Abstract
The end replication problem hypothesis proposes that the ends of linear DNA cannot be replicated completely during lagging strand DNA synthesis. Although the idea has been widely accepted for explaining telomere attrition during cell proliferation, it has never been directly demonstrated. In order to take a biochemical approach to understand how linear DNA ends are replicated, we have established a novel in vitro linear simian virus 40 DNA replication system. In this system, terminally biotin-labeled linear DNAs are conjugated to avidin-coated beads and subjected to replication reactions. Linear DNA was efficiently replicated under optimized conditions, and replication products that had replicated using the original DNA templates were specifically analyzed by purifying bead-bound replication products. By exploiting this system, we showed that while the leading strand is completely synthesized to the end, lagging strand synthesis is gradually halted in the terminal ∼500-bp region, leaving 3′ overhangs. This result is consistent with observations in telomerase-negative mammalian cells and formally demonstrates the end replication problem. This study provides a basis for studying the details of telomere replication.
Semiconservative DNA replication is achieved by a harmonious cooperation between two distinct modes of DNA synthesis, leading and lagging strand DNA syntheses. These processes have been extensively characterized in vitro using the simian virus 40 (SV40) DNA replication system, the results of which are summarized as follows (reviewed in reference 37). In leading strand synthesis, the direction of DNA synthesis is the same as that of the replication fork movement. Consequently, leading strand synthesis, performed by DNA polymerase δ and ɛ, is processive. On the other hand, in lagging strand synthesis, DNA is synthesized in a direction opposite to replication fork movement, as short pieces that are eventually ligated to form a continuous DNA strand. This process involves repeated synthesis of RNA primers (∼10 nucleotides [nt]), which are elongated into Okazaki fragments. The RNA primer synthesis and the initial phase of DNA elongation (up to 40 nt) are carried out by the DNA polymerase α-primase complex, which produces RNA-DNA primers. Subsequently, a DNA polymerase switch takes place, and the RNA-DNA primer is elongated by DNA polymerase δ to the full length of its respective Okazaki fragment. Finally, consecutive Okazaki fragments are ligated by removal of RNA primers to form an uninterrupted progeny strand.
In replication of linear DNA molecules, the 3′ end of the nascent strand is synthesized by leading strand synthesis, whereas the 5′ end is synthesized by lagging strand synthesis. In the early 1970s, it was first suggested that the lagging strand synthesis of linear DNA templates would be incomplete for two reasons (28, 39). First, there is no known mechanism that ensures priming of the most distal Okazaki fragment synthesis from the very end of the template molecule. Accordingly, the template sequence between the end and the most distal Okazaki fragment would not be replicated. Second, there is no known mechanism for the most distal RNA primer to be replaced by DNA. Consequently, the sequence of the most distal RNA primer would be lost in the daughter strand. In contrast to lagging strand synthesis, leading strand synthesis is thought to continue to the very end of the template molecule.
The native ends of linear chromosomes are called telomeres. Telomeres contain the ends of linear genomic DNA. All studied eukaryotic cells maintain linear chromosomes and thus have telomeres (reviewed in reference 19). Telomere lengths are reduced as cells proliferate (14, 15). In most eukaryotes, a specialized reverse transcriptase called telomerase is responsible for counteracting this progressive telomere size reduction during cell growth by synthesizing telomeric DNA de novo (13, 25). The telomere reduction rates have been estimated to vary between organisms. For example, in a telomerase-negative Saccharomyces cerevisiae mutant, telomeres were reduced at an approximate rate of 3 bp per generation (33). In contrast to this relatively small reduction rate in yeast, telomeres in telomerase-negative human diploid cells are lost at rates of ∼50 to 200 bp per cell division (reviewed in reference 18). A similar estimate was obtained in vivo using TERC (this gene encodes the mouse telomerase template RNA, mTR) knockout mice (1). These telomere shortenings in telomerase-negative cells are generally attributed to the end replication problem. However, inability of DNA polymerases to replicate the end of the linear DNA molecule during lagging strand synthesis has not been directly demonstrated. Therefore, we do not know the extent of the end replication problem caused by the DNA replication machinery per se. Could the end replication problem be caused by priming failure of the Okazaki fragment at the extreme end, failure of the most distal RNA primer to be replaced by DNA, or both? What might be the reason for the large difference of telomere reduction rates per division between yeast and mammalian cells? To answer these questions, it is necessary to have a biochemical system in which the end replication problem hypothesis can be directly analyzed. Therefore, we developed a novel in vitro system for linear SV40 DNA replication. Results show that the end replication problem indeed occurs in this simple system and may solely explain the telomere reduction rates reported in human cells.
MATERIALS AND METHODS
Preparation of cytoplasmic S100 extracts, SV40 T-ag, and plasmid DNAs used for in vitro replication reactions.
Preparation of cytoplasmic extracts from 293 cells and immunoaffinity purification of SV40 T antigen (T-ag) from recombinant SV40 T-ag-expressing baculovirus-infected insect cells were performed as described previously (5). Plasmid DNAs used for replication reactions were prepared by purifying closed circular plasmid DNA by equilibrium centrifugation in CsCl-ethidium bromide (EtBr) gradients.
In vitro replication of linear DNA.
Linearized plasmid filled in with biotinylated deoxynucleoside triphosphates (dNTPs) was prepared as follows. pSVO11, a plasmid carrying an origin for SV40 replication (29), was first digested with BsrFI. BsrFI cuts the plasmid at a single site, which is located approximately opposite the origin. BsrFI produces a 4-nt 5′ protruding end, which was filled in with dGTP and biotinylated dCTP (Pharmacia) using Klenow enzyme. Biotin-labeled pUC19 was also prepared as described above. Biotin-labeled pSVO10 was digested with AflII and filled in with dATP and biotin-labeled dUTP. Biotin-labeled DNA was then bound to avidin-coated beads (MagneSphere Magnetic Paramagnetic Particles; Promega). By this procedure, more than 50% of DNA was labeled with biotin and captured on beads (data not shown). DNA captured on the beads was subjected to an in vitro replication reaction, basically under the conditions described (36). Products obtained from in vitro replication were purified by phenol extraction and chloroform extraction followed by ethanol precipitation and subjected to the subsequent analysis. Biotin-labeled DNAs captured on beads were recovered efficiently (more than 75% was recovered) by this method.
Analysis of replication intermediates by neutral-neutral 2D gel electrophoresis.
In order to detect intermediates in the replication reactions, we employed neutral-neutral two-dimensional (2D) gel electrophoresis (4). Samples obtained from circular DNA replication were digested with the restriction enzymes indicated in the figure legends before loading on the gels. The first dimension was run in 0.45% agarose in 1× TBE without EtBr, and the second was run in 1.2% agarose in 1× TBE with EtBr. The gel was photographed, dried on a Whatman 3MM paper, and exposed to X-ray film.
Analysis of replication products by denaturing gel electrophoresis.
Products obtained from in vitro replication performed in the presence of [α-32P]dATP were purified as outlined above. The purified DNA was either treated with or without λ exonuclease (GIBCO), exonuclease III (Takara), and exonuclease I (New England BioLabs). Exonuclease III treatment was performed for 1.5 or 3 min at 15°C, and λ exonuclease treatment was performed for 1 or 2 min at 15°C. DNA fragments were further digested with restriction enzymes and separated by 6% denaturing acrylamide gel electrophoresis. To calculate the efficiency of lagging strand synthesis as shown in Fig. 5B, the radioactivity level of each band in the acrylamide gels was quantified by an imaging analyzer.
FIG. 5.
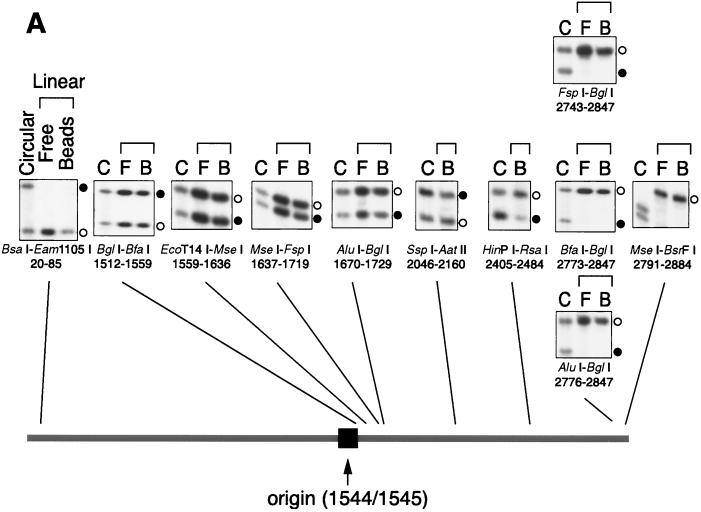
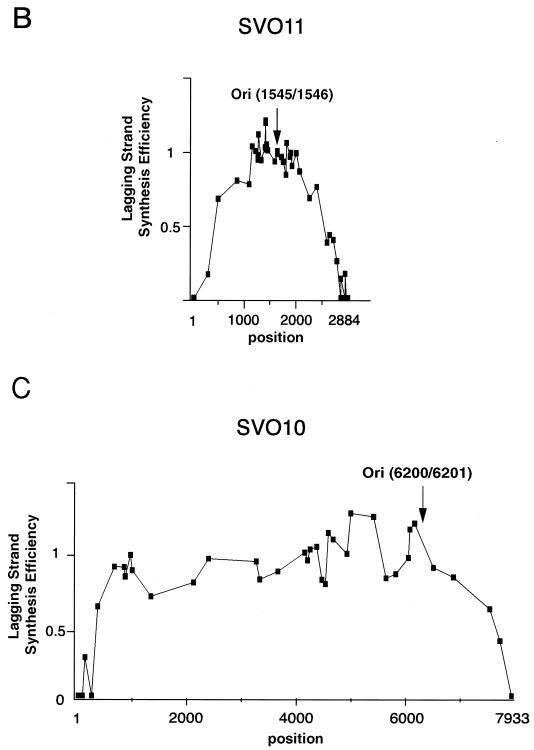
Relative efficiencies of local leading and lagging strand syntheses in the linear DNA replication system. (A) Labeled replication products from circular (lanes C), and linear (lanes F) pSVO11 in solution, and from pSVO11-beads (lanes B), were digested with the combinations of restriction enzymes indicated. The samples were run in 6% denaturing acrylamide gels and autoradiographed. Signals deriving from leading strand syntheses are shown by open circles, and those from lagging strand syntheses are shown by filled circles. Fragments are presented in the same order as their restriction sites are located along the linear pSVO11 DNA. The replication origin is shown by an arrow. Since linear DNA was digested with BsrFI and subsequently filled in by the Klenow enzyme, whereas circular DNA was only digested with BsrFI, there is a small difference in fragment size at both ends, as seen in panel MseI-BsrFI. (B) Relative efficiencies of lagging versus leading strand synthesis were calculated for different regions on the linear pSVO11 molecule. Experiments similar to those in panel A were done extensively, thus covering many regions of the replication products derived from pSVO11-beads and circular pSVO11. Nascent lagging and leading strand fragments for each restriction fragment were inferred from their expected DNA lengths. Then, the lagging strand intensity was divided by the leading strand intensity (Ilag/Ilead) for each restriction fragment. Because the circular pSVO11 products are completely replicated, the corresponding values serve as a reference for the variation in labeling efficiencies caused by nucleotide compositions. Ilag/Ilead was divided by this factor to compensate for the labeling efficiencies before plotting. The position of the SV40 replication origin is shown by an arrow. Most values are means of at least two replicate experiments. (C) Similar analyses were done for the pSVO10 molecule, which is a 7,933-bp SV40 origin-containing plasmid. Most values are means of at least two replicate experiments.
In-gel Southern hybridization.
Strand-specific probes originating from positions 1 to 197 and 2681 to 2884 of pSVO11 were prepared as follows. PCR primers were synthesized with sense and antisense sequences derived from positions 2681 to 2700 (primer A; ATG AAG CCA TAC CAA ACG ACG AGC G) and 178 to 197 (primer B; CAA TCT AAA GTA TAT ATG AGT AAA C), respectively. Primer A was phosphorylated using T4 polynucleotide kinase. PCR was performed against circular pSVO11 with phosphorylated primer A and nonphosphorylated primer B to obtain double-stranded products. The products were then treated with λ exonuclease to digest phosphorylated strands to completion. To prepare replication products for in-gel Southern hybridization, in vitro replication was performed without α-32P-labeled dNTPs. In-gel Southern hybridization under native conditions was performed as described previously (26). Strand specific probes originating from positions 1 to 197 and 2681 to 2884 of pSVO11 (prepared as described above) were used as probes for the hybridizations.
RESULTS
Requirements for SV40 replication origin-containing linear DNA to incorporate dNMPs in vitro.
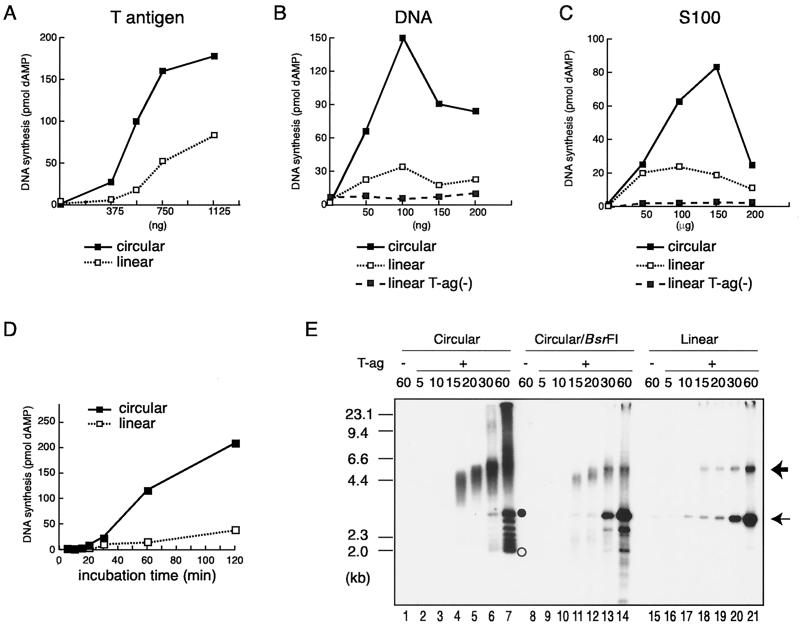
To study the end-replication problem in vitro, we first examined whether linear DNA could be replicated in the in vitro SV40 DNA replication system. In order to establish optimal conditions for linear DNA replication, we examined deoxyribonucleoside 5′-monophosphate (dNMP) incorporation levels under various amounts of purified SV40 T-ag, S100 cell extracts derived from 293 cells, and circular or linearized purified pSVO11 DNA. pSVO11 is a 2,880-bp plasmid containing the SV40 replication origin (29). The BsrFI restriction site, positioned approximately opposite to the origin, was used for linearization of the plasmid (see Fig. 4). Replication reactions were carried out in the presence of [α-32P]dATP to monitor the incorporation of dNMPs into synthesized DNA. As shown in Fig. 1A to C, linear pSVO11 significantly incorporated [32P]dAMP in an SV40 T-ag-dependent manner, although in agreement with previous studies (21, 34, 43) the efficiencies were lower than those of circular pSVO11 under all conditions examined. Interestingly, the optimum conditions for the incorporation of dNMPs into circular and linear pSVO11 were different. For example, increased amounts of S100 extracts resulted in more [32P]dAMP incorporation for circular pSVO11, except at the highest S100 concentration (200 μg of S100 extract/25 μl of reaction mixture) (Fig. 1C). In contrast, incorporation to linear pSVO11 reached a plateau with relatively small amounts of S100 extracts (50 μg of S100 extract/25 μl of reaction mixture) (Fig. 1C). Increased amounts of T-ag resulted in more [32P]dAMP incorporation for both linear and circular pSVO11, but this effect was more profound for the linear DNA (Fig. 1A). In addition, due to the SV40 T-ag-independent incorporation of dNMPs, it was hard to detect SV40 T-ag-dependent DNA synthesis when more than 100 ng of DNA was included in the 25 μl reaction (Fig. 1B and data not shown). Accordingly, under optimal conditions for the linear DNA (750 ng of SV40 T-ag, 50 ng of pSVO11 DNA, and 100 μg of S100 extract per 25 μl of reaction mixture), which are significantly different from those for the circular DNA, 36 pmol of dAMP was incorporated in the linear DNA in a 25-μl reaction mixture during the 2-h incubation (Fig. 1D). Although this value was still lower (1/5.6) than the corresponding value for circular pSVO11 (202 pmol), the incorporation level was 45 times higher than that of a T-ag-minus sample, suggesting that this incorporation was the result of replication. As will be described below, T-ag-dependent DNA synthesis of linear DNA fulfills characteristics for DNA replication products. Therefore, we concluded that linear DNA can be replicated significantly in the SV40 origin-based in vitro system; hereafter in this work we will refer to these DNA synthesis reactions simply as “DNA replication.” In the following experiments, we used the DNA replication conditions optimized for linear DNA molecules.
FIG. 4.
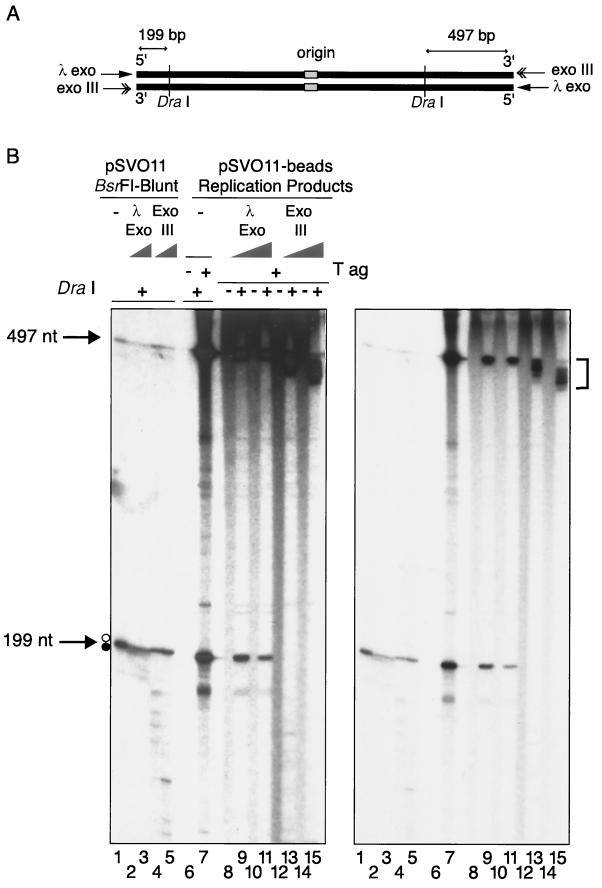
Analysis of terminal restriction fragments from replicated linear DNAs. (A and B) The bound fraction of pSVO11-bead replication products was purified and treated with either λ exonuclease or exonuclease III. To know the exonuclease digestion rates, we treated a separately prepared 199-bp terminal fragment with these exonucleases and found that under the employed conditions, approximately 100 nt is digested from the ends, albeit relatively asymmetrically (data not shown). After the digestion, a half aliquot of the DNA was further treated with DraI, which produces 199- and 497-bp fragments from the left and right arms of the DNA, respectively (A). Samples were run in a 6% denaturing acrylamide gel, dried, and autoradiographed. Heavily and lightly exposed autoradiographs of the same gel are shown. Control pSVO11 DNA was digested with BsrFI, and the two ends were filled-in with dNTPs. The resultant blunt-ended linear pSVO11 was first treated with either λ exonuclease or exonuclease III, followed by DraI digestion. The products were first dephosphorylated by alkaline phosphatase at their 5′ ends and then labeled by T4 polynucleotide kinase and [γ-32P]ATP. DraI digests DNA at a TTT/AAA site, leaving blunt ends. Therefore, the two 199-nt and 497-nt fragment strands have the same nucleotide lengths (arrows). However, because of the effect of different base compositions on migration rates, two distinct 199-nt single-stranded DNA bands are visible in lane 1. The upper and lower bands (marked by open and filled circles, respectively) of the 199-nt doublet were completely digested by λ exonuclease and exonuclease III, respectively (lanes 2 to 5). The 199- and 197-nt bands were detected in pSV011-band replication products. These two bands were resistant to λ exonuclease (lanes 9 and 11). In contrast, the 199-nt band was completely digested, and the 497-nt band was significantly trimmed by exonuclease III (lanes 13 and 15; shorter-sized 497-nt bands are indicated by a bracket). These results indicate that the observed 497- and 199-nt bands were derived solely from a strand whose 3′ ends correspond to nascent radiolabeled DNA ends. Several extra bands were observed in lane 7. We do not know the precise origin of these signals. However, because they are both λ exonuclease and exonuclease III sensitive, it is likely they represent unligated lagging strand DNA molecules derived from internal template regions. It seemed that λ exonuclease had reached the DraI site on the template (cold) strand of some molecules, because the signal intensity of the 199-nt band decreased after the λ exonuclease treatment.
FIG. 1.
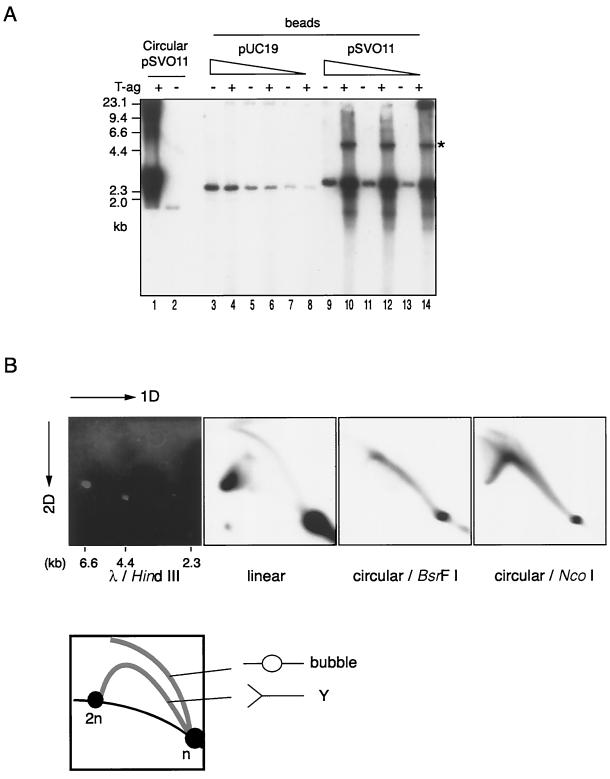
In vitro replication of circular and linear pSVO11 DNAs. (A to C) Titrations of the amounts of SV40 T-ag, template DNA, and cytosol S100 extracts derived from 293 cells in DNA synthesis from circular and linear pSVO11 DNA templates. Reactions were conducted using varied amounts of the immunoaffinity-purified SV40 T-ag (A), pSVO11 template (B), and 293 cell S100 (C) extracts, as indicated in the figure. Other components in the reaction mixture (25 μl each) were 50 ng of DNA template and 80 μg of S100 extracts (A), 600 ng of T-ag and 80 μg of S100 extracts (B), and 50 ng of DNA template and 600 ng of T-ag (C). The samples were incubated at 37°C for 1.5 h. The average incorporation of [32P]dAMP (in picomoles) under each condition was measured. (D) Kinetics of DNA synthesis. Reaction mixtures (25 μl each) containing 50 ng of either circular or linearized pSVO11 DNA, 750 ng of T-ag, and 100 μg of cytosol S100 extracts were incubated for the times indicated, and the average incorporation of [32P]dAMP (in picomoles) was measured. (E) Gel electrophoresis analysis of the DNA products obtained from circular and linear pSVO11 template DNAs. One-fifth (for 20 to 60 min) or 1/2.5 (for 5 to 15 min) of each product obtained in the reactions containing the linear pSVO11 templates and 1/10 (20 to 60 min) or 1/5 (5 to 15 min) of each product obtained in the reactions containing the circular pSVO11 templates were run in a 0.8% agarose gel. For circular DNA reactions, the products were analyzed either with (Circular/BsrFI) or without (Circular) BsrFI restriction digestion prior to gel loading. Following electrophoresis, the gel was dried and autoradiographed. λ DNA digested with HindIII was used as a size marker. The closed and open circles indicate the positions of relaxed and supercoiled circular DNAs, respectively. The thin arrow marks the position of full-length linear DNA products. The linear dimer DNA products potentially produced by the endogenous DNA ligase activity present in S100 extracts are indicated by the thick arrow.
We next analyzed the replication products by agarose gel electrophoresis. As shown in Fig. 1E, an apparently full-length product was detected in both the circular and linear pSVO11 reactions (Fig. 1E). These signals were significantly stronger in T-ag-plus samples than in T-ag-minus samples, indicating that most are replication products. The template plasmid DNA used in this study was prepared from a dam+ (DNA adenine methyltransferase gene) strain. Accordingly, the GATC sequence on both strands of the original template DNA was methylated and sensitive to DpnI digestion. When the semiconservative replication happens, one expects the DNA product to become resistant to DpnI digestion, since the nascent strand is not methylated and DpnI fails to digest the hemimethylated GATC sequence. We found that the labeled DNA products derived from linear templates as well as circular templates were resistant to DpnI digestion (data not shown). This result further supported the idea that the DNA products were synthesized by replication reaction. Finally, the full-length products were detected with similar time courses in both the circular and linear pSVO11 reactions. These results suggested that although the efficiencies of DNA replication were lower for the linear DNA (Fig. 1D), the processivity of DNA synthesis in the linear DNA replication was comparable to that of the circular DNA replication. The weak signals observed in the T-ag-minus linear DNA sample are most likely derived from DNA repair reactions (Fig. 1E, lane 15). This signal was higher than those found in T-ag-minus circular DNA samples (Fig. 1E, lanes 1 and 8), suggesting that linear DNA molecules are prone to repair reactions at their ends. We also noticed a signal, the size of which is approximately double that of the original template DNA, that appeared as the linear DNA replication reactions proceeded (Fig. 1E, lanes 18 to 21). This molecular species is linear DNA (see Fig. 3B) and probably is produced by an endogenous DNA ligase activity present in the S100 extracts.
FIG. 3.
DNA replication of linear DNA molecules bound to beads. (A) One-dimensional gel electrophoresis of DNA products. Circular pSVO11, linear bead-captured pUC19 (pUC19-beads) and linear bead-captured pSVO11 (pSVO11-beads) were subjected to the replication reaction. Circular DNA was replicated in solution as described in the legend to Fig. 1. For pUC19-beads and pSVO11-beads, three batches of beads that had been incubated with three different amounts of DNA (60, 20, and 6.6 ng, indicated as the graded triangles) were used as templates. In a separate experiment, it was confirmed that the volume of beads used in these experiments had the capacity to bind these amounts of DNA. In the pSVO11-bead reactions, DNA products liberated from beads were discarded, while the bound DNA fractions (bound fraction) were collected and further analyzed. Aliquots of the DNA products were run on an agarose gel as described in the legend to Fig. 1. Reactions were assembled with (+) or without (−) T-ag. (B) 2D gel electrophoresis of DNA products. The labeled DNA obtained from circular pSVO11 and pSVO11-beads (bound fraction) was assayed by neutral-neutral 2D gel electrophoresis assay (4). λ DNA digested with HindIII was run in parallel and is shown in the leftmost panel to serve as a marker for the linear DNA species. The bound fraction obtained from the pSVO11-bead reaction was run in the second panel. DNA products obtained from circular pSVO11 were digested with either BsrFI or NcoI prior to gel loading. Both enzymes digest the circular DNA products at a single site. The BsrFI site is positioned approximately opposite the replication origin, whereas the NcoI site neighbors the origin. Typical Y arcs were observed for both BsrFI-digested and NcoI-digested circular pSVO11 products (the third and fourth panels). Although bubble-form intermediates, and not Y-form intermediates, were expected to comprise the majority of DNA species in the BsrFI-digested circular pSVO11 products, a bubble arc was not observed in the autoradiograph (the third panel). It has been reported that for an unknown reason, restriction enzymes disrupt bubble structures (3). Typical Y and bubble arcs were observed for the bound fraction of the pSVO11-bead reaction (the second panel). A schematic representation of the relative positions of linear DNA, bubble arcs, and Y arcs is shown below the electrophoresis data. n and 2n represent the positions of full-sized and double-sized linear DNA molecules. The strong signal found at the 2n position of linear species in the pSVO11-bead products probably represents the linear dimer, which is also denoted by an asterisk in panel A, in addition to replication intermediates.
In vitro replication of linear DNA captured on beads.
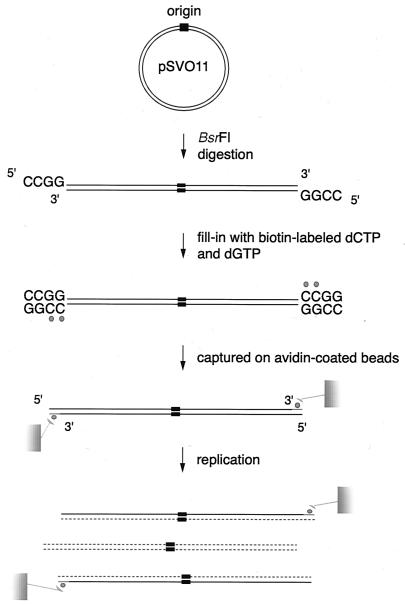
SV40 DNA replication in vitro can proceed for multiple rounds of replication (34). To know if the end replication problem occurs during linear DNA replication, it is essential to examine DNA replication products that have undergone a known number (preferably, a single round) of replications of the original DNA template. Towards this goal, we have developed a novel system to monitor linear DNA replication in vitro, in which the template DNA is conjugated to beads (Fig. 2). pSVO11 is first digested with BsrFI to produce linear DNA whose two ends are 3′ recessive (BsrFI digests an R/CCGGY sequence, where R and Y represent any purines and pyrimidines, respectively). Subsequently, both ends are filled in by the Klenow enzyme using biotinylated dCTP, and the DNAs are captured on avidin-coated beads (pSVO11-beads). The beads are used as templates for the in vitro replication reactions as described in Materials and Methods. In preliminary experiments, we found that irrespective of the DNA concentrations we used, both ends of linear DNA were conjugated simultaneously to the beads (data not shown). In this context, daughter DNA molecules replicated using the original template are expected to remain bound to the beads. In contrast, daughter DNA molecules that have undergone multiple rounds of replication using nascent DNA as templates will likely have lost biotinylated nucleotide and be dissociated from the beads and released into the aqueous phase. Consequently, by purifying DNA bound to the beads after replication reactions, we can recover the replication products that arose only from the original templates.
FIG. 2.
Method to analyze DNA products that have undergone a single round of replication reaction on an original DNA strand template. Plasmid DNA is linearized by BsrFI producing two ends with 5′-protruding 5′-CCGG-3′ sequences. The 3′-recessive ends are subsequently filled in with biotinylated dCTPs and dGTPs by the Klenow enzyme. The resultant DNA molecules possess two biotinylated, blunt ends. Both ends are conjugated to avidin beads. When these bead-captured linear DNAs are used as templates for in vitro DNA replication, their daughter DNA molecules remain bound to the beads, whereas daughter DNA molecules that have been replicated using nascent DNA strands as templates are liberated to the aqueous phase. By simply collecting the DNA molecules bound to the beads after replication, it is possible to analyze the products of a single round of DNA replication on the original DNA strand. Labeled biotin is represented by the small filled circles. Nascent DNA is shown by the dotted lines.
As shown in Fig. 3A, when the bead-captured pUC19 DNA lacking the SV40 replication origin (pUC19-beads) was used as a template in replication reactions, T-ag-dependent incorporation of [32P]dAMP was not observed. On the other hand, when pSVO11-beads were used in the reactions, T-ag-dependent incorporation was observed. If the observed labeling of DNA was the result of DNA replication, fragments proximal to the replication origin were expected to be labeled earlier than distal fragments (29). With this in mind, we digested the labeled DNA products derived from pSVO11-beads with restriction enzymes, ran the products in a gel, and examined the time course of [32P]dAMP incorporation into restriction fragments that were positioned at various distances from the replication origin. In DNA products obtained from the pSVO11-beads and circular pSVO11, origin-proximal fragments were selectively labeled in the earlier stages of DNA synthesis, whereas origin-distal fragments appeared in the later stages (data not shown). This result supports the notion that the labeled DNA products resulted from DNA replication and not from nonspecific DNA synthesis, such as that in DNA repair reactions.
To confirm that the reaction was indeed DNA replication, we employed a neutral-neutral 2D gel electrophoresis assay that exhibits replication intermediates as the slower-migrating species in the second dimension of the electrophoresis (4). The labeled DNA products obtained from circular pSVO11 and pSVO11-beads were subjected to 2D gel electrophoresis (Fig. 3B). As expected, DNA products obtained from circular pSVO11 templates were observed as slow-migrating DNA replication intermediates (Fig. 3B, right two panels). The bound fraction of pSVO11-bead products showed two slow-migrating species which were interpreted as a Y arc and a bubble arc (Fig. 3B, second panel; the relative positions of these two arcs are illustrated below the autoradiographs). Taken together, the T-ag-dependency, the earlier labeling of origin-proximal fragments, and the 2D gel migration pattern characteristic of replication intermediates strongly indicate that the pSVO11-bead reactions are DNA replication.
Leading strand but not lagging strand is synthesized to the end.
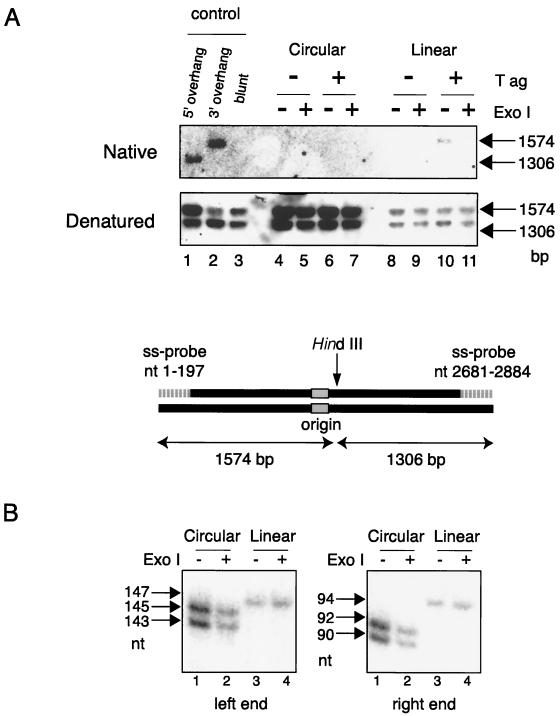
Using the linear DNA replication system described above, we next analyzed whether or not linear DNA was replicated to the end. pSVO11-bead replication reactions were performed with [α-32P]dATP, and the replicated DNA (bound fraction) was digested with DraI, which produces 199- and 497-bp fragments derived from the bead-linked ends of the left and right arms, respectively (in the orientation illustrated in Fig. 4A). After digestion, the size of nascent terminal fragments was examined by denaturing polyacrylamide gel electrophoresis. As shown in Fig. 4B, both the 199- and 497-nt labeled fragments were detected in a T-ag-dependent manner (compare lanes 6 and 7). We then treated DraI-digested samples with NaOH to remove RNA primers that may exist at the 5′ ends of nascent lagging stands. However, there was no detectable difference in the length of the 199- and 497-nt fragments between NaOH-treated and untreated samples (data not shown). This result indicated that the labeled 199- and 497-nt fragments do not contain species with alkali-labile RNA regions and suggested that they did not represent the DNA strands produced by the lagging strand synthesis. To test this hypothesis, we treated the products with λ exonuclease, a 5′-to-3′ exonuclease, or exonuclease III, a 3′-to-5′ exonuclease, prior to restriction digestion (Fig. 4A). Since the replication products subjected to the analyses used the original (cold) DNA strands bound to the beads as templates, they are hybrids of an original template strand and a radiolabeled nascent strand. Thus, radiolabeled nascent strands positioned 3′ to the origin are synthesized solely by leading strand synthesis, and those 5′ to the origin are synthesized by lagging strand synthesis. Consequently, λ exonuclease specifically digests the labeled nascent lagging strand signals from the 5′ end, whereas exonuclease III specifically digests the labeled nascent leading strand signals from the 3′ end. When replication products were treated with exonuclease III, the 199-nt fragment was not detected, and the 497-nt fragment became significantly shorter, indicating that both fragments are susceptible to exonuclease III (Fig. 4B, lanes 13 and 15). In contrast, when products were treated with λ exonuclease, no significant change in migration rates was observed, indicating that both fragments are resistant to λ exonuclease (lanes 9 and 11). It was possible that this λ exonuclease-resistance was caused by residual RNA primers (which was not detected by the NaOH-treatment experiments) present at the 5′ ends of the lagging strand fragments. If this had been the case, however, at least some fractions of the fragments should have been resistant to exonuclease III, which we did not observe. Therefore, the 199- and 497-nt labeled single-stranded fragments present in lane 7 are terminal fragments synthesized by leading strand synthesis. Because these labeled nascent leading strand DraI-digested fragments (Fig. 4B, lane 7) were precisely the same length as those of the control terminal fragments (Fig. 4B, lanes 2 and 3), we concluded that leading strand synthesis occurred completely to the very ends of the template DNA, thus likely producing blunt ends. Lagging strand synthesis was apparently defective since we did not observe NaOH or λ exonuclease-sensitivity or exonuclease III-resistance of the radiolabeled fragments. The lagging strand synthesis may have ended before the two terminal DraI regions were reached. Alternatively, the synthesis may have been halted at various points in these regions, giving rise to products of different lengths that resulted in smears of labeled DraI fragments. An experiment presented in the following section suggests the latter possibility.
Lagging strand synthesis is halted within 500 bp from the end.
The results described above suggest that lagging strand synthesis was not completed to the terminal regions in the linear DNA replication system. We therefore analyzed relative efficiencies of lagging strand versus leading strand synthesis in more detail. Towards this goal, we exploited restriction length polymorphisms of two strands of restriction fragments. For example, when a DNA region is digested by a restriction enzyme leaving a 4-nt 5′ overhang at one end and by another enzyme leaving a 4-nt 3′ overhang at the other end, one would expect to see an 8-nt difference between the longer and shorter strands. We thus performed double restriction digestions of labeled replication products with different combinations of restriction enzymes and ran the digested fragments in denaturing polyacrylamide gels. By comparing the intensities of the two distinctly migrating labeled strands derived from the same restriction fragments, we were able to estimate the relative DNA synthesis efficiencies of the two strands. These analyses were performed throughout the length of the linear DNA, and typical results are shown in Fig. 5A. The band intensities could be influenced by relative densities of the nucleotide used for labeling (adenine in this case). Results obtained similarly for circular pSVO11 products (completely replicated in both leading and lagging strand syntheses) served as a good reference in accounting for this sequence-specific variation in labeling efficiencies. When band intensities were roughly examined, the circular pSVO11 products apparently gave rise to approximately the same band intensities for the two strands in all the restriction fragments examined (Fig. 5A). In contrast, fragments derived from the pSVO11-bead products varied in relative intensity of the two strand signals. Intensities from the lagging and leading strand syntheses were approximately the same for restriction fragments derived from the site neighboring the replication origin (see, for example, the MseI-FspI fragment in Fig. 5A). Interestingly, however, nascent lagging strands derived from restriction fragments close to the two ends of the pSVO11-beads showed highly reduced intensities compared to corresponding leading strands (see, for example, the BsaI-Eam1105I and FspI-BglI fragments in Fig. 5A). To qualify the observations more accurately, we first measured the intensities of leading and lagging strand bands and calculated their relative intensities by dividing intensity values of the lagging strand by those of the leading strand. A similar calculation was done for the corresponding strands derived from circular pSVO11 products. Finally, to compensate for sequence-specific variations in labeling efficiencies, values obtained from the pSVO11-bead products were normalized to those obtained from circular pSVO11 products. The relative lagging versus leading strand synthesis efficiencies for the various restriction fragments obtained from the pSVO11-bead replication system are plotted in Fig. 5B. Results showed that leading and lagging strand synthesis efficiencies are approximately equal for internal fragments derived from regions greater than ∼0.5 kb from both ends. In contrast, in the two end-proximal 0.5 kb regions, the closer the examined fragment was positioned to the ends, the less efficiently lagging strand synthesis occurred compared to the leading strand synthesis. Indeed, the nascent lagging strand synthesis was not detected as discrete bands for the two terminal fragments BsaI-Eam1105 I (20 to 85 bp away from the left end) and AluI-BglI (38 to 109 bp away from the right end).
The inefficient lagging strand synthesis may have been caused not because these fragments are close to ends but because these fragments are distant from the replication origin. To exclude this possibility, we examined a second 7,929-bp plasmid, pSVO10, containing the SV40 replication origin. We linearized pSVO10 such that the position of the origin was not at the center but relatively close to one end of the molecule. Again, fragments derived from the internal region (>0.5 kb from both ends) showed approximately equal replication efficiencies between the leading and lagging strand syntheses (Fig. 5C). In contrast, lagging strand synthesis in the terminal 0.5-kb regions decreased in efficiency towards the ends. Importantly, the efficiency of lagging strand synthesis apparently depends on the absolute distance of the fragment from the ends of the template, rather than on the distance to the replication origin (note that the left arm end is more distant from the origin than the right arm end). Therefore, we concluded that lagging strand synthesis is compromised in terminal regions of linear DNA.
One concern about the bead-conjugated DNA replication system was that the DNA replication complex may have been excluded from the terminal regions because of steric hindrance caused by the proximity to the bead surface. However, such a spatial constraint appears not to be the case, because we observed similar inefficient lagging strand synthesis in the DNA replication of free linear pSVO11 in solution (for example, compare F and B lanes of BsaI-Eam1105I and FspI-BglI fragments in Fig. 5A, where F and B are results of free and conjugated pSVO11, respectively). Thus, we believe that the bead-conjugated replication system accurately reflects natural replication reactions in solution.
Lagging strand synthesis leaves 3′-overhanging ends, and leading strand synthesis leaves blunt ends.
The results shown above suggest that ends produced by lagging strand synthesis have single-stranded 3′ overhangs, whereas ends produced by leading strand synthesis are blunt. However, end structures may be modified by postreplication mechanisms, such as 5′ exonucleases, as has been suggested in vivo (24, 41). To analyze the structure of the ends more accurately, we applied a nondenaturing in-gel hybridization technique, which is commonly used to detect single-stranded overhangs at telomeres in vivo (40). Replication reactions of circular pSVO11 or pSVO11-beads were carried out with cold dNTPs. The circular pSVO11 products were digested with BsrFI (which was used to linearize pSVO11 on beads) and HindIII. The pSVO11-bead products were digested with HindIII only. These digests were run in agarose gels and hybridized with a mixture of strand-specific probes corresponding to the upper strands of the fragments from nt 1 to 197 and nt 2681 to 2884 (Fig. 6A). The probe for nt 1 to 197 was expected to hybridize with a 3′-protruding 1,574-bp BsrFI-HindIII fragment, whereas the probe for nt 2681 to 2884 was anticipated to bind with a 5′-protruding 1,306-bp BsrFI-HindIII fragment. The predictions were confirmed by the results shown in Fig. 6A, lanes 1 to 3, where BsrFI-HindIII fragments artificially modified to possess 5′, 3′, and blunt ends manifested the expected hybridization pattern. Under these conditions, the digests of circular pSVO11 products did not hybridize with either probe, as expected (Fig. 6A, lanes 4 to 7). On the other hand, the 1,574-bp HindIII fragment derived from pSVO11-bead products hybridized with the nt 1 to 197 upper-strand probe, indicating that at least a fraction of replicated DNAs had 3′-overhanging left ends (Fig. 6A, lane 10). These 3′ overhangs were not accidentally formed during preparation of the DNA materials, since prior treatment of the replication product with exonuclease I, which specifically removes single-stranded 3′-overhangs, abolished the signal (Fig. 6A, lane 11). Furthermore, the signal was observed only in samples that were incubated with T-ag during the replication reactions (Fig. 6A, lanes 8 and 9), indicating that the 3′ overhangs we observed resulted from DNA replication. In contrast, the 1,306-bp HindIII fragment of pSVO11-bead products failed to hybridize with the nt 2681 to 2884 upper-strand probe (Fig. 6A, lane 10), suggesting that the right end did not have a 5′ overhang. In a comparable experiment performed using a mixture of two lower-strand-derived probes, a similar conclusion was obtained: At least a fraction of replicated molecules possessed 3′-overhanging right ends, and there was no indication that left ends had 5′ overhangs (data not shown).
FIG. 6.
Analysis of the end structure of replicated DNAs. (A) Nondenaturing hybridization analysis of the replicated products. The circular pSVO11 (Circular) and pSVO11-bead (Linear) replication reactions were carried out in the absence (−) or presence (+) of SV40 T-ag with cold dNTPs. The replication products were treated (+) or not treated (−) with exonuclease I. pSVO11 products were subsequently digested with BsrFI (used to prepare pSVO11-beads) and HindIII, while pSVO11-bead products were digested with HindIII only. The approximate positions of the HindIII site on the linearized pSVO11 are shown. The restriction digests were run in an agarose gel and then subjected to in-gel hybridization with strand-specific probes. A mixture of two strand-specific probes, corresponding to the upper strands of the regions from nt 1 to 197 and nt 2681 to 2884, was used. Control DNA BsrFI-HindIII fragments possessing 5′-protruding (5′ overhang), 3′-protruding (3′ overhang), or blunt BsrFI sites were run in parallel. The 5′-protruding and 3′-protruding DNAs were prepared by treating the end-filled DNA with exonuclease III and λ exonuclease, respectively. (B) The ends containing the terminal nascent leading strand do not have 3′ overhangs. In vitro replication was performed with circular pSVO11 and pSVO11-beads in the presence of [α-32P]dATP. The products were treated with (+) or without (−) exonuclease I, followed by restriction enzyme digestion. The circular pSVO11 products were digested with BsrFI and MseI. The pSVO11-bead products were digested with MseI only. By MseI digestion, 147- and 94-nt signals deriving from both ends are expected as a result of leading strand synthesis for pSVO11-bead products without exonuclease I treatment. The digests were run in a 6% denaturing acrylamide gel and then autoradiographed. The reason for the observed difference in size between circular and pSVO11-bead replication products is described in the legend to Fig. 5A.
These results can be most simply explained by assuming that lagging strand synthesis left 3′-overhanging ends due to the end replication problem, while leading strand synthesis left blunt ends. To confirm this possibility more vigorously, we investigated the origin of the 3′-overhanging ends. Circular pSVO11 and pSVO11-beads were replicated in the presence of [α-32P]dATP. The circular pSVO11 products were linearized with BsrFI and digested with MseI (Fig. 6B, lanes 1 and 2). The pSVO11-bead products were digested with MseI only, producing two end-derived fragments of 145 and 92 bp (Fig. 6B, lanes 3 and 4). These digests were run in a denaturing polyacrylamide gel and then autoradiographed. The terminal radiolabeled fragments derived from pSVO11-beads were solely derived from leading strand synthesis and have 3′-OHs at their ends (as shown in Fig. 4). If these ends had been modified to possess 3′ overhangs by a postreplication mechanism, such as 5′ exonuclease activity that digests 5′ ends of the cold template strand, treatment of the products with 3′-overhang-specific exonuclease I would digest the labeled strand. However, this was not the case as no size difference was observed between terminally labeled leading strands incubated with or without the exonuclease I (Fig. 6B, lanes 3 and 4). Therefore, we concluded that the 3′-overhanging ends detected in Fig. 6A were derived by lagging strand synthesis, and most probably occurred due to the end replication problem. On the other hand, because the terminal nascent leading strand fragment showed the same length as its template fragment (Fig. 4), ends produced by leading strand synthesis remain blunt following the replication process.
DISCUSSION
Since its first proposal in the early 1970s, the end replication problem hypothesis has provided a possible explanation for a large body of puzzling biological phenomena, such as the limited capacity of normal cells to proliferate (cellular senescence) (2) and the chromosomal instabilities in cancerous cells (11). However, no in vitro system explored thus far has been suitable for direct analysis of replication at linear DNA ends. In this study, we have established a novel in vitro linear SV40 DNA replication system. Although it is generally believed that the SV40-based system replicates linear DNA very poorly (21), we have found that linear DNA can be efficiently replicated under optimized conditions. Taking advantage of SV40's efficient replication reactions in vitro, we were able to show the presence of the end replication problem at molecular levels.
Linear DNA in vitro replication system.
We first examined replication efficiencies of circular and linear DNAs under a variety of conditions. In agreement with previous reports, under all conditions examined, linear DNA templates incorporated dNMPs less effectively than circular DNAs. One remarkable finding was that optimum conditions for replication reactions of circular and linear DNAs were different. We needed to add relatively large amounts of T-ag and relatively small amounts of S100 extracts and template DNAs to obtain maximal replication efficiencies for linear DNA (750 ng of SV40 T-ag, 50 ng of pSVO11 DNA, and 100 μg of S100 extract per 25 μl of reaction mixture, for example). This reaction composition is different from that found in typical replication reactions of circular DNAs. We speculate that linear DNA replicated very poorly in the SV40-based system, because previous experiments were done under conditions optimal for circular DNA but not for linear DNA. Recently, it has been reported that when linear double-stranded DNA was preincubated with S100 extracts, efficiencies of the SV40 in vitro-replication system were reduced, probably due to activation of endogenous DNA-dependent protein kinase (DNA-PK) by the linear DNA molecules (38). However, our system does not include this preincubation step and, thus, is considerably different from the reported system. Furthermore, increasing the amount of template DNA did not profoundly reduce the replication efficiencies (Fig. 1B). Therefore, we think that the effect of DNA-PK activation, if any, is not significant in our system.
In order to analyze changes at the ends of linear DNA precisely, we developed a novel system in which linear DNAs conjugated to beads were used as templates. By purifying DNA still attached to beads after replication reactions, it was possible to isolate products that had replicated from original templates. Three lines of evidence support the notion that DNA replication indeed occurs in this system. First, DNA synthesis reactions depended on the presence of the SV40 replication origin on the DNA template, as well as on the presence of T-ag in the reaction. Second, typical replication intermediates, revealed as the bubble and the Y arcs, were observed following 2D gel electrophoresis of the products. Finally, the time course of DNA synthesis of fragments located at different distances from the origin supported the expectation of propagation of DNA synthesis occurring from the origin towards the ends.
End replication problem.
From the analysis of linear DNA replication products, we showed that leading strands are synthesized completely to their end, while lagging strand synthesis leaves nascent DNAs incomplete at their 5′ ends, thus, for the first time, formally demonstrating the end replication problem. SV40 T-ag is a 3′-to-5′ helicase and forms a hexameric ring-shaped complex (reviewed in reference 7). It is believed that the T-ag hexamer moves ahead of replication forks along the leading strand template. Therefore, in the leading strand synthesis, T-ag may continue to unwind DNA until the very end of the linear leading strand template to assure the completion of DNA synthesis. In vivo, DNA replication-coupled helicases may execute a similar function in place of T-ag, although the identity of such helicases remains unknown.
As DNA polymerization progressed towards the ends, lagging strand synthesis occurred less efficiently. By plotting the relative replication efficiencies for lagging and leading strand syntheses, we specified that lagging strand synthesis is gradually halted within the most distal ∼500-bp regions. Two molecular mechanisms have been proposed for the end replication problem. The first idea suggests an inability for the most distal RNA primer to be replaced by DNA, explaining only an ∼10-nt (the length of one RNA primer) loss from the nascent DNA end. Since we found that lagging strand synthesis fails to replicate an average length of ∼250 nt (roughly 25-fold the size of a primer) at ends of linear DNA templates, it is highly unlikely that the lack of end primer replacement is the sole basis for the end replication problem. Accordingly, the second hypothesis, which predicts an inability for DNA polymerase α-primase to initiate lagging strand synthesis from the very end of linear DNA, should be a major cause of the end replication problem.
Typical Okazaki fragments formed in the in vitro SV40 DNA replication system are ∼200 to 500 nt long (for example, see reference 23). A consensus sequence for initiating the primer synthesis on the SV40 genome has been reported (6). However, the consensus is very short and was calculated to be present every 19 nt on average. Therefore, priming events in lagging strand synthesis of SV40 DNA apparently happen in a relatively random manner. If such random priming holds true for the end replication of linear DNA, it could explain why the most distal ∼500-bp region of linear DNA suffers from incomplete lagging strand synthesis. This is because since two neighboring Okazaki fragments need to be ligated to form a continuous nascent DNA strand, a priming event should happen at least once per 500 nt (the maximum Okazaki fragment length). Therefore, in lagging strand synthesis, any region of >500 bp interior to the DNA ends is expected to be replicated at a probability close to 1, whereas the very ends will be replicated at a probability near 0. Because the priming happens randomly, any point in the terminal 500-bp region will be replicated at a probability proportional to its distance from the ends. These predictions are consistent with the observation in this study that replication efficiency decreased towards the ends of linear DNA (Fig. 5B and C). However, the experimental resolution was not high enough to vigorously test this prediction, and thus further study is necessary.
It is possible that the 3′-terminal single-stranded regions resulting from the incomplete lagging strand synthesis may be reprimed by the polymerase α-primase complex and be modified in a postreplication manner. However, we think that this possibility is unlikely, because it is known that the single-stranded DNA-binding protein RPA is present abundantly in the SV40 replication system and suppresses such nonspecific DNA synthesis from single-stranded regions (8).
Vertebrate telomeres consist of tandem repeats of six nucleotides (T2AG3) (10, 27). The T2AG3 strand is oriented in a direction of its 3′ end being toward the telomeric ends and is replicated by the leading strand synthesis. The complementary C3TA2 repeat is replicated by the lagging strand synthesis. We also examined in vitro replication reactions of linear DNA molecules having a telomeric repeat sequence at one end (R. Ohki and F. Ishikawa, unpublished data). We found that the telomeric end possessed 400- to 500-nt single-stranded T2AG3 repeats after replication, but no single-stranded C3TA2-repeat was detected. The single-stranded T2AG3 repeats were found specifically at telomeric ends that had been synthesized by the lagging strand synthesis, but not at those synthesized by the leading strand synthesis. This result indicated that the leading strand was synthesized completely to the ends, but the lagging strand was not. Therefore, the end replication problem happens similarly at both telomeric and unique DNA ends in our system.
These results, however, were not expected straightforwardly. Telomeric DNA replication may be unique in a variety of ways during and after replication. For example, a primase activity that specifically synthesizes telomeric C-rich strands using single-stranded telomeric G-rich strand, has been observed in Oxytricha nova (47). We think it is unlikely that any telomere-specific trans-acting factor provided by the S100 extract would have been sufficient enough to react with the substrates, because telomeric DNAs existed abundantly in our in vitro replication system (∼3 × 1010 ends/25 μl of reaction mixture). Therefore, it is difficult to characterize trans-acting factors in the present system. cis elements present on telomeric DNA, however, may have modified the replication reactions. For example, single-stranded G-rich sequences are known to form specialized tertiary structures called G quartets (17, 32, 42). The G-quartet structure is a poor substrate for telomerase (48). It is possible that during the T-ag-induced melting of double-stranded telomeric DNA in the replication reaction, the G-rich strand forms a G-quartet structure, which does not serve as an efficient template for the DNA replication. In addition, it is known that mammalian DNA polymerase α-primase preferentially initiates priming at or near pyrimidine-rich (more than six consecutive pyrimidines) sequences (9, 35). This is clearly not the case in mammalian telomeric G-rich strands since they have only two thymines in one repeat. Thus, it is possible that mammalian primase does not initiate the lagging strand synthesis efficiently. However, as far as within the limitation of experimental sensitivity, we found that telomeric ends were replicated in a similar manner as unique ends (R. Ohki and F. Ishikawa, unpublished data). Therefore, telomeric DNA sequences appear not to have significant effects on the general DNA replication machinery. Indeed, using purified DNA polymerase α-primase and synthetic oligonucleotide templates containing G-rich telomeric repeats, it has been shown that RNA priming happens at the 3′-thymidine (underlined) of the template 5′TTAGGG3′ sequence in vitro (30). However, the question of whether lagging strand synthesis occurs at telomeric repeats as efficiently as at unique sequences remains to be answered.
Telomere structures in vivo
In 1981, it was first discovered that both ends of linear gene-sized minichromosomes present in macronuclei of hypotrichous ciliates possess 12- to 16-nt GT-rich 3′ overhangs (20). In the yeast S. cerevisiae, it was demonstrated that >30-nt G-rich overhangs (G tails) are present on telomeres at the end of S phase (40). In addition, from an analysis of linear minichromosomes in these yeasts, it was concluded that both ends of one linear DNA molecule possess G tails (41). In contrast, the precise structure of telomeric DNA termini in higher eukaryotes is not well understood. In human cells, relatively long (100- to 300-nt) G tails reside at telomeres throughout the cell cycle for virtually all types of cells examined, including telomerase-positive transformed cells, telomerase-negative normally dividing cells, and dormant cells (24, 26, 44). These observations suggest that G tails are general features of eukaryotic telomeres, a hypothesis consistent with the idea that telomere single-stranded DNA-binding proteins may play an important role in telomere protection (22). However, the question of whether both ends of one linear DNA molecule in higher eukaryote chromosomes simultaneously have similar G tails or not, remains controversial. In one study, it was concluded that the two ends of a single chromosome have relatively long G tails (24). The end replication problem does not explain the presence of G tails at both ends. Moreover, deletions of genes essential for telomerase activity did not significantly affect the G-tail structure in yeast and mice, indicating that G tails occur independent of telomerase (12, 16, 46). Accordingly, it has been suggested that some modification mechanisms, such as C-rich strand-specific 5′ exonucleases, might be responsible for the formation of G tails (24, 41). In contrast, another study argued that in normal human fibroblasts, a single chromosome has one telomere with a long G tail (200 ± 75 nt) and one with a short G tail or a blunted end (44). Very recently, it was found that only a half fraction of the total telomeres in plant cells possess detectable G-tails, suggesting that two ends of a single chromosome have different telomeric structures (31). Since the end replication problem produces one blunt end and one 3′-overhanging end in a single replicated linear DNA (Fig. 6), it is possible that this mechanism is the primary cause of the “asymmetrical telomeres” observed in mammalian and plant cells. A postreplication mechanism (such as C-strand-specific 5′ exonucleases) may form short G tails at telomeres that were originally blunt immediately after replication, although the presence of a short G tail at this end has not been proven yet. In this scenario, telomere attrition during cell proliferation is mostly caused by the end replication problem (44, 45). In this study, we report for the first time the magnitude of the end replication problem caused by the general replication machinery. The estimated DNA loss at the DNA ends replicated by the lagging strand synthesis (250 nt on average) remarkably agrees with the estimated values in a previous study proposing the “asymmetrical telomere model” (44, 45). Because our present experimental system does not address the effects of telomerespecific trans-acting factors, the present study does not provide evidence supporting specifically either of the two current models concerning the structure of human telomeric ends. In yeast, the nascent asymmetrical telomeres produced by the replication machinery may be postreplicationally modified into “symmetrical telomeres” by virtue of telomerase-associated activities, such as DNA polymerase α and C-strand-specific 5′ exonucleases. Different postreplicational activities may explain the different telomere attrition rates between telomerase-negative yeast and mammalian cells. Alternatively, yeast may have specialized factors associated with the replication machinery, which minimizes effects of the end replication problem. A system including recombinant proteins of molecules involved in these telomere-specific reactions will provide a good opportunity for us to study the mechanistic details of telomere maintenance in human cells.
ACKNOWLEDGMENTS
We are grateful to K. Pepin (Tokyo Institute of Technology) for critical reading of and comments on the manuscript. The excellent secretarial work of F. Nishizaki, K. Saito, and K. Yokoyama is also acknowledged.
This work was supported by a grant-in-aid from the Organization for Pharmaceutical Safety and Research of Japan and a grant-in-aid for Cancer Research from the Ministry of Education, Cultures, Sports, Science, and Technology of Japan.
REFERENCES
- 1.Blasco M A, Lee H W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 2.Bodnar A G, Ouellette M, Frolkis M, Holt S E, Chiu C P, Morin G B, Harley C B, Shay J W, Lichtsteiner S, Wright W E. Extension of life-span by introduction of telomerase into normal human cells. Science. 1998;279:349–352. doi: 10.1126/science.279.5349.349. [DOI] [PubMed] [Google Scholar]
- 3.Braguglia D, Heun P, Pasero P, Duncker B P, Gasser S M. Semi-conservative replication in yeast nuclear extracts requires Dna2 helicase and supercoiled template. J Mol Biol. 1998;281:631–649. doi: 10.1006/jmbi.1998.1973. [DOI] [PubMed] [Google Scholar]
- 4.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 5.Brush G S, Kelly T J, Stillman B. Identification of eukaryotic DNA replication proteins using simian virus 40 in vitro replication system. Methods Enzymol. 1995;262:522–548. doi: 10.1016/0076-6879(95)62043-5. [DOI] [PubMed] [Google Scholar]
- 6.Bullock P A, Tevosian S, Jones C, Denis D. Mapping initiation sites for simian virus 40 DNA synthesis events in vitro. Mol Cell Biol. 1994;14:5043–5055. doi: 10.1128/mcb.14.8.5043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock P A. The initiation of simian virus 40 DNA replication in vitro. Crit Rev Biochem Mol Biol. 1997;32:503–568. doi: 10.3109/10409239709082001. [DOI] [PubMed] [Google Scholar]
- 8.Collins K L, Kelly T J. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol Cell Biol. 1991;11:2108–2115. doi: 10.1128/mcb.11.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davey S K, Faust E A. Murine DNA polymerase α fills gaps to completion in a direct assay. J Biol Chem. 1990;265:4098–4104. [PubMed] [Google Scholar]
- 10.de Lange T, Shiue L, Myers R M, Cox D R, Naylor S L, Killery A M, Varmus H E. Structure and variability of human chromosome ends. Mol Cell Biol. 1990;10:518–527. doi: 10.1128/mcb.10.2.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Lange T, DePinho R A. Unlimited mileage from telomerase? Science. 2000;283:947–949. doi: 10.1126/science.283.5404.947. [DOI] [PubMed] [Google Scholar]
- 12.Dionne I, Wellinger R J. Cell cycle-regulated generation of single-stranded G-rich DNA in the absence of telomerase. Proc Natl Acad Sci USA. 1996;93:13902–13907. doi: 10.1073/pnas.93.24.13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greider C W. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- 14.Harley C B, Futcher A B, Greider C W. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 15.Hastie N D, Dempster M, Dunlop M G, Thompson A M, Green D K, Allshire R C. Telomere reduction in human colorectal carcinoma and with ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 16.Hemann M T, Greider C W. G-strand overhangs on telomeres in telomerase-deficient mouse cells. Nucleic Acids Res. 1999;27:3964–3969. doi: 10.1093/nar/27.20.3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson E, Hardin C C, Walk S K, Tinoco I, Jr, Blackburn E H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine-guanine base pairs. Cell. 1987;51:899–908. doi: 10.1016/0092-8674(87)90577-0. [DOI] [PubMed] [Google Scholar]
- 18.Huffman K E, Levene S D, Tesmer V M, Shay J W, Wright W E. Telomere shortening is proportional to the size of the G-rich telomeric 3′-overhang. J Biol Chem. 2000;275:19719–19722. doi: 10.1074/jbc.M002843200. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa F, Naito T. Why do we have linear chromosomes? A matter of Adam and Eve. Mutat Res. 1999;434:99–107. doi: 10.1016/s0921-8777(99)00017-8. [DOI] [PubMed] [Google Scholar]
- 20.Klobutcher L A, Swanton M T, Donini P, Prescott D M. All gene-sized DNA molecules in four species of hypotrichs have the same terminal sequence and an unusual 3′ terminus. Proc Natl Acad Sci USA. 1981;78:3015–3019. doi: 10.1073/pnas.78.5.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li J, Kelly T. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lingner J, Cooper J P, Cech T R. Telomerase and DNA end replication: no longer a lagging strand problem? Science. 1995;269:1533–1534. doi: 10.1126/science.7545310. [DOI] [PubMed] [Google Scholar]
- 23.Mackenney V J, Barnes D E, Lindahl T. Specific function of DNA ligase I in simian virus 40 DNA replication by human cell-free extracts is mediated by the amino-terminal non-catalytic domain. J Biol Chem. 1997;272:11550–11556. doi: 10.1074/jbc.272.17.11550. [DOI] [PubMed] [Google Scholar]
- 24.Makarov V L, Hirose Y, Langmore J P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 25.McEachern M J, Krauskopf A, Blackburn E H. Telomeres and their control. Annu Rev Genet. 2000;34:331–358. doi: 10.1146/annurev.genet.34.1.331. [DOI] [PubMed] [Google Scholar]
- 26.McElligott R, Wellinger R J. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moyzis R K, Buckingham J M, Cram L S, Dani M, Deaven L L, Jones M D, Meyne J, Ratliff R L, Wu J R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc Natl Acad Sci USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olovnikov A M. A theory of marginotomy. J Theor Biol. 1973;41:181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 29.Prelich G, Stillman B. Coordinated leading and lagging strand synthesis during SV40 DNA replication in vitro requires PCNA. Cell. 1988;53:117–126. doi: 10.1016/0092-8674(88)90493-x. [DOI] [PubMed] [Google Scholar]
- 30.Reveal P M, Henkels K M, Turchi J J. Synthesis of the mammalian telomere lagging strand in vitro. J Biol Chem. 1997;272:11678–11681. doi: 10.1074/jbc.272.18.11678. [DOI] [PubMed] [Google Scholar]
- 31.Riha K, McKnight T D, Fajkus J, Vyskot B, Shippen D E. Analysis of the G-overhang structures on plant telomeres: evidence for two distinct telomere architectures. Plant J. 2000;23:633–641. doi: 10.1046/j.1365-313x.2000.00831.x. [DOI] [PubMed] [Google Scholar]
- 32.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 33.Singer M S, Gottschling D E. TLC1: template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 34.Stillman B, Gerard R D, Guggenheimer R A, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki M, Savoysky E, Izuta S, Tatebe M, Okajima T, Yoshida S. RNA priming coupled with DNA synthesis on natural template by calf thymus DNA polymerase α-primase. Biochemistry. 1993;32:12782–12792. doi: 10.1021/bi00210a030. [DOI] [PubMed] [Google Scholar]
- 36.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. II. Switching of DNA polymerase α and δ during initiation of leading and lagging strand synthesis. J Biol Chem. 1991;266:1961–1968. [PubMed] [Google Scholar]
- 37.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 38.Wang Y, Zhou X-Y, Wang H, Huq M S, Iliakis G. Roles of replication protein A and DNA-dependent protein kinase in the regulation of DNA replication following DNA damage. J Biol Chem. 1999;274:22060–22064. doi: 10.1074/jbc.274.31.22060. [DOI] [PubMed] [Google Scholar]
- 39.Watson J D. The origin of concatemeric T7 DNA. Nat New Biol. 1972;239:197–201. doi: 10.1038/newbio239197a0. [DOI] [PubMed] [Google Scholar]
- 40.Wellinger R J, Wolf A J, Zakian V A. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- 41.Wellinger R J, Ethier K, Labrecque P, Zakian V A. Evidence for a new step in telomere maintenance. Cell. 1996;85:423–433. doi: 10.1016/s0092-8674(00)81120-4. [DOI] [PubMed] [Google Scholar]
- 42.Williamson J R, Raghuraman M K, Cech T R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 43.Wobbe C R, Dean F, Weissbach L, Hurwitz J. In vitro replication of duplex circular DNA containing the simian virus 40 DNA origin site. Proc Natl Acad Sci USA. 1985;82:5710–5714. doi: 10.1073/pnas.82.17.5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright W E, Tesmer V M, Huffman K E, Levene S D, Shay J W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wright W E, Tesmer V M, Liao M L, Shay J W. Normal human telomeres are not late replicating. Exp Cell Res. 1999;251:492–499. doi: 10.1006/excr.1999.4602. [DOI] [PubMed] [Google Scholar]
- 46.Yuan X, Ishibashi S, Hatakeyama S, Saito M, Nakayama J, Nikaido R, Haruyama T, Watanabe Y, Iwata H, Iida M, Sugimura H, Yamada N, Ishikawa F. Presence of telomeric G-strand tails in the telomerase catalytic subunit TERT knockout mice. Genes Cells. 1999;4:563–572. doi: 10.1046/j.1365-2443.1999.00284.x. [DOI] [PubMed] [Google Scholar]
- 47.Zahler A M, Prescott D M. DNA primase and the replication of the telomeres in Oxytricha nova. Nucleic Acids Res. 1989;17:6299–6317. doi: 10.1093/nar/17.15.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zahler A M, Williamson J R, Cech T R, Prescott D M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]