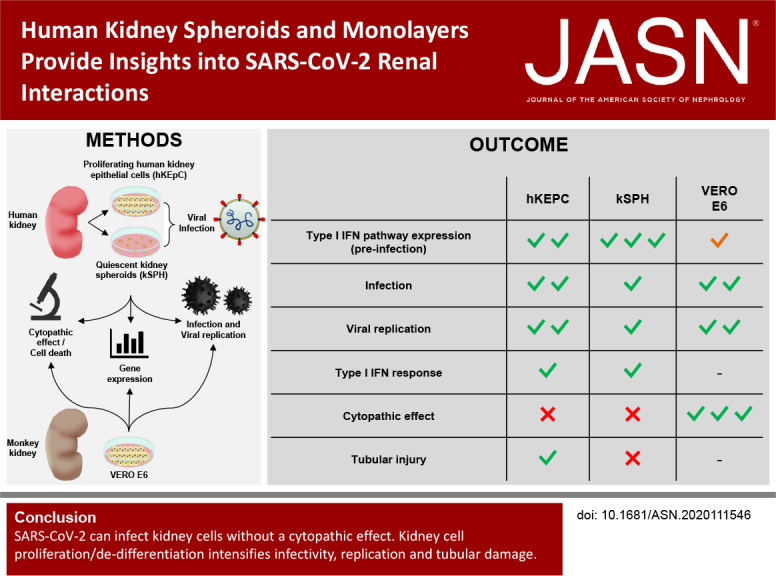
Significance Statement
Coronavirus disease 2019 (COVID-19) commonly results in AKI, but it is unknown whether AKI in patients with COVID-19 stems from direct kidney infection or indirect mechanisms. The authors explored the effects of infection with the causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), both in monolayers of primary human kidney cells and in kidney spheroids, a three-dimensional model recapitulating human kidneys in cell composition and function. They demonstrated that the virus effectively infects and replicates in human tubular epithelial cells, but does not cause cytopathic effects. They also provide molecular evidence that activation of a type 1 IFN response may be the underlying mechanism of resistance to SARS-CoV-2–related cytopathic damage. In all, the findings indicate minimal—if any—direct tubular damage by SARS-CoV-2.
Keywords: renal tubular epithelial cells, COVID-19, kidney spheroids, acute kidney injury, interferon pathway, SARS-CoV-2, cytopathic damage, kidney stem cells, renal progenitors, human tubular kidney cells
Visual Abstract
Abstract
Background
Although coronavirus disease 2019 (COVID-19) causes significan t morbidity, mainly from pulmonary involvement, extrapulmonary symptoms are also major componen ts of the disease. Kidney disease, usually presenting as AKI, is particularly severe among patients with COVID-19. It is unknown, however, whether such injury results from direct kidney infection with COVID-19’s causative virus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), or from indirect mechanisms.
Methods
Using ex vivo cell models, we sought to analyze SARS-CoV-2 interactions with kidney tubular cells and assess direct tubular injury. These models comprised primary human kidney epithelial cells (derived from nephrectomies) and grown as either proliferating monolayers or quiescent three-dimensional kidney spheroids.
Results
We demonstrated that viral entry molecules and high baseline levels of type 1 IFN–related molecules were present in monolayers and kidney spheroids. Although both models support viral infection and replication, they did not exhibit a cytopathic effect and cell death, outcomes that were strongly present in SARS-CoV-2–infected controls (African green monkey kidney clone E6 [Vero E6] cultures). A comparison of monolayer and spheroid cultures demonstrated higher infectivity and replication of SARS-CoV-2 in actively proliferating monolayers, although the spheroid cultures exhibited high er levels of ACE2. Monolayers exhibited elevation of some tubular injury molecules—including molecules related to fibrosis (COL1A1 and STAT6) and dedifferentiation (SNAI2)—and a loss of cell identity, evident by reduction in megalin (LRP2). The three-dimensional spheroids were less prone to such injury.
Conclusions
SARS-CoV-2 can infect kidney cells without a cytopathic effect. AKI-induced cellular proliferation may potentially intensify infectivity and tubular damage by SARS-CoV-2, suggesting that early intervention in AKI is warranted to help minimize kidney infection.
The recent pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which is responsible for coronavirus disease 2019 (COVID-19), has infected >110 million people and resulted in >2,500,000 deaths globally, as of July 18, 2020.1 Although its main manifestations are pulmonary (e.g., pneumonia and acute respiratory distress syndrome), other organs have been shown to be involved, one of the most clinically significant of which being the kidney.2–5 The spectrum of kidney involvement in COVID-19 is wide and includes AKI, hematuria, proteinuria, and metabolic acidosis.6 There is currently an ongoing debate surrounding the incidence and the mechanism, of kidney damage in COVID-19. In particular, it is unknown whether SARS-CoV-2–associated kidney disease results from direct kidney infection or from indirect mechanisms. The latter potentially include endothelial dysfunction with microvascular damage,7 cytokine storm,8 and other nonspecific etiologies (e.g., hypotension and sepsis) that characterize patients who are severely ill and mechanically ventilated. Indeed, both clinical and molecular studies surrounding COVID-19–related kidney injury reported highly discordant results. With regard to the clinical presentation, although early studies from China reported low (0.5%–15%) AKI incidence in patients who were hospitalized,4,9,10 later studies in the United States pointed toward a much higher incidence of 37%–57%.2,3 Similarly, attempts to uncover the mechanisms underlying renal involvement in COVID-19 yielded contradictory results. Supporting direct renal damage, several groups have identified alleged viral particles within renal tubules via electron microscopy11–13 and positive immunostaining for the virus. Very recently, SARS-CoV-2 was isolated from an autopsied kidney, and was shown to infect and replicate in nonhuman kidney cells.14 Moreover, human angiotensin-converting enzyme 2 (ACE2), the SARS-CoV-2 receptor,15 has been shown to be highly expressed in renal tubular cells,16,17 suggesting potential tropism for the kidney. In contrast, other groups showed that the antibody used to detect the virus was nonspecific,18 and called into question the nature of the electron microscopy–detected particles, claiming they are, in fact, also nonspecific.19,20 Accordingly, several studies either failed to detect viral RNA in the urine, or detected it in a very small number of patients.21–23 Hence, it is unknown whether SARS-CoV-2 can infect and replicate in kidney cells. More importantly, the mere presence of the virus in kidney cells does not equate to kidney damage, which is even more difficult to study in patients. The main hurdle toward answering these key questions is the lack of a human disease model that recapitulates the infective processes using human renal cells, and allows us to examine the viral effect on the kidney in a direct and prospective manner. Previously, we developed an in vitro, three-dimensional culture platform of human kidney epithelial cells, termed kidney spheroids (kSPH), which enables the maintenance of slowly proliferating renal epithelial cell identity that recapitulates the native adult kidney in terms of gene and surface marker expression.24,25 Herein, we queried the clinical data of the first 36 consecutive patients critically ill with COVID-19 arriving at Sheba Medical Center (SMC) and found that all patients who developed AKI also had a second condition that could explain AKI, apart from the SARS-CoV-2 infection itself. Then, to analyze the effects of SARS-CoV-2, we established adult kSPH and monolayers as reliable models for studying host-pathogen relationships in human coronavirus-229E (HCoV-229E) and SARS-CoV-2. Importantly, human adult kidney (hAK) cells harbor predetermined elevated levels of type 1 IFN molecules compared with Vero E6 kidney epithelial cells, which may account for relative protection. We go on to show that, although no cytopathic effect/cell death were noted whatsoever in human kidney cultures, proliferating cultures were more prone to infection and to exhibit upregulation of molecular markers of tubular injury. These data undermine the ability of Vero E6 kidney epithelial cells to serve as a reliable model of kidney–SARS-CoV-2 interactions. Ex vivo cell behavior may predict specific vulnerability of kidney tubular cells to SARS-CoV-2 after cell proliferation induced by acute tubular necrosis.
Methods
Study Design and Participants
We conducted a single-center, retrospective cohort study of patients who were critically ill and hospitalized in the SMC dedicated intensive care unit for patients with COVID-19 (coronavirus critical care unit; CCCU) during March to June 2020. This study was approved by the SMC Institutional Review Board (7230-20-SMC).
The diagnosis of COVID-19 was determined on the basis of a positive nasopharyngeal swab taken upon admission to the hospital. Positive specimens taken outside the hospital were confirmed through repeat testing before admission to the CCCU. Patients who were critically ill with COVID-19 were defined as those with respiratory failure necessitating ventilation, shock, or organ failure requiring treatment and surveillance in the intensive care unit.
Data Collection and Definitions
We used SMC’s computerized system to identify all patients admitted to the CCCU during the study period. Next, we searched the electronic medical records of these patients and extracted their baseline characteristics—including age at admission, sex, body mass index, and smoking status—and any significant comorbidity (e.g., diabetes mellitus, systemic hypertension, CKD, chronic lung disease, obesity [defined as a body mass index >30 kg/m2], cardiovascular disease, or active cancer). In addition, we extracted their continuous vital sign measurements, chronic medications, laboratory results, and medical order lists. We consolidated documented patients’ notes, daily follow-up examinations, and assessments to compile a comprehensive and thorough dataset. Specifically, we examined BP measurements, use of crystalloid fluids and vasopressors, hourly and daily urine output, and feeding status during the days previous to the diagnosis of AKI, where available.
AKI was defined according to the Kidney Disease Improving Global Outcomes clinical practice guidelines.26 Sepsis was defined retrospectively as a combination of positive blood culture (bacterial or fungal) and clinical finding suggestive of organ failure, as supported by the 2016 Society of Critical Care Medicine task force recommendations.27 Cardiovascular collapse was defined as shock secondary to pump failure (acute myocardial infarction, fulminant viral myocarditis, uncompensated heart failure, or massive pulmonary embolism). Hypertension was defined according to the 2017 American College of Cardiology/American Heart Association Hypertension Guideline.28 Hypotension was defined as systolic BP <90 mm Hg and/or diastolic BP <60 mm Hg. Rhabdomyolysis was defined as a creatine phosphokinase level gr eater than five times the upper limit of normal. Urinary retention was diagnosed using ultrasonographic evaluation or a volume of >500 ml in the urinary bag after catheterization.
Human Fetal Kidneys, hAKs, and Human Adult Lungs
Human fetal kidneys (hFKs) were collected from elective abortions. The fetal gestational age ranged from 15 to 19 weeks. hAK samples were retrieved from the borders of renal cell carcinoma tumors from patients who underwent partial nephrectomy. Lung tissue was obtained from human lung resections. hFKs, hAKs and lung tissues were handled within 1 hour after the curettage or resection procedures, respectively. All studies were approved by the local ethics committees, and informed consent was provided by the legal guardians of the patients involved, according to the declaration of Helsinki. Collected tissues were cut into 0.5-cm cubes using sterile surgical scalpels. hAK and hFK cells were grown as previously described,25,29 using serum-containing medium (SCM; Iscove’s modified Dulbecco’s medium; Biological Industries, Kibbutz Beit-Haemek, Israel) supplemented with 10% FBS (Invitrogen), 1% 100 M penicillin streptomycin, 1% l-glutamine (both purchased from Biological Industries), 100 ng/ml EGF, 100 ng/ml basic fibroblast growth factor, and 10 ng/ml stem cell factor (all growth factors were purchased from PeproTech Asia).
kSPH Formation
Human kidney epithelial cells (hKEpC) or hFK cells were grown in two dimensions (2D). Upon reaching a confluency of 80%–100%, the cells were harvested and seeded on plates precoated with poly(2-hydroxyethyl methacrylate) (Sigma-Aldrich), in serum-free medium (SFM), at a concentration of 5.5 × 104–13 × 104 cells/ml. SFM comprised N2 medium (Biological Industries) supplemented with 1% 100 M penicillin streptomycin, 1% l-glutamine, 0.4% B27 supplement (Gibco), 4 µg/ml heparin sodium (IntraMed), 1% nonessential amino acids, 1% sodium pyruvate, 0.2% Chemically Defined Lipid Concentrate (a ll purchased from Invitrogen), 2.4 mg/ml glucose, 0.4 mg/ml transferrin, 10 mg/ml insulin, 38.66 µg/ml putrescine, 0.04% sodium selenite, 12.6 µg/ml progesterone (all from Sigma-Aldrich), 10 ng/ml fibroblast growth factor, and 20 ng/ml EGF. kSPH were photographed using Nikon Eclipse TS100 and Nikon Digital Sight cameras. After 7–10 days in culture, kSPH were collected and dissociated by incubation with TrypLE (Gibco) for 10 minutes. kSPH-derived cells were then counted for further experimentation. For generating agarose blocks and RNA, medium containing 2 million kSPH cells was collected and centrifuged, and the kSPH pellet was retrieved.
Cell Lines and Viruses
Vero E6 cells (CRL-1586; ATCC) were grown in growth medium (DMEM containing 10% FBS, MEM nonessential amino acids, 2 mM l-glutamine, 100 U/ml penicillin, 0.1 mg/ml streptomycin, 12.5 U/ml nystatin; all from Biological Industries). Cells were cultured at 37°C with 5% carbon dioxide and at 95% humidity.
SARS-CoV-2 (GISAID accession number EPI_ISL_406862) was kindly provided by The Bundeswehr Institute of Microbiology (Munich, Germany). Virus stocks were propagated (4 passages) and titered on Vero E6 cells. All handling of SARS-CoV-2 was conducted in a Biosafety Level 3 facility, in accordance with the biosafety guidelines of the Israel Institute for Biologic Research.
In Vitro Viral Infection Procedure
Adherent Cells
Primary cultures of hAK-derived cells (hKEpC) were cultured on six-well plates. On the day of infection, wells were washed with PBS and 200 μl of viral solution (2 × 107 viral copies of HCOV-229E or SARS-CoV-2 in SCM with 2% FBS and without antibiotics) was added. Media (200 μl) without virus was added to control wells. Plates were then incubated at 35°C (for HCOV-229E) or 37°C (for SARS-CoV-2) for 2 hours. After incubation, wells were washed twice with PBS and growth media was added (SCM). After 48 hours, cells were harvested, 100 μl of the supernatant was removed, and 200 μl TRIzol was added for RNA analysis. Cell pellets were suspended in 300 μl TRIzol for RNA analysis.
Spheroid Cells
Primary cultures of adult human kidney–derived spheroids (kSPH) were cultured on six-well plates coated with poly(2-hydroxyethyl methacrylate). On the day of infection, cells were collected and centrifuged. The remaining medium was discarded and the pellet was washed with PBS and centrifuged again. The pellet was then suspended with either 200 μl of viral solution (2 × 107 viral copies in SFM without antibiotics) or 200 μl of medium without virus for the control wells. Tubes were then incubated at 35°C (for HCOV-229E) or 37°C (for SARS-CoV-2) for 2 hours. After incubation, cells were centrifuged and washed with PBS, and the pellet was returned its original well and fresh SFM was added. Cells were collected and centrifuged, and 100 μl of the supernatant was removed, and 200 μl TRIzol was added for RNA analysis. Cell pellets were suspended using 300 μl TRIzol for RNA analysis.
RNA Isolation and Quantitative PCR
Total RNA was prepared using the Direct-zol RNA MiniPrep Kit (Zymo Research) according to the manufacturer’s instructions. Real-time quantitative PCR(qPCR) was carried out as previously described,30 using the following primers:
IRF7, forward, GCTGGACGTGACCATCATGTA; reverse, GGGCCGTATAGGAACGTGC;
USP18, forward, AACGTGCCCTTGTTTGTCCAA; reverse, GAGTCCTTCACCCGGATCGTA;
CCL5, forward, CCAGCAGTCGTCTTTGTCAC; reverse, CTCTGGGTTGGCACACACTT;
ACTB, forward, CCACCATGTACCCTGGCATT; reverse, CGGACTCGTCATACTCCTGC;
ACE2, forward, CATTGGAGCAAGTGTTGGATCTT; reverse, GAGCTAATGCATGCCATTCTCA;
TMPRSS2, forward, CAAGTGCTCCAACTCTGGGAT; reverse, AACACACCGATTCTCGTCCTC;
BSG, forward, GAAGTCGTCAGAACACATCAACG; reverse, TTCCGGCGCTTCTCGTAGA; and
GAPDH, forward, ACAGTCAGCCGCATCTTCTT; reverse, GTTAAAAGCAGCCCTGGTGA.
qPCR primers and probes for the detection of SARS-CoV-2 N1 were obtained using the 2019-nCoV CDC Emergency Use Authorization Kit (catalog number 10006606; Integrated DNA Technologies).
Immunohistochemical and Immunofluorescence Staining of Paraffin-Embedded Grafts
Paraffin sections (5 μm) were pretreated using OmniPrep Solution (Zytomed Systems) in 95°C for 1 hour according to the manufacturer’s protocol. For immunohistochemistry (IHC), blocking was performed using CAS‐Block solution (Invitrogen) for 20 minutes, followed by 1 hour of incubation at room temperature with the primary antibodies anti-ACE2 (15348) and anti-CD147 (666; both from Abcam). Samples were incubated with secondary antibodies (ImmPRESS anti-mouse/rabbit reagent peroxidase; Vector Laboratories, Burlingame, CA) for 30 minutes at room temperature, and detected using the ImmPACT DAB kit (Vector Laboratories), according to the manufacturer’s protocol. Hematoxylin was used for counterstaining. For immunofluorescence, sections were blocked with CAS‐Block solution (Invitrogen) for 1 hour, and then incubated with the primary antibodies biotinylated lotus tetragonolobus lectin (Vector Laboratories), anti-EMA (247M-98; Cell Marque), and anti-CD13 (108382; abcam). For the detection of SARS-CoV-2, we used hyperimmune rabbit serum from intravenous SARS-CoV-2–infected rabbit. Sections were then washed and incubated with secondary antibodies for 1 hou r, and mounting medium containing 4′,6-diamidino-2-phenylindole (Southern Biotech) was applied. Photos were obtained using Olympus BX51TF Fluorescence Microscope with an Olympus DP72 camera and cellSens standard software.
Grafting into Mice
A total of 1 × 106–2 × 106 kSPH were suspended within Matrigel (BD Biosciences) and implanted into the subcutaneous space of 8- to 10-week-old NOD-SCID mice. Grafts were harvested 14 days after injection, fixed overnight in 4% buffered paraformaldehyde, and then embedded in paraffin.
RNA-Sequencing Analysis
To compare expression levels of nephron segment markers between Vero E6 cells and hKEpC, the average of log2(transcripts per million [TPM]+1) values was calculated for known markers of each denoted nephron segment’s ortholog. TPM values for Vero E6 cells were derived from RNA-sequencing (RNA-seq) data generated for Vero E6 cells by Ogando et al.31 (GSE153940; three control samples). TPM values for hKEpC were obtained from RNA-seq data of 2D epithelial cells derived from adult kidney generated by Harari-Steinberg et al.25 (GSE141257; four Adherent samples).
Statistical Analysis
Summary statistics for the study group were expressed as mean (SD) or percentage. The Fisher exact test was used to compare categoric variables, and the matched two-sample t test was used for comparison of metric continuous variables. The analysis of viral load in the medium over time was carried out via a one-way ANOVA with post hoc Tukey post hoc correctio n. A two-sided P value <0.05 was considered statistically significant.
Results
kSPH Express the SARS-CoV-2 Receptors ACE2, TMPRSS2, and CD147
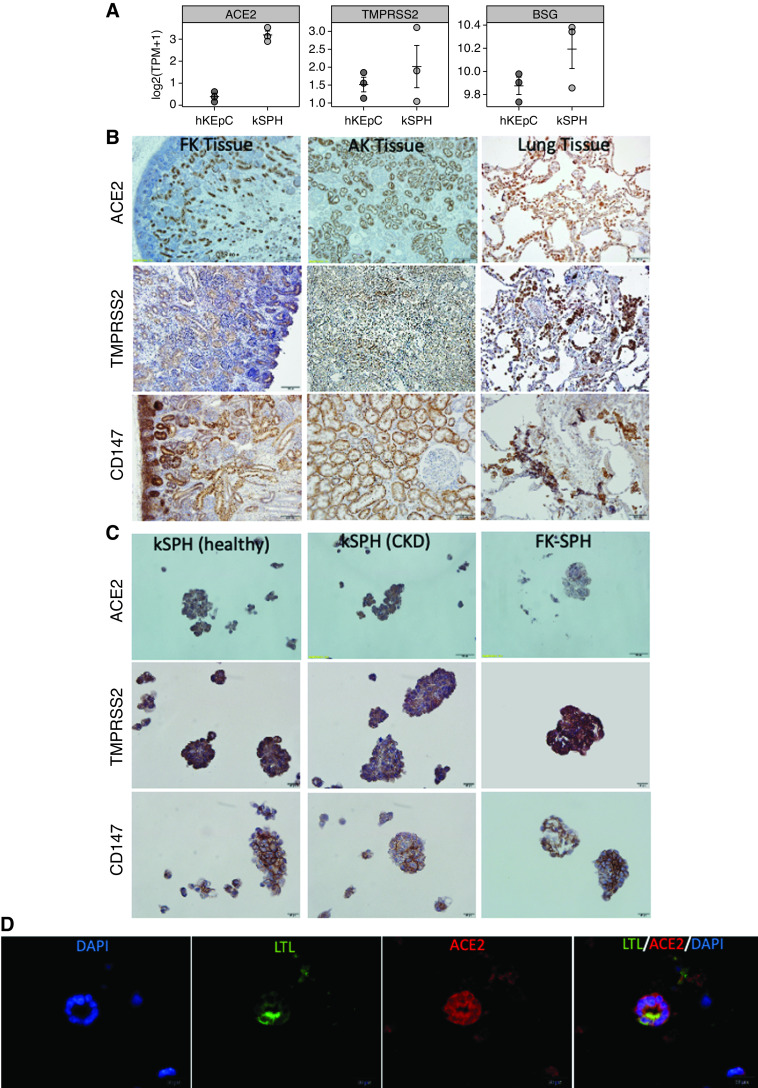
To assess the interactions between SARS-CoV-2 and the human kidney, we sought to evaluate the effect of SARS-CoV-2 infection in an in vitro model of the human kidney. To this end, we decided to use kSPH, which were previously shown to recapitulate the epithelial phenotypes and function of human kidneys.25 Initially, to determine whether the kSPH could serve as a model to study the host-pathogen interaction of the human kidney and SARS-CoV-2, we assessed whether they express the surface markers thought to serve as receptors for SARS-CoV-2: ACE2,32 TMPRSS2,33 and BSG (CD147).34 Interestingly, kSPH expressed significantly higher levels of both ACE2 and BSG, and similar levels of TMPRSS2, compared with primary 2D hKEpC, as assessed by RNA-seq (Figure 1A).
Figure 1.
kSPH express the SARS-CoV-2 receptors. (A) Relative gen e expression, shown as TPM of ACE2, TMPRSS2, and BSG in hKEpC, grown in a monolayer, and kSPH. (B) Immunostaining of fetal kidney (FK) and adult kidney (AK) and lung tissues for ACE2, TMPRSS2, and CD147. (C) Immunostaining of kSPH derived from the adult kidneys of a healthy donor (left panel), a patient with CKD (middle panel), and from fetal kidney (right panel) for ACE2, TMPRSS2, and CD147. (D) kSPH were subcutaneously injected into NOD-SCID mice and the grafts were analyzed after 14 days for the formation of kidney tubuloids. Shown is immunofluorescence staining for the proximal tubule marker lotus tetragonolobus lectin (LTL; green) and ACE2 (red). DAPI, 4′,6-diamidino-2-phenylindole. (Scale bar: B: 100μm, C: 20μm, D: 20μm).
IHC staining of both adult and fetal human kidneys confirmed robust and widespread tubular expression of ACE2, CD147, and TMPRSS2 at the protein level, which was comparable with their expression in lung tissue (Figure 1B). Notably, proteins were localized in renal tubules, and absent from glomeruli, as previously described.16 Confirming their resemblance to native adult kidney tissue, kSPH exhibited strong expression of ACE2, CD147, and TMPRSS2 at the protein level, regardless of whether the donor was healthy or a patient with CKD (Figures 1C). Interestingly, kSPH generated from fetal kidneys exhibited lower expression levels of ACE2 (Figure 1C). Lastly, we injected kSPH subcutaneously into NOD-SCID mice, thereby generating human tubular structures, as previously described.25 Staining of the tubular structures for the proximal tubule marker lotus tetragonolobus lectin and ACE2 demonstrated proximal tubules expressing ACE2 (Figure 1D), consistent with the in vitro observations. Taken together, these results demonstrate that kSPH resemble the native kidney in the expression of the SARS-CoV-2 receptors.
SARS-CoV-2 Infects kSPH and Monolayers
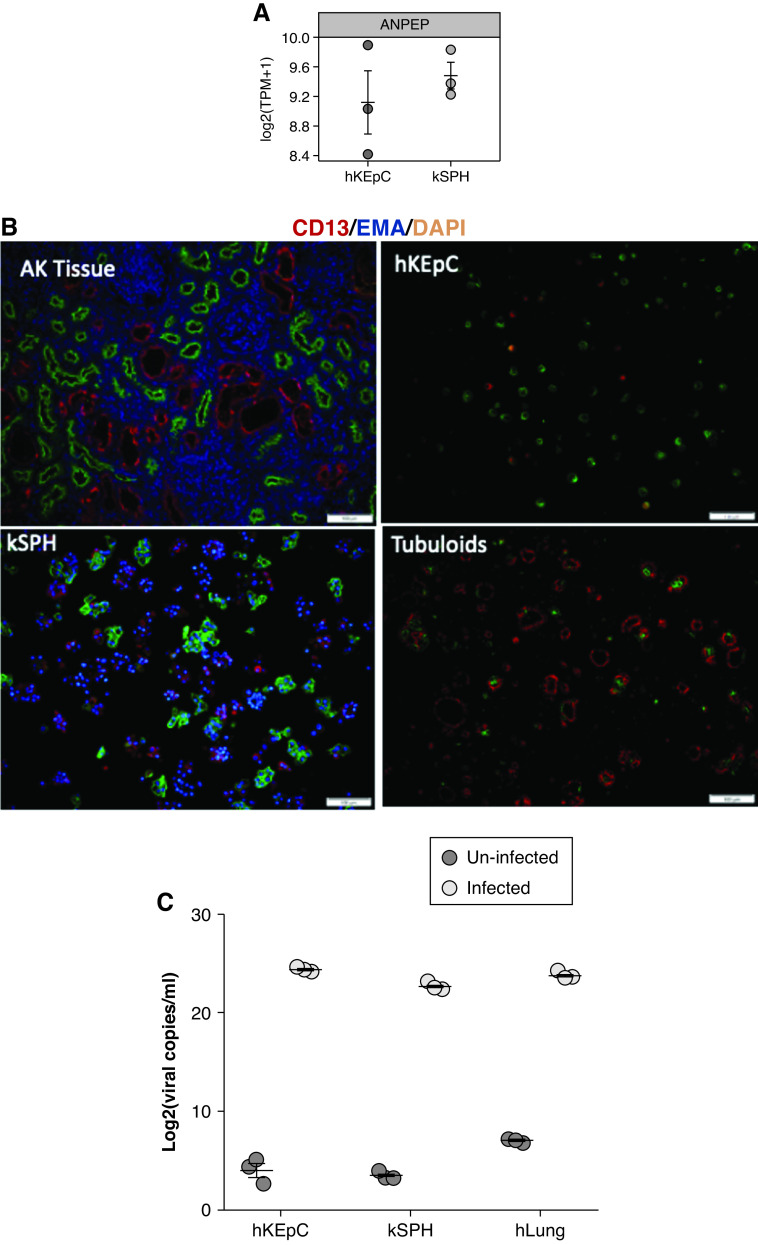
Subsequently, we sought to establish a direct link between SARS-CoV-2 infection and the kidney damage seen in COVID-19, by testing whether SARS-CoV-2 can infect primary human kidney cells and kSPH, and whether this results in a cytopathic effect. Initially, as a proof of concept, we evaluated whether hKEpC and kSPH could be infected by HCoV-229E, which is a different strain of coronavirus and a common cause of viral upper respiratory tract infections.35 Transcriptomic data demonstrated strong expression of the HCoV-229E receptor CD13 (ANPEP) in both hKEpC and kSPH (Figure 2A), which was confirmed at the protein level via immunofluorescence staining (Figure 2B) adult kidney tissue, hKEpC, kSPH, and in vivo tubular structures derived thereof. Hence, we incubated hKEpC and kSPH with HCoV-229E for 48 hours and assessed the level of infection, as indicated by the viral copies in the medium, with primary passage 1 human lung cells serving as a positive control. qPCR demonstrated high levels of viral load in all cell types, with hKEpC and kSPH showing comparable levels of infection as human lung cells (Figure 2C).
Figure 2.
HCOV-229E infects hKEpC and kSPH. (A) hKEpC and kSPH express CD13 (ANPEP), which is the receptor for HCOV-229E. (B) CD13 immunostaining of adult kidney tissue, hKEpC, kSPH, and the in vivo tubuloids derived thereof. (C) After infection with HCOV-229E, hKEpC and kSPH express the viral RNA at similar levels as human lung cells (hLung). EMA, Epithelial Membrane Antigen; DAPI, 4′,6-diamidi no-2-phenylindole. (Scale bar: B: 100μm).
Having shown that hKEpC and kSPH effectively facilitate infection of HCoV-229E, we next attempted to infect both types of cultures with SARS-CoV-2, quantifying the level of infection according to the total number of plaque-forming unit equivalents in both medium and cells, as measured by qPCR 48 hours postinfection (Figure 3A). Interestingly, both hKEpC and kSPH demonstrated a significant increase in viral RNA in both the cells and medium (Figure 3A). In addition, we measured viral levels in the culture medium of hKEpC, kSPH, and Vero E6 cells, comparing total plaque-forming units in uninfected cells versus infected cells at day 0 and day 1. We detected a significant increase in viral levels at 24 hours in all cell types (Figure 3B). Notably, these infection levels are similar (kSPH) or higher (hKEpC) than those that are reported to result in functional cytopathic effects in other cell types.36 We next tested subgenomic RNA to verify virus replication. We could see high levels of viral subgenomic RNA in the infected group, in both hKEpC and kSPH (Figure 3C), confirming viral infection. For SARS-CoV-2 detection by immunostaining, we used hyperimmune rabbit serum from intravenous SARS-CoV-2–infected rabbit. We could see the presence of the virus in both hKEpC and in kSPH, and in the Vero E6 cells that served as a positive control (Figure 3D).
Figure 3.
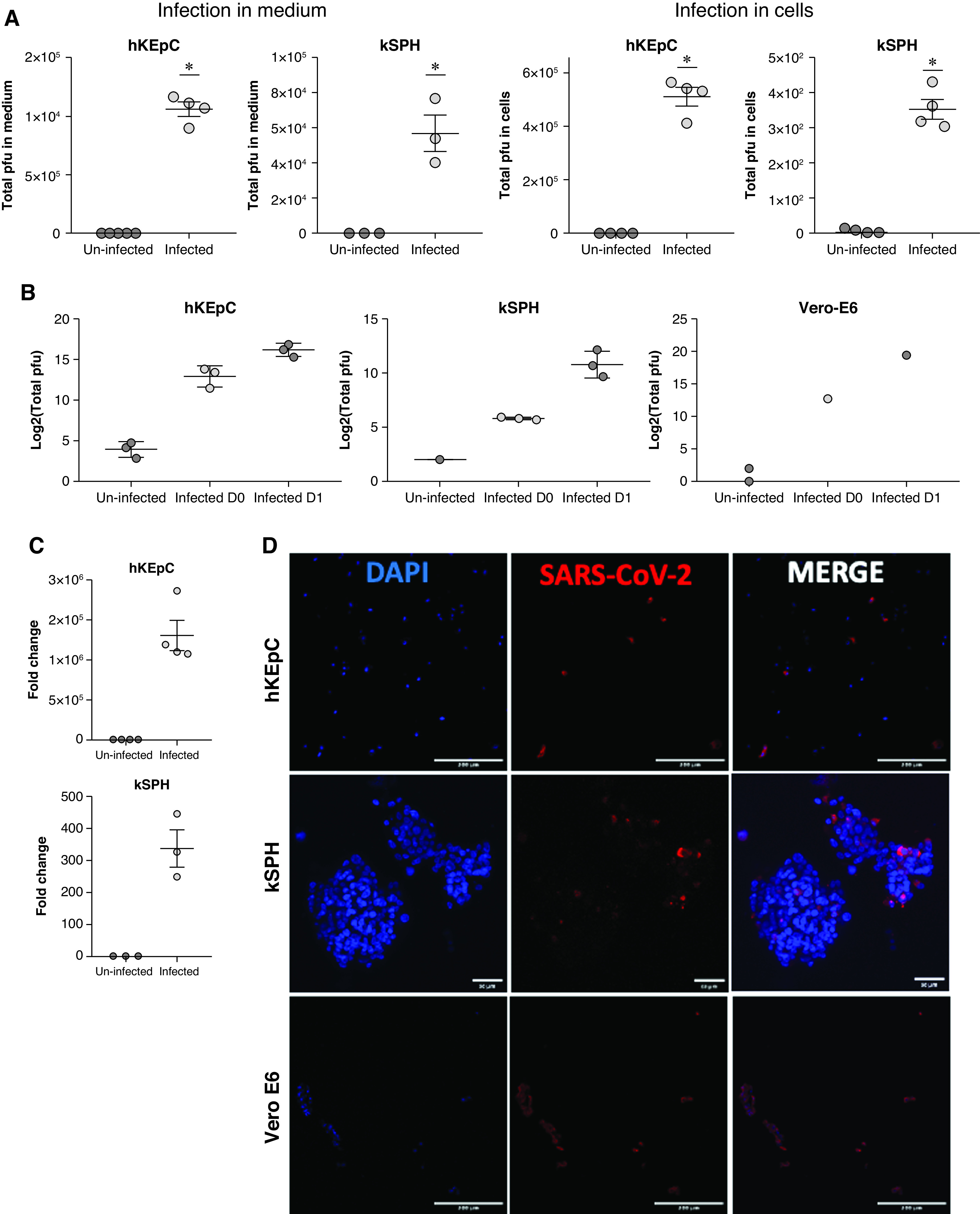
SARS-CoV-2 infects hKEpC and kSPH. (A) Quantification of the viral load in the medium and in hKEpC and kSPH 48 hours after infection with SARS-CoV-2. (B) Quantification of the viral load in the medium at the day of infection and 24 hours postinfection. (C) Real-time PCR of viral subgenomic RNA 48 hours postinfection in hKEpC and kSPH. (D) Detection of SARS-CoV-2 in hKEpC, kSPH, and Vero E6 cells by immunostaining using hyperimmune rabbit serum from intravenous SARS-CoV-2–infected rabbit. *P<0.05. D0, day 0; D1, day 1; DAPI, 4′,6-diamidino-2-phenylindole; pfu, plaque-forming units. ( Scale bar: D: 200μm).
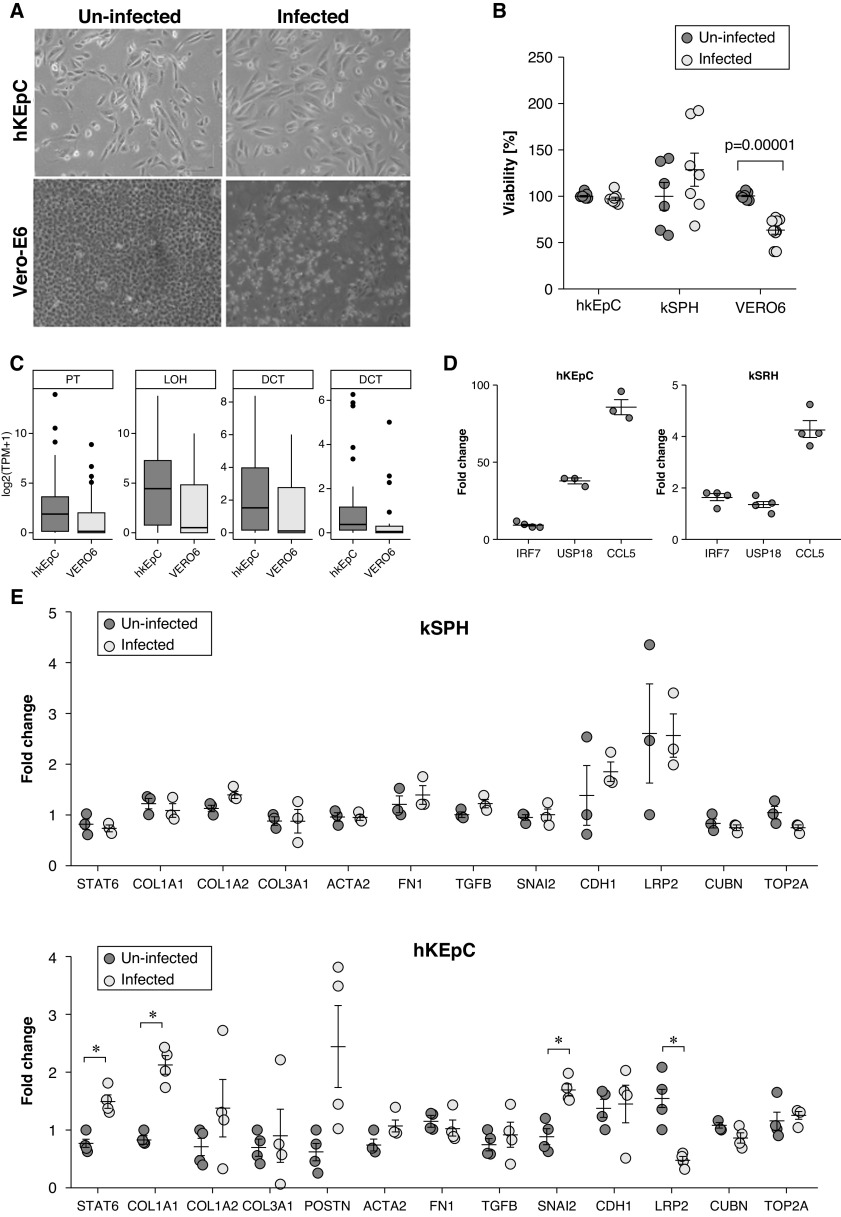
SARS-CoV-2 Does Not Result in a Cytopathic Effect but Can Induce Changes in Kidney Injury Molecules
Nonetheless, although SARS-CoV-2 infection of Vero E6 resulted in a clear cytopathic effect, both hKEpC and kSPH did not demonstrate any recognizable morphologic change, as observed by light microscopy, even when infected using very high viral titers (moi=4–8; Figure 4A). This manifestation was also observed when we infected the cells for a longer time, 72 hours, and tested cell viability. These results demonstrate that, although infected Vero E6 cells show significantly decreased viability, hKEpC and kSPH show no difference in viability upon infection (Figure 4B). Importantly, although Vero E6 cells were originally derived from monkey kidney, expression of various kidney-specific markers was negligible compared with native human kidney, indicating a very low resemblance to primary human kidney cells (Figure 4C). In addition, we could see upregulation of IRF7, USP18, and CCL5, genes that are activated during viral infection,37–39 especially in hKEpC (Figure 4D). Subsequently, to assess whether detrimental effects might be evident at the molecular level, we tested the effect of the infection on the expression of genes related to tubular damage. Thus, we carried out qPCR analysis of genes related to fibrosis and those implicated in the tubular response to damage, which involves dedifferentiation and epithelial-mesenchymal transitio n.40 In kSPH, we did not detect significant changes, implying that during a steady state, as reflected in the quiescent kSPH, no molecular damage takes place as a result of infection. Because COVID-19 often results in AKI, during which kidney cells enter a proliferative state, we next tested whether 2D hKEpC, which are more proliferative and thus better resemble the injured kidney, would show molecular evidence of damage. Indeed, we found an increase in several fibrosis-related genes (STAT6 and COL1A1), and an increase in the SNAI2 gene, suggestive of epithelial-mesenchymal transition and kidney tubular injury (Figure 4E upper graph). Accordingly, we detected downregulation of the proximal tubule marker LRP2, implying transient loss of epithelial identity (i.e., dedifferentiation), typical of AKI. Taken together, these results indicate that, although the steady-state kidney allows viral entry and proliferation, this does not result in cellular damage. In contrast, once in a more proliferative state, as occurs during AKI, viral replication is augmented and, accordingly, molecular evidence of damage can be seen.
Figure 4.
SARS-CoV-2 elicits molecular changes associated with kidney injury in hKEpC and kSPH, without evidence of a cytopathic effect. (A) Morphology of SARS-CoV-2–infected hKEpC and Vero E6 cells versus uninfected cells. (B) Viability of hKEpC, kSPH, and Vero E6 cells 72 hours postinfection compared with uninfected cells. (C) Box plots representing mean expression levels (log2[TPM+1]) of nephron segment markers in Vero E6 cells and hKEpC, on the basis of RNA-seq data. Nephron segment genes are downregulated in Vero E6 cells in comparison with their human orthologs, hKEpC. (D) Relative expression of type 1 IFN response–related genes in infected hKEpC versus kSPH. (E) Relative expression of genes related to kidney tubular damage. CD, collecting duct; DCT, distal convoluted tubule; LOH, loop of Henle; PT, proximal tubule.
Infection with SARS-CoV-2 Results in Activation of the Type 1 IFN Response Pathway in Human Kidney Cells
Subsequently, we were interested in exploring the molecular mechanisms that endow human kidney cells with resistance to SARS-CoV-2–related cytopathic damage. Because recent reports have demonstrated that type 1 IFN response plays a pivotal role in the immune response of patients with COVID-19,41–43 we compared the activity of this pathway in human kidney cells (hKEpC and kSPH) and Vero E6 cells, which demonstrate significant cytopathic effects upon infection with SARS-CoV-2.
Intriguingly, we found significant activation of multiple genes associated with the activation of the type 1 IFN reaction in hKEpC and kSPH, compared with Vero E6 cells (Figure 5), including IRF7, IRF8, and STAT2.
Figure 5.
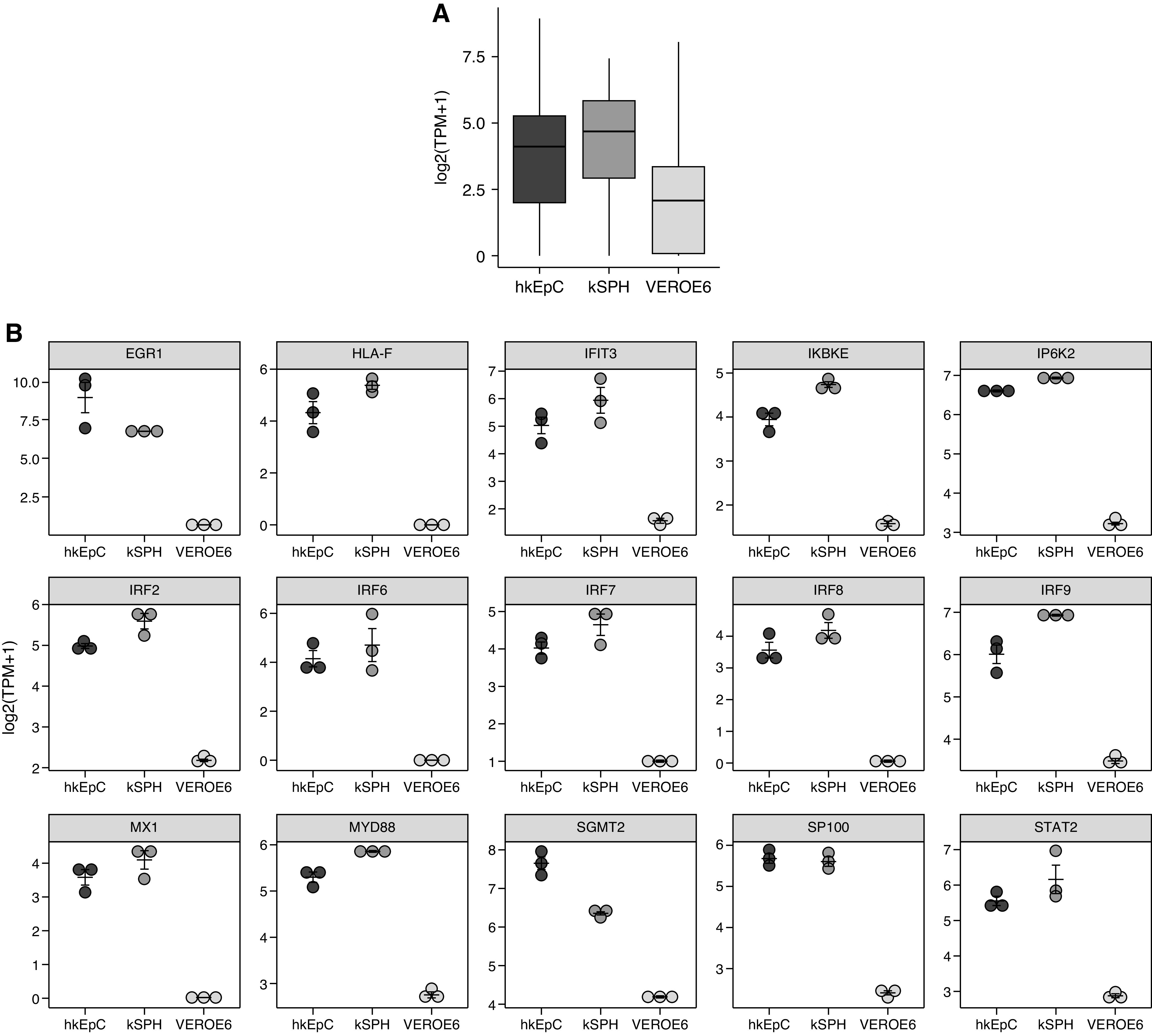
SARS-CoV-2 activates Type 1 IFN response in human kidney cells. (A) Box plots showing mean expression levels (log2[TPM+1]) of genes associated with the Gene Ontology term “response to type 1 IFN” (0034340) in Vero E6 cells, hKEpC, and kSPH from RNA-seq. These genes are downregulated in Vero E6 cells in comparison with their human counterparts, hKEpC and kSPH. (B) Representation of 15 genes showing the highest levels of expression in hKEpC, kSPH, and Vero E6 cells.
Clinical Data Do Not Support a Major Role for Primary Kidney Damage in COVID-19–Induced AKI
To assess how our in vitro findings correlate with clinical findings, we analyzed a cohort of 36 patients, representing all patients with COVID-19 admitted to the CCCU at our institution between March and June 2020. Supplemental Table 1 shows the baseline characteristics of these patients. The mean±SD age of the cohort was 60.9±14.9 years, and most patients were male (83.3%). All patients were mechanically ventilated, and 47.3% died during their hospitalization. Approximately two thirds (61.1%) of patients (22 of 36) had AKI during their hospitalization, after a median of 4 days. Of these patients, 36.4% (eight of 22) had AKI upon admission, and 65.6% (14 of 22 patients) developed AKI during hospitalization (Supplemental Table 2). In an attempt to differentiate AKI resulting from direct viral injury and AKI secondary to other factors, we queried the medical records for possible AKI-causing insults, other than COVID-19 infection, preceding the appearance of AKI. These included prolonged hypotension, volume depletion, shock (septic [not related to COVID-19], cardiogenic, obstructive, hemorrhagic), exposure to nephrotoxic drugs, rhabdomyolysis, and postrenal obstruction. Interestingly, all eight patients presenting with AKI at admission had documented hypovolemia and dehydration in the days before hospitalization. Similarly, within the AKI group, most patients had documented hypotension before renal function deterioration (86.4%, 19 of 22 patients). Of the patients with AKI, 27.3% (six of 22) developed AKI within a few hours of intubation. Moreover, 40.1% of patients (nine of 22) had sepsis diagnosed before AKI, and 27.3% (six of 22) had cardiovascular collapse before AKI (Supplemental Table 2). Taken together, in our cohort of patients critically ill with COVID-19, the development of AKI was invariably preceded by a clear etiologic factor, other than COVID-19 infection. Collectively, this points against direct viral infection as the reason for AKI among these patients, which is in line with previous reports and the in vitro results, although we note that the cohort size is too small, and the determination of AKI etiology too subjective, to draw definite conclusions.
Discussion
Although the hallmark of the current COVID-19 pandemic is its pulmonary involvement, various organ systems have been shown to be implicated as well. With an incidence of 8%–9%, according to two recent meta-analyses,44,45 AKI is a particularly enigmatic facet of COVID-19, with the mechanism of injury representing an unresolved issue.
One of the main obstacles toward deciphering the underlying mechanisms of kidney damage in COVID-19 is the need for a reliable cellular model that can allow direct assessment of infection on hAK cells. Notably, most studies used different cell lines, both human and nonhuman, which yielded conflicting results. For instance, although infection of the monkey-derived cell lines Vero and Vero E6 resulted in a cytopathic effect,46,47 the canine MDCK47 and human 29348 cell lines were unaffected. Similarly, a recent study, which reported the isolation of SARS-CoV-2 from an autopsied kidney, relied on primate rather than human kidney cells to test the replicative capacity of the virus.14
Importantly, both monolayer hKEpC and three-dimensional kSPH support infection and replication of SARS-CoV-2. However, this does not result in a cytopathic effect nor profound cell damage, which is clearly evident in Vero E6 cells. We found high baseline levels of type 1 IFN response–related molecules associated with hKEpC and kSPH transcriptomes before infection compared with those in Vero E6 ce lls. The type 1 IFN response has been suggested as a cellular means to combat viral-induced cytopathy42,49 and, therefore, this observation may set the basis for the level of cytopathic/cell damage response in human kidney cultures once infected. Altogether, these data, showing a lack of profound cellular effects, are in line with our clinical information gathered from the first 36 patients treated in the intensive care unit (see Supplemental Table 2), which demonstrates that AKI occurs with secondary etiologies.
Further work will be needed to determine why, despite effective infection and replication of SARS-CoV-2 in human kidney cells, no cytopathic damage is observed. Notably, similar levels of viral titers have been shown to induce significant cytopathic effects in other organs, such as the heart36; however, successful infection of human lung organoids, modeling the primary site of viral infection, both in vitro and in vivo, was not reported to lead to a cytopathic effect.50 As suggested, one possible explanation is a stronger and more effective activation of antiviral cellular pathways, such as the IFN signaling pathway, which has recently been suggested to have potent activity against SARS-CoV-2.42,43,49 In addition, given the significant vascular damage observed in COVID-19 patients, it would be intersting to assess the effects of the virus using more complex models, harboribg both renal and vascular compartments (DOI: https://doi.org/10.1681/ASN.2019050508).
Nevertheless, after viral infection, molecular assessment of the injury molecules induced during AKI—such as STAT6, COL1A1, and SNAI2—showed significant elevation, whereas LRP2 was diminished, indicating that more subtle injury may occur once the virus infects and replicates. These changes were most apparent in hKEpC cultures. Interestingly, close comparison of kSPH and monolayers revealed the latter to support higher infectivity/replication and a more pronounced elevation of molecules associated with SARS-CoV-2 infection, such as IRF7, USP18, and CCL5.
kSPH and hKEpC represent two opposing sides of a spectrum.25 In hKEpC, the dissociated human kidney tissue, once placed in culture and grown as monolayer, displays a loss of quiescence, proliferation, and partial dedifferentiation—a cascade of events that occurs in AKI.
In contrast, kSPH more closely resemble the cellular composition of the renal tubular compartments, as previously shown by transcriptomics and protein analysis.24,25 Similar to the native adult kidney, kSPH contain low-cycling epithelial cells. Although they express a high level of viral entry molecules and support infection and replication, evidence of significant elevation of AKI-related tubular injury molecules was not apparent. Thus, the dichotomy between culture models may hint toward a possible scenario in which direct viral infection does not pose significant damage for renal tubules, but once AKI-induced cell proliferation takes place due to a variety of other etiologies, viral infection and replication may intensify damage, leading to further dedifferentiation, loss of cell identity, and a profibrotic state.
Finally, the ability to generate similar cultures from urine could allow this approach to be used for personalized and precision medicine, by culturing the patient’s own cells in a noninvasive manner. This could help to answer some of the outstanding questions surrounding renal involvement in COVID-19, with regards to age, sex and ethnic background and the effects of pre-exisiting CKD.
Disclosures
B. Dekel repor ts having ownership interest in KidneyCure as a founder and shareholder; having patents and inventions with SMC; and serving as a scientific advisor for, or editorial member of, Stem Cells Translational Medicine, Stem Cell Reports and the UK Regenerative Medicine Platform. All remaining authors have nothing to disclose.
Funding
This study was supported by The Lisa and David Pulver Family Foundation, Israel Science Foundation grant 2071/17, Israel Innovation Authority KAMIN grant 61910, and The Euro-Asian Jewish Congress.
Supplementary Material
Footnotes
D.O. and O.P contributed equally to this work.
Published online ahead of print. Publication date available at www.jasn.org.
Supplemental Material
This article contains the following supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2020111546/-/DCSupplemental.
Supplemental Table 1. Patient characteristics.
Supplemental Table 2. Comparison of clinical characteristics of COVID-19 patients with and without AKI.
References
- 1.World Health Organization: WHO Coronavirus (COVID-19) dashboard. Available at: https://covid19.who.int/. Accessed March 11, 2021
- 2.Fisher M, Neugarten J, Bellin E, Yunes M, Stahl L, Johns TS, et al.: AKI in hospitalized patients with and without COVID-19: A comparison study. J Am Soc Nephrol 31: 2145–2157, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch JS, Ng JH, Ross DW, Sharma P, Shah HH, Barnett RL, et al.; Northwell COVID-19 Research Consortium; Northwell Nephrology COVID-19 Research Consortium:. Acute kidney injury in patients hospitalized with COVID-19. Kidney Int 98: 209–218, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al.; China Medical Treatment Expert Group for Covid-19:. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382: 1708–1720, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richardson S, Hirsch JS, Narasimhan M, Crawford JM, McGinn T, Davidson KW, et al.; the Northwell COVID-19 Research Consortium:. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 323: 2052–2059, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gupta A, Madhavan MV, Sehgal K, Nair N, Mahajan S, Sehrawat TS, et al.: Extrapulmonary manifestations of COVID-19. Nat Med 26: 1017–1032, 2020 [DOI] [PubMed] [Google Scholar]
- 7.Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al.: Endothelial cell infection and endotheliitis in COVID-19. Lancet 395: 1417–1418, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, Liu J, Zhang D, Xu Z, Ji J, Wen C: Cytokine storm in COVID-19: The current evidence and treatment strategies. Front Immunol 11: 1708, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, Luo R, Wang K, Zhang M, Wang Z, Dong L, et al.: Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int 97: 829–838, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al.: Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 395: 1054–1062, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farkash EA, Wilson AM, Jentzen JM: Ultrastructural evidence for direct renal infection with SARS-CoV-2. J Am Soc Nephrol 31: 1683–1687, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kissling S, Rotman S, Gerber C, Halfon M, Lamoth F, Comte D, et al.: Collapsing glomerulopathy in a COVID-19 patient. Kidney Int 98: 228–231, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su H, Yang M, Wan C, Yi LX, Tang F, Zhu HY, et al.: Renal histopathological analysis of 26 postmortem findings of patients with COVID-19 in China. Kidney Int 98: 219–227, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun F, Lütgehetmann M, Pfefferle S, Wong MN, Carsten A, Lindenmeyer MT, et al.: SARS-CoV-2 renal tropism associates with acute kidney injury. Lancet 396: 597–598, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al.: A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579: 270–273, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang YM, Zhang H: Genetic roadmap for kidney involvement of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Clin J Am Soc Nephrol 15: 1044–1046, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh M, Bansal V, Feschotte C: A single-cell RNA expression map of human coronavirus entry factors. Cell Rep 32: 108175, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Larsen CP, Bourne TD, Wilson JD, Saqqa O, Sharshir MA: Collapsing glomerulopathy in a patient with COVID-19. Kidney Int Rep 5: 935–939, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller SE, Brealey JK: Visualization of putative coronavirus in kidney. Kidney Int 98: 231–232, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Calomeni E, Satoskar A, Ayoub I, Brodsky S, Rovin BH, Nadasdy T: Multivesicular bodies mimicking SARS-CoV-2 in patients without COVID-19. Kidney Int 98: 233–234, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al.: Detection of SARS-CoV-2 in different types of clinical specimens. JAMA 323: 1843–1844, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bwire GM, Majigo MV, Njiro BJ, Mawazo A: Detection profile of SARS-CoV-2 using RT-PCR in different types of clinical specimens: A systematic review and meta-analysis. J Med Virol 93: 719–725, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ling Y, Xu SB, Lin YX, Tian D, Zhu ZQ, Dai FH, et al.: Persistence and clearance of viral RNA in 2019 novel coronavirus disease rehabilitation patients. Chin Med J (Engl) 133: 1039–1043, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buzhor E, Harari-Steinberg O, Omer D, Metsuyanim S, Jacob-Hirsch J, Noiman T, et al.: Kidney spheroids recapitulate tubular organoids leading to enhanced tubulogenic potency of human kidney-derived cells. Tissue Eng Part A 17: 2305–2319, 2011 [DOI] [PubMed] [Google Scholar]
- 25.Harari-Steinberg O, Omer D, Gnatek Y, Pleniceanu O, Goldberg S, Cohen-Zontag O, et al.: Ex vivo expanded 3D human kidney spheres engraft long term and repair chronic renal injury in mice. Cell Rep 30: 852–869.e4, 2020 [DOI] [PubMed] [Google Scholar]
- 26.KDIGO Clinical Practice Guideline for Acute Kidney Injury, 2012. Avalable at: https://kdigo.org/wp-content/uploads/2016/10/KDIGO-2012-AKI-Guideline-English.pdf. Accessed May 1, 2021
- 27.Rhodes A, Evans LE, Alhazzani W, Levy MM, Antonelli M, Ferrer R, et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock: 2016. Crit Care Med 45: 486–552, 2017. 28098591 [DOI] [PubMed] [Google Scholar]
- 28.Whelton PK, Carey RM, Aronow WS, Casey DE, Collins KJ, Himmelfarb CD, et al.: 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APha/ASH/ASPC/NMA/PCNA Guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines [published correction appears in Hypertension 71: e136–e139, 2018]. Hypertension 71: 1269–1324, 2018. 29133354 [Google Scholar]
- 29.Harari-Steinberg O, Metsuyanim S, Omer D, Gnatek Y, Gershon R, Pri-Chen S, et al.: Identification of human nephron progenitors capable of generation of kidney structures and functional repair of chronic renal disease. EMBO Mol Med 5: 1556–1568, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Metsuyanim S, Harari-Steinberg O, Buzhor E, Omer D, Pode-Shakked N, Ben-Hur H, et al.: Expression of Stem Cell Markers in the Human Fetal Kidney 2009. Available at: 10.1371/journal.pone.0006709. Accessed May 1, 2021 10.1371/journal.pone.0006709 [DOI] [PMC free article] [PubMed]
- 31.Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, Limpens RWAL, van der Meer Y, Caly L, et al.: SARS-coronavirus-2 replication in Vero E6 cells: Replication kinetics, rapid adaptation and cytopathology. J Gen Virol 101: 925–940, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D: Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181: 281–292.e6, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al.: SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271–280.e8, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang K, Chen W, Zhang Z, Deng Y, Lian JQ, Du P, et al.: CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal Transduct Target Ther 5: 283, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Hoek L: Human coronaviruses: What do they cause? Antivir Ther 12[4 Pt B]: 651–658, 2007 [PubMed] [Google Scholar]
- 36.Sharma A, Garcia G, Wang Y, Plummer JT, Morizono K, Arumugaswami V, et al.: Human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Reports Med, 1: 100052, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Au WC, Moore PA, LaFleur DW, Tombal B, Pitha PM: Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem 273: 29210–29217, 1998 [DOI] [PubMed] [Google Scholar]
- 38.Ritchie KJ, Hahn CS, Kim KL, Yan M, Rosario D, Li L, et al.: Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat Med 10: 1374–1378, 2004 [DOI] [PubMed] [Google Scholar]
- 39.Crawford A, Angelosanto JM, Nadwodny KL, Blackburn SD, Wherry EJ: A role for the chemokine RANTES in regulating CD8 T cell responses during chronic viral infection. PLoS Pathog 7: e1002098, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar S: Cellular and molecular pathways of renal repair after acute kidney injury. Kidney Int 93: 27–40, 2018 [DOI] [PubMed] [Google Scholar]
- 41.Hassan AO, Case JB, Winkler ES, Thackray LB, Kafai NM, Bailey AL, et al.: A SARS-CoV-2 infection model in mice demonstrates protection by neutralizing antibodies. Cell 182: 744–753.e4, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, et al.: Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11: 3810, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lokugamage KG, Hage A, de Vries M, Valero-Jimenez AM, Schindewolf C, Dittmann M, et al.: Type I interferon susceptibility distinguishes SARS-CoV-2 from SARS-CoV. J Virol 94: e01410–e01420, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hansrivijit P, Qian C, Boonpheng B, Thongprayoon C, Vallabhajosyula S, Cheungpasitporn W, et al.: Incidence of acute kidney injury and its association with mortality in patients with COVID-19: A meta-analysis. J Investig Med 68: 1261–1270, 2020 [DOI] [PubMed] [Google Scholar]
- 45.Chen YT, Shao SC, Hsu CK, Wu IW, Hung MJ, Chen YC: Incidence of acute kidney injury in COVID-19 infection: A systematic review and meta-analysis. Crit Care 24: 346, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, et al.: Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A 117: 7001–7003, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barr IG, Rynehart C, Whitney P, Druce J: SARS-CoV-2 does not replicate in embryonated hen’s eggs or in MDCK cell lines. Euro Surveill 25: 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chu H, Chan JF-W, Yuen TT-T, Shuai H, Yuan S, Wang Y, et al.: Comparative tropism, replication kinetics, and cell damage profiling of SARS-CoV-2 and SARS-CoV with implications for clinical manifestations, transmissibility, and laboratory studies of COVID-19: An observational study. Lancet Microbe 1: e14–e23, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xia H, Cao Z, Xie X, Zhang X, Chen JYC, Wang H, et al.: Evasion of type I interferon by SARS-CoV-2. Cell Rep 33: 108234, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Han Y, Duan X, Yang L, Nilsson-Payant BE, Wang P, Duan F, et al.: Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 589: 270–275, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.