Abstract
AU-rich elements (ARE) present in the 3′ untranslated regions of many cytokines and immediate-early genes are responsible for targeting the transcripts for rapid decay. We present evidence from cotransfection experiments in NIH 3T3 cells that two signaling pathways, one involving phosphatidylinositol 3-kinase (PI3-K), and one involving the p38 mitogen-activated protein kinase (MAPK), lead to stabilization of interleukin-3 mRNA in parallel. Stabilization mediated by either of the two pathways was antagonized by tristetraprolin (TTP), an AU-binding protein known to promote constitutive decay of ARE-containing transcripts. Remarkably, the stabilizing AU-binding protein HuR, in collaboration with p38 MAPK but not with PI3-K, could overcome the destabilizing effect of TTP. These data argue that the stabilizing kinases PI3-K and p38 MAPK do not act through direct inactivation of TTP but via activating pathway-specific stabilizing AU-binding proteins. Our data suggest an integrated model of mRNA turnover control, where stabilizing (HuR) and destabilizing (TTP) AU-binding proteins compete and where the former are under the positive control of independent phosphokinase signaling pathways.
Stabilization of short-lived mRNAs of cytokines and proto-oncogenes is an important aspect of gene expression and plays a role in cell activation and oncogenesis (6, 13, 26, 30, 37, 44, 45). This regulation involves AU-rich elements (ARE) located in the 3′ untranslated region (UTR) that direct deadenylation followed by rapid degradation of mRNA (4–6, 31, 35, 42). Stabilization of ARE-containing transcripts can be achieved by upstream signals, such as allergic stimulation in mast cells, elevation of intracellular Ca2+, or protein kinase C activation by tetradecanoyl phorbol acetate (TPA) (11, 44, 45); activation of T cells by anti-CD3 and anti-CD28 antibodies (21); or overexpression of the AU-binding protein (AUBP) HuR in various cultured cells (8, 15, 19, 27). While the mechanisms by which upstream signals regulate the mRNA decay machinery remain to be elucidated, the involvement of protein kinases and phosphatases has been suggested through the use of specific inhibitors which destabilize various cytokine mRNAs (1, 9, 18, 20, 26, 33, 47). Direct evidence for a kinase pathway regulating cytokine mRNA turnover has recently been obtained by cotransfection experiments. We showed that c-jun N-terminal kinase (JNK) is involved in interleukin-3 (IL-3) mRNA turnover control in mast cells (24), and Chen et al. (3) demonstrated that the same pathway regulates IL-2 mRNA in T cells. Moreover, Winzen et al. (43) reported that the p38 mitogen-activated protein kinase (MAPK) pathway signals for cytokine-induced mRNA stabilization via MAPK-activated protein kinase 2 (MK2).
Obvious candidate targets of these kinase pathways are AUBPs, several of which have now been cloned (41) and linked to specific functions. Overexpression of the ELAV protein Hel-N1 stabilized the ARE-containing glucose transporter 1 mRNA (15). Transfection of HuR, closely related to Hel-N1, led to stabilization of c-fos and granulocyte-macrophage colony-stimulating factor ARE reporter transcripts, as well as of vascular endothelial growth factor mRNA (8, 19, 27) and of mRNA from cyclins A and B1 (38, 39). More recently, another AUBP, termed tristetraprolin (TTP), was identified and shown to promote ARE-dependent decay. This discovery followed the observation that TTP−/− mice had high systemic levels of tumor necrosis factor alpha, indicating a role for TTP in a constitutive default degradation pathway (2). In cellular mutants with a specific defect in ARE-dependent degradation, TTP could restore rapid degradation in two complementation groups (36). The third AUBP with an established role is AUF1 (41). Loflin et al. (22) have recently shown that overexpressed AUF1 in erythroleukemia cells antagonized the stabilizing effect of hemin on ARE-containing reporter transcripts.
Here, we have used an NIH 3T3 cell-based transient-transfection system to analyze three protein kinase pathways for the potential to stabilize IL-3 mRNA. We demonstrate that phosphatidylinositol 3-kinase (PI3-K) and p38 MAPK independently stabilize IL-3 mRNA. Cotransfection experiments with AUBPs showed that the protein kinases do not act by inactivating the destabilizing function of TTP and revealed a synergism between HuR and p38 MAPK. Based on our results, we present an integrated working model of mRNA turnover control involving AUBPs, protein phosphokinases, and phosphatases.
MATERIALS AND METHODS
Materials.
Reagents were purchased or obtained from the following sources: N,N-bis(2-hydroxy-ethyl)-2-amino-ethanesulfonic acid and actinomycin D (actD) from Calbiochem; anti-FLAG M2 monoclonal antibody from Eastman Kodak Company; and anti-mouse immunoglobulin G (H+L) AP conjugate and Western Blue stabilized substrate for alkaline phosphatase from Promega. The recombinant glutathione S-transferase (GST) fusion proteins GST–c-jun (1–79) and GST-ATF2 (1–254), containing the first 79 and 254 amino acids of c-jun and ATF2, respectively, were purified as described previously (12).
Plasmid construction.
Mxh-IL-3-wt and Mx-IL-3-ΔAU (35), pMxh-β-IL3(UTR)wt, pMxh-β-IL3(UTR)ΔAU, SRα3-MEK6DD, pCMV-M2-JNK-APF, pCMV-M2-p38-AGF, pGEX-c-jun (1–79), and pGEX-ATF2 (1–254) have been described previously (24). pEF-Bos-rCD2-p110 and pcDNA-MKK7D were generously provided by D. A. Cantrell (28, 29) and K. R. Chien (40), respectively. pTet-Myc-over-HuR (27) was kindly provided by A.-B. Shyu.
To generate myc- and His-tagged HuR, an HuR cDNA fragment was first prepared by EcoRV digestion of plasmid pTet-Myc-over-HuR and then ligated in frame into the EcoRV site of pcDNA 3.1-myc/his.B (Invitrogen). The stop codon of HuR was then removed by QuickChange site-directed mutagenesis (Stratagene GMBH), and the final plasmid was referred to as pcDNA-HuR-myc/His.B. Construction of pcDNA-mTTP-myc/His.A was described previously (36). pcDNA-mTTPC139R-myc/His.A was constructed by QuickChange site-directed mutagenesis.
Cell culture and transfection.
Mouse NIH 3T3 B2A2 cells (46) were kindly provided by A.-B. Shyu. The cells were maintained in complete Iscove's modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum (FCS) at 37°C in an atmosphere of 7% CO2 and were passaged when they reached confluency. The cells were seeded at a density of 2 × 106/10-cm-diameter dish in IMDM with 10% FCS 18 to 20 h before transfection by the calcium phosphate method (32). Transfection mixtures for each plate contained 1 to 8 μg of Mxh-IL-3-wt, 1 μg of Mx-IL-3-ΔAU reporter plasmid, or 6 μg of Mxh-βglobin-IL-3(UTR) reporter plasmid and 0.5 to 5 μg of test plasmid. The hygromycin B phosphotransferase (hph) cDNA under control of simian virus 40 regulatory regions on the reporter plasmids was used as an internal control for transfection efficiency. After exposure to the plasmid precipitates for 12 to 16 h, the cells were washed twice with Ca2+-Mg2+-free phosphate-buffered saline and cultured in IMDM containing 0.5% FCS for 22 to 24 h before the addition of actD. RNA was extracted after various time intervals. Note that different amounts of reporter plasmids were used in order to give a comparable reporter signal on the Northern blot, as a result of which the hph signals vary among different experimental groups.
Northern blot analysis.
Total cytoplasmic RNA was isolated by the method of Gough (10), and hybridizations were performed as described previously (26). 32P-labeled antisense RNA probes were synthesized using an SP6 transcription kit (Boehringer). The following riboprobes were used: pSP65-multi-CSF, containing a 633-bp XbaI-SpeI 3′ UTR fragment of IL-3 cDNA; pGEM-hph, containing a 96-bp EcoRI-PstI fragment of hph cDNA as described previously (24); and pSP73-βglobin, containing an 86-bp EcoRI-BglII fragment. Quantification of the mRNA signals was performed with a PhosphorImager (Molecular Dynamics) using ImageQuant software as described previously (1). The reporter mRNA signals were normalized to the respective hph reference signal after subtraction of the filter background. In all the plotted graphics, each point represents the average value from three transfection experiments. Standard errors are included unless they were too small to be displayed on the semilogarithmic plots.
Immunoblotting.
Cell extracts were prepared by lysing cells in 250 μl of extraction buffer (120 mM NaCl, 50 mM Tris [pH 8.0], 20 mM NaF, 1 mM benzamidine, 1 mM EDTA, 1 mM EGTA, 1 mM PPi, 30 mM 4-nitrophenyl phosphate disodium salt hexahydrate, 1% NP-40, and 0.1 M phenylmethylsulfonyl fluoride), as described previously (24). Extracts (15 μl) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrophoretically transferred to an Immobilon P membrane (Millipore), and the membrane was incubated overnight with the corresponding first antibody at 4°C with gentle agitation after being blocked with 5% skim milk. The protein was decorated with an anti-mouse alkaline phosphatase-conjugated secondary antibody and detected using Western Blue stabilized substrate. Detection of tagged TTP has been described elsewhere (36).
In vitro kinase assay.
For kinase assays, 100 μg of lysates from cells transfected with M2-JNK or M2-p38 MAPK was immunoprecipitated with 5 μl of M2 monoclonal antibody (7). The immunoprecipitates were then assayed for kinase activity using GST–c-Jun (1–79) or GST-ATF2 (1–254) fusion protein, respectively, as the substrate (24).
RESULTS
Regulated mRNA decay in an NIH 3T3 cell transient-transfection system.
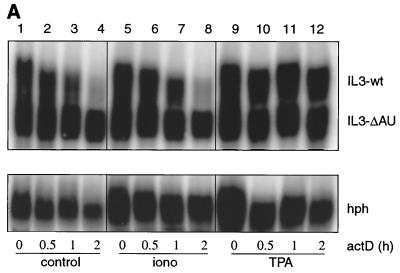
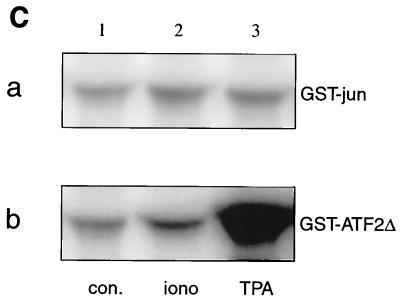
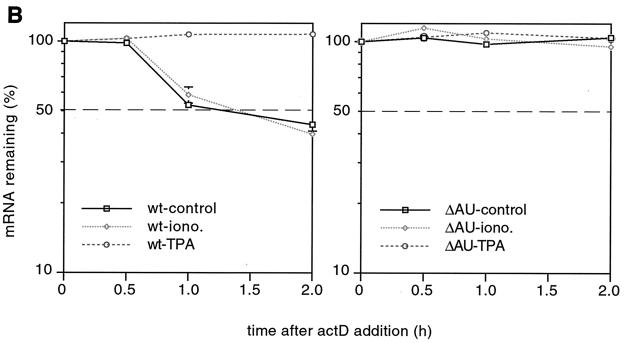
To extend our studies of the regulation of IL-3 mRNA turnover by signal transduction pathways, we established a transient-transfection system where the ARE-dependent turnover of reporter transcripts and its sensitivity to upstream signals could be evaluated. As the mast cell lines used in our previous work showed poor transfection efficiency, we switched to NIH 3T3 cells, a cell line that has been successfully used for studying mRNA turnover by this technique (27, 32, 46). IL-3 genes, both wild type (wt) and with the ARE deletion (ΔAU), served as reporter genes. They were under the control of the Moloney retroviral long terminal repeat (Mx), referred to as Mxh-IL-3-wt and Mx-IL-3-ΔAU, respectively (35). The reporter plasmids also included the hph cDNA serving as a control for transfection efficiency and loading. Figure 1A shows a typical transient-transfection experiment in which Mxh-IL-3-wt was cotransfected with Mx-IL-3-ΔAU. Decay was measured in the presence of actD over 2 h. Similar to actD chase experiments in mast cells (35), IL-3-wt mRNA had a relatively short half-life, but the form with ARE deleted was stable (Fig. 1A, lanes 1 to 4). TPA stabilized the wt transcripts but did not increase the abundance of IL-3-ΔAU (lanes 9 to 12). Ionomycin, in contrast to its stabilizing effect in mast cells, was inactive (lanes 5 to 8). Quantification of the data is shown in Fig. 1B. In vitro kinase assays revealed that ionomycin activated neither JNK nor p38 MAPK (Fig. 1C, lane 2), while TPA activated p38 MAPK with very little, if any, effect on JNK activity (Fig. 1C, lane 3). As this system showed rapid and regulatable ARE-dependent mRNA turnover, it appeared to provide a suitable tool to study the corresponding signaling pathways.
FIG. 1.
Stability of IL-3 transcripts in a transient-transfection assay. (A) NIH 3T3 B2A2 cells were cotransfected with 3 μg of Mxh-IL-3-wt and 1 μg of Mx-IL-3-ΔAU, the latter plasmid carrying a deletion of the ARE. hph served to monitor transfection efficiency and control for loading. Forty-eight hours after transfection, actD (5 μg/ml) was added for the indicated time in the absence (control; lanes 1 to 4) or presence of 2 μM ionomycin (iono; lanes 5 to 8) or 20 ng of TPA/ml (lanes 9 to 12). (B) Quantification of the signal intensities shown in panel A by PhosphorImager, with the hph-normalized values at time zero taken as 100%. Each point represents the average of three transfection experiments. (C) In vitro kinase assays. Cells were either untreated (con.; lane 1) or stimulated with 2 μM ionomycin (lane 2) or 20 ng of TPA/ml (lane 3) for 20 min. Then lysates were prepared, and 100 μg was subjected to in vitro JNK (a) or p38 MAPK (b) assay using either GST-jun (1–79) or GST-ATF2 (1–254), respectively, as the substrate.
PI3-K and p38 MAPK pathways independently stabilize IL-3 mRNA.
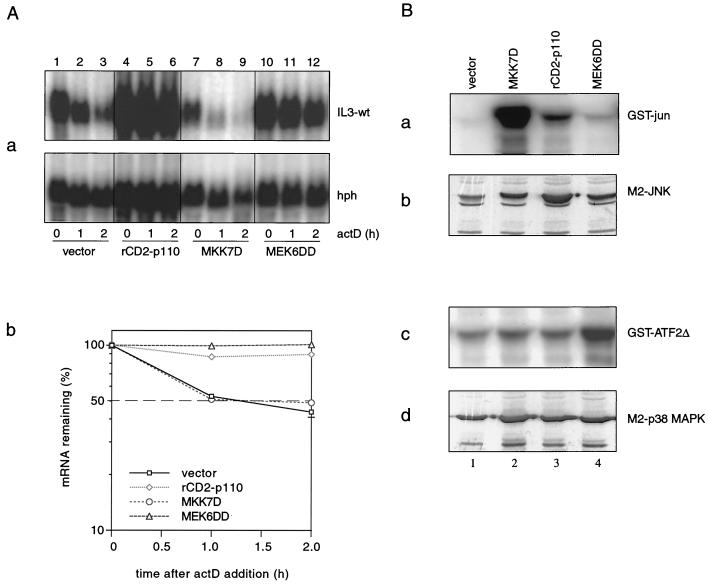
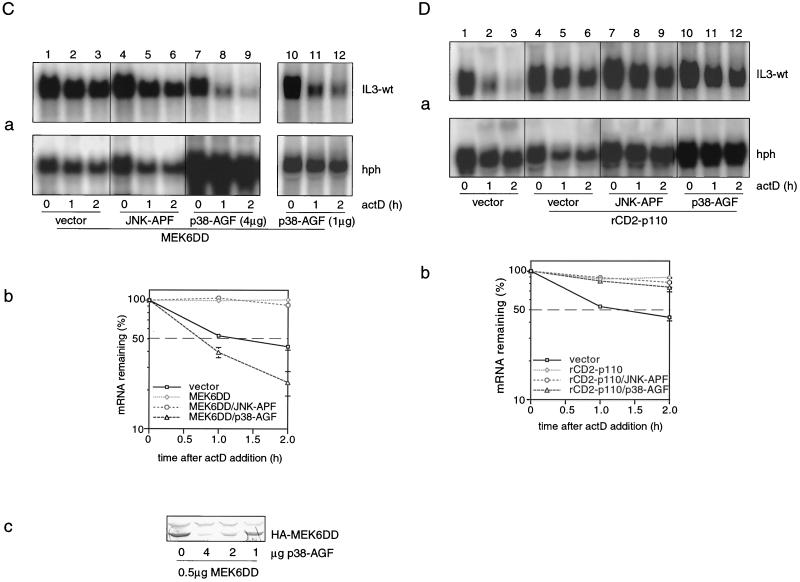
We first wished to determine whether individual signaling components from established pathways, such as PI3-K, JNK, or p38 MAPK, would be sufficient for stabilizing IL-3 mRNA. The wt IL-3 reporter construct was cotransfected with either rat (rCD2)-p110, MKK7D, or MEK6DD, constitutive active forms of PI3-K, MKK7, and MEK6, respectively (29, 34, 40). rCD2-p110 is a chimeric molecule in which the cytoplasmic domain of the rCD2 cell-surface antigen has been replaced with p110α, the catalytic subunit of PI3-K that is activated by plasma-membrane localization (28, 29). The membrane-bound rCD2-p110 induces accumulation of PI(3,4)P2 and PI(3,4,5)P3 equivalent to levels seen after mitogenic stimulation (29). While JNK activation by PI3-K has been reported (14, 23), MEK6 and MKK7 are specific upstream activators of p38 MAPK and JNK, respectively (25, 34). Immune complex kinase assays confirmed that MKK7 and MEK6 specifically activate JNK and p38 MAPK, respectively (Fig. 2B, lanes 2 and 4, respectively), whereas PI3-K weakly activates JNK without any effect on p38 MAPK activity in NIH 3T3 cells (Fig. 2B, lane 3). The effect of rCD2-p110, MKK7D, or MEK6DD on the stability of wt IL-3 mRNA was then assessed in the decay assay. As shown in Fig. 2A, ectopic expression of either rCD2-p110 or MEK6DD alone was able to antagonize IL-3-wt mRNA decay (lanes 4 to 6 and 10 to 12), indicating that both the PI3-K and the p38 MAPK pathways are involved in mRNA stabilization. The strong JNK activator MKK7D did not exhibit any stabilizing effect (lanes 7 to 9). This is noteworthy in view of the fact that JNK mediates ionomycin-induced IL-3 mRNA stabilization in mast cells (24). Thus, the pattern seen in NIH 3T3 cells is more closely related to that in HeLa cells, where a recent study showed that p38 MAPK controls ARE-mediated mRNA decay (43).
FIG. 2.
PI3-K and p38 MAPK are independently involved in IL-3 mRNA stabilization. (A) Effect of rCD2-p110, MKK7D, or MEK6DD on wt IL-3 mRNA decay. Three micrograms of the reporter plasmid Mxh-IL-3-wt was cotransfected with 2 μg of vector (lanes 1 to 3), rCD2-p110 (lanes 4 to 6), or MKK7D (lanes 7 to 9) or 0.5 μg of MEK6DD (lanes 10 to 12). Decay assays were performed as described in the legend to Fig. 1. Shown is a representative result from three independent experiments. For quantification, the averages of three transfection experiments are plotted in graph b. (B) Stimulation of JNK or p38 activity by MKK7, PI3-K, or MEK6. Cells were cotransfected with 2 μg of either M2-JNK (a and b) or M2-p38 (c and d) in combination with 2 μg of vector (lane 1), MKK7D (lane 2), rCD2-p110 (lane 3), or MEK6DD (lane 4). One hundred micrograms of the lysates was used for immunoprecipitation with the M2 monoclonal antibody and subjected to in vitro JNK (a) or p38 kinase (c) assay using GST–c-jun (1–79) or GST-ATF2 (1–254), respectively, as the substrate. The expression of M2-JNK (b) and M2-p38 (d) was analyzed by Western blotting using anti-M2. (C and D) Effect of JNK-APF or p38-AGF on MEK6DD- or rCD2-p110-mediated stabilization. Mxh-IL-3-wt reporter plasmid was transfected alone (D, panel a, lanes 1 to 3) or in combination with MEK6DD (C, panel a, lanes 1 to 12) or rCD2-p110 (D, panel a, lanes 4 to 12) in the absence (C, panel a, lanes 1 to 3; D, panel a, lanes 4 to 6) or presence of JNK-APF (C, panel a, lanes 4 to 6; D, panel a, lanes 7 to 9), 4 μg of p38-AGF (C, panel a, lanes 7 to 9; D, panel a, lanes 10 to 12), or 1 μg of p38-AGF (C, panel a, lanes 10 to 12). A decay assay was performed 2 days after transfection as described for panel A. Graph b shows quantification of signal intensities using the average of three transfection experiments. (C) Panel c reveals the expression of MEK6DD by Western blot analysis using the anti-hemagglutinin monoclonal antibody 12CA5.
The fact that expression of either PI3-K or MEK6DD led to stabilization of IL-3 mRNA (Fig. 2A) despite their different specificities (Fig. 2B) suggested that they may operate in parallel. To test this hypothesis, MEK6DD or rCD2-p110 was cotransfected with the dominant-negative construct JNK-APF or p38-AGF. While coexpression of p38-AGF strongly interfered with MEK6DD-induced IL-3 mRNA stabilization (Fig. 2C, panel a, compare lanes 7 to 9 with lanes 1 to 3), it did not significantly affect rCD2-p110-mediated stabilization (Fig. 2D, panel a, compare lanes 10 to 12 with lanes 4 to 6). No significant inhibitory effect was observed when JNK-APF was cotransfected with MEK6DD (Fig. 2C, panel a, lanes 4 to 6) or rCD2-p110 (Fig. 2D, panel a, lanes 7 to 9). To rule out the possibility that the inhibitory effect on MEK6DD-mediated IL-3 mRNA stabilization by p38-AGF is not due to unexpected transcriptional effects on MEK6DD, the expression of MEK6DD in the absence or presence of 4 μg of p38-AGF was determined by Western blot analysis using the monoclonal antihemagglutinin antibody 12CA5. As shown in Fig. 2C, panel c, cotransfection of 4 μg of p38-AGF strongly suppresses the expression of MEK6DD. However, when p38-AGF was titrated to 1 μg, where no significant inhibition on MEK6DD expression was observed (Fig. 2C, panel c), the MEK6DD-mediated mRNA stabilization was still antagonized (Fig. 2C, panel a, lanes 10 to 12). These observations indicate that MEK6 affects IL-3 mRNA stability in NIH 3T3 cells through the p38 MAPK pathway, whereas PI3-K stabilizes through effectors other than JNK or p38 MAPK. Thus, PI3-K and p38 MAPK represent two independent pathways that operate in parallel to control IL-3 mRNA turnover in NIH 3T3 cells.
Role of HuR and TTP.
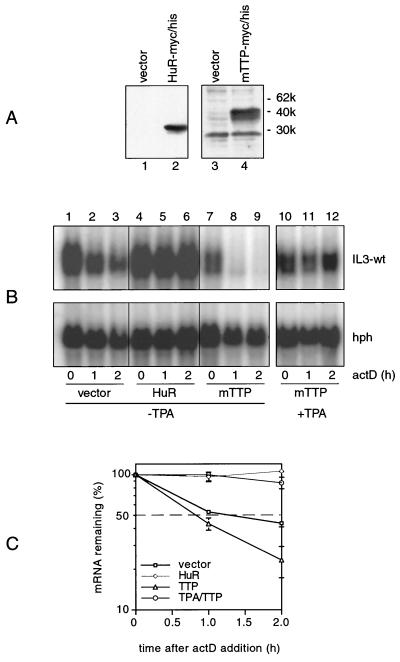
Next, we wished to study whether and how these pathways interact with downstream candidate AUBPs, and we concentrated on HuR and TTP. The genes for both were transfected together with the IL-3 wt reporter. Expression was analyzed by Western blotting and is shown in Fig. 3A. Their possible effect on decay was explored in the absence or presence of stabilizing TPA. In the absence of TPA, ectopic expression of HuR led to stabilization of IL-3 reporter transcripts (Fig. 3B, lanes 4 to 6). In contrast, TTP accelerated the decay rate, an observation frequently made in repeated experiments (Fig. 3B, lanes 7 to 9). Of note, TTP was unable to antagonize mRNA stabilization induced by TPA (Fig. 3B, lanes 10 to 12).
FIG. 3.
Role of HuR and TTP in IL-3 mRNA turnover in vivo. (A) Expression of myc-tagged HuR (lane 2) and TTP (lane 4) detected by Western blotting using monoclonal anti-myc antibody 9E10. (B) Cells were transfected with Mxh-IL-3-wt reporter plasmid alone (lanes 1 to 3) or with 1 μg of HuR (lanes 4 to 6) or 0.3 μg of TTP (lanes 7 to 12). The effect of the ectopically expressed AUBPs on constitutive IL-3 mRNA decay (lanes 1 to 9) or on TPA-induced stabilization (lanes 10 to 12) was assessed in a decay assay. Note that 2 μg of TTP was transfected for the Western blot analysis shown in panel A, whereas only 0.3 μg was used in all other decay assays, since 2 μg of TTP decreased the IL-3 mRNA signal to undetectable levels while TTP was not detectable by Western blotting when only 0.3 μg was transfected (data not shown). (C) Quantification of signal intensities, using the average of three transfection experiments.
Functional competition experiments.
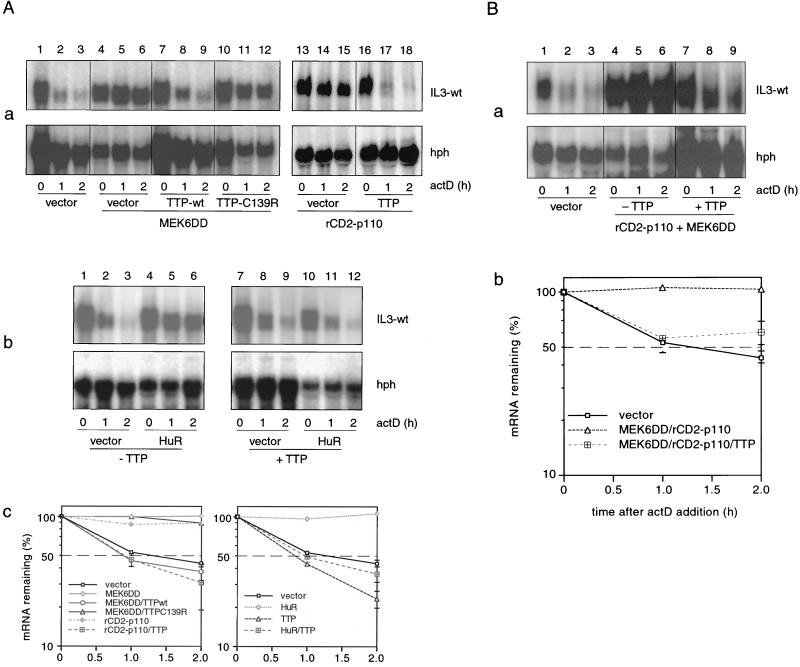
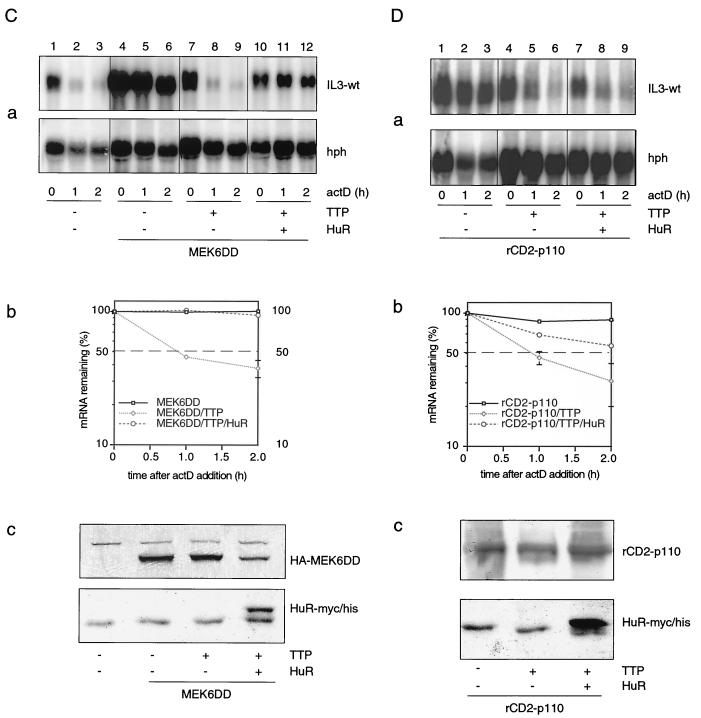
The inability of TTP to antagonize the stabilizing effect of TPA suggested that one of the TPA-activated signaling pathways may inactivate the destabilizing function of TTP, perhaps by direct phosphorylation. We therefore asked whether PI3-K or p38 MAPK may analogously stabilize IL-3 mRNA by inactivating the destabilizing function of TTP. We cotransfected the IL-3-wt reporter with rCD2-p110 or MEK6DD in the absence or presence of TTP, as indicated in Fig. 4A, blot a, and performed decay assays to see whether stabilization or decay would prevail. Strikingly, both PI3-K- and p38 MAPK-mediated stabilization was strongly overruled by ectopic expression of TTP (Fig. 4A, panel a, compare lanes 7 to 9 and 4 to 6, as well as 16 to 18 and 13 to 15). The TTP zinc finger mutant C139R, which is unable to bind the ARE (16), failed to antagonize the stabilizing effect of p38 MAPK (Fig. 4A, panel a, lanes 10 to 12). These results rule out the possibility that PI3-K or MEK6DD stabilizes IL-3 mRNA through functional inactivation of TTP, and in particular that TTP is a direct substrate of these enzymes. We next examined whether HuR-mediated stabilization would also be overcome by TTP and performed similar functional competition experiments. Again, destabilizing TTP could override HuR (Fig. 4A, panel b). Lastly, we also examined whether transfection of both PI3-K and MEK6DD would cooperatively antagonize TTP. Remarkably, the destabilizing effect of TTP could even override a combination of the two stabilizing kinases (Fig. 4B).
FIG. 4.
Mutual interactions of upstream kinases and AUBPs in regulating IL-3 mRNA turnover. (A) Effect of TTP on PI3-K-, p38 MAPK-, or HuR-mediated stabilization. Cells were transfected with Mxh-IL-3-wt reporter plasmid alone (a and b, lanes 1 to 3) or together with active kinases (a, lanes 4 to 18) or stabilizing HuR (b, lanes 4 to 6 and 10 to 12) in the absence (a, lanes 1 to 6 and 13 to 15; b, lanes 1 to 6) or presence of 0.3 μg of TTP-wt (a, lanes 7 to 9 and 16 to 18; b, lanes 7 to 12) or 1 μg of TTP-C139R (a, lanes 10 to 12). Shown in graphs c are the averages of three transfection experiments. (B) Effect of TTP (lanes 7 to 9) on stabilization mediated by a combination of rCD2-p110 with MEK6DD (lanes 4 to 9). The experiment was performed as described for panel A, and the plasmids were transfected as indicated in panel a. The averages of three transfection experiments are shown in graph b. (C and D) mRNA decay assay was performed using cells transfected with Mxh-IL-3-wt alone (C, panel a, lanes 1 to 3) or in combination with MEK6DD (C, panel a, lanes 4 to 12) or rCD2-p110 (D, panel a, lanes 1 to 9) in the absence (C, panel a, lanes 4 to 9; D, panel a, lanes 1 to 6) or presence of HuR (C, panel a, lanes 10 to 12; D, panel a, lanes 7 to 9). In addition, TTP was cotransfected in lanes 7 to 12 (C, panel a) or lanes 4 to 9 (D, panel a). Shown is one representative result from three transfection experiments. Quantification of the signal intensities using the average of three transfection experiments is shown in graph b. Panels c show the expression of MEK6DD and HuR (C) or rCD2-p110 and HuR (D) by Western blotting. rCD2-p110 expression was detected with anti-rCD2 antibody.
Our data described above showed that the TTP effect prevailed over individual stabilizing kinases and HuR but not over the pharmacological stabilizer TPA. This could mean that TPA-induced signal amplification, a characteristic of kinase cascades, generated sufficient stabilizing capacity to override the limiting destabilization activity of TTP. With this possibility in mind, we tested whether PI3-K or p38 MAPK, when combined with HuR, could antagonize TTP and performed transfections of TTP with combinations of HuR and the two kinase genes. While ectopically expressed TTP again antagonized MEK6DD (Fig. 4C, panel a, compare lanes 7 to 9 and lanes 4 to 6) or rCD2-p110-mediated stabilization (Fig. 4D, panel a, compare lanes 4 to 6 and lanes 1 to 3), it was striking that TTP did not have a significant destabilizing effect when MEK6DD was cotransfected with HuR (Fig. 4C, panel a, lanes 10 to 12). In contrast, the combination of rCD2-p110 with HuR (Fig. 4D, panel a, lanes 7 to 9) was still antagonized by TTP. To ensure that the antagonizing effects of various effectors on mRNA stability was not due to transcriptional effects such as promoter competition, we performed Western blots for control. As shown in Fig. 4C, panel c and 4D, panel c, no inhibitory effect of TTP or HuR on the expression of MEK6DD or rCD2-p110 was evident. While TTP was not detectable by Western blotting at the concentration used for transfection, the data in Fig. 4D, panel a, lanes 7 to 9, rule out the possibility that HuR downregulates TTP expression, as decay still prevailed. Taken together, these data reinforced the idea that PI3-K and MEK6DD stabilize IL-3 mRNA not by inactivation of TTP but rather by activating downstream stabilizing targets. In addition, these results allowed the functional assignment of HuR to the p38 MAPK pathway, while a corresponding PI3-K-specific AUBP remains to be identified.
ARE as sufficient regulatory target.
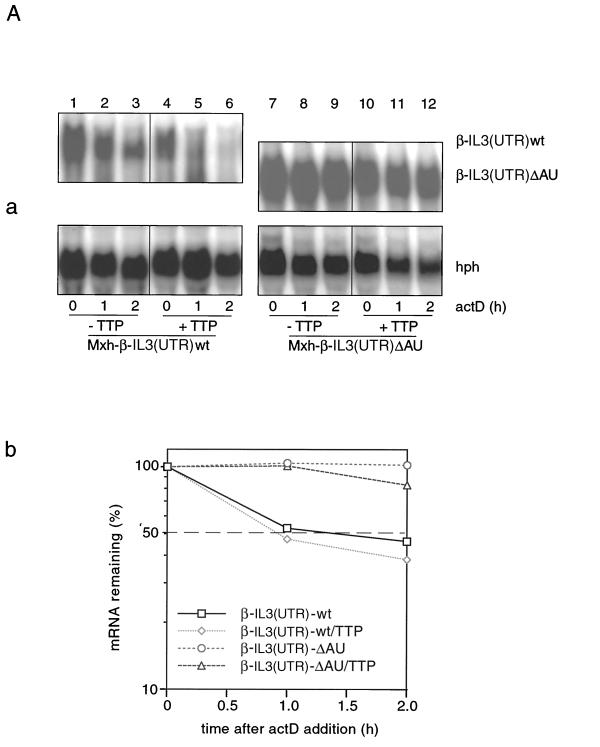
The ARE in the 3′ UTR of IL-3 is both necessary and sufficient for directing rapid decay of IL-3 transcripts (35). Moreover, it is the cis element through which the JNK pathway exerts its regulatory effect on IL-3 mRNA turnover in mast cells (24). We therefore wished to determine whether the 3′ UTR of IL-3 mRNA is also sufficient as the cis target for the regulation observed in this study. First, we ensured that TTP binds the ARE from IL-3, as should be expected (2). This was verified by corresponding gel mobility shift assays (data not shown). Next, we performed an experiment similar to that described above using a β-globin reporter construct carrying the 3′ UTR of IL-3 with (wt) or without (ΔAU) the ARE. As shown in Fig. 5A, the reporter transcripts with an ARE deletion were stable and insensitive to TTP (Fig. 5A, panel a, lanes 7 to 12), in contrast to transcripts carrying the ARE (Fig. 5A, panel a, lanes 1 to 6). These data, in analogy with data from tumor necrosis factor alpha (2), demonstrated that the ARE from IL-3 is the cis element required for TTP-mediated rapid decay.
FIG. 5.
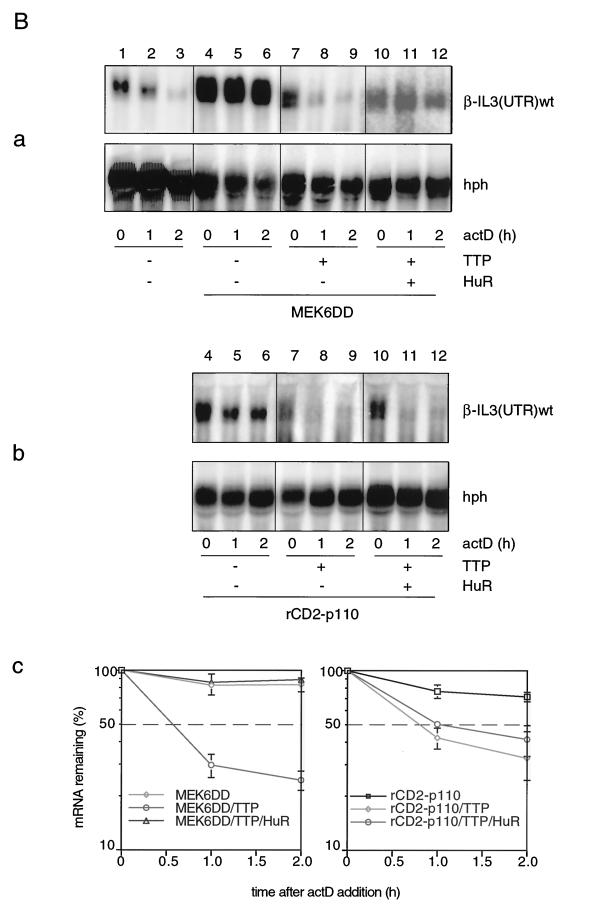
ARE is the cis element through which IL-3 mRNA turnover is regulated by PI3-K or p38 MAPK involving AUBPs. (A) Effect of TTP on the decay of β-globin reporter transcripts. Cells were transfected with Mxh-β-IL3(UTR)wt (lanes 1 to 6) or Mxh-β-IL3(UTR)ΔAU (lanes 7 to 12) in the absence (lanes 1 to 3 and 7 to 9) or presence (lanes 4 to 6 and 10 to 12) of 0.3 μg of TTP. (B) The experiment was performed as described in the legend to Fig. 4C except that Mxh-β-IL3(UTR)wt was used as the reporter construct instead of Mxh-IL-3-wt. Shown in panel A, graph b, and panel B, graph c are the average values taken from three transfection experiments.
Finally we wished to examine whether the conclusions drawn above would also hold true with the β-globin reporter constructs. Results comparable to those with the IL-3 reporter mRNA were obtained, as shown in Fig. 5B. Again, TTP dramatically antagonized MEK6DD- or rCD2-p110-mediated stabilization (Fig. 5B, panels a and b, compare lanes 7 to 9 and lanes 4 to 6) but did not significantly inhibit the stabilizing effect mediated by a combination of MEK6DD with HuR (Fig. 5B, panel a, lanes 10 to 12). Furthermore, stabilization by a combination of rCD2-p110 with HuR (Fig. 5B, panel b, lanes 10 to 12) was again antagonized by TTP. Taken together, these results demonstrate that the ARE-containing 3′ UTR of IL-3 is necessary and sufficient to confer the pattern of regulation shown in this study.
DISCUSSION
Stabilization of short-lived ARE-containing mRNAs is a physiological mechanism to amplify and fine tune gene expression in response to extracellular stimuli. While the pathways regulating transcription are well described, less is known about how signaling is connected to the stabilization of short-lived transcripts and their translation. It is thought that AUBPs serve as adapter proteins linking upstream signaling elements to mRNA and a yet-to-be-identified RNase. In this work, we provide firm evidence that two signal transduction pathways, the PI3-K and the p38 MAPK pathways, are independently implicated in turnover control of IL-3 mRNA. This links mRNA stabilization within a single cell to more than one receptor system. While mRNA stabilization by p38 MAPK has previously been reported in HeLa cells (43), the involvement of PI3-K is a novel finding. By using an NIH 3T3 cell-based transient-transfection system, where multiple collaborating and antagonistic components could be evaluated, we assigned HuR to the p38 MAPK pathway and showed that the 3′ UTR of IL-3 contains the necessary and sufficient cis element.
In our previous work with mast cells, using a dominant-negative construct of JNK, we provided evidence that the JNK pathway mediates ionomycin-induced IL-3 mRNA stabilization (24). Pharmacological data argued that JNK alone is not sufficient and that collaborating pathways might be required as well. In fact, kinase inhibitors with different specificities, i.e., wortmannin (PI3-K and JNK), SB202198 (JNK and p38 MAPK), or cyclosporine A (p38 MAPK), antagonized stabilization by ionomycin. It thus appeared that at least in mast cells coordinated signaling by more than one pathway is necessary to maintain transcript stability (24). In contrast, the present data argue that TPA-induced IL-3 mRNA stabilization in NIH 3T3 fibroblasts employs several redundant pathways, as none of the inhibitors alone (wortmannin, cyclosporine A, or SB202198) could antagonize TPA-induced stabilization (data not shown). The existence of redundant and independent stabilizing pathways was supported by two lines of evidence. The first came from experiments using constitutively activated kinase genes and corresponding dominant-negative mutants. While MEK6DD stabilized IL-3 reporter transcripts via the p38 MAPK pathway (Fig. 2C, panel a), PI3-K led to stabilization independently of JNK or p38 MAPK (Fig. 2D, panel b), implying that the two pathways control IL-3 mRNA turnover in parallel. The second piece of evidence is provided by functional-competition experiments in which MEK6DD, but not PI3-K, could synergize with the stabilizing AUBP HuR to counteract TTP-mediated destabilization (Fig. 4C and 5B), indicating that the two kinase pathways stabilize IL-3 mRNA via different downstream targets. In view of the fact that JNK was reported to be under the control of PI3-K in both HeLa and mast cells (14, 23), it is important to point out that dominant-negative JNK failed to antagonize PI3-K-induced stabilization in NIH 3T3 cells (Fig. 2C, panel b). Moreover, TPA failed to activate JNK appreciably, and the constitutive active JNK-specific activator MKK7 failed to stabilize transcripts in NIH 3T3 cells (Fig. 2A). Thus, JNK does not appear to be involved in the mRNA stabilization pathways in NIH 3T3 cells, which is in contrast to its effect in mast cells and Jurkat T cells (3, 24) but consistent with result from HeLa cells (43). These observations suggest that pathways stabilizing mRNA operate in a cell-type-specific fashion, like the pathways controlling other cellular functions.
In recent years, much has been learned about the roles of trans-acting AUBPs in ARE-directed mRNA turnover (41). HuR, AUF1, and TTP have been assigned a functional role in vivo (2, 8, 15, 17, 19, 22, 27). In agreement with these reports, we demonstrate here that HuR and TTP function as trans-acting stabilizing and destabilizing AUBPs, respectively. Interestingly, TTP also antagonized stabilization brought about by HuR in cotransfection experiments (Fig. 4A, panel b) under conditions where TTP was expressed at a much lower level than HuR (data not shown). Furthermore, TTP antagonized both stabilizing kinases (Fig. 4A, panel a), even when they were transfected in combination (Fig. 4B). This rules out a model where constitutive decay, brought about in resting cells by TTP, is overcome following functional inactivation of the destabilizing TTP by the PI3-K or p38 MAPK pathway. Instead, the data favor a model where destabilizing TTP is competed out by a stabilizing AUBP that needs prior activation by an upstream signaling pathway. Here we provide functional evidence that for the p38 MAPK pathway the AUBP is HuR, as their combination overcame TTP-mediated decay (Fig. 4C, panel a, and 5B, panel a). A stabilizing AUBP activated by the PI3-K pathway remains to be identified. In view of the fact that HuR is not reported to be a phosphoprotein, a critical question that remains is whether HuR is the direct downstream target of the p38 MAPK pathway or whether HuR remains unphosphorylated but cooperates with a p38 MAPK target. A tryptic peptide map of HuR in the presence and absence of MEK6DD should help to resolve this issue.
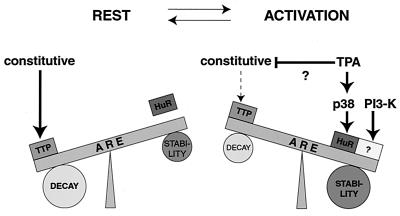
The data presented here are summarized in a model (Fig. 6) that integrates our view of how IL-3 mRNA turnover is regulated by exogenous signals. (i) ARE-directed IL-3 mRNA turnover is under the control of a number of trans-acting AUBPs, including stabilizing HuR and destabilizing TTP. The destabilizing function of TTP is, under resting conditions, dominant over the stabilizing function of HuR, which results in constitutive rapid mRNA decay. (ii) HuR and perhaps other yet-to-be-identified AUBPs are under the control of multiple parallel signal transduction pathways, which allows a variety of stimuli and receptor systems to affect mRNA decay. TPA acts as a pleiotropic upstream activator. (iii) Stabilization of IL-3 mRNA could be achieved by two basically independent mechanisms, either by inactivation of destabilizing TTP or by activation of stabilizing AUBPs such as HuR, which in turn antagonize TTP. Both the PI3-K and p38 MAPK pathways stabilize IL-3 mRNA by the second mechanism. Evidence for the existence of the first mechanism has not been obtained so far, but a TPA-induced stabilizing pathway targeting TTP remains a possibility. An important task will be to study whether the postulated activation of HuR involves phosphorylation and, if so, how phosphorylation affects its function in ARE-mediated mRNA turnover control.
FIG. 6.
Integrated model of ARE-mediated mRNA turnover control. IL-3 mRNA turnover is under the control of both stabilizing (HuR) and destabilizing (TTP) AUBPs. Under resting conditions, the destabilizing function of TTP is dominant and responsible for constitutive rapid decay of the transcripts. Upon stimulation, independent signaling pathways become activated and lead either to activation of the stabilizing AUBPs or inactivation of destabilizing AUBPs, as a result of which degradation of the transcripts is blocked. The question marks indicate that the element is hypothetical.
ACKNOWLEDGMENTS
We thank L. Brennan and A. Wyss for their comments on the manuscript, A.-B. Shyu for the NIH 3T3 B2A2 cell line and pTet-myc-over-HuR plasmid, D. A. Cantrell for the rCD2-p110 plasmid, and K. R. Chien for the pcDNA-MKK7D plasmid. We also thank S. Degen for constructing the TTP mutant, B. Gross for performing FACS analysis, and M. Colombi for valuable help in preparing the figures.
This work was supported by grant 31-40816.94 of the Schweizerische Nationalfonds to C.M.
REFERENCES
- 1.Banholzer R, Nair A P K, Hirsch H H, Ming X-F, Moroni C. Rapamycin destabilizes IL-3 mRNA in autocrine tumor cells by a mechanism requiring an intact 3′-untranslated region. Mol Cell Biol. 1997;17:3254–3260. doi: 10.1128/mcb.17.6.3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carballo E, Lai W S, Blackshear P J. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 3.Chen C-Y, Gatto-Konczak F D, Wu Z, Karin M. Stabilization of interleukin-2 mRNA by the c-jun NH2-terminal kinase pathway. Science. 1998;280:1945–1949. doi: 10.1126/science.280.5371.1945. [DOI] [PubMed] [Google Scholar]
- 4.Chen C-Y A, Shyu A-B. AU-rich elements: characterization and importance in mRNA degradation. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 5.Chen C-Y A, Shyu A-B. Selective degradation of early-response-gene mRNAs: functional analyses of sequence features of the AU-rich elements. Mol Cell Biol. 1994;14:8471–8482. doi: 10.1128/mcb.14.12.8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C-Y A, Xu N, Shyu A-B. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derijard B, Raingeaud J, Barrett T, Wu I H, Han J, Ulevitch R J, Davis R J. Independent human MAP-kinase signal transduction pathways defined by MEK and MKK isoforms. Science. 1995;267:682–685. doi: 10.1126/science.7839144. . (Erratum, 269:17, 1995.) [DOI] [PubMed] [Google Scholar]
- 8.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorospe M, Wang X, Holbrook N J. p53-dependent elevation of p21Waf1 expression by UV light is mediated through mRNA stabilization and involves a vanadate-sensitive regulatory system. Mol Cell Biol. 1998;18:1400–1407. doi: 10.1128/mcb.18.3.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gough N M. Rapid quantitative preparation of cytoplasmic RNA from small numbers of cells. Anal Biochem. 1988;173:93–95. doi: 10.1016/0003-2697(88)90164-9. [DOI] [PubMed] [Google Scholar]
- 11.Hahn S, Moroni C. Modulation of cytokine expression in PB-3c mastocytes by IBMX and PMA. Lymphokine Cytokine Res. 1994;13:247–252. [PubMed] [Google Scholar]
- 12.Hibi M, Lin A, Smeal T, Minden A, Karin M. Identification of an oncoprotein- and UV-responsive protein kinase that binds and potentiates the c-Jun activation domain. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 13.Hirsch H H, Nair A P K, Backenstoss V, Moroni C. Interleukin-3 mRNA stabilization by a trans-acting mechanism in autocrine tumors lacking interleukin-3 gene rearrangements. J Biol Chem. 1995;270:20629–20635. doi: 10.1074/jbc.270.35.20629. [DOI] [PubMed] [Google Scholar]
- 14.Ishizuka T, Terada N, Gerwins P, Hamelmann E, Oshiba A, Fanger G R, Johnson G L, Gelfand E W. Mast cell tumor necrosis factor a production is regulated by MEK kinases. Proc Natl Acad Sci USA. 1997;94:6358–6363. doi: 10.1073/pnas.94.12.6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jain R G, Andrews L G, McGowan K M, Pekala P H, Keene J D. Ectopic expression of Hel-N1, an RNA-binding protein, increases glucose transporter (GLUT1) expression in 3T3–L1 adipocytes. Mol Cell Biol. 1997;17:954–962. doi: 10.1128/mcb.17.2.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai W S, Carballo E, Strum J R, Kennington E A, Phillips R S, Blackshear P J. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laroia G, Cuesta R, Brewer G, Schneider R J. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 18.Lee J C, Laydon J T, McDonnell P C, Gallagher T F, Kumar S, Green D, McNulty D, Blumenthal M J, Heys J R, Landvatter S W, Strickler J E, McLaughlin M M, Siemens I R, Fisher S M, Livi G P, White J R, Adams J L, Young P R. A protein kinase involved in the regulation of inflammatory cytokine biosynthesis. Nature. 1994;372:739–746. doi: 10.1038/372739a0. [DOI] [PubMed] [Google Scholar]
- 19.Levy N S, Chung S, Furneaux H, Levy A P. Hypoxic stabilization of vascular endothelial growth factor mRNA by the RNA-binding protein HuR. J Biol Chem. 1998;273:6417–6423. doi: 10.1074/jbc.273.11.6417. [DOI] [PubMed] [Google Scholar]
- 20.Lieberman A P, Pitha P M, Shin M L. Poly(A) removal is the kinase-regulated step in tumor necrosis factor mRNA decay. J Biol Chem. 1992;267:2123–2126. [PubMed] [Google Scholar]
- 21.Lindsten T, June C H, Ledbetter J A, Stella G, Thompson G B. Regulation of lymphokine messenger RNA stability by a surface-mediated T Cell activation pathway. Science. 1989;244:339–343. doi: 10.1126/science.2540528. [DOI] [PubMed] [Google Scholar]
- 22.Loflin P, Chen C Y, Shyu A B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Logan S K, Falasca M, Hu P, Schlessinger J. Phosphatidylinositol 3-kinase mediated epidermal growth factor-induced activation of the c-jun N-terminal kinase signaling pathway. Mol Cell Biol. 1997;17:5784–5790. doi: 10.1128/mcb.17.10.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ming X-F, Kaiser M, Moroni C. c-jun N-terminal kinase is involved in AUUUA-mediated interleukin-3 mRNA turnover in mast cells. EMBO J. 1998;17:6039–6048. doi: 10.1093/emboj/17.20.6039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moriguchi T, Toyoshima F, Masuyama N, Hanafusa H, Gotoh Y, Nishida E. A novel SAPK/JNK kinase, MKK7, stimulated by TNFα and cellular stresses. EMBO J. 1997;16:7045–7053. doi: 10.1093/emboj/16.23.7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nair A P K, Hahn S, Banholzer R, Hirsch H H, Moroni C. Cyclosporin A inhibits growth of autocrine tumour cell lines by destabilizing interleukin-3 mRNA. Nature. 1994;369:239–242. doi: 10.1038/369239a0. [DOI] [PubMed] [Google Scholar]
- 27.Peng S S, Chen C Y, Xu N, Shyu A B. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reif K, Lucas S, Cantrell D. A negative role for phosphoinositide 3-kinase in T-cell antigen receptor function. Curr Biol. 1997;7:285–293. doi: 10.1016/s0960-9822(06)00151-5. [DOI] [PubMed] [Google Scholar]
- 29.Reif K, Nobes C D, Thomas G, Hall A, Cantrell D A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- 30.Shaw G, Kamen R. A conserved AU sequence from the 3′untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell. 1986;46:659–667. doi: 10.1016/0092-8674(86)90341-7. [DOI] [PubMed] [Google Scholar]
- 31.Shyu A-B, Belasco J G, Greenberg M E. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 32.Shyu A-B, Greenberg M E, Belasco J G. The c-fos transcript is targeted for rapid decay by two distinct mRNA degradation pathways. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 33.Sirenko O I, Lofquist A K, DeMaria C T, Morris J S, Brewer G, Haskill J S. Adhesion-dependent regulation of an A + U-rich element-binding activity associated with AUF1. Mol Cell Biol. 1997;17:3898–3906. doi: 10.1128/mcb.17.7.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stein B, Brady H, Yang M X, Young D B, Barbosa M S. Cloning and characterization of MEK6, a novel member of the mitogen-activated protein kinase kinase cascade. J Biol Chem. 1996;271:11427–11433. doi: 10.1074/jbc.271.19.11427. [DOI] [PubMed] [Google Scholar]
- 35.Stoecklin G, Hahn S, Moroni C. Functional hierarchy of AUUUA motifs in mediating rapid interleukin-3 mRNA decay. J Biol Chem. 1994;269:28591–28597. [PubMed] [Google Scholar]
- 36.Stoecklin G, Ming X-F, Looser R, Moroni C. Somatic mRNA turnover mutants implicate tristetraprolin in the interleukin-3 mRNA degradation pathway. Mol Cell Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thorens B, Mermod J J, Vassalli P. Phagocytosis and inflammatory stimuli induce GM-CSF mRNA in macrophages through posttranscriptional regulation. Cell. 1987;48:671–679. doi: 10.1016/0092-8674(87)90245-5. [DOI] [PubMed] [Google Scholar]
- 38.Wang W, Caldwell M C, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. EMBO J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang W, Furneaux H, Cheng H, Caldwell M C, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Su B, Sah V P, Brown J H, Han J, Chien K R. Cardiac hypertrophy induced by mitogen-activated protein kinase kinase 7, a specific activator for c-Jun NH2-terminal kinase in ventricular muscle cells. J Biol Chem. 1998;273:5423–5426. doi: 10.1074/jbc.273.10.5423. [DOI] [PubMed] [Google Scholar]
- 41.Wilson G M, Brewer G. The search for trans-acting factors controlling messenger RNA decay. Prog Nucleic Acid Res Mol Biol. 1999;62:257–291. doi: 10.1016/s0079-6603(08)60510-3. [DOI] [PubMed] [Google Scholar]
- 42.Wilson T, Treismann R. Removal of poly(A) and consequent degradation of c-fos mRNA facilitated by 3′ AU-rich sequences. Nature. 1988;336:396–399. doi: 10.1038/336396a0. [DOI] [PubMed] [Google Scholar]
- 43.Winzen R, Kracht M, Ritter B, Wilhelm A, Chen C Y, Shyu A B, Muller M, Gaestel M, Resch K, Holtmann H. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 1999;18:4969–4980. doi: 10.1093/emboj/18.18.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wodnar-Filipowicz A, Heusser C, Moroni C. Production of the haemopoietic growth factors GM-CSF and interleukin-3 by mast cells in response to IgE receptor mediated activation. Nature. 1989;339:150–152. doi: 10.1038/339150a0. [DOI] [PubMed] [Google Scholar]
- 45.Wodnar-Filipowicz A, Moroni C. Regulation of interleukin 3 mRNA expression in mast cells occurs at the posttranscriptional level and is mediated by calcium ions. Proc Natl Acad Sci USA. 1990;87:777–781. doi: 10.1073/pnas.87.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu N, Loflin P, Chen C Y, Shyu A B. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 1998;26:558–565. doi: 10.1093/nar/26.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang L, Steussy C N, Fuhrer D K, Hamilton J, Yang Y-C. Interleukin-11 mRNA stabilization in phorbol ester-stimulated primate bone marrow stromal cells. Mol Cell Biol. 1996;16:3300–3307. doi: 10.1128/mcb.16.7.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]