae are coded by distinct genes that are differentially controlled. J Biol Chem. 1991;266:15602–15607. [PubMed] [Google Scholar]
- 14.Fosset M, Muir L W, Nielsen L D, Fischer E H. Purification and properties of yeast glycogen phosphorylase a and b. Biochemistry. 1971;10:4105–4113. doi: 10.1021/bi00798a015. [DOI] [PubMed] [Google Scholar]
- 15.Francois J, Parrou J L. Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev. 2001;25:125–145. doi: 10.1111/j.1574-6976.2001.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 16.Francois J M, Thompson-Jaeger S, Skroch J, Zellenka U, Spevak W, Tatchell K. GAC1 may encode a regulatory subunit for protein phosphatase type 1 in Saccharomyces cerevisiae. EMBO J. 1992;11:87–96. doi: 10.1002/j.1460-2075.1992.tb05031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Funakoshi T, Matsuura A, Noda T, Ohsumi Y. Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene. 1997;192:207–213. doi: 10.1016/s0378-1119(97)00031-0. [DOI] [PubMed] [Google Scholar]
- 18.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gasch A P, Spellman P T, Kao C M, Carmel-Harel O, Eisen M B, Storz G, Botstein D, Brown P O. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geddes R. Glycogen: a metabolic viewpoint. Biosci Rep. 1986;6:415–428. doi: 10.1007/BF01116132. [DOI] [PubMed] [Google Scholar]
- 21.Geddes R, Harvey J D, Wills P R. The molecular size and shape of liver glycogen. Biochem J. 1977;163:201–209. doi: 10.1042/bj1630201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilboe D P, Larson K L, Nuttall F Q. Radioactive method for the assay of glycogen phosphorylases. Anal Biochem. 1972;47:20–27. doi: 10.1016/0003-2697(72)90274-6. [DOI] [PubMed] [Google Scholar]
- 23.Hardie D G, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 24.Hardy T A, Huang D Q, Roach P J. Interactions between cAMP-dependent and SNF1 protein kinases in the control of glycogen accumulation in Saccharomyces cerevisiae. J Biol Chem. 1994;269:27907–27913. [PubMed] [Google Scholar]
- 25.Hardy T A, Roach P J. Control of yeast glycogen synthase-2 by COOH-terminal phosphorylation. J Biol Chem. 1993;268:23799–23805. [PubMed] [Google Scholar]
- 26.Huang D, Farkas I, Roach P J. Pho85p, a cyclin-dependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4357–4365. doi: 10.1128/mcb.16.8.4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang D, Moffat J, Wilson W A, Moore L, Cheng C, Roach P J, Andrews B. Cyclin partners determine Pho85 protein kinase substrate specificity in vitro and in vivo: control of glycogen biosynthesis by Pc18 and Pc110. Mol Cell Biol. 1998;18:3289–3299. doi: 10.1128/mcb.18.6.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang K P, Cabib E. Yeast glycogen synthetase in the glucose 6-phosphate-dependent form. II. The effect of proteolysis. J Biol Chem. 1974;249:3858–3861. [PubMed] [Google Scholar]
- 29.Hwang P K, Tugendreich S, Fletterick R J. Molecular analysis of GPH1, the gene encoding glycogen phosphorylase in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:1659–1666. doi: 10.1128/mcb.9.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnston M. Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet. 1999;15:29–33. doi: 10.1016/s0168-9525(98)01637-0. [DOI] [PubMed] [Google Scholar]
- 30a.Kaffman A, Herskowitz I, Tjian R, O'Shea E K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263:1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- 30b.Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–1513. doi: 10.1083/jcb.150.6.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones, Webb E, Webb G C, Hiller M A. Biogenesis and function of the yeast vacuoles. In: Pringle J R, Jones E W, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 3. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 363–470. [Google Scholar]
- 32.Kim J, Dalton V M, Eggerton K P, Scott S V, Klionsky D J. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim J, Klionsky D J. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- 34.Klionsky D J, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Kronstad J, De Maria A D, Funnell D, Laidlaw R D, Lee N, de Sa M M, Ramesh M. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch Microbiol. 1998;170:395–404. doi: 10.1007/s002030050659. [DOI] [PubMed] [Google Scholar]
- 36.Lang T, Schaeffeler E, Bernreuther D, Bredschneider M, Wolf D H, Thumm M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998;17:3597–3607. doi: 10.1093/emboj/17.13.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lillie S H, Pringle J R. Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol. 1980;143:1384–1394. doi: 10.1128/jb.143.3.1384-1394.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin K, Hwang P K, Fletterick R J. Mechanism of regulation in yeast glycogen phosphorylase. J Biol Chem. 1995;270:26833–26839. doi: 10.1074/jbc.270.45.26833. [DOI] [PubMed] [Google Scholar]
- 39.Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene. 1997;192:245–250. doi: 10.1016/s0378-1119(97)00084-x. [DOI] [PubMed] [Google Scholar]
- 40.Measday V, Moore L, Retnakaran R, Lee J, Donoviel M, Neiman A M, Andrews B. A family of cyclin-like proteins that interact with the Pho85 cyclin-dependent kinase. Mol Cell Biol. 1997;17:1212–1223. doi: 10.1128/mcb.17.3.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Melcher M L, Thorner J. Identification and characterization of the CLK1 gene product, a novel CaM kinase-like protein kinase from the yeast Saccharomyces cerevisiae. J Biol Chem. 1996;271:29958–29968. doi: 10.1074/jbc.271.47.29958. [DOI] [PubMed] [Google Scholar]
- 42.Mu J, Cheng C, Roach P J. Initiation of glycogen synthesis in yeast—requirement of multiple tyrosine residues for function of the self-glucosylating Glg proteins in vivo. J Biol Chem. 1996;271:26554–26560. doi: 10.1074/jbc.271.43.26554. [DOI] [PubMed] [Google Scholar]
- 43.Ni H T, Laporte D C. Response of a yeast glycogen synthase gene to stress. Mol Microbiol. 1995;16:1197–1205. doi: 10.1111/j.1365-2958.1995.tb02342.x. [DOI] [PubMed] [Google Scholar]
- 44.Noda T, Matsuura A, Wada Y, Ohsumi Y. Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1995;210:126–132. doi: 10.1006/bbrc.1995.1636. [DOI] [PubMed] [Google Scholar]
- 45.Parrou J L, Francois J. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem. 1997;248:186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 46.Parrou J L, Teste M A, Francois J. Effects of various types of stress on the metabolism of reserve carbohydrates in Saccharomyces cerevisiae: genetic evidence for a stress-induced recycling of glycogen and trehalose. Microbiology. 1997;143:1891–1900. doi: 10.1099/00221287-143-6-1891. [DOI] [PubMed] [Google Scholar]
- 47.Preiss J, Walsh D A. The comparative biochemistry of glycogen and starch. In: Ginsburg V, Robbins P, editors. Biology of carbohydrates. Vol. 1. New York, N.Y: Wiley; 1981. pp. 99–134. [Google Scholar]
- 48.Pugh T A, Shah J C, Magee P T, Clancy M J. Characterization and localization of the sporulation glucoamylase of Saccharomyces cerevisiae. Biochim Biophys Acta. 1989;994:200–209. doi: 10.1016/0167-4838(89)90294-x. [DOI] [PubMed] [Google Scholar]
- 49.Rothman-Denes L B, Cabib E. Glucose 6-phosphate dependent and independent forms of yeast glycogen synthetase. Their properties and interconversions. Biochemistry. 1971;10:1236–1242. doi: 10.1021/bi00783a021. [DOI] [PubMed] [Google Scholar]
- 50.Rothman-Denes L B, Cabib E. Two forms of yeast glycogen synthetase and their role in glycogen accumulation. Proc Natl Acad Sci USA. 1970;66:967–974. doi: 10.1073/pnas.66.3.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sikorski R S, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Silljé H H W, Ter Schure E G, Rommens A J M, Huls P G, Woldringh C L, Verkleij A J, Boonstra J, Verrips C T. Effects of different carbon fluxes on G1 phase duration, cyclin expression, and reserve carbohydrate metabolism in Saccharomyces cerevisiae. J Bacteriol. 1997;179:6560–6565. doi: 10.1128/jb.179.21.6560-6565.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol. 1992;119:301–311. doi: 10.1083/jcb.119.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanida I, Mizushima N, Kiyooka M, Ohsumi M, Ueno T, Ohsumi Y, Kominami E. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Mol Biol Cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas J A, Schlender K K, Larner J. A rapid filter paper assay for UDP glucose-glycogen glucosyltransferase, including an improved biosynthesis of UDP-14C-glucose. Anal Biochem. 1968;25:486–499. doi: 10.1016/0003-2697(68)90127-9. [DOI] [PubMed] [Google Scholar]
- 55a.Thompson-Jaeger S, Francois J, Gaughram J P, Tatchell K. Delection of SNF1 alleles affects the nutrient response of yeast and resembles mutations which activate the adenylate cyclase pathway. Genetics. 1991;129:697–706. doi: 10.1093/genetics/129.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Toh-e A, Tanaka K, Uesono Y, Wickner R B. PHO85, negative regulator of the PHO system, is a homolog of the protein kinase gene, CDC28, of Saccharomyces cerevisiae. Mol Gen Genet. 1988;214:162–164. doi: 10.1007/BF00340196. [DOI] [PubMed] [Google Scholar]
- 57.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 58.Uetz P, Giot L, Cagney G, Mansfield T A, Judson R S, Knight J R, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg J M. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 59.Vivier M A, Lambrechts M G, Pretorius I S. Coregulation of starch degradation and dimorphism in the yeast Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1997;32:405–435. doi: 10.3109/10409239709082675. [DOI] [PubMed] [Google Scholar]
- 60.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 61.Yang R, Chun K T, Wek R C. Mitochondrial respiratory mutants in yeast inhibit glycogen accumulation by blocking activation of glycogen synthase. J Biol Chem. 1998;273:31337–31344. doi: 10.1074/jbc.273.47.31337. [DOI] [PubMed] [Google Scholar]