Abstract
Sip4 is a Zn2Cys6 transcriptional activator that binds to the carbon source-responsive elements of gluconeogenic genes in Saccharomyces cerevisiae. The Snf1 protein kinase interacts with Sip4 and regulates its phosphorylation and activator function in response to glucose limitation; however, evidence suggested that another kinase also regulates Sip4. Here we examine the role of the Srb10 kinase, a component of the RNA polymerase II holoenzyme that has been primarily implicated in transcriptional repression but also positively regulates Gal4. We show that Srb10 is required for phosphorylation of Sip4 during growth in nonfermentable carbon sources and that the catalytic activity of Srb10 stimulates the ability of LexA-Sip4 to activate transcription of a reporter. Srb10 and Sip4 coimmunoprecipitate from cell extracts and interact in two-hybrid assays, suggesting that Srb10 regulates Sip4 directly. We also present evidence that the Srb10 and Snf1 kinases interact with different regions of Sip4. These findings support the view that the Srb10 kinase not only plays negative roles in transcriptional control but also has broad positive roles during growth in carbon sources other than glucose.
The transcriptional activator Sip4 of Saccharomyces cerevisiae belongs to a family of activators with a Zn2Cys6 binuclear cluster DNA-binding domain, which includes Gal4, Hap1, Leu3, Put3, and others (20, 22). Sip4 was identified by its two-hybrid interaction with the Snf1 protein kinase of the glucose signaling pathway (17, 37). Sip4 binds to the carbon source-responsive elements (CSRE) (28) in the promoters of gluconeogenic genes (34) and also has an inhibitory effect on glucose depletion-dependent invasive growth (6). Both the expression and the function of Sip4 are regulated in response to glucose. Transcription of SIP4 is repressed by glucose (17, 34), and RNA levels increase during the diauxic shift and sporulation (3, 8).
The physical interaction of Sip4 with the Snf1 protein kinase reflects a role of Snf1 in regulating Sip4 function. In response to glucose limitation, Sip4 is rapidly phosphorylated and its ability to activate transcription is rapidly upregulated; both processes depend on Snf1 kinase activity (17). Biochemical and genetic evidence indicates that a specific β subunit of the Snf1 kinase, Gal83, mediates the physical and functional interaction of the kinase with Sip4 (33). These findings show that Snf1 modulates the activity of Sip4 in response to glucose, but it has not been demonstrated that Sip4 is a direct target of Snf1. Moreover, both in vitro and in vivo studies indicate that another kinase besides Snf1 contributes to phosphorylation of Sip4 (33).
Several lines of evidence suggested the Srb10 (Ssn3, Ume5) kinase as a candidate. Srb10 and its cyclin, Srb11 (Ssn8), are associated with the RNA polymerase II holoenzyme, as are their mammalian homologs, cyclin-dependent kinase 8 (cdk8) and cyclin C (15, 18, 19). This kinase phosphorylates the carboxy-terminal domain (CTD) of the polymerase, thereby inhibiting transcription (10). Genetic studies implicated Srb10 in transcriptional repression of genes that are regulated by glucose repression, meiotic development, mating type, and heat shock (1, 4, 13, 15, 30, 32, 36; for a review, see reference 2). A negative role for Srb10 in response to carbon source availability was further supported by microarray analysis: in glucose-grown cells, Srb10 negatively regulates 173 genes, including 75 that are induced during the diauxic shift (12). Moreover, Srb10 protein levels are depleted during the diauxic shift (12), and Srb11 levels are reduced during growth on nonfermentable carbon sources (5).
Studies of GAL gene regulation, however, indicate that the regulatory roles of Srb10 in response to a carbon source are not limited to repression. Srb10 also has a positive role in induction of the GAL genes (1, 11, 15, 18), and Srb10 has been shown to regulate the activator Gal4. Srb10 phosphorylates Gal4 on Ser699, and this phosphorylation is required for full induction of transcription (11, 23, 27). The activity of Srb10 does not appear to be regulated by galactose, and Sadowski and colleagues have proposed that Srb10 communicates signals regarding the physiological state of the cell to Gal4 during its interaction with the RNA polymerase II holoenzyme.
These findings that Srb10 phosphorylates an activator in the zinc cluster family, together with evidence that Snf1 interacts with Srb10 (14), prompted us to investigate the relationship between Srb10 and Sip4. We show that phosphorylation of Sip4 in vivo depends on the Srb10 kinase and that Srb10 stimulates activation of a reporter by LexA-Sip4. We further show that Srb10 interacts with Sip4 in the two-hybrid system and that the two proteins coimmunoprecipitate from cell extracts. We also examine the relationship of Srb10 and Snf1 with respect to their interactions with Sip4. Our findings, together with previous work on Gal4, support the view that the Srb10 kinase not only plays negative roles in transcriptional control but also has broad positive roles during growth in carbon sources other than glucose.
MATERIALS AND METHODS
Strains and genetic methods.
S. cerevisiae strains used are listed in Table 1. Transformation and other genetic manipulations were done by standard procedures (24). Cultures were grown in synthetic complete (SC) medium lacking appropriate supplements to select for plasmids.
TABLE 1.
S. cerevisiae strains
| Strain | Genotypea |
|---|---|
| MCY3605b | MATaura3-52 his3Δ200 leu2-3,112 |
| MCY3634b | MATasrb10Δ::HIS3 ura3-52 his3Δ200 leu2-3,112 |
| MCY3694b | MATα srb10-D290A ura3-52 his3Δ200 leu2Δ1 trp1Δ63 |
| CTY10-5d | MATagal4 gal80 URA3::lexAop-lacZ his3Δ200 leu2Δ1 ade2-101 trp1-901 |
| MCY3691 | CTY10-5d srb10Δ::HIS3 |
| MCY4024 | CTY10-5d gal83Δ::TRP1 |
| W303-1A | MATatrp1-1 ura3-1 his3-11,15 leu2-3,112 ade2-1 can1-100 |
srb10Δ::HIS3 is the allele ssn3Δ1::HIS3 (15).
Strain has the S288C genetic background.
Plasmids.
To construct pSK37, we digested pACTII (16) with HindIII to remove the GAD sequence and inserted a BamHI linker. The BamHI-EcoRV fragment of pSK37 was then replaced with that of pACTII to restore the ADH1 terminator, yielding pSK134. pSK135 and pSK136 contain the BglII-SalI fragment from pSK84 (14) and pSK85, respectively, cloned into pSK134 and express the triple hemagglutin (HA) epitope-tagged proteins HA-Srb10 and HA-Srb10D290A from the ADH1 promoter. pSK85 contains the BamHI fragment of pSK74 (13) subcloned into pWS93 (29). pSK45 expresses Srb10 from the vector pSK37. pOV8 and pOV21 express LexA-Snf1 and LexA-Sip4, respectively, from the ADH1 promoter of vector pBTM116 (gift of Stan Fields, University of Washington, Seattle). pPL50 and pPL54 express LexA-Sip4 and LexA-Sip4(1–690) from the ADH1 promoter of vector pLexA(1–202)+PL (26). pSK33, pOV48, and pRJ55 express LexA-Srb10, LexA-Cat8(1–1203), and LexA-Snf1, respectively, from pLexA(1–202)+PL. LexA-Srb10 has kinase function. pPL69 expresses full-length GAD-Sip4 (17), and other plasmids expressing partial GAD-Sip4 fusions are also derivatives of pACTII. pOV47 is derived from pACTII and expresses GAD-Cat8, which functions for activation of a CSRE-lacZ reporter and also complements cat8 for growth on ethanol.
β-Galactosidase assays.
Transformants were patched onto selective SC medium plus 2% glucose and grown for 1 or 2 days, and filter lift assays for blue color were performed as described previously (37). For quantitative assays, cells were grown to exponential phase in selective SC medium containing 2% glucose unless otherwise noted. β-Galactosidase activity was assayed in permeabilized cells and expressed in Miller units.
Coimmunoprecipitation assays and immunoblot analysis.
Preparation of protein extracts and immunoprecipitation were carried out as described previously (33). Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were analyzed by immunoblotting with monoclonal anti-LexA (Clontech), polyclonal anti-LexA (gift of C. Denis, University of New Hampshire, New Hampshire, Conn.), or monoclonal anti-HA antibody 12CA5. Antibodies were detected by enhanced chemiluminescence with ECL Plus reagents (Amersham). Extracts were also prepared by boiling cells and vortexing with glass beads as described previously (33), and proteins were analyzed by immunoblotting as described above.
RESULTS
Phosphorylation of Sip4 requires the Srb10 kinase.
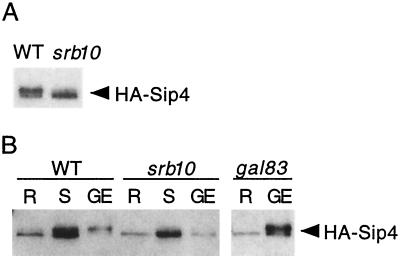
We tested the role of Srb10 in the phosphorylation of Sip4 in two different S. cerevisiae strain backgrounds. First we examined HA-Sip4, expressed from its own promoter, in wild-type and srb10Δ mutant cells of the S288C genetic background. Protein extracts were prepared from cells grown in glycerol plus ethanol, separated by SDS-PAGE, and immunoblotted with anti-HA antibody (Fig. 1A). In the wild type, HA-Sip4 migrated as a doublet, and the species previously shown to correspond to phosphorylated HA-Sip4 (17) was prominent. In contrast, in the srb10Δ mutant the major species comigrated with unphosphorylated HA-Sip4, although more slowly migrating material was faintly detectable above the major band. We next examined HA-Sip4 in wild-type CTY10-5d and an isogenic srb10Δ mutant (Fig. 1B). Cells were grown to mid-log phase in glucose and then shifted to glycerol plus ethanol for 4 h. After the shift, phosphorylated HA-Sip4 was easily detected in the wild type, but very little was present in the mutant. Cultures were also grown in glycerol plus ethanol with results similar to those described above (only 40% as much protein was loaded for this growth condition).
FIG. 1.
Srb10 is required for phosphorylation of Sip4 in vivo. (A) Strains used were MCY3605 (wild type [WT]) and MCY3634 (srb10Δ) expressing HA-Sip4 from its native promoter on pPL76 (the same as pHA-Sip4 [17]). Cultures were grown in 2% glycerol plus 3% ethanol. Extracts were prepared by the boiling method, and proteins were separated by SDS–6% PAGE, immunoblotted, and detected with anti-HA. (B) Strains were CTY10-5d (WT) and its derivatives, MCY3691 (srb10Δ) and MCY4024 (gal83Δ), expressing HA-Sip4 from pOV64, a LEU2-marked derivative of pPL76 (33). Cultures were grown to mid-log phase in 2% glucose (R, glucose-repressed) and shifted (S) to 2% glycerol plus 3% ethanol for 4 h, or cells were grown in 2% glycerol plus 3% ethanol (GE). For the cells grown in GE, only 40% as much protein was loaded for the WT and srb10Δ samples, whereas the full amount was loaded for the gal83Δ sample. Immunoblot analysis was done as described above. Similar results were obtained with a second set of srb10Δ and gal83Δ transformants.
We also included in this experiment a gal83Δ mutant derivative of CTY10-5d. The Gal83 β subunit of the Snf1 kinase mediates both physical and functional interaction with Sip4 (33); moreover, Gal83 is the only β subunit that is localized to the nucleus and Sip4 is a nuclear protein (35). Previously we showed that a gal83Δ mutant exhibits no change in mobility of HA-Sip4 within 8 h of a shift from glucose to ethanol (33). Here we found that HA-Sip4 became modified after long-term growth in glycerol plus ethanol (Fig. 1B). These findings suggest that the Srb10-dependent modification of Sip4 is delayed, but not abolished, in the absence of Gal83-mediated Snf1 kinase activity. The relevant Gal83-mediated phosphorylation event may not occur at all in the gal83Δ mutant; alternatively, Snf1 containing one of the other β subunits, or no β subunit, may function inefficiently such that phosphorylation is delayed after a shift but is achieved during long-term growth conditions. These possibilities cannot be easily distinguished because a snf1 mutant cannot grow on nonfermentable carbon sources.
Thus, both Snf1 and Srb10 kinase activity is required for rapid modification of HA-Sip4 after a shift to a nonfermentable carbon source, while Srb10 remains important during long-term growth in glycerol plus ethanol.
Transcriptional activation by LexA-Sip4 is reduced in an srb10 mutant.
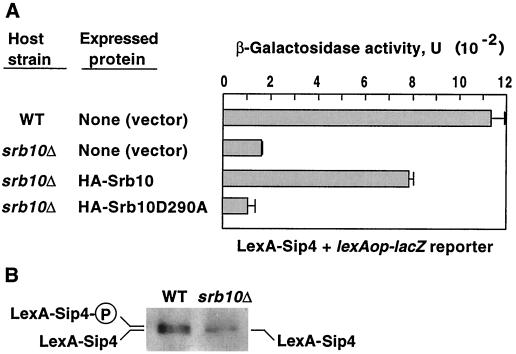
Previous studies showed that the phosphorylation of Sip4 correlates with upregulation of its ability to activate transcription (17, 33). To test the role of Srb10 in transcriptional activation by Sip4, we assayed the ability of LexA-Sip4 to activate a lacZ reporter with LexA sites in wild-type CTY10-5d and its srb10Δ derivative (Fig. 2A). Transformants expressing LexA-Sip4 were grown in glycerol plus ethanol and assayed for β-galactosidase activity. Activity was sevenfold higher in wild-type cells than in srb10Δ cells (Fig. 2A). Immunoblot analysis showed that levels of unphosphorylated LexA-Sip4 were comparable, but no phosphorylated LexA-Sip4 was detected in the srb10Δ cells (Fig. 2B), consistent with the view that the phosphorylated species is responsible for most of the activation. In parallel, srb10Δ transformants expressing LexA-Sip4 and either HA-Srb10 or the kinase-dead mutant HA-Srb10D290A (13) were assayed, and activity was similarly 7.3-fold higher in cells expressing the active Srb10 than in those expressing the mutant kinase (Fig. 2A). Thus, the Srb10 catalytic activity stimulates transcriptional activation by LexA-Sip4. Evidence that an srb10Δ mutation does not impair the function of every activator (14) supports the specificity of this interaction.
FIG. 2.
Srb10 affects transcriptional activation by LexA-Sip4. Strains were wild-type (WT) CTY10-5d and its srb10Δ derivative MCY3691 expressing LexA-Sip4 from pOV21. Strains also expressed HA-Srb10 or the kinase-dead mutant HA-Srb10D290A, as indicated, from pSK135 or pSK136 or carried the vector pSK134 so that values could be compared. Cells were grown in SC-Trp-Leu containing 2% glycerol plus 2% ethanol into early stationary phase and then diluted in fresh medium to an optical density at 600 nm of 0.2 and allowed to grow for 24 h. (A) Values for β-galactosidase activity are averages for three transformants. Error bars are shown. (B) Protein extracts were prepared from transformants expressing LexA-Sip4 and vector pSK134 by the boiling method. Proteins were separated by SDS–8% PAGE, immunoblotted, and probed with anti-LexA. Lines indicate phosphorylated (P) and unphosphorylated species. Immunoblot analysis of transformants expressing LexA-Sip4 and HA-Srb10 or HA-Srb10D290A with both anti-LexA and anti-HA confirmed the expression of both tagged proteins (data not shown). WT, wild type.
Sip4 coimmunoprecipitates with Srb10.
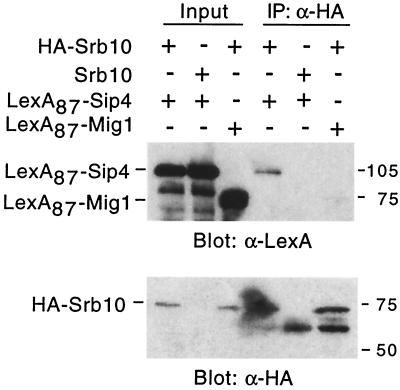
To assess the physical interaction of Sip4 and Srb10, we tested for their coimmunoprecipitation from cell extracts (Fig. 3). A wild-type strain was transformed with plasmids expressing a LexA DNA-binding domain fusion to Sip4 (LexA87-Sip4) and either HA-Srb10 or untagged Srb10. HA-Srb10 was immunoprecipitated with monoclonal HA antibody, and the precipitates were resolved by SDS-PAGE and subjected to immunoblot analysis with anti-LexA antibody. LexA87-Sip4 coprecipitated with HA-Srb10; only a fraction of the protein coprecipitated, consistent with the idea that Sip4 has a functional interaction with Srb10 but is not stably associated with the RNA polymerase II holoenzyme. In control experiments, LexA87-Sip4 did not coimmunoprecipitate with the untagged Srb10, and LexA87-Mig1 did not coprecipitate with HA-Srb10.
FIG. 3.
Coimmunoprecipitation of Srb10 and Sip4. Strain W303-1A expressed HA-Srb10, Srb10, LexA87-Sip4, and LexA87-Mig1, as indicated, from pSK135, pSK45, pPL49 (the same as pLexA-Sip4 [17]), and pLexA-Mig1 (31). Protein extracts were prepared from cells grown to mid-log phase in 2% glucose, and proteins (11 μg) were immunoprecipitated (IP) with monoclonal anti (α)-HA antibody. Both the input proteins (1.2 μg) and the precipitates were separated by SDS-PAGE and immunoblotted with α-LexA. Immunoprecipitation of HA-Srb10 was confirmed by immunoblot analysis with α-HA.
Srb10 interacts with Sip4 in the two-hybrid system.
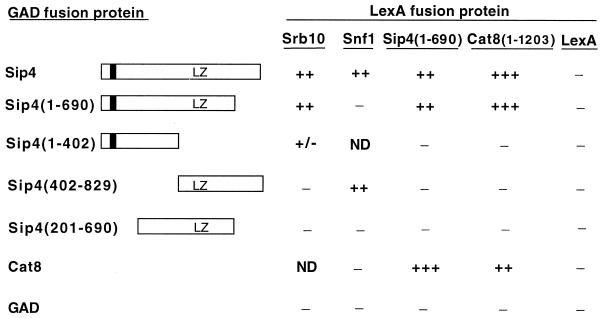
To confirm the interaction of Srb10 and Sip4 in vivo, we tested for two-hybrid interaction between LexA-Srb10 and a Gal4 activation domain (GAD) fusion to Sip4 (Fig. 4). A filter assay for β-galactosidase activity produced a strong blue color, and quantitative assays of glucose-grown cells showed high-level activity (76 U; average for four transformants). Activity in control transformants was low (0.3 U for LexA plus GAD-Sip4 and 1.6 U for LexA-Srb10 plus GAD). We were unable to detect interaction during growth in glycerol plus ethanol, but GAD-Sip4 levels were much lower than in glucose-grown cells (data not shown), probably because the ADH1 promoter is less active (7).
FIG. 4.
Two-hybrid interactions of Sip4, Srb10, and Cat8. Strain CTY10-5d was transformed with plasmids expressing the indicated GAD-Sip4 and LexA fusion proteins. LexA proteins were expressed from pSK33, pPL54, pOV48, pOV8, and pRJ55. The open bar represents Sip4 sequence (residues 1 to 829); the dark bar represents Zn2Cys6 zinc cluster (residues 45 to 76); LZ indicates a leucine zipper motif beginning at residue 503. Transformants were grown on medium containing 2% glucose, and two-hybrid interaction was monitored using the filter assay for blue color. Color is represented as follows: +++, strong blue; ++, moderate blue; +/−, very light blue; −, white. ND, not determined. LexA-Sip4(1–690) also interacted with GAD-Cat8(1–1203), and full-length LexA-Sip4 alone generated a light blue color but nonetheless clearly interacted with both GAD-Sip4 and GAD-Cat8 (residues 1 to 1433). The interaction of LexA-Snf1 with GAD-Sip4 and GAD-Sip4(402–829) has been reported previously (17, 33).
To determine the region of Sip4 required for this two-hybrid interaction, we tested a series of GAD fusions to different Sip4 sequences (Fig. 4), which were all expressed at similarly high levels as judged by immunoblots (two transformants tested for each; data not shown). Notable features of Sip4 include the N-terminal Zn2Cys6 binuclear cluster and a leucine zipper motif, comprising one isoleucine (residue 503) and four leucines. In filter assays for interaction with LexA-Srb10, GAD-Sip4(1–690) produced blue color of the same intensity as the full-length GAD-Sip4 containing all 829 residues. A much weaker blue color was detected with GAD-Sip4(1–402), and no blue color developed with GAD-Sip4(402–829) or GAD-Sip4(201–690).
Previous studies indicated that different Sip4 sequences are required for interaction with the Snf1 kinase. Residues 402 to 829 mediate the two-hybrid interaction of Sip4 with Snf1 (17), and these residues also suffice for interaction in vitro with the conserved ASC domain of the β subunit of Snf1 (33). In accord with these findings, a LexA fusion to the ASC sequence interacted with GAD-Sip4(402–829) in two-hybrid assays (data not shown), whereas no interaction was detected between LexA-Snf1 and GAD-Sip4(1–690) (Fig. 4). Consistent with the distinct sequence requirements for interaction of Sip4 with the Snf1 and Srb10 kinases, the interaction of GAD-Sip4 with LexA-Snf1 was as strong in an srb10Δ mutant as in the wild type (data not shown).
Sip4 interacts with itself and with Cat8.
Members of the zinc cluster transcriptional activator family have been shown to bind to DNA as dimers (22). Thus, it seemed likely that Sip4 forms homodimers and possible that Sip4 forms heterodimers with Cat8, another zinc cluster activator that binds to the CSRE and has a major role in the activation of gluconeogenic genes (9, 21, 25). We addressed this issue because of the further possibility that Srb10 has a role in dimerization.
To test two-hybrid interactions, we used the truncated proteins LexA-Sip4(1–690) and LexA-Cat8(1–1203), which do not activate transcription alone. In filter assays, interaction between Sip4 and Cat8 was easily detected (Fig. 4). Moreover, LexA-Sip4(1–690) interacted more strongly with full-length GAD-Cat8 (residues 1 to 1433) than with GAD-Sip4, and conversely, LexA-Cat8(1–1203) interacted more strongly with GAD-Sip4 than with GAD-Cat8.
Some members of the family contain dimerization domains located immediately C-terminal to the zinc cluster (22), which in Sip4 comprises residues 45 to 76. To assess the sequence requirements for dimerization, we tested different GAD-Sip4 fusions (Fig. 4). Both LexA-Sip4(1–690) and LexA-Cat8(1–1203) interacted with GAD-Sip4(1–690), but neither interacted with GAD-Sip4(1–402). LexA-Sip4(1–402) also did not interact with GAD-Sip4 or GAD-Cat8 (data not shown). The data suggest that the region of Sip4 containing the leucine zipper is necessary, but not sufficient, for dimerization; however, it is possible that truncation at residue 402 somehow impairs interaction.
The pattern of two-hybrid interactions between Srb10 and Sip4 sequences is consistent with a role for Srb10 in dimerization. To examine this possibility, we assayed β-galactosidase activity in transformants of CTY10-5d and its srb10Δ derivative that expressed LexA-Sip4(1–690) plus either GAD-Sip4 or GAD-Cat8. Values were the same within a factor of 2 in glucose-grown wild-type and mutant transformants (80 and 150 U, respectively, for GAD-Sip4, and 510 and 380 U, respectively, for GAD-Cat8; values are averages for three or four transformants, and immunoblot analysis showed similar levels of LexA-Sip4). In addition, no effect of srb10Δ was observed after growth of transformants in glycerol plus ethanol for 16 h (data not shown). Thus, no role of the Srb10 kinase was apparent, although it remains possible that Srb10 affects the dimerization of the native proteins.
Assays for phosphorylation of Sip4 by the Srb10 kinase.
The evidence that Srb10 interacts physically with Sip4 and is required for phosphorylation of Sip4 in vivo suggested that Sip4 is a direct target of the kinase. However, Srb10 could also affect the activity of another kinase towards Sip4. In an attempt to distinguish between these two possibilities, we used several assays to detect phosphorylation of Sip4 by Srb10 in vitro. Because Sip4 and Srb10 coprecipitate (see Fig. 3), we immunoprecipitated HA-Sip4 from an srb10Δ mutant expressing Srb10, kinase-dead Srb10D290A, or no Srb10 and incubated the immunoprecipitates with [γ-32P]ATP. In all cases HA-Sip4 was phosphorylated, indicating that another kinase also coprecipitated; this other kinase is not Snf1, because HA-Sip4 was similarly phosphorylated when precipitated from an snf1Δ srb10Δ mutant (data not shown). It is possible that phosphorylation by Srb10 was obscured by the other kinase. Three other assays showed no phosphorylation of Sip4 in vitro. In the first assay, we immunoprecipitated HA-Sip4(1–690), which does not coprecipitate detectable kinase activity, and added GST-Srb10 purified from a strain overexpressing Srb11. In the second assay, we immunoprecipitated HA-Srb10 or HA-Srb10D290A from an srb10 mutant that also overexpressed Lex87-Sip4 to assay phosphorylation of the coprecipitating Lex87-Sip4. Finally, we immunoprecipitated HA-Srb10 or HA-Srb10D290A from a strain overexpressing Srb11 and added bacterially expressed GST-Sip4(401–829), which contains a region of Sip4 that coprecipitates a kinase from yeast cell extracts (33). None of these assays provided any evidence for Srb10-dependent phosphorylation of Sip4 in vitro; however, these assays also do not exclude the possibility that Srb10 directly phosphorylates Sip4 in vivo.
DISCUSSION
We have examined the interaction, both functional and physical, of the Srb10 kinase with the transcriptional activator Sip4. We show that the Srb10 kinase activity is required for phosphorylation of Sip4 during growth on nonfermentable carbon sources and that the Srb10 catalytic activity stimulates transcriptional activation by LexA-Sip4. Two lines of evidence support the view that Sip4 and Srb10 interact physically in vivo: the two proteins coimmunoprecipitate from cell extracts, and they interact in two-hybrid assays. These findings strongly suggest that the regulatory effects of Srb10 are direct, but it remains unclear whether Srb10 phosphorylates Sip4 or regulates the activity of another associated kinase.
What is the functional significance of the Srb10-dependent phosphorylation of Sip4 during growth in nonfermentable carbon sources? The reduced phosphorylation of Sip4 in srb10 mutants correlates with the reduced ability of Sip4 to activate transcription in the mutant; similarly, the delay in phosphorylation of Sip4 in a gal83 mutant correlates with a delay in transcriptional activation (33). A simple model is that phosphorylation potentiates the activator function of Sip4, as is the case for Gal4 (11). Alternatively, it is possible that phosphorylation is a functionally neutral consequence of the interaction of Sip4 with the holoenzyme during activation; in that case, the reduced activation by LexA-Sip4 in the srb10Δ mutant could reflect compromised recruitment of the holoenzyme due to loss of the physical interaction between Sip4 and Srb10. We do not favor this explanation, because activation was also reduced in the presence of catalytically inactive Srb10 protein. Identification and mutation of specific phosphorylation sites will be required to address this issue.
The Srb10 kinase has been primarily implicated in transcriptional repression. Previous studies have documented not only specific negative regulatory effects on diverse genes (1, 2, 4, 13, 15, 30, 32, 36), including repression of 173 genes during growth in glucose (12), but also general inhibitory effects on the function of the RNA polymerase II holoenzyme (10). Until the present study, the only clear example of positive regulatory action by Srb10 concerned the GAL genes. Genetic evidence indicated that Srb10 contributes to transcriptional activation of GAL genes (1, 15, 18), and detailed molecular studies showed that full induction requires phosphorylation of the activator Gal4 by Srb10 (11, 23, 27). Our evidence for interaction of Srb10 with Sip4, an activator of gluconeogenic genes, supports the view that Srb10 has broad positive regulatory effects under conditions other than growth in glucose.
It is worth noting that protein levels of the Srb10 kinase subunits decrease during the diauxic shift (Srb10 [12]) and during growth on nonfermentable carbon sources (Srb11 [5]). These decreases are easily reconciled with roles of the kinase in glucose repression. However, decreased protein levels are not incompatible with roles in transcriptional activation under these conditions. It is even possible that physical interaction with Sip4 stabilizes Srb10 during growth under conditions where the Srb10 kinase is otherwise unstable, that is, Sip4 “reserves” a fraction of Srb10 for the transcription of gluconeogenic genes during growth in a gluconeogenic carbon source.
Sadowski and colleagues have proposed that the Srb10 kinase communicates signals regarding the physiological state of the cell to gene-specific activators during their interaction with the RNA polymerase II holoenzyme. Specifically, they proposed that the activity of Srb10 towards Gal4 is inhibited in the absence of a fermentable carbon source (11, 23). Our results are consistent with this general idea, with the proviso that the activity of the Srb10 kinase towards Sip4 must be inhibited by glucose or stimulated by nonfermentable carbon sources.
It is not yet clear whether Srb10 directly phosphorylates Sip4, as is the case for Gal4, or rather stimulates the activity of another kinase towards Sip4. Although Srb10 is required for phosphorylation of Sip4 during growth in nonfermentable carbon sources in vivo, we were unable to detect Srb10-dependent phosphorylation of Sip4 in various in vitro assays. However, during its physiological function as a transcriptional activator, Sip4 binds DNA at a site near a promoter and is thereby positioned near the Srb10 kinase, which is associated with the RNA polymerase II holoenzyme; perhaps DNA binding of Sip4 is a prerequisite for phosphorylation by Srb10. It is also possible that some other factor, missing from the in vitro assays, is required for phosphorylation of Sip4 by Srb10. Thus, these in vitro assays do not exclude the possibility that Srb10 directly phosphorylates Sip4 in vivo.
Whether or not Sip4 proves to be a direct target of Srb10, another kinase may be involved in regulating Sip4. Phosphorylation of Sip4 in vivo was greatly diminished, but not totally abolished, in an srb10 mutant, and another kinase besides Srb10 or Snf1 coprecipitated with Sip4 and phosphorylated it in vitro. Such another kinase may be regulated by Srb10 in vivo.
It is possible that the Srb10 and Snf1 kinases are functionally interconnected with respect to their effects on Sip4. Previous studies indicated that some fraction of the Snf1 protein in the cell is associated with Srb10 (14). The present findings that distinct Sip4 sequences are required for interaction with Srb10 and Snf1 suggest that the two kinases interact independently with Sip4, but the resulting physical proximity may facilitate functional interactions between these kinases. The Snf1 kinase activity is required for the rapid phosphorylation of Sip4 when cells are shifted to glucose-limiting conditions, but it may not be important for the phosphorylation of Sip4 during long-term growth on glycerol and ethanol. One interpretation of these results is that the Snf1 kinase activity facilitates the rapid Srb10-dependent modification of Sip4. It is possible that Snf1 directly phosphorylates Sip4 and thereby improves recognition by Srb10 or that Snf1 somehow stimulates the catalytic activity of Srb10 when both kinases are associated with Sip4.
The regulatory relationship of Srb10 and Sip4 may be even more complex, as evidence suggests that Srb10 not only positively regulates Sip4 function in nonfermentable carbon sources but also negatively regulates SIP4 gene expression in glucose. Transcription of SIP4 is repressed by glucose and increases during the diauxic shift (8, 17, 34), and Srb10 has a role in glucose repression of similarly regulated genes (no data were reported for SIP4) (12). We found that during growth in glucose, the srb10Δ mutation increases expression of both HA-Sip4 and a SIP4-lacZ promoter fusion (17) in strains of the S288C genetic background (O. Vincent, unpublished results). However, srb10Δ had no effect on HA-Sip4 levels in strain CTY10-5d (Fig. 1). These results show that Srb10 contributes to glucose repression of SIP4 in certain genetic backgrounds.
Finally, we found that Sip4 interacts with itself in two-hybrid assays and also interacts with Cat8, another zinc cluster activator that binds to the CSRE (21, 25). Genetic analysis has shown that Cat8 is the major activator for gluconeogenic genes; mutation of CAT8 has much more severe consequences than does mutation of SIP4, in part because expression of SIP4 requires Cat8 (9, 34). These results suggest that Sip4 may function primarily as a heterodimer with Cat8.
ACKNOWLEDGMENTS
We thank Pascale Lesage for plasmids.
This work was supported by Public Health Service grants GM34095 and GM47259 from the National Institutes of Health (NIH) to M.C. R.T. also received support from NIH training grant GM08224.
REFERENCES
- 1.Balciunas D, Ronne H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995;23:4421–4425. doi: 10.1093/nar/23.21.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson M. Genetics of transcriptional regulation in yeast: connection to the RNA polymerase II CTD. Annu Rev Cell Dev Biol. 1997;13:1–23. doi: 10.1146/annurev.cellbio.13.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 4.Cooper K F, Mallory M J, Smith J B, Strich R. Stress and developmental regulation of the yeast C-type cyclin Ume3p (Srb11p/Ssn8p) EMBO J. 1997;16:4665–4675. doi: 10.1093/emboj/16.15.4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper K F, Mallory M J, Strich R. Oxidative stress-induced destruction of the yeast C-type cyclin Ume3p requires phosphatidylinositol-specific phospholipase C and the 26S proteasome. Mol Cell Biol. 1999;19:3338–3348. doi: 10.1128/mcb.19.5.3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen P J, Sprague G F., Jr Glucose depletion causes haploid invasive growth in yeast. Proc Natl Acad Sci USA. 2000;97:13619–13624. doi: 10.1073/pnas.240345197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denis C L, Ferguson J, Young E T. mRNA levels for the fermentative alcohol dehydrogenase of Saccharomyces cerevisiae decrease upon growth on a nonfermentable carbon source. J Biol Chem. 1983;258:1165–1171. [PubMed] [Google Scholar]
- 8.DeRisi J L, Iyer V R, Brown P O. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 9.Hedges D, Proft M, Entian K-D. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengartner C J, Myer V E, Liao S-M, Wilson C J, Koh S S, Young R A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol Cell. 1998;2:43–53. doi: 10.1016/s1097-2765(00)80112-4. [DOI] [PubMed] [Google Scholar]
- 11.Hirst M, Kobor M S, Kuriakose N, Greenblatt J, Sadowski I. GAL4 is regulated by the RNA polymerase II holoenzyme-associated cyclin-dependent protein kinase SRB10/CDK8. Mol Cell. 1999;3:673–678. doi: 10.1016/s1097-2765(00)80360-3. [DOI] [PubMed] [Google Scholar]
- 12.Holstege F C P, Jennings E G, Wyrick J J, Lee T I, Hengartner C J, Green M R, Golub T R, Lander E S, Young R A. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 13.Kuchin S, Carlson M. Functional relationships of Srb10-Srb11 kinase, carboxy-terminal domain kinase CTDK-I, and transcriptional corepressor Ssn6-Tup1. Mol Cell Biol. 1998;18:1163–1171. doi: 10.1128/mcb.18.3.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchin S, Treich I, Carlson M. A regulatory shortcut between the Snf1 protein kinase and RNA polymerase II holoenzyme. Proc Natl Acad Sci USA. 2000;97:7916–7920. doi: 10.1073/pnas.140109897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuchin S, Yeghiayan P, Carlson M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc Natl Acad Sci USA. 1995;92:4006–4010. doi: 10.1073/pnas.92.9.4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Legrain P, Dokhelar M-C, Transy C. Detection of protein-protein interactions using different vectors in the two-hybrid system. Nucleic Acids Res. 1994;22:3241–3242. doi: 10.1093/nar/22.15.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lesage P, Yang X, Carlson M. Yeast SNF1 protein kinase interacts with SIP4, a C6 zinc cluster transcriptional activator: a new role for SNF1 in the glucose response. Mol Cell Biol. 1996;16:1921–1928. doi: 10.1128/mcb.16.5.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liao S-M, Zhang J, Jeffery D A, Koleske A J, Thompson C M, Chao D M, Viljoen M, van Vuuren H J J, Young R A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature. 1995;374:193–196. doi: 10.1038/374193a0. [DOI] [PubMed] [Google Scholar]
- 19.Maldonado E, Shiekhattar R, Sheldon M, Cho H, Drapkin R, Rickert P, Lees E, Anderson C W, Linn S, Reinberg D. A human RNA polymerase II complex associated with SRB and DNA-repair proteins. Nature. 1996;381:86–89. doi: 10.1038/381086a0. [DOI] [PubMed] [Google Scholar]
- 20.Pan T, Coleman J E. GAL4 transcription factor is not a “zinc finger” but forms a Zn(II) binding site with the DNA-binding domain of the GAL4 transcription factor. Proc Natl Acad Sci USA. 1989;86:3145–3149. doi: 10.1073/pnas.86.9.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahner A, Hiesinger M, Schuller H. Deregulation of gluconeogenic structural genes by variants of the transcriptional activator Cat8p of the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;34:146–156. doi: 10.1046/j.1365-2958.1999.01588.x. [DOI] [PubMed] [Google Scholar]
- 22.Reece R J, Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 23.Rohde J R, Trinh J, Sadowski I. Multiple signals regulate GAL transcription in yeast. Mol Cell Biol. 2000;20:3880–3886. doi: 10.1128/mcb.20.11.3880-3886.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rose M D, Winston F, Hieter P. Methods in yeast genetics, a laboratory course manual. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 25.Roth S, Schuller H-J. Cat8 and Sip4 mediate regulated transcriptional activation of the yeast malate dehydrogenase gene MDH2 by three carbon source-responsive promoter elements. Yeast. 2001;18:151–162. doi: 10.1002/1097-0061(20010130)18:2<151::AID-YEA662>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 26.Ruden D M, Ma J, Li Y, Wood K, Ptashne M. Generating yeast transcriptional activators containing no yeast protein sequences. Nature. 1991;350:250–252. doi: 10.1038/350250a0. [DOI] [PubMed] [Google Scholar]
- 27.Sadowski I, Costa C, Dhanawansa R. Phosphorylation of Gal4p at a single C-terminal residue is necessary for galactose-inducible transcription. Mol Cell Biol. 1996;16:4879–4887. doi: 10.1128/mcb.16.9.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schöler A, Schüller H-J. A carbon source-responsive promoter element necessary for activation of the isocitrate lyase gene ICL1 is common to genes of the gluconeogenic pathway in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3613–3622. doi: 10.1128/mcb.14.6.3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song W, Carlson M. Srb/mediator proteins interact functionally and physically with transcriptional repressor Sfl1. EMBO J. 1998;17:5757–5765. doi: 10.1093/emboj/17.19.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strich R, Slater M R, Esposito R E. Identification of negative regulatory genes that govern the expression of early meiotic genes in yeast. Proc Natl Acad Sci USA. 1989;86:10018–10022. doi: 10.1073/pnas.86.24.10018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vallier L G, Carlson M. Synergistic release from glucose repression by mig1 and ssn mutations in Saccharomyces cerevisiae. Genetics. 1994;137:49–54. doi: 10.1093/genetics/137.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vincent O, Carlson M. Gal83 mediates the interaction of the Snf1 kinase complex with the transcription activator Sip4. EMBO J. 1999;18:6672–6681. doi: 10.1093/emboj/18.23.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vincent O, Carlson M. Sip4, a Snf1 kinase-dependent transcriptional activator, binds to the carbon source-responsive element of gluconeogenic genes. EMBO J. 1998;17:7002–7008. doi: 10.1093/emboj/17.23.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent O, Townley R, Kuchin S, Carlson M. Subcellular localization of the Snf1 kinase is regulated by specific β subunits and a novel glucose signaling mechanism. Genes Dev. 2001;15:1104–1114. doi: 10.1101/gad.879301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wahi M, Johnson A D. Identification of genes required for α2 repression in Saccharomyces cerevisiae. Genetics. 1995;140:79–90. doi: 10.1093/genetics/140.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang X, Hubbard E J A, Carlson M. A protein kinase substrate identified by the two-hybrid system. Science. 1992;257:680–682. doi: 10.1126/science.1496382. [DOI] [PubMed] [Google Scholar]