Abstract
Improved long-term kidney allograft survival is largely related to better outcomes at 12 months, in association with declining acute rejection rates and more efficacious immunosuppression. Finding the right balance between under- and overimmunosuppression or rejection versus immunosuppression toxicity remains one of transplant’s holy grails. In the absence of precise measures of immunosuppression burden, transplant clinicians rely on nonspecific, noninvasive tests and kidney allograft biopsy generally performed for cause. This review appraises recent advances of conventional monitoring strategies and critically examines the plethora of emerging tests utilizing tissue, urine, and blood samples to improve upon the diagnostic precision of allograft surveillance.
Keywords: transplantation, transplant outcomes, molecular genetics, immunosuppression, biopsy, kidney transplantation series
Introduction
Long-term improvement in kidney transplant outcomes is mostly related to better 1-year allograft survival (1–3), concomitant with declining acute rejection under contemporary immunosuppression largely comprising T cell–depleting induction and tacrolimus-mycophenolic acid-corticosteroid maintenance therapy (2). Between years 2 and 10 post-transplant, allograft attrition rates approximate 5%–7% per year (1). Allograft failure occurs from immunologic or nonimmunologic causes, whereas immunosuppression toxicity affects patient morbidity and mortality. Optimizing transplant outcomes is a central recipient care tenet, commencing with pretransplant immunologic assessment. Without a post-transplant measure of immunosuppression burden or risk of rejection versus overimmunosuppression, clinicians rely on nonspecific markers and indication biopsies for guidance, with allograft histology or transplant failure the gold standard against which nonspecific tests are measured. Noninvasive monitoring tools are being developed for use in several contexts, including as diagnostic, prognostic, and/or predictive markers (Table 1). We herein review conventional tools used for these assessments and highlight emerging tissue, urine, and blood biomarkers aimed at improving precision.
Table 1.
Context of use of biomarkers in kidney transplantation
| Test | Commercially Available | Biomarker Subtype | ||||
| Susceptibility | Diagnostic | Prognostic | Serial Monitoring | Reference Nos. Cited | ||
| Pretransplant DSA | + | + | + | 4–6 | ||
| Eplet mismatch | + | + | + | 7–13 | ||
| Serum creatinine/GFR | + | + | + | + | + | 20,21 |
| UP/Cr | + | + | + | + | + | 22–24 |
| Tacrolimus TTR/CV | + | + | P | P | + | 27–31 |
| EBV NAT | + | + | + | 23,38,42 | ||
| BKV NAT | + | + | + | + | 39–41,43–46 | |
| De novo DSA | + | + | + | P | P | 7,13,32–37 |
| Surveillance biopsy | + | P | + | + | 50–62 | |
| Pretransplant ELISPOT | − | P | P | 16–18 | ||
| MMDX | + | P | + | P | 64–67 | |
| GoCAR 13-gene set | − | P | P | P | 69 | |
| Urine chemokines | − | P | P | P | P | 25,70–72 |
| Blood gene profiling | + | P | P | P | P | 54,75 |
| kSORT | − | P | P | P | P | 73,74 |
| dd-cfDNA | + | P | P | P | P | 77–83 |
DSA, donor-specific antibody; +, established use for biomarker test; UP/Cr, urine protein-urine creatinine ratio; TTR/CV, time in therapeutic ratio/coefficient of variation; P, potential use for biomarker pending further validation studies; EBV, Epstein–Barr virus; NAT, nucleic acid testing; BKV, BK virus; ELISPOT, enzyme-linked immune absorbent spot; MMDX, microarray-based molecular diagnostic system; GoCAR, Study of the Genomics of Chronic Allograft Rejection; kSORT, kidney Solid Organ Response Test; dd-cfDNA, donor-derived cellfree DNA.
Pretransplant Immunologic Risk Assessment
Preexisting Donor-Specific Antibody and Histocompatibility Leukocyte Antigen Epitope Mismatch
Detectable preexisting donor-specific antibody (DSA), ascertained at transplant, is associated with chronic allograft failure (4). Patients with pretransplant DSA and positive crossmatches have high rates of persistent or recurrent DSA post-transplant. Despite desensitization protocols, antibody-mediated rejection occurs in approximately 50% of patients with persistent DSA post-transplant (5,6).
Alloantibodies to HLA engage the HLA molecule’s epitope. Epitope binding affinity is determined by an eplet, a single or small number of polymorphic amino acids near the HLA surface that influence antibody specificity and immunologic risk (7). A seminal 286-patient study demonstrated that locus-specific epitope mismatch correlated more robustly with de novo DSA formation than traditional DR/DQ mismatch (8). Epitope mismatch has since been correlated with rejection and allograft loss when combined with nonadherence (9) and with immunosuppression minimization (10). More recent application of HLA-DR/DQ single-molecular eplet mismatch further improved correlation with de novo DSA and facilitated stratification of recipients into low–, intermediate–, and high–alloimmune risk categories (11). Molecular mismatch risk category was associated with de novo DSA, antibody- and T cell–mediated rejection, and graft loss, findings since validated elsewhere (12,13). Molecular mismatch risk stratification should be easily implementable in HLA laboratories, with potential to positively affect post-transplant outcomes.
Nonhistocompatibility Leukocyte Antigen Mismatching and Nonhistocompatibility Leukocyte Antigen Antibodies
Recently, donor/recipient non-HLA allelic mismatches associating with antibody-mediated rejection were identified through exome sequencing of mononuclear cell DNA (14). Genes for kidney and blood vessel cell surface proteins incurred risk independent of HLA mismatch. In an unrelated study, the presence pretransplant of non-HLA antibodies targeting glomerular endothelium (MHC class 1–related chain A, endothelin 1 type A, and angiotensin type 1 receptor) has been associated with early post-transplant microvascular injury resembling antibody-mediated rejection (15). These preliminary observations indicate that non-HLA–related biomarkers are promising tools to assess post-transplant immunologic risk of potential candidates.
Enzyme-Linked Immune Absorbent Spot
Enzyme-linked immune absorbent spot (ELISPOT) quantitatively measures the cytokine secreting cell frequency. ELISPOT assays for IFN-γ identify effector memory T cells pretransplant when mixed with T cell–depleted donor cells. Pretransplant ELISPOT positivity correlated with rejection in higher-risk populations (16) and during tacrolimus withdrawal in lower-risk patients (17). The association of pretransplant ELISPOT with rejection is tempered by antithymocyte globulin therapy. A recent analysis found a rejection rate of 15% in ELISPOT-positive patients versus 3% in negative patients (18). Among ELISPOT-positive patients who received antithymocyte globulin induction, there was no rejection. In Clinical Trials in Organ Transplantation 01 (CTOT-01), pretransplant ELISPOT similarly predicted allograft function only in the nondepleting induction subgroup (17).
Peripheral Blood Gene Signature
Investigators from the Study of the Genomics of Chronic Allograft Rejection (GoCAR) recently performed whole-blood RNA sequencing in patients pretransplant, developing a 23-gene set that predicted early T cell rejection and associated with late graft loss (19). Further validation of this signature is underway.
Conventional Post-Transplant Monitoring
Allograft Function
Studies demonstrate that 1-year kidney transplant function predicts longer-term failure (20,21), a risk that is greater with worsening albuminuria (22–24). Kidney Disease Improving Global Outcomes (KDIGO) guidelines recommend ongoing serum creatinine and urine protein monitoring, with dysfunction episodes evaluated by allograft ultrasound (25). Allograft biopsy is indicated when diagnosis is uncertain, for evaluating proteinuria, or where histologic findings will affect treatment.
Drug-Level Monitoring
Although drug monitoring for tacrolimus, cyclosporin, sirolimus, and everolimus is routine, the utility of mycophenolic acid levels is not established (26). Trough calcineurin inhibitor levels correlate fairly well with total drug exposure. Out of range levels may signify nonadherence; underdosing; formulation change; or unrecognized drug-drug, food-drug, or gut-drug interactions, providing opportunity for patient counseling or regimen adjustment before rejection or toxicity ensues.
Tacrolimus-level variability over time predicts underdosing/nonadherence and rejection (27–30). High intrapatient tacrolimus-level variability is associated with interstitial fibrosis/tubular atrophy (IFTA) (27), rejection, and allograft failure (28). In one study, recipients with de novo DSA had higher proportions of tacrolimus levels <5 ng/ml; moreover, levels were significantly lower in the 6 months preceding de novo DSA detection than at earlier time points (29).
Time in therapeutic range, typically used in anticoagulation management, is a newer concept in transplantation. Time in therapeutic range—a calculation of the percentage of time a level is within the predefined target range in individual patients—has been applied to tacrolimus therapy (30). On the basis of target levels 5–10 ng/ml within the first post-transplant year, time in therapeutic range <60% associates with de novo DSA and rejection risk by 12 months and allograft loss by 5 years post-transplant. Analysis incorporating both time in therapeutic range and coefficient of variation suggests the immunologic risk associated with high intrapatient tacrolimus-level variation is due to low time in therapeutic range rather than variability in and of itself (31). Use of these simple tracking tools is an actionable strategy to monitor medication nonadherence, optimize dosing, and improve outcomes.
Donor-Specific Antibody
De novo DSA develops in 15%–20% of patients within the first few post-transplant years (7,13). One study monitored de novo DSA over a mean of 6.2 years in 315 nonsensitized patients, demonstrating that its presence associated with HLA-DR mismatch and patient nonadherence and adversely affected 10-year allograft survival (7). Other DSA risk factors include immunosuppression minimization, DQ mismatching, and early T cell–mediated rejection (32), with mostly class 2 DSA identified in this latter setting (33,34).
In patients with antibody-mediated rejection, de novo DSA appears to have greater negative effect than preexisting DSA. In a 205-patient antibody-mediated rejection cohort evenly divided between recipients with pretransplant DSA and de novo DSA, antibody-mediated rejection occurred earlier in the latter group (median 85 versus 1437 days). Patients with de novo DSA demonstrated more proteinuria, greater class 2 antibody, higher antibody titers, more frequent transplant glomerulopathy, and worse allograft survival at 8 years (35).
Transplantation Society guidelines recommend DSA monitoring in the setting of recipients with pretransplant DSA (36), immunosuppression reduction, patient nonadherence, or a rejection episode occurrence, with close allograft function surveillance when detected (37). Transplant biopsy, similarly recommended upon DSA detection, notably has no evidence grade. In this setting, we believe biopsy may have diagnostic utility, although acknowledge absence of data demonstrating the procedure results in improved outcomes.
Viral Screening
Through the interplay between antiviral and alloimmune responses, viruses result from overimmunosuppression but may also trigger rejection. Virus-specific T cells crossreactive to alloantigen have been demonstrated in the circulation of Epstein–Barr virus (EBV)– and/or cytomegalovirus-infected patients (38). BK virus (BKV) impairs allograft function through cytopathic injury yet is associated with de novo DSA and rejection (39,40). Mechanisms linking BKV to alloimmunity are unclear but include immunosuppression reduction or heterologous immunity. Supporting the latter, BKV nephropathy allograft biopsies have been shown to simultaneously contain both BKV-reactive and alloreactive T cell clones (41).
These viral infections typically occur within 6 months post-transplant. Except for pre-emptive treatment strategies or guiding therapy in infected individuals, no cytomegalovirus screening recommendations exist (23). KDIGO guidelines suggest EBV nucleic acid testing in EBV-seronegative recipients of EBV-seropositive kidneys once in week 1, monthly for 3–6 months, then quarterly until 12 months post-transplant, and when treating rejection (23). Although American Society of Transplantation (AST) Infectious Diseases Community of Practice (IDCOP) guidelines suggest more frequent testing (42), both guidelines advocate immunosuppression reduction for worsening EBV viremia.
BK viremia and BKV nephropathy prevalence rates are 10%–30% and 2%, respectively (43). Routine screening enables BKV detection before nephropathy affects kidney function (44). KDIGO suggests monthly monitoring through 6 months and then quarterly for 6 months. Because BK viral loads >10,000 copies per milliliter predict BKV nephropathy (45), KDIGO recommends this level as a threshold above which immunosuppression should be reduced. Recent AST IDCOP guidelines recommend monthly testing until 9 months and then quarterly until 24 months because 30% of BKV infections occur beyond 6 months post-transplant (46). Because intragraft BKV replication may be focal, up to one third of BKV-infected biopsy samples may test negative; this false-negative rate declines as viral loads exceed six log10 copies per milliliter (46). We recommend allograft biopsy in recipients with both BK viremia and either new allograft dysfunction or another abnormal diagnostic biomarker.
Torque Tino Virus—An Emerging Immunostat?
Torque teno virus (TTV) is an apathogenic virus with no known therapies (47). In the transplant context, a recent study reported that around 43 days prebiopsy, patients with rejection had lower blood TTV levels than nonrejecting patients (48). A subsequent prospective, observational study incorporated TTV viral load measurements weekly initially and then quarterly until 12 months post-transplant (49). Torque Teno viral load peaked 3 months post-transplant; thereafter, each TTV viral load log increase associated with a 22% lower rejection odds and an 11% greater odds for another infection. Viral loads between 1 × 106–108 copies per milliliter were identified as the “sweet spot” for optimally minimized risk for rejection and infection. Although further validation is required, TTV represents another potential immune status monitoring tool.
Surveillance Biopsies
Historically performed in “high-risk” patients, the rationale for surveillance kidney transplant biopsy is determination of “subclinical rejection” described in cyclosporin-treated patients, where 30% of recipients biopsied by protocol early post-transplant displayed histologic tubulitis despite stable laboratory values (50,51). A similar study subsequently conducted in tacrolimus-MMF-prednisone–treated patients observed subclinical rejection rates <5%, with no differences in patient/allograft survival or IFTA at 2 years post-transplant (52,53). The investigators concluded there was no benefit to surveillance biopsies in patients receiving this immunosuppression regimen.
Recent surveillance biopsy studies have focused on subclinical borderline T cell–mediated rejection, detected in approximately 25%–40% of participants (54–56). Follow-up biopsies have demonstrated histologic progression despite treatment (54,55). Although these data may support surveillance biopsy (and potentially, rebiopsy when subclinical injury is detected), it should be borne in mind that consensus around the histologic definition of borderline T cell rejection (57) and effectiveness of treatment in this setting is not established.
Surveillance biopsies may have value in patients undergoing major immunosuppression modification. Heilman et al. (58) correlated 1- or 4-month post-transplant biopsy findings with a 12-month biopsy in 256 recipients who underwent rapid corticosteroid withdrawal. Although 6% developed overt rejection by 12 months, early surveillance biopsy revealed subclinical rejection or inflammation in 27%. Both subclinical rejection and inflammation predicted greater IFTA at 12 months. Another study randomized low–immunologic risk recipients to continued tacrolimus versus conversion to sirolimus at 3 months post-transplant (59). Despite similar kidney function, 24-month surveillance biopsies showed more subclinical inflammation and IFTA in the sirolimus arm.
A recent US transplant center survey found that surveillance biopsies were performed by 38 of 83 (46%) responding centers; 20 centers biopsied all patients, whereas 18 were more selective (60). This increased biopsy rate compared with a prior survey (61) may reflect currently perceived need for histology in lieu of reliable immune monitoring tools. An analysis that examined complications after 2514 kidney allograft biopsies observed fewer major complications in surveillance than for-cause procedures (0.3% versus 3%), with surgical intervention undertaken in 0.6% and no attributable graft losses or death (62). These findings support that surveillance kidney transplant biopsies are safe.
Limitations of Conventional Monitoring and Biopsy
Serum creatinine, widely available, inexpensive, and with rapid turnaround, is neither specific nor sensitive, and often, it is a late injury indicator. Kidney biopsy is costly, sampling error prone, limited by subjective interpretation, and inconvenient, and it carries some risk. Surveillance biopsies are lower yield than indication biopsies because many patients with unremarkable histology will be biopsied. Moreover, optimal timing and surveillance biopsy frequency are unknown. Finally, although identifying morphologic changes that predict outcomes, surveillance biopsies have yet to result in interventions established to improve outcomes.
These collective limitations, coupled with technological advancement, have spawned interest in finding more specific, noninvasive tissue, urine, and blood biomarkers. An important premise for seeking status quo alternatives is that allograft injury and damage are driven by subclinical, often repetitive events (63), underscoring the need for scalable monitoring strategies.
Novel Tissue Diagnostics
Halloran et al. (64) analyzed molecular allograft rejection phenotypes by measuring mRNA transcripts in biopsy tissue. Comparing T cell–mediated and antibody-mediated rejection biopsies with normal histology revealed that prominent transcripts for both rejection types were induced by IFN-γ. Effector T cell and myeloid cell expressions were T cell–mediated rejection specific, whereas transcripts for natural killer cell localization and endothelial injury were unique to antibody-mediated rejection. There was some transcript overlap between rejecting and nonrejecting allografts (65), but generally strong associations of transcripts with rejection and rejection subtypes were preserved in validation testing.
Application of this “microarray-based molecular diagnostic system” (MMDX) was demonstrated in a multicenter study (66). MMDX defined antibody-mediated rejection in 41% of biopsies where it was not reported originally and revealed antibody-mediated rejection transcript signaling in C4d-positive and -negative biopsies. The MMDX system has been used to analyze biopsies with inflammation in areas of IFTA, revealing predominant antibody-mediated rather than T cell–mediated rejection (67). Recently, the Banff Working Group described a 770 biopsy tissue–derived gene panel using a Nano-String platform. Genes were categorized by host organ transplant responses, including rejection, tolerance, drug toxicity, and viral infection, with plans for future multicenter validation using a commercially available assay (68).
The GoCAR study utilized mRNA microarray analysis to predict fibrosis progression at 1 year from biopsy tissue collected at 3 months post-transplant (69). Using the Chronic Allograft Damage Index score, 12-month biopsies with scores of > 2 were analyzed, identifying a gene set that correlated with fibrosis. After application to 3-month biopsies in cases where fibrosis worsened by month 12, a 13–gene set panel was derived that predicted subsequent fibrosis, independent of clinicopathologic variables.
Novel Noninvasive Biomarkers
Ideal diagnostic biomarkers should identify patients with high disease probability (high positive predictive value). Prognostic and predictive biomarkers can then be applied to determine both patients at high risk for a bad prognosis and predicted treatment benefit. Assays should provide greater lead time for detecting and monitoring allograft injury, be reproducible, have a low coefficient of variation, have rapid turnaround, and permit cost-effective surveillance. Moreover, they could provide dynamic analyses of allograft and immune status, enabling precise risk stratification, with potential to guide need for biopsy or immunosuppression modification.
Urinary Biomarkers
Chemokines CXCL9 and CXCL10 recruit effector T cells in response to IFN-γ and have been identified as urinary markers of acute rejection in separate clinical trials. In CTOT-04, Suthanthiran et al. (70) analyzed urinary mRNA collected serially from 485 kidney recipients within the first post-transplant year. A three-gene signature was derived using CD3ε, CXCL10, and 18S ribosomal RNA that discriminated T cell–mediated rejection from no rejection. Analysis of urine samples from rejecters showed increased chemokine gene expression as early as 120 days before biopsy.
In CTOT-01, Hricik et al. (71) analyzed protocol and for-cause biopsies in 255 first kidney recipients. Rejection was present in 33% of indication biopsies, and urinary CXCL9 protein could detect rejection. CXCL9 also correlated with inflammation and was elevated up to 30 days before rejection. For the entire cohort, CXCL9 at 6 months correlated with rejection and functional deterioration by 24 months post-transplant.
In CTOT-09, tacrolimus withdrawal resulted in urinary CXCL9 elevation in six of 14 patients; four had rejection, five had de novo DSA, and two had BKV (25). Urinary CXCL9 alone could not differentiate BKV from rejection. Prospective trials investigating urinary chemokine monitoring and outcomes are planned (72).
Blood Biomarkers
To date, three blood biomarker assays have been evaluated in the post-transplant setting: kidney Solid Organ Response Test (kSORT), whole-genome peripheral blood gene expression profiling, and donor-derived cellfree DNA (cfDNA). One caveat with these assays in their quest to supplant allograft biopsy is that histology continues to serve as their rejection “gold standard.”
Kidney Solid Organ Response Test.
This whole blood–derived molecular assay uses quantitative PCR to detect a 17-gene panel. The Assessment of Acute Rejection Trial study, involving 436 patients, collected aggregated blood samples in a cross-sectional manner and matched them with contemporaneous kidney allograft biopsy. The kSORT analysis suite, an algorithm that used varying numbers of smaller subsets of the gene panel, differentiated rejection from no rejection on the basis of patterns consistent with increased inflammation or immune quiescence (73). Study limitations included lack of serial blood samples, cohort heterogeneity, and inability to distinguish T cell–mediated from antibody-mediated rejection. A real-world, retrospective, multicenter study comprising 1763 samples from 1134 patients could not validate kSORT for detecting rejection in the first post-transplant year (74). Observational and interventional studies are ongoing.
Peripheral Blood Gene Expression Profiling.
Most established is a DNA microarray-based gene expression test that identifies 57 classifier genes that distinguish subclinical acute rejection (with stable allograft function) from histologic quiescence. In the pivotal study, where subclinical acute rejection was the primary end point, patients were followed for 24 months with gene profiling paired with surveillance biopsies between 2 and 6 months, at 12 and 24 months, and with for-cause biopsies (54). The subclinical acute rejection incidence was 42%, although was mostly “borderline” T cell rejection. At the optimal diagnostic threshold, the sensitivity, specificity, positive predictive value, and negative predictive value of the biomarker for subclinical acute rejection were 64%, 87%, 61%, and 88%, respectively. Secondarily, investigators observed that the biomarker profile that correlated with subclinical acute rejection also associated with worse transplant outcomes after 24 months. Real-world experience in centers not using surveillance biopsies reflects similar performance characteristics in confirming immune quiescence (75).
A peripheral blood 17-gene signature using targeted RNA expression has been reported from GoCAR that associates with rejection on 3-month surveillance biopsy, although it requires further validation (76).
Donor-Derived Cellfree DNA.
Cellfree DNA (cfDNA) is nonencapsulated circulating DNA. Degrading into approximately 166-base nucleosomal units, cfDNA has a half-life around 30 minutes. Donor-derived cfDNA are DNA fragments in recipient circulation originating from donor tissue injury. The most common detection approach is on the basis of detecting differences in highly homozygous single-nucleotide polymorphisms with high allelic frequency between donor and recipient. Normally a miniscule fraction of total cfDNA, donor-derived cfDNA increases with increasing allograft injury. Levels are elevated very early post-transplant after ischemia-reperfusion injury, declining to baseline steady state more rapidly in living than deceased donor recipients (77).
The prospective, observational Diagnosing Active Rejection in Kidney Transplant Recipients (DART) trial investigated donor-derived cfDNA as a rejection marker (78). At a 1% diagnostic cutoff, donor-derived cfDNA levels measured concomitantly with for-cause biopsy differentiated active rejection from no rejection, outperforming serum creatinine. Donor-derived cfDNA performed most robustly for discriminating antibody-mediated rejection from no antibody-mediated rejection, with sensitivity, specificity, positive predictive, and negative predictive values of 81%, 83%, 44%, and 96%, respectively. Data, frequently single center, with different donor-derived cfDNA assays from the United States, Europe, and Australia have since been reported. Most (77,79–81), although not all (82), studies have confirmed DART’s observation. Some assays have additionally shown promise for T cell–mediated rejection (79,83) and ATN (77). Reported differences in performance characteristic between assays, and between studies with the same assay, are likely related to differences in diagnostic thresholds used or different histologic classifications (Banff 2013 versus 2017). Although attention has focused on donor-derived cfDNA’s ability to discriminate rejection from nonrejection, it most plausibly reflects overall injury burden regardless of triggering cause. Better-quality data are required to evaluate donor-derived cfDNA’s potential for diagnosing and predicting rejection and other allograft injury, as well as postintervention surveillance.
Where to from Here?
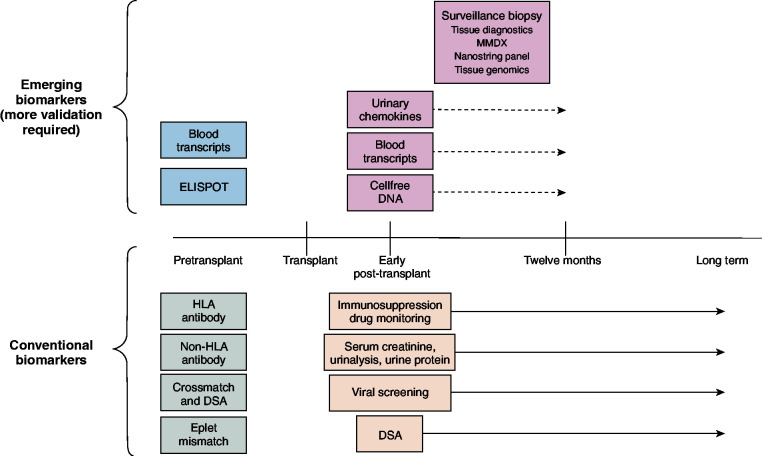
At present, the published quality of evidence regarding emerging biomarkers is variable, the clinical framework for their use is not clearly established (Table 2), and they have not been independently validated. Despite these limitations, costly commercially available biomarkers are now widely used in various unproven contexts. All demonstrate a strong negative predictive value, indicating potential to avoid unnecessary biopsies; however, they perform less well in identifying patients at risk for a poor outcome (lower positive predictive value). Furthermore, studies have not compared these biomarkers against most conventional approaches or against one another. Table 3 highlights knowledge gaps related to optimal use of novel biomarkers, including testing frequency and timely resulting as well as other logistic and technical considerations. Finally, it is necessary to determine whether they are cost effective in low– and high–immunologic risk recipients alike. Figure 1 depicts hypothetical time frames for using these biomarkers in practice assuming their clinical utility was established.
Table 2.
Conventional and emerging biomarkers used in kidney transplant
| Test | Suggested Testing Frequency | Potential Benefit | Limitation(s) | Comment |
| Conventional | ||||
| UP/Cr | 1, 3 mo, then quarterly | Low cost, actionable surveillance test |
|
Opportunity for antiproteinuric therapy/targeted therapy |
| Tacrolimus TTR/CV | Ongoing | Actionable, low cost, monitor nonadherence |
|
Can incorporate into electronic medical record |
| EBV NAT | Monthly for 6–12 mo | IS management guide in EBV+/− recipients |
|
Reduce IS for rising viral load |
| BKV NAT | 1, 3, 6, 12 mo | Guide IS management |
|
Reduce IS for rising viral load |
| DSA | 1, 3, 6, 12 mo | Risk stratification/guide IS management |
|
Opportunity to enroll in contemporary clinical trials |
| Emerging | ||||
| Surveillance biopsy | Once in first 3 mo? | Risk stratification, relatively safe |
|
Possible use in conversion/minimization regimens, patients with DSA, clinical trials |
| MMDX | TBD | Discriminates rejection types, with improved consistency |
|
Consider for patients being biopsied where histology findings are unclear to guide therapy |
| Urine chemokines | TBD | Predicts rejection, reproducible |
|
|
| ELISPOT | Prior to transplant | Risk stratification |
|
May be more practical in live donor recipients |
| Blood gene profiling | TBD | High NPV for subclinical rejection |
|
Consider where 2- to 3-d result delay will not affect management plan |
| kSORT | TBD | May risk stratify rejection |
|
Lack of benefit in recent large, multicenter, retrospective study |
| dd-cfDNA | TBD | High NPV for rejection |
|
Consider for ruling out AMR, where 2- to 3-d result delay will not affect management plan |
UP/Cr, urine protein-urine creatinine ratio; TTR/CV, time in therapeutic ratio/coefficient of variation; EBV, Epstein–Barr virus; NAT, nucleic acid testing; IS, immunosuppression; BKV, BK virus; DSA, donor-specific antibody; MMDX, microarray-based molecular diagnostic system; TBD, to be determined; ELISPOT, enzyme-linked immune absorbent spot; NPV, negative predictive value; kSORT, kidney Solid Organ Response Test; dd-cfDNA, donor-derived cellfree DNA; AMR, antibody-mediated rejection.
Table 3.
Information gaps with novel tests to monitor kidney transplant health
| Current Knowledge Gaps |
| Utility for diagnosis versus surveillance |
| Optimal use of each test |
| Timing and frequency of testing |
| Diagnostic thresholds |
| Measure of immunosuppression |
| Rejection subtype/severity |
| Discriminating allograft injury not due to rejection |
| Recurrent disease |
| BK nephropathy |
| Other infection (CMV, adenovirus, bacterial, etc.) |
| Drug toxicity |
| De novo disease |
| Donor quality parameters |
| Comparative performance of each test |
| Combined use of tests |
| Which |
| When |
| How |
| Logistic and technologic considerations |
| Timely turnaround |
| Scalability |
| Access to all patients |
| Changes in the Banff classification |
| Cost-benefit versus conventional testing |
CMV, cytomegalovirus.
Figure 1.
Schematic depicting potential time frames for using conventional and emerging transplant biomarkers. Conventional testing is shown below the timeline with pretransplant (grey) and post-transplant (tan) testing. Novel and experimental tests are shown above the timeline with pretransplant (blue) and post-transplant (light red) testing. DSA, donor-specific antibody; ELISPOT, enzyme-linked immune absorbent spot; MMDX, microarray-based molecular diagnostic system.
Kidney transplantation’s successful evolution has created a high bar for biomarkers to overcome. Outcomes have improved with conventional monitoring, and death accounts for most allograft loss. Moreover, acute rejection rates are <10% (2), while subclinical acute rejection is relatively infrequent and its treatment benefit unproven (43,44). Moving beyond measuring serum creatinine, immunosuppression drug levels and indication biopsy will require large, prospective, observational and interventional randomized controlled studies to demonstrate that these promising biomarkers result in improved outcomes in order to justify their incorporation into routine post-transplant care.
Disclosures
J.J. Augustine reports employment with the Cleveland Clinic Foundation and Cleveland Veterans Affairs Medical Center. R.D. Bloom reports employment with the University of Pennsylvania; consultancy agreements with Veloxis Pharmaceuticals; receiving research funding from CareDx, Veloxis, and Vitaeris; receiving honoraria from Veloxis; receiving royalties from UpToDate; serving as a scientific advisor or member of Allovir, CareDx, Natera, Paladin Labs, and Veloxis; and serving on the Editorial Board of American Journal of Kidney Diseases.
Funding
None.
Footnotes
Published online ahead of print. Publication date available at www.cjasn.org.
References
- 1.Lamb KE, Lodhi S, Meier-Kriesche HU: Long-term renal allograft survival in the United States: A critical reappraisal. Am J Transplant 11: 450–462, 2011 [DOI] [PubMed] [Google Scholar]
- 2.Hart A, Smith JM, Skeans MA, Gustafson SK, Wilk AR, Castro S, Foutz J, Wainright JL, Snyder JJ, Kasiske BL, Israni AK: OPTN/SRTR 2018 annual data report: Kidney. Am J Transplant 20[Suppl s1]: 20–130, 2020 [DOI] [PubMed] [Google Scholar]
- 3.Poggio ED, Augustine JJ, Arrigain S, Brennan DC, Schold JD: Long-term kidney transplant graft survival-making progress when most needed [published online ahead of print December 21, 2020]. Am J Transplant 10.1111/ajt.16463 [DOI] [PubMed] [Google Scholar]
- 4.Terasaki PI, Ozawa M: Predicting kidney graft failure by HLA antibodies: A prospective trial. Am J Transplant 4: 438–443, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Sharif A, Kraus ES, Zachary AA, Lonze BE, Nazarian SM, Segev DL, Alachkar N, Arend LJ, Bagnasco SM, Racusen LC, Montgomery RA: Histologic phenotype on 1-year posttransplantation biopsy and allograft survival in HLA-incompatible kidney transplants. Transplantation 97: 541–547, 2014 [DOI] [PubMed] [Google Scholar]
- 6.Vo AA, Aubert O, Haas M, Huang E, Zhang X, Choi J, Peng A, Najjar R, Sethi S, Ammerman N, Lim K, Jordan SC: Clinical relevance of posttransplant DSAs in patients receiving desensitization for HLA-incompatible kidney transplantation. Transplantation 103: 2666–2674, 2019 [DOI] [PubMed] [Google Scholar]
- 7.Wiebe C, Gibson IW, Blydt-Hansen TD, Karpinski M, Ho J, Storsley LJ, Goldberg A, Birk PE, Rush DN, Nickerson PW: Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant. Am J Transplant 12: 1157–1167, 2012 [DOI] [PubMed] [Google Scholar]
- 8.Wiebe C, Pochinco D, Blydt-Hansen TD, Ho J, Birk PE, Karpinski M, Goldberg A, Storsley LJ, Gibson IW, Rush DN, Nickerson PW: Class II HLA epitope matching-A strategy to minimize de novo donor-specific antibody development and improve outcomes. Am J Transplant 13: 3114–3122, 2013 [DOI] [PubMed] [Google Scholar]
- 9.Wiebe C, Nevins TE, Robiner WN, Thomas W, Matas AJ, Nickerson PW: The synergistic effect of class II HLA epitope-mismatch and nonadherence on acute rejection and graft survival. Am J Transplant 15: 2197–2202, 2015 [DOI] [PubMed] [Google Scholar]
- 10.Hricik DE, Formica RN, Nickerson P, Rush D, Fairchild RL, Poggio ED, Gibson IW, Wiebe C, Tinckam K, Bunnapradist S, Samaniego-Picota M, Brennan DC, Schröppel B, Gaber O, Armstrong B, Ikle D, Diop H, Bridges ND, Heeger PS; Clinical Trials in Organ Transplantation-09 Consortium: Adverse outcomes of tacrolimus withdrawal in immune-quiescent kidney transplant recipients. J Am Soc Nephrol 26: 3114–3122, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiebe C, Kosmoliaptsis V, Pochinco D, Gibson IW, Ho J, Birk PE, Goldberg A, Karpinski M, Shaw J, Rush DN, Nickerson PW: HLA-DR/DQ molecular mismatch: A prognostic biomarker for primary alloimmunity. Am J Transplant 19: 1708–1719, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senev A, Coemans M, Lerut E, Van Sandt V, Kerkhofs J, Daniëls L, Driessche MV, Compernolle V, Sprangers B, Van Loon E, Callemeyn J, Claas F, Tambur AR, Verbeke G, Kuypers D, Emonds MP, Naesens M: Eplet mismatch load and de novo occurrence of donor-specific anti-HLA antibodies, rejection, and graft failure after kidney transplantation: An observational cohort study. J Am Soc Nephrol 31: 2193–2204, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis S, Wiebe C, Campbell K, Anobile C, Aubrey M, Stites E, Grafals M, Pomfret E, Nickerson P, Cooper JE: Adequate tacrolimus exposure modulates the impact of HLA class II molecular mismatch: A validation study in an American cohort. Am J Transplant 21: 322–328, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pineda S, Sigdel TK, Chen J, Jackson AM, Sirota M, Sarwal MM: Novel non-histocompatibility antigen mismatched variants improve the ability to predict antibody-mediated rejection risk in kidney transplant. Front Immunol 8: 1687, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delville M, Lamarthée B, Pagie S, See SB, Rabant M, Burger C, Gatault P, Giral M, Thaunat O, Arzouk N, Hertig A, Hazzan M, Matignon M, Mariat C, Caillard S, Kamar N, Sayegh J, Westeel PF, Garrouste C, Ladrière M, Vuiblet V, Rivalan J, Merville P, Bertrand D, Le Moine A, Duong Van Huyen JP, Cesbron A, Cagnard N, Alibeu O, Satchell SC, Legendre C, Zorn E, Taupin JL, Charreau B, Anglicheau D: Early acute microvascular kidney transplant rejection in the absence of anti-HLA antibodies is associated with preformed IgG antibodies against diverse glomerular endothelial cell antigens. J Am Soc Nephrol 30: 692–709, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Augustine JJ, Siu DS, Clemente MJ, Schulak JA, Heeger PS, Hricik DE: Pre-transplant IFN-gamma ELISPOTs are associated with post-transplant renal function in African American renal transplant recipients. Am J Transplant 5: 1971–1975, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Hricik DE, Augustine J, Nickerson P, Formica RN, Poggio ED, Rush D, Newell KA, Goebel J, Gibson IW, Fairchild RL, Spain K, Iklé D, Bridges ND, Heeger PS; CTOT-01 Consortium: Interferon gamma ELISPOT testing as a risk-stratifying biomarker for kidney transplant injury: Results from the CTOT-01 multicenter study. Am J Transplant 15: 3166–3173, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gandolfini I, Crespo E, Baweja M, Jarque M, Donadei C, Luque S, Montero N, Allesina A, Perin L, Maggiore U, Cravedi P, Bestard O: Impact of preformed T-cell alloreactivity by means of donor-specific and panel of reactive T cells (PRT) ELISPOT in kidney transplantation. PLoS One 13: e0200696, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W, Yi Z, Wei C, Keung KL, Sun Z, Xi C, Woytovich C, Farouk S, Gallon L, Menon MC, Magee C, Najafian N, Samaniego MD, Djamali A, Alexander SI, Rosales IA, Smith RN, O’Connell PJ, Colvin R, Cravedi P, Murphy B: Pretransplant transcriptomic signature in peripheral blood predicts early acute rejection. JCI Insight 4: e127543, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kasiske BL, Israni AK, Snyder JJ, Skeans MA; Patient Outcomes in Renal Transplantation (PORT) Investigators: The relationship between kidney function and long-term graft survival after kidney transplant. Am J Kidney Dis 57: 466–475, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Schnitzler MA, Johnston K, Axelrod D, Gheorghian A, Lentine KL: Associations of renal function at 1-year after kidney transplantation with subsequent return to dialysis, mortality, and healthcare costs. Transplantation 91: 1347–1356, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Naesens M, Lerut E, Emonds MP, Herelixka A, Evenepoel P, Claes K, Bammens B, Sprangers B, Meijers B, Jochmans I, Monbaliu D, Pirenne J, Kuypers DR: Proteinuria as a noninvasive marker for renal allograft histology and failure: An observational cohort study. J Am Soc Nephrol 27: 281–292, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam NN, Tonelli M, Lentine KL, Hemmelgarn B, Ye F, Wen K, Klarenbach S: Albuminuria and posttransplant chronic kidney disease stage predict transplant outcomes. Kidney Int 92: 470–478, 2017 [DOI] [PubMed] [Google Scholar]
- 24.Weiner DE, Park M, Tighiouart H, Joseph AA, Carpenter MA, Goyal N, House AA, Hsu CY, Ix JH, Jacques PF, Kew CE, Kim SJ, Kusek JW, Pesavento TE, Pfeffer MA, Smith SR, Weir MR, Levey AS, Bostom AG: Albuminuria and allograft failure, cardiovascular disease events, and all-cause death in stable kidney transplant recipients: A cohort analysis of the FAVORIT trial. Am J Kidney Dis 73: 51–61, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kidney Disease: Improving Global Outcomes (KDIGO) Transplant Work Group: KDIGO clinical practice guideline for the care of kidney transplant recipients, 2009. Available at: https://kdigo.org/guidelines/transplant-recipient. Accessed February 23, 2021 [Google Scholar]
- 26.Gaston RS, Kaplan B, Shah T, Cibrik D, Shaw LM, Angelis M, Mulgaonkar S, Meier-Kriesche HU, Patel D, Bloom RD: Fixed- or controlled-dose mycophenolate mofetil with standard- or reduced-dose calcineurin inhibitors: The Opticept trial. Am J Transplant 9: 1607–1619, 2009 [DOI] [PubMed] [Google Scholar]
- 27.Vanhove T, Vermeulen T, Annaert P, Lerut E, Kuypers DRJ: High intrapatient variability of tacrolimus concentrations predicts accelerated progression of chronic histologic lesions in renal recipients. Am J Transplant 16: 2954–2963, 2016 [DOI] [PubMed] [Google Scholar]
- 28.Taber DJ, Su Z, Fleming JN, McGillicuddy JW, Posadas-Salas MA, Treiber FA, Dubay D, Srinivas TR, Mauldin PD, Moran WP, Baliga PK: Tacrolimus trough concentration variability and disparities in African American kidney transplantation. Transplantation 101: 2931–2938, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wiebe C, Rush DN, Nevins TE, Birk PE, Blydt-Hansen T, Gibson IW, Goldberg A, Ho J, Karpinski M, Pochinco D, Sharma A, Storsley L, Matas AJ, Nickerson PW: Class II eplet mismatch modulates tacrolimus trough levels required to prevent donor-specific antibody development. J Am Soc Nephrol 28: 3353–3362, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davis S, Gralla J, Klem P, Tong S, Wedermyer G, Freed B, Wiseman A, Cooper JE: Lower tacrolimus exposure and time in therapeutic range increase the risk of de novo donor-specific antibodies in the first year of kidney transplantation. Am J Transplant 18: 907–915, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis S, Gralla J, Klem P, Stites E, Wiseman A, Cooper JE: Tacrolimus intrapatient variability, time in therapeutic range, and risk of de novo donor-specific antibodies. Transplantation 104: 881–887, 2020 [DOI] [PubMed] [Google Scholar]
- 32.Zhang R: Donor-specific antibodies in kidney transplant recipients. Clin J Am Soc Nephrol 13: 182–192, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.El Ters M, Grande JP, Keddis MT, Rodrigo E, Chopra B, Dean PG, Stegall MD, Cosio FG: Kidney allograft survival after acute rejection, the value of follow-up biopsies. Am J Transplant 13: 2334–2341, 2013 [DOI] [PubMed] [Google Scholar]
- 34.Chemouny JM, Suberbielle C, Rabant M, Zuber J, Alyanakian MA, Lebreton X, Carmagnat M, Pinheiro N, Loupy A, Van Huyen JP, Timsit MO, Charron D, Legendre C, Anglicheau D: De novo donor-specific human leukocyte antigen antibodies in nonsensitized kidney transplant recipients after T cell-mediated rejection. Transplantation 99: 965–972, 2015 [DOI] [PubMed] [Google Scholar]
- 35.Aubert O, Loupy A, Hidalgo L, Duong van Huyen JP, Higgins S, Viglietti D, Jouven X, Glotz D, Legendre C, Lefaucheur C, Halloran PF: Antibody-mediated rejection due to preexisting versus de novo donor-specific antibodies in kidney allograft recipients. J Am Soc Nephrol 28: 1912–1923, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tait BD, Süsal C, Gebel HM, Nickerson PW, Zachary AA, Claas FH, Reed EF, Bray RA, Campbell P, Chapman JR, Coates PT, Colvin RB, Cozzi E, Doxiadis II, Fuggle SV, Gill J, Glotz D, Lachmann N, Mohanakumar T, Suciu-Foca N, Sumitran-Holgersson S, Tanabe K, Taylor CJ, Tyan DB, Webster A, Zeevi A, Opelz G: Consensus guidelines on the testing and clinical management issues associated with HLA and non-HLA antibodies in transplantation. Transplantation 95: 19–47, 2013 [DOI] [PubMed] [Google Scholar]
- 37.Schinstock CA, Mannon RB, Budde K, Chong AS, Haas M, Knechtle S, Lefaucheur C, Montgomery RA, Nickerson P, Tullius SG, Ahn C, Askar M, Crespo M, Chadban SJ, Feng S, Jordan SC, Man K, Mengel M, Morris RE, O’Doherty I, Ozdemir BH, Seron D, Tambur AR, Tanabe K, Taupin J-L, O’Connell PJ: Recommended treatment for antibody-mediated rejection after kidney transplantation: The 2019 expert consensus from the Transplantation Society Working Group. Transplantation 104: 911–922, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heutinck KM, Yong SL, Tonneijck L, van den Heuvel H, van der Weerd NC, van der Pant KA, Bemelman FJ, Claas FH, Ten Berge IJ: Virus-specific CD8(+) T cells cross-reactive to donor-alloantigen are transiently present in the circulation of kidney transplant recipients infected with CMV and/or EBV. Am J Transplant 16: 1480–1491, 2016 [DOI] [PubMed] [Google Scholar]
- 39.Elfadawy N, Flechner SM, Schold JD, Srinivas TR, Poggio E, Fatica R, Avery R, Mossad SB: Transient versus persistent BK viremia and long-term outcomes after kidney and kidney-pancreas transplantation. Clin J Am Soc Nephrol 9: 553–561, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawinski D, Forde KA, Trofe-Clark J, Patel P, Olivera B, Goral S, Bloom RD: Persistent BK viremia does not increase intermediate-term graft loss but is associated with de novo donor-specific antibodies. J Am Soc Nephrol 26: 966–975, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zeng G, Huang Y, Huang Y, Lyu Z, Lesniak D, Randhawa P: Antigen-specificity of T cell infiltrates in biopsies with T cell-mediated rejection and BK polyomavirus viremia: Analysis by next generation sequencing. Am J Transplant 16: 3131–3138, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Allen UD, Preiksaitis JK; AST Infectious Diseases Community of Practice: Post-transplant lymphoproliferative disorders, Epstein-Barr virus infection, and disease in solid organ transplantation: Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 33: e13652, 2019 [DOI] [PubMed] [Google Scholar]
- 43.Hirsch HH, Randhawa P; AST Infectious Diseases Community of Practice: BK polyomavirus in solid organ transplantation. Am J Transplant 13[Suppl 4]: 179–188, 2013 [DOI] [PubMed] [Google Scholar]
- 44.Hirsch HH, Vincenti F, Friman S, Tuncer M, Citterio F, Wiecek A, Scheuermann EH, Klinger M, Russ G, Pescovitz MD, Prestele H: Polyomavirus BK replication in de novo kidney transplant patients receiving tacrolimus or cyclosporine: A prospective, randomized, multicenter study. Am J Transplant 13: 136–145, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Randhawa P, Brennan DC: BK virus infection in transplant recipients: An overview and update. Am J Transplant 6: 2000–2005, 2006 [DOI] [PubMed] [Google Scholar]
- 46.Hirsch HH, Randhawa PS; AST Infectious Diseases Community of Practice: BK polyomavirus in solid organ transplantation: Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin Transplant 33: e13528, 2019 [DOI] [PubMed] [Google Scholar]
- 47.Tyschik EA, Rasskazova AS, Degtyareva AV, Rebrikov DV, Sukhikh GT: Torque teno virus dynamics during the first year of life. Virol J 15: 96, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strassl R, Schiemann M, Doberer K, Görzer I, Puchhammer-Stöckl E, Eskandary F, Kikic Ž, Gualdoni GA, Vossen MG, Rasoul-Rockenschaub S, Herkner H, Böhmig GA, Bond G: Quantification of Torque Teno virus viremia as a prospective biomarker for infectious disease in kidney allograft recipients. J Infect Dis 218: 1191–1199, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Doberer K, Schiemann M, Strassl R, Haupenthal F, Dermuth F, Görzer I, Eskandary F, Reindl-Schwaighofer R, Kikić Ž, Puchhammer-Stöckl E, Böhmig GA, Bond G: Torque teno virus for risk stratification of graft rejection and infection in kidney transplant recipients: A prospective observational trial. Am J Transplant 20: 2081–2090, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rush DN, Henry SF, Jeffery JR, Schroeder TJ, Gough J: Histological findings in early routine biopsies of stable renal allograft recipients. Transplantation 57: 208–211, 1994 [DOI] [PubMed] [Google Scholar]
- 51.Rush D, Nickerson P, Gough J, McKenna R, Grimm P, Cheang M, Trpkov K, Solez K, Jeffery J: Beneficial effects of treatment of early subclinical rejection: A randomized study. J Am Soc Nephrol 9: 2129–2134, 1998 [DOI] [PubMed] [Google Scholar]
- 52.Rush DN, Cockfield SM, Nickerson PW, Arlen DJ, Boucher A, Busque S, Girardin CE, Knoll GA, Lachance JG, Landsberg DN, Shapiro RJ, Shoker A, Yilmaz S: Factors associated with progression of interstitial fibrosis in renal transplant patients receiving tacrolimus and mycophenolate mofetil. Transplantation 88: 897–903, 2009 [DOI] [PubMed] [Google Scholar]
- 53.Rush D, Arlen D, Boucher A, Busque S, Cockfield SM, Girardin C, Knoll G, Lachance JG, Landsberg D, Shapiro J, Shoker A, Yilmaz S: Lack of benefit of early protocol biopsies in renal transplant patients receiving TAC and MMF: A randomized study. Am J Transplant 7: 2538–2545, 2007 [DOI] [PubMed] [Google Scholar]
- 54.Friedewald JJ, Kurian SM, Heilman RL, Whisenant TC, Poggio ED, Marsh C, Baliga P, Odim J, Brown MM, Ikle DN, Armstrong BD, Charette JI, Brietigam SS, Sustento-Reodica N, Zhao L, Kandpal M, Salomon DR, Abecassis MM; Clinical Trials in Organ Transplantation 08 (CTOT-08): Development and clinical validity of a novel blood-based molecular biomarker for subclinical acute rejection following kidney transplant. Am J Transplant 19: 98–109, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nankivell BJ, Agrawal N, Sharma A, Taverniti A, P’Ng CH, Shingde M, Wong G, Chapman JR: The clinical and pathological significance of borderline T cell-mediated rejection. Am J Transplant 19: 1452–1463, 2019 [DOI] [PubMed] [Google Scholar]
- 56.Seifert ME, Agarwal G, Bernard M, Kasik E, Raza SS, Fatima H, Gaston RS, Hauptfeld-Dolejsek V, Julian BA, Kew CE, Kumar V, Mehta S, Ong S, Rosenblum F, Towns G, Mannon RB: Impact of subclinical borderline inflammation on kidney transplant outcomes. Transplant Direct 7: e663, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nankivell BJ, P’Ng CH, Chapman JR: Does tubulitis without interstitial inflammation represent borderline acute T cell mediated rejection? Am J Transplant 19: 132–144, 2019 [DOI] [PubMed] [Google Scholar]
- 58.Heilman RL, Devarapalli Y, Chakkera HA, Mekeel KL, Moss AA, Mulligan DC, Mazur MJ, Hamawi K, Williams JW, Reddy KS: Impact of subclinical inflammation on the development of interstitial fibrosis and tubular atrophy in kidney transplant recipients. Am J Transplant 10: 563–570, 2010 [DOI] [PubMed] [Google Scholar]
- 59.de Sandes-Freitas TV, Felipe CR, Campos ÉF, de Lima MG, Soares MF, de Franco MF, Aguiar WF, Tedesco-Silva H, Medina-Pestana JO: Subclinical lesions and donor-specific antibodies in kidney transplant recipients receiving tacrolimus-based immunosuppressive regimen followed by early conversion to sirolimus. Transplantation 99: 2372–2381, 2015 [DOI] [PubMed] [Google Scholar]
- 60.Lee DM, Abecassis MM, Friedewald JJ, Rose S, First MR: Kidney graft surveillance biopsy utilization and trends: Results from a survey of high-volume transplant centers. Transplant Proc 52: 3085–3089, 2020 [DOI] [PubMed] [Google Scholar]
- 61.Mehta R, Cherikh W, Sood P, Hariharan S: Kidney allograft surveillance biopsy practices across US transplant centers: A UNOS survey. Clin Transplant 31: e12945, 2017 [DOI] [PubMed] [Google Scholar]
- 62.Morgan TA, Chandran S, Burger IM, Zhang CA, Goldstein RB: Complications of ultrasound-guided renal transplant biopsies. Am J Transplant 16: 1298–1305, 2016 [DOI] [PubMed] [Google Scholar]
- 63.Naesens M, Khatri P, Li L, Sigdel TK, Vitalone MJ, Chen R, Butte AJ, Salvatierra O, Sarwal MM: Progressive histological damage in renal allografts is associated with expression of innate and adaptive immunity genes. Kidney Int 80: 1364–1376, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Halloran PF, Venner JM, Madill-Thomsen KS, Einecke G, Parkes MD, Hidalgo LG, Famulski KS: Review: The transcripts associated with organ allograft rejection. Am J Transplant 18: 785–795, 2018 [DOI] [PubMed] [Google Scholar]
- 65.Halloran PF, Venner JM, Famulski KS: Comprehensive analysis of transcript changes associated with allograft rejection: Combining universal and selective features. Am J Transplant 17: 1754–1769, 2017 [DOI] [PubMed] [Google Scholar]
- 66.Halloran PF, Pereira AB, Chang J, Matas A, Picton M, De Freitas D, Bromberg J, Serón D, Sellarés J, Einecke G, Reeve J: Microarray diagnosis of antibody-mediated rejection in kidney transplant biopsies: An international prospective study (INTERCOM). Am J Transplant 13: 2865–2874, 2013 [DOI] [PubMed] [Google Scholar]
- 67.Halloran PF, Matas A, Kasiske BL, Madill-Thomsen KS, Mackova M, Famulski KS: Molecular phenotype of kidney transplant indication biopsies with inflammation in scarred areas. Am J Transplant 19: 1356–1370, 2019 [DOI] [PubMed] [Google Scholar]
- 68.Mengel M, Loupy A, Haas M, Roufosse C, Naesens M, Akalin E, Clahsen-van Groningen MC, Dagobert J, Demetris AJ, Duong van Huyen JP, Gueguen J, Issa F, Robin B, Rosales I, Von der Thüsen JH, Sanchez-Fueyo A, Smith RN, Wood K, Adam B, Colvin RB: Banff 2019 Meeting Report: Molecular diagnostics in solid organ transplantation: Consensus for the Banff Human Organ Transplant (B-HOT) gene panel and open source multicenter validation. Am J Transplant 20: 2305–2317, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.O’Connell PJ, Zhang W, Menon MC, Yi Z, Schröppel B, Gallon L, Luan Y, Rosales IA, Ge Y, Losic B, Xi C, Woytovich C, Keung KL, Wei C, Greene I, Overbey J, Bagiella E, Najafian N, Samaniego M, Djamali A, Alexander SI, Nankivell BJ, Chapman JR, Smith RN, Colvin R, Murphy B: Biopsy transcriptome expression profiling to identify kidney transplants at risk of chronic injury: A multicentre, prospective study. Lancet 388: 983–993, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Suthanthiran M, Schwartz JE, Ding R, Abecassis M, Dadhania D, Samstein B, Knechtle SJ, Friedewald J, Becker YT, Sharma VK, Williams NM, Chang CS, Hoang C, Muthukumar T, August P, Keslar KS, Fairchild RL, Hricik DE, Heeger PS, Han L, Liu J, Riggs M, Ikle DN, Bridges ND, Shaked A; Clinical Trials in Organ Transplantation 04 (CTOT-04) Study Investigators: Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med 369: 20–31, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hricik DE, Nickerson P, Formica RN, Poggio ED, Rush D, Newell KA, Goebel J, Gibson IW, Fairchild RL, Riggs M, Spain K, Ikle D, Bridges ND, Heeger PS; CTOT-01 consortium: Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant 13: 2634–2644, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ho J, Sharma A, Kroeker K, Carroll R, De Serres S, Gibson IW, Hirt-Minkowski P, Jevnikar A, Kim SJ, Knoll G, Rush DN, Wiebe C, Nickerson P: Multicentre randomised controlled trial protocol of urine CXCL10 monitoring strategy in kidney transplant recipients. BMJ Open 9: e024908, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Roedder S, Sigdel T, Salomonis N, Hsieh S, Dai H, Bestard O, Metes D, Zeevi A, Gritsch A, Cheeseman J, Macedo C, Peddy R, Medeiros M, Vincenti F, Asher N, Salvatierra O, Shapiro R, Kirk A, Reed EF, Sarwal MM: The kSORT assay to detect renal transplant patients at high risk for acute rejection: Results of the multicenter AART study [published erratum appears in PLoS Med 12: e1001790, 2015]. PLoS Med 11: e1001759, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Loon E, Giral M, Anglicheau D, Lerut E, Dubois V, Rabeyrin M, Brouard S, Roedder S, Spigarelli MG, Rabant M, Bogaerts K, Naesens M, Thaunat O: Diagnostic performance of kSORT, a blood-based mRNA assay for noninvasive detection of rejection after kidney transplantation: A retrospective multicenter cohort study. Am J Transplant 21: 740–750, 2021. 10.1111/ajt.16179 [DOI] [PubMed] [Google Scholar]
- 75.Peddi VR, Patel PS, Schieve C, Rose S, First MR: Serial peripheral blood gene expression profiling to assess immune quiescence in kidney transplant recipients with stable renal function. Ann Transplant 25: e920839, 2020. 10.12659/AOT.920839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W, Yi Z, Keung KL, Shang H, Wei C, Cravedi P, Sun Z, Xi C, Woytovich C, Farouk S, Huang W, Banu K, Gallon L, Magee CN, Najafian N, Samaniego M, Djamali A, Alexander SI, Rosales IA, Smith RN, Xiang J, Lerut E, Kuypers D, Naesens M, O’Connell PJ, Colvin R, Menon MC, Murphy B: A peripheral blood gene expression signature to diagnose subclinical acute rejection. J Am Soc Nephrol 30: 1481–1494, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oellerich M, Shipkova M, Asendorf T, Walson PD, Schauerte V, Mettenmeyer N, Kabakchiev M, Hasche G, Gröne HJ, Friede T, Wieland E, Schwenger V, Schütz E, Beck J: Absolute quantification of donor-derived cell-free DNA as a marker of rejection and graft injury in kidney transplantation: Results from a prospective observational study. Am J Transplant 19: 3087–3099, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, Matas AJ, Mehta S, Mannon RB, Sharfuddin A, Fischbach B, Narayanan M, Jordan SC, Cohen D, Weir MR, Hiller D, Prasad P, Woodward RN, Grskovic M, Sninsky JJ, Yee JP, Brennan DC; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators: Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol 28: 2221–2232, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sigdel TK, Archila FA, Constantin T, Prins SA, Liberto J, Damm I, Towfighi P, Navarro S, Kirkizlar E, Demko ZP, Ryan A, Sigurjonsson S, Sarwal RD, Hseish SC, Chan-On C, Zimmermann B, Billings PR, Moshkevich S, Sarwal MM: Optimizing detection of kidney transplant injury by assessment of donor-derived cell-free DNA via massively multiplex PCR. J Clin Med 8: 19, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Whitlam JB, Ling L, Skene A, Kanellis J, Ierino FL, Slater HR, Bruno DL, Power DA: Diagnostic application of kidney allograft-derived absolute cell-free DNA levels during transplant dysfunction. Am J Transplant 19: 1037–1049, 2019 [DOI] [PubMed] [Google Scholar]
- 81.Huang E, Sethi S, Peng A, Najjar R, Mirocha J, Haas M, Vo A, Jordan SC: Early clinical experience using donor-derived cell-free DNA to detect rejection in kidney transplant recipients. Am J Transplant 19: 1663–1670, 2019 [DOI] [PubMed] [Google Scholar]
- 82.Gielis EM, Ledeganck KJ, Dendooven A, Meysman P, Beirnaert C, Laukens K, De Schrijver J, Van Laecke S, Van Biesen W, Emonds MP, De Winter BY, Bosmans JL, Del Favero J, Abramowicz D: The use of plasma donor-derived, cell-free DNA to monitor acute rejection after kidney transplantation. Nephrol Dial Transplant 35: 714–721, 2020 [DOI] [PubMed] [Google Scholar]
- 83.Stites E, Kumar D, Olaitan O, John Swanson S, Leca N, Weir M, Bromberg J, Melancon J, Agha I, Fattah H, Alhamad T, Qazi Y, Wiseman A, Gupta G: High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant 20: 2491–2498, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]