Abstract
During T-cell activation, c-Myb is induced upon interleukin 2 (IL-2) stimulation and is required for correct proliferation of cells. In this paper, we provide evidence that IL-2-mediated induction of the c-myb gene occurs via the phosphoinositide 3-kinase (PI3K) signaling pathway, that protein kinase B (PKB) is the principal transducer of this signal, and that activation of the c-myb promoter can be abolished by deletion of conserved E2F and NF-κB binding sites. We show that Myb is required to protect activated peripheral T cells from bcl-2-independent apoptosis and that overexpression of oncogenic v-Myb is antiapoptotic. Overexpression of a Myb dominant-negative transgene abrogates PKB-mediated protection from apoptosis. Taken together, these results suggest that induction of c-myb transcription is an important downstream event for PKB-mediated protection of T cells from programmed cell death.
Interleukin 2 (IL-2) regulates the survival, proliferation, and differentiation of mature T cells and is responsible for their progression from G1 to S phase following antigenic activation (17). Studies of the molecules involved in signaling from the IL-2 receptor (IL-2R) have shown that activation of phosphoinositide 3-kinase (PI3K) and its downstream effector, protein kinase B (PKB), appear to be most important for the survival functions mediated by IL-2 (reviewed in reference 19). Proapoptotic proteins which can be phosphorylated and inhibited by PKB include BAD (20, 22), human caspase 9 (16), and the forkhead family of transcription factors (12). PKB can also cause stimulation of NF-κB activity by up-regulating IκB degradation via phosphorylation of IκB Kinase and by affecting NF-κB itself (29, 31, 33, 43, 52), thereby allowing the transcription of genes involved in promoting survival, such as the bcl-2 homologue bfl-1 (78). In addition to the forkhead and NF-κB families, E2F-mediated transcription can also be activated by the hyperphosphorylation and subsequent inactivation of retinoblastoma protein (Rb) in response to signals from PI3K and its downstream effectors, PKB and p70S6 kinase (10, 11, 25). Recently, overexpression of activated PKB in transgenic mice has been shown to enhance the resistance of both thymocytes and T cells to challenges with apoptotic stimuli and to promote survival following antigenic activation (29).
The transcription factors activated by PI3K and PKB are of great interest in the IL-2 response, as they regulate the genes responsible for determining whether activated T cells survive, proliferate, or die. We have been studying a candidate PI3K-regulated transcription factor, c-Myb. c-Myb is one of three mammalian Myb proteins, all of which are transcription factors implicated in the regulation of proliferation, differentiation, and apoptosis (reviewed in reference 42). During T-cell activation, cell cycle progression in response to IL-2R signaling is associated with a sixfold induction of c-myb expression, with the highest levels seen around late G1 (57). Both c-Myb and its DNA binding activity are similarly up-regulated in response to IL-2 stimulation (77). c-myb is predominantly regulated by an attenuation block in the first intron of the gene (7, 71), and IL-2 treatment releases this block, allowing full-length c-myb mRNA to be transcribed (51). IL-2 mediated release of c-myb attenuation can be inhibited by rapamycin (51), which interferes with signals downstream of PI3K, suggesting a role for the PI3K pathway in c-myb regulation.
Myb proteins play an important role at a number of points in T-cell development. c-myb is absolutely required for early thymopoiesis (1), and it is also required for IL-2-mediated progression out of G1 phase during T-cell activation (24). Transgenic expression of oncogenic v-Myb leads to T-cell lymphomas (5), whereas mice expressing a dominant-negative Myb (MEnT) during T-cell development suffer a partial block in early thymopoiesis and have a proliferative defect in more mature cells (4). Thymocytes and resting splenocytes from MEnT mice are more susceptible to apoptosis than normal controls, implying that Myb proteins can act as survival factors during T-cell development. In the T-cell line EL4, expression of an inducible version of MEnT causes down-regulation of bcl-2 and apoptosis (62), and we and others have shown that the bcl-2 gene is a direct target of v-, c-, and B-Myb (23, 26, 62). More recently, the link between Myb proteins and apoptosis has been substantiated in a number of other cell types (for example, see reference 74).
Given that c-myb lies downstream of IL-2 and that Myb proteins can protect from cell death, we were interested to determine the precise means by which IL-2 acts to up-regulate c-myb, whether this involves signaling via PI3K, and if Myb proteins can affect the antiapoptotic signal from PI3K. We show here that the c-myb promoter can be activated by PI3K and PKB and that this activation requires conserved E2F and NF-κB sites. We demonstrate that activation of the endogenous c-myb gene in response to IL-2 stimulation can be blocked by chemical inhibitors of PI3K and NF-κB. Blocking Myb function in activated T cells results in a fivefold increase in apoptosis which cannot be rescued by bcl-2, while overexpressing v-Myb can protect activated cells from death. When MEnT transgenic mice are crossed to transgenic animals expressing activated PKB, the survival advantage conferred by the activated PKB during T-cell activation is abolished. These data show that maintenance of c-myb expression is dependent on signals from PI3K and define c-Myb as an important downstream effector of the PKB survival signal.
MATERIALS AND METHODS
IL-2 signaling experiments.
Spleens were disaggregated and single-cell suspensions were cultured at 1 × 106 cells/ml in 10% CO2. Activation medium was Dulbecco's modified Eagle's medium (Gibco) with 5% heat-inactivated fetal calf serum (Gibco), 2 mM l-glutamine, 1 mM sodium pyruvate (Gibco), 1 mM nonessential amino acids (Gibco), 20 ng of monothioglycerol (Sigma) per ml, and 1.25 μg of concanavalin A (ConA) (Sigma) per ml. After 72 h a sample of cells was analyzed by flow cytometry following staining with antibodies against T-cell receptor (TCR) β chain (phycoerythrin conjugated, clone H57-597; Pharmingen) and CD25 (fluorescein isothiocyanate [FITC] conjugated, clone 7D4; Pharmingen) and 7-amino-actinomycin D (7-AAD) (Sigma) to check activation and proliferation of cultures, and an RNA sample was taken. The remaining cells were washed twice in Dulbecco's modified Eagle's medium and then starved of ConA for 16 h. The cells were then treated with inhibitors for 1 h prior to restimulation and continuously thereafter with 10 ng of recombinant human IL-2 (AMS Biotechnology) per ml, and RNA samples were taken using RNAzol B (Biogenesis Ltd) at the times indicated in the figures. B6.1 cells were cultured as described in reference 55, with 10 ng of recombinant human IL-2/ml. For induction experiments, subconfluent cells were starved for 16 h in medium lacking IL-2. Cells were then treated with inhibitors for 1 h prior to IL-2 treatment and continuously thereafter. RNA was harvested at the times indicated in the figures. The following inhibitors were used: 50 μM LY294002 (Biomol), 50 μM PD98059 (New England Biolabs), and 25 μM pyrrolidine dithiocarbamate (PDTC) (Sigma). c-myb, c-jun, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were monitored in RNase protection assays (see below).
Western blotting.
Spleens were disaggregated and cultured as described above. After 48 h, some cells were harvested (activated population), and the remainder were starved of ConA for 24 h and either harvested immediately (starved population) or following stimulation with 10 ng of human IL-2/ml for 5 h (IL-2 population). Cells were washed twice with phosphate-buffered saline (PBS); harvested in 200 μl of lysis buffer (10 mM Tris-HCl [pH 7.4], 20% glycerol, 0.2 mM EDTA, 300 mM NaCl, 25 mM KCl, 5 μg of leupeptin/ml, 5 μg of pepstatin/ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM dithiothreitol) on ice for 30 min. The lysate was centrifuged for 10 min at 17,900 × g at 4°C. Supernatants were stored at −80°C. A total of 60 μg of total protein from each sample was run in a sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and blotted onto polyvinylidene difluoride membrane, pretreated with TBST (TBS containing 0.1% Tween 20) and 5% dry milk, and incubated for 1 h at room temperature (RT). Mouse monoclonal anti-c-myb (clone 1-1; Upstate Biotechnology) was used at 1 μg/ml. After the washes, goat anti-mouse immunoglobulin G (IgG)-horseradish peroxidase (Amersham) was used in TBST and 5% dry milk and incubated for 1 h at RT. Subsequent washes were done with TBST. The proteins were then visualized using an ECL kit (Amersham). Rabbit anti-mouse PKB (clone 9916; New England Biolabs) with anti-rabbit IgG-horseradish peroxidase as secondary antibody was used to visualize PKB as a loading control.
Transient transfections.
NIH 3T3 cells were transfected using Lipofectamine (Gibco) as described in reference 62, except that a total of 2 μg of DNA was used. RNA was harvested after 72 h using RNAzol B (Biogenesis Ltd). All experiments were done a minimum of five times and representative data are shown.
RNase protection assays.
Probes were produced by in vitro transcription using T7 or SP6 RNA polymerases (Promega) and [α-32P] riboUTP (Amersham). Samples were digested with 5 U of RNase ONE (Promega) and prepared for electrophoresis according to the manufacturer's instructions. Details of probe construction are available upon request. The c-myb probe is full length, 350 bp, and protects 254-bp; the c-jun probe is full length, 330 bp, and protects 180-bp; the GAPDH probe (Pharmingen) is full length, 125 bp, and protects 97 bp; the c-myb-βG reporter probe is full-length, 355 bp, and protects 315 bp; and the c-myb-αG reporter probe is full length, 105 bp, and protects 96 bp.
Constructs.
Expression vectors were as follows: the activated PI3K construct is rCD2p110 (49); activated PKB is gagPKB (13); kinase-dead dominant-negative PKB is HA-Akt(K179A) (32); RasV12, RasV12S35, and RasV12C40 are described elsewhere (30); and RasV12A38 is described in reference 50. E2F1 and E1A vectors were gifts of X. Lu and S. Mittnacht, respectively, and the IκBα vector was a gift of R. Hay. The c-myb-βG reporter construct was made by excision of a 2.3-kb EcoRI/NcoI fragment of murine c-myb genomic DNA (from the ATG start site [+1] to −2.3 kb) from plasmid PKR4A-R3 (72). Following end-repair of the EcoRI site, this fragment was cloned into SmaI/NcoI-digested pβ128 (73). The NcoI religation regenerates the ATG start codon in the same position as that of the human βG gene contained in pβ128. This reporter was mutagenized using the Sculptor in vitro mutagenesis kit from Amersham Pharmacia Biotech. All mutants were sequenced. Oligonucleotides used for mutagenesis were as follows, with base changes shown in bold type: E2F, GGACACTCCCCCTCCATACAAATCTGGCGCCCCTGC; NF-κB, GAGGTTTGGACACTGAGCCTCCCGCCAAATC; and GAL4, GGACACTCCCCCTCCTACTGTCCTCCGAGCGGAGTCTGGCGCCCCTGCAGTGC. The c-myb-αG reporter contains a 6.6-kb EcoRI/SpeI fragment from plasmid PKR4A-R3 carrying the same 2.3-kb upstream sequence as c-myb-βG, followed by the 5′ end of the c-myb gene and ending at a SpeI site 185 bp from the 3′ end of intron 1. This fragment was ligated into pSKII (Stratagene) along with a 700-bp SmaI/PstI fragment from the human α-globin gene containing plasmid HPα2 (17), which carries the 3′ end of the gene, beginning at the SmaI site (+120) within intron 1. Full details of plasmid construction are available on request.
Bandshifts.
Nuclear extracts from activated splenocytes cultured for 3 days with ConA were made exactly as described previously (6). Extracts were used at a concentration of 3.5 μg/μl. Jurkat plus TPA plus CI extract was purchased from Geneka Biotechnology Inc. and was used at a concentration of 5 μg/μl. In vitro translation was carried out using the TnT system (Promega). E2F and DP1 plasmid templates were a gift of N. Jones. E2F bandshifts were carried out as described previously (6). A total of 10 μg of nuclear extract or 3 μl of in vitro-translated protein was incubated with 1 ng of radiolabeled probe for 30 min at RT. Complexes were resolved on a 5% nondenaturing polyacrylamide gel (14:1 acrylamide:bisacrylamide) in 0.5× TBE. NF-κB bandshifts were carried out as described elsewhere (34). Ten micrograms of nuclear extract or 7 ng of purified p50 and p65, or a total of 14 ng of p50 plus p65 (2) was incubated with 1 ng of radiolabeled probe for 20 min at RT. Purified IκBα (gift of R. Hay) was added to some reaction mixtures at the indicated concentrations. Gel conditions were as for E2F. Rabbit polyclonal antibodies against p50 and p65 were gifts of R. Hay, and the anti-c-rel rabbit polyclonal antibody was a gift of S. Goodbourn. Antibodies against p52 and RelB were purchased from Santa Cruz Biotechnology. All antibodies were added 20 min before the labeled probe. Probes were annealed, end repaired with [α-32P]dCTP (Amersham) and avian myeloblastosis virus reverse transcriptase (Pharmacia), and purified on MicroSpin G25 columns (Pharmacia) prior to use. Oligonucleotide sequences were as follows (with base changes in bold type and binding consensus sequences for the E2F and NF-κB transcription factors underlined): wild-type E2F oligonucleotide, 5′ GGGAGGGGCGCCAGATTTGGCGGGAGGGGGAGT 3′; mutant E2F oligonucleotide 5′ GGGAGGGGCGCCAGATTTGTATGGAGGGGGAGT 3′; wild-type NF-κB oligonucleotide, 5′ GGGGGAGTGTCCAAACC 3′; and mutant NF-κB oligonucleotide, 5′ GGGGGTGTGTGGTAAACC 3′.
Survival assays.
The vMyb4 transgenic mice are described in (5), the MEnT mice in (4), and the Eμ-bcl-2-25 mice in (59). All these lines are on a C57B10 background. The gag-PKB transgenic line B6/PKB is described in (29), and is on a C57BL6 background. Mice were between 5 and 10 weeks of age when sacrificed. Following disaggregation, splenocytes were cultured for 3 days in conA. For all experiments, cells were stained with allophyocyanin-conjugated anti-TCR-β (Pharmingen) and phycoerythrin-conjugated anti-CD25 (Pharmingen). For detection of intracellular bcl-2 levels, cells were then washed in PBS and permeabilized in PBS–0.03% saponin–0.1% sodium azide. Cells were then stained with 20 μl of FITC-conjugated anti-mouse bcl-2 (clone 3F11; Pharmingen) or with 20 μl of FITC-conjugated IgG1 control antibody (Pharmingen) for 30 min. After washing, they were analyzed on a Becton Dickinson FACScalibur flow cytometer using Cellquest software. For detection of apoptosis, cells stained as above for TCR-β and CD25 were washed in PBS and resuspended in 500 μl of annexin V binding buffer (Nexins Research), 0.5 μl of annexin- V-FITC Conjugate (250 mg/ml stock; Nexins Research), and 0.5 μl of 7-AAD (1 mg/ml stock) for 15 min on ice prior to flow cytometry. All mouse experiments were done at least three times.
RESULTS
The c-myb gene lies downstream of PI3K in the IL-2 response.
Human thymic blast cells induce c-myb in response to IL-2 stimulation, and this induction can be prevented by rapamycin (51). As rapamycin is an indirect inhibitor of p70S6 kinase (p70S6K), which lies downstream of PI3K, we sought to determine whether the PI3K pathway was required for IL-2 induction of c-myb mRNA during the process of T-cell activation.
To simulate antigen activation of T cells in tissue culture, splenic T cells were isolated and treated with ConA. This treatment cross-links the TCR, resulting in expression of IL-2 and the high-affinity αβγ-IL-2R and progression into the cell cycle. T cells were activated and allowed to proliferate for 3 days and then were arrested in G1 by overnight incubation in medium devoid of IL-2. Following this synchronization, an average of 89% of cells were in G1/G0 (data not shown). Then, IL-2 was added to the medium, and cells were assessed for progression into the cell cycle and expression of c-myb in the presence and absence of various inhibitors of downstream IL-2 signaling pathways. After 18 h of IL-2 treatment in the absence of inhibitors, the percentage of cells in S, G2, and M phases had increased to 27% (data not shown). RNase protection assays showed that c-myb was induced 3.5-fold on average (three experiments) by 4 h after IL-2 treatment and that expression was maintained up to 24 h later (Fig. 1A, compare the first lane with the next four). Western blotting of starved and IL-2-stimulated extracts demonstrated that c-Myb protein was also induced after 5 h of IL-2 treatment, in comparison to PKB, whose levels are known to remain constant (29) (Fig. 1B) These data are in good agreement with those reported previously (45, 57, 77). However, when the PI3K inhibitor LY294002 was added to the culture, c-myb expression was not induced (Fig. 1A, IL-2 + LY), showing that during T-cell activation IL-2-mediated activation of c-myb expression is dependent upon signaling via the PI3K pathway. In contrast, c-myb was expressed normally if IL-2-stimulated cultures were treated with the MEK inhibitor PD98059 (Fig. 1A, IL-2 + PD), indicating that the MAP kinase pathway is not involved in regulation of c-myb. We wished to explore the induction of c-myb in more detail, and for this we turned to the cytotoxic T-cell line B6.1 (55). In contrast to primary T cells, B6.1 cells are a homogeneous population and are less susceptible to apoptosis in the absence of IL-2. They can be more efficiently arrested if starved of IL-2 for 16 h (an average of 93.2% of cells are in G0/G1; data not shown), and they will then proliferate in response to IL-2 addition. Furthermore, c-myb expression is correctly regulated during the IL-2 response (21). The upper panel of Fig. 1C shows that B6.1 cells growing exponentially contain c-myb mRNA, and this was greatly reduced upon IL-2 starvation (compare first two lanes). When IL-2 was added back, c-myb was reexpressed within 3 h (Fig. 1C, lane 3), and its induction was inhibited by LY294002 (Fig. 1C, compare third and fourth lanes) and unaffected by the MEK inhibitor PD98059 (44) (Fig. 1C, fifth lane). In contrast to c-myb, transcription of c-jun was almost completely inhibited by PD98059 and only slightly inhibited by LY294002 (Fig. 1C, right panel).
FIG. 1.
c-myb is activated by PI3K following IL-2 stimulation. T cells were starved for 16 h and stimulated with 10 ng of IL-2/ml in the presence or absence of various inhibitors. After the times indicated (in hours), total RNA was obtained from cells and c-myb, c-jun, and GAPDH expression was analyzed in RNase protection assays. (A) ConA-activated splenocytes were starved and stimulated with IL-2 ± 50 μM LY294002 or 50 μM PD98059. (B) ConA-activated splenocytes were starved and then stimulated with IL-2 for 5 h. Extracts were blotted and probed with anti-c-Myb antibody (upper lanes) or anti-PKB antibody as a loading control (lower lanes). (C) B6.1 cells were starved and stimulated with IL-2 ± 50 μM LY294002 or 50 μM PD98059. (D) B6.1 cells were starved and stimulated with IL-2 ± 25 μM PDTC.
NF-κB transcription factors are induced during T-cell activation by both TCR signaling (reviewed in reference 15) and perhaps by IL-2R activation (3; but see reference 28), and they have recently been shown to be activated in response to PI3K signaling (29, 31, 33, 43, 52). The c-myb promoter contains a well-conserved NF-κB binding site (see below), so we investigated whether PDTC, an inhibitor of NF-κB DNA binding (54), could also prevent IL-2 stimulation of c-myb in B6.1 cells. Figure 1D demonstrates that PDTC severely affected the IL-2 signal to c-myb but that it could not affect induction of c-jun, which is not regulated by NF-κB factors. Taken together, these data show that IL-2-stimulated transcription of the c-myb gene requires activation of PI3K and the presence of NF-κB transcription factors.
PI3K regulates the c-myb promoter.
Expression of c-myb is regulated at the levels of both transcription initiation and attenuation in the first intron (7, 71, 72). It is known that IL-2 signaling leads to an increase in c-myb transcription and release of attenuation, but how this might happen is unclear (51). The polymerase-pausing mechanism by which attenuation occurs has been proposed to be a function of the interactions that RNA polymerase makes with transcription factors at the promoter (76). A more processive polymerase can be generated when transcription factors such as E2F or p53 are present; other activators such as Sp1, although able to increase initiation, have no effect on elongation (9). Therefore, given the importance of the promoter as a regulator of both initiation and elongation, we chose to analyze the c-myb promoter in detail.
The c-myb promoter contains a region of 119 bp (−304 to −185) which is conserved between humans, mice, and chickens (66) and is therefore likely to contain regulatory sequences of importance. Within this region (Fig. 2A), there is a putative NF-κB site (−266 to −256) and an E2F site (−278 to −271). To examine whether the c-myb promoter could respond to PI3K signals, we made a reporter construct in which 2.3 kb of the murine c-myb upstream sequence, including the 119-bp conserved region, was linked to the human β-globin gene. This construct (c-myb-βG) was transfected into NIH 3T3 cells either alone or together with the effector plasmids detailed below. Seventy-two hours after transfection, RNA was harvested and hybridized in RNase protection assays to probes recognizing the βG sequences or, as an internal control, endogenous GAPDH.
FIG. 2.
The c-myb promoter is activated by PI3K and PKB. (A) Schematic of the murine c-myb promoter. E2F and NF-κB sites are underlined and shown in bold, and the putative SP1 site is overlined. (B) NIH 3T3 cells were transfected with the c-myb-βG reporter either alone or in concert with effector plasmids encoding activated PI3K, activated PKB, or dominant-negative PKB. Transcription from c-myb-βG and endogenous GAPDH was analyzed in RNase protection assays. (C) NIH 3T3 cells were transfected with c-myb-βG either alone or with the Ras effector mutants shown (see text). RNA analysis was as described above. (D) NIH 3T3 cells were transfected with c-myb-αG either alone or together with effector plasmids encoding activated PI3K or activated PKB.
In the first set of experiments, we tested whether the c-myb promoter could be activated by PI3K or its downstream effector kinase, PKB. We cotransfected c-myb-βG into NIH 3T3 cells in the presence or absence of plasmids encoding either activated PI3K (49) or activated (13) or dominant-negative (32) PKB. Both activated PI3K and PKB reproducibly up-regulated c-myb-βG to levels about fourfold over baseline (Fig. 2B, lanes 1 to 5). Dominant-negative PKB completely inhibited activation by PI3K (Fig. 2B, compare lanes 5 and 6), implying that it is the major downstream component of the PI3K signal to the c-myb promoter.
In a complementary approach, we also tested the ability of c-myb-βG to respond to plasmids encoding effector mutants of the Ras oncoprotein. RasV12 is an activated oncogenic form of Ras (75) and can stimulate multiple pathways, including the Raf/mitogen-activated protein (MAP) kinase pathway and the PI3K pathway. RasV12C40 can switch on PI3K but not Raf (30), whilst RasV12S35 activates Raf but not PI3K (50). Finally, RasV12A38 is inactive (50). As shown in Fig. 2C, oncogenic RasV12 strongly activated the c-myb promoter (lane 4), as did the PI3K effector RasV12C40 (lane 3). The Raf effector RasV12S35 could not stimulate the promoter above baseline levels (lane 2), and the inactive RasV12A38 mutant appeared to repress even baseline promoter activity (lane 5). Therefore, our transient-transfection data confirm that the c-myb promoter, and hence initiation of transcription of c-myb, is responsive to the PI3K pathway but not to activation of the Raf/MAP kinase cascade.
To examine whether attenuation of c-myb transcription could be relieved by PI3K signaling, we constructed a second reporter plasmid, c-myb-αG, which contains the −2.3-kb region of the c-myb promoter followed by the 5′ end of the c-myb gene, extending through exon 1 into intron 1 beyond the attenuation region. This was fused to the 3′ end of the human α-globin gene, from midway through intron 1 to the poly(A) site (17). This reporter should generate a hybrid c-myb-αG mRNA which is susceptible to regulation by attenuation in its first intron. Baseline levels of hybrid mRNA were detected when the reporter alone was transfected into NIH 3T3 cells, but cotransfection of either activated PI3K or activated PKB led to production of reporter mRNA (Fig. 2D, compare lane 1 with lanes 2 and 3), implying that attenuation was being relieved. To prove definitively that PI3K and PKB were relieving attenuation, we attempted to perform runoff analyses in NIH 3T3 cells transiently transfected with the reporter and effector plasmids, but unfortunately these experiments proved to be technically unfeasible.
Conserved E2F and NF-κB sites are important for c-myb promoter activity.
In order to define which region(s) of the c-myb promoter transduced the PI3K signal, we first had to find which transcription factors were required for promoter activity. We decided to analyze the conserved E2F and NF-κB sites in the promoter (Fig. 2A). To determine whether E2F and NF-κB factors could bind to these sites, we performed bandshifts with proteins made in vitro and also with activated T-cell nuclear extracts. Figure 3A shows that an oligonucleotide carrying the E2F site from the c-myb promoter bound in vitro-translated E2F1/DP1 (lane 1) and also a number of species in activated T-cell extracts (lane 3) which could be competed away with an excess of unlabeled wild-type oligonucleotide (lane 4). E2F1/DP1 could not bind to the site when it was mutated to the sequence that was shown in transient-transfection assays to severely reduce promoter activity (lane 2). Similar analysis of the NF-κB site demonstrated that purified p65 but not p50 protein could bind to the site (Fig. 3B, lanes 2 and 3). In addition to binding p65 homodimers, when equimolar amounts of p65 and p50 were added together the NF-κB site could bind a heterodimeric complex (Fig. 3B, lane 4), which could be supershifted by addition of an anti-p65 antibody (data not shown) and also an anti-p50 antibody (Fig. 3B, lane 1). Binding activity was reduced to nil upon addition of increasing amounts of purified IκBα (Fig. 3B, lanes 5, 6, and 7; 1, 5, and 10 ng, respectively), which is known to interfere with site recognition by NF-κB factors (2). In activated T-cell extracts, the NF-κB site recognized two bands (Fig. 3C, lane 1), the lower of which was nonspecific. The slower-migrating specific band could be efficiently competed with excess unlabeled wild-type oligonucleotide (lane 2) but only slightly by an excess of an unlabeled oligonucleotide in which the NF-κB site had been mutated (lane 3). Addition of both anti-p50 and anti-c-Rel antibodies supershifted small amounts of bound complex (lanes 5 and 9). Both the primary complex and these supershifted bands were dependent on the presence of nuclear extract (data not shown). Preimmune serum (lane 4) or antibodies against the NF-κB family members p65, p52, and RelB had no effect on complex mobility (lanes 6 to 8). Therefore, the E2F and NF-κB sites are able to bind their cognate proteins, both in vitro and in activated T-cell extracts.
FIG. 3.
E2F and NF-κB proteins bind to the c-myb promoter. (A) Bandshifts with an E2F probe from the c-myb promoter or a mutant thereof. Lanes 1 and 2, in vitro-translated E2F1 and DP1; lanes 3 and 4, activated T-cell extracts. (B) Bandshifts with an NF-κB probe from the c-myb promoter. Purified NF-κB proteins and IκBα were added as detailed in Materials and Methods. (C) Bandshifts with the c-myb NF-κB probe and activated T-cell extracts. Unlabeled competitor oligonucleotides and antibodies were added as shown.
Having established that the E2F and NF-κB sites were genuine, we looked at their functional significance. To analyze the E2F site, we cotransfected the c-myb-βG reporter construct into NIH 3T3 cells together with effector plasmids encoding E2F1 or E1A. Transcription from c-myb-βG was enhanced between 4- and 20-fold (in 10 separate experiments) by expression of either E2F1 (Fig. 4A, compare lanes 1 and 2) or E1A (Fig. 4A, compare lanes 3 and 4). Mutation of the E2F site in c-myb-βG resulted in the promoter being severely inhibited (Fig. 4B, compare lanes 1 and 2). Therefore, addition of extra E2F1, or E1A-mediated release of the cells' own supplies of E2F factors, results in the c-myb promoter being activated, and the conserved E2F site is needed for proper function of the promoter.
FIG. 4.
The E2F and NF-κB sites are required for basal and PI3K-activated transcription from the c-myb promoter. Transfections into NIH 3T3 cells and RNase protection analyses were performed as described above. (A) Cotransfection of c-myb-βG and E1A or E2F expression vectors results in up-regulation of c-myb-βG. (B) Individual or double mutation of the E2F and NF-κB sites results in loss of activity of the c-myb-βG reporter. (C) Lanes 1 to 3, mutation of the E2F site to a GAL4 site causes loss of PI3K responsiveness. Lanes 4 to 6, mutation of the E2F site to a GAL4 site and inhibition of NF-κB activity causes complete loss of PI3K inducibility.
To determine whether the c-myb promoter requires the conserved NF-κB site for activity, we mutated the site in c-myb-βG and tested the mutant promoter in transient-transfection assays. Loss of the NF-κB site resulted in a 50% reduction in promoter activity (Fig. 4B, compare lanes 1 and 3). A promoter carrying a double mutation of both the E2F and NF-κB sites was almost completely nonfunctional (Fig. 4B, lane 4), illustrating the importance of both of these sites. In summary then, the conserved E2F and NF-κB sites in the c-myb promoter can dictate whether the 2.3 kb of the c-myb upstream sequence included in our reporter construct is transcriptionally active.
PI3K and PKB require the E2F and NF-κB sites to activate the c-myb promoter.
Having shown that the E2F and NF-κB sites are essential components of the c-myb promoter, we wished to see whether they were also required for PI3K activation of c-myb transcription. As the E2F site is required for basal promoter activity, we were unable simply to delete it and assay the promoter for PI3K-mediated activation. Therefore, we replaced it with a site for GAL4 in our c-myb-βG reporter construct to make c-myb-GAL4-βG. c-myb-GAL4-βG has little or no activity except when GAL4 is present to support transcription (Fig. 4C, compare lanes 1 and 2). We assayed c-myb-GAL4-βG for its responsiveness to PI3K signals and found that it could not be appreciably superactivated by cotransfection of activated PI3K with GAL4 (lane 3). We did observe that the baseline activity of c-myb-GAL4-βG could still be augmented by activated PI3K (compare lanes 4 and 5), indicating that some PI3K responsiveness had been retained. However, this responsiveness was abolished if c-myb-GAL4-βG was cotransfected with activated PI3K in the presence of IκBα, which sequesters and inactivates NF-κB family members (lane 6). Taken together, these data suggest that the c-myb promoter responds to a PI3K signal which is transduced via PKB and that full promoter activation requires the presence of the conserved E2F and NF-κB sites.
Myb proteins protect activated T cells from death.
PKB is an important survival kinase, and it has been shown to protect against cell death in a number of circumstances, including following T-cell activation; Myb proteins are also antiapoptotic (see the introduction). To determine whether c-Myb is a downstream effector of PKB-mediated survival following IL-2 signaling, we decided to explore the survival function of c-Myb in activated T cells by using two lines of transgenic mice, vMyb4 and MEnT. vMyb4 animals express the v-Myb oncoprotein in their T cells and develop lymphomas with a latency of over a year (5), and MEnT transgenic mice express a dominant interfering Myb protein, also in a T-cell-specific fashion (4). MEnT consists of the Myb DNA binding domain fused to the Drosophila Engrailed repressor domain, and it efficiently and specifically represses Myb target genes (4, 56). Previously, our laboratory has shown that the thymocytes and resting splenocytes of MEnT mice have enhanced susceptibility to apoptosis and that this phenotype can be partially rescued by overexpression of bcl-2 (62).
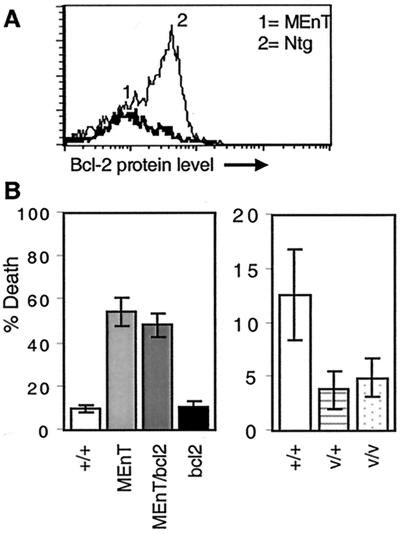
To see whether Myb proteins affect apoptosis following T-cell activation and induction of the IL-2 signaling cascade, splenocytes from MEnT and vMyb4 transgenic animals, together with nontransgenic controls, were isolated and activated by 3 days of ConA treatment. Cells were then harvested and analyzed by flow cytometry. Activated T cells were identified by their expression of αβTCR and CD25, the IL-2Rα chain. First, we determined whether expression of the Myb target gene bcl-2, which is normally upregulated from very low levels during T-cell activation (69), was affected in MEnT mice. Figure 5A shows bcl-2 protein levels in activated T cells and demonstrates that there is little or no expression in MEnT cells (peak 1), relative to nontransgenic controls (peak 2). We then assessed the amount of cell death occurring following T-cell activation by annexin V and 7-AAD staining of αβTCR+ CD25+ cells. MEnT T cells were far more susceptible to death than nontransgenic controls; an average of 57% of MEnT cells had died, in contrast to 11% of nontransgenic cells (Fig. 5B, left panel). Conversely, expression of v-myb was antiapoptotic in activated T cells. Only around 5% of cells were dead in v-Myb4 transgenics, in contrast to 12% of nontransgenic control cells (Fig. 5B, right panel).
FIG. 5.
Myb is a survival factor during T-cell activation. Splenocytes were activated with ConA and assessed for bcl-2 levels and apoptosis as described in the text. Activated T cells were gated for expression of αβTCR and CD25. (A) Activated MEnT splenocytes have reduced bcl-2 expression. Peak 1, ConA-stimulated MEnT splenocytes; peak 2, ConA-stimulated nontransgenic splenocytes. (B) Left panel: MEnT causes enhanced apoptosis during T-cell activation. Right panel: v-Myb protects activated cells from apoptosis. The percentages of dead and dying cells in the cultures were quantitated by annexin V and 7-AAD staining. Error bars indicate the standard deviation from the mean of at least three separate experiments.
One of the characteristics of death following T-cell activation is that it cannot be rescued by overexpression of bcl-2 (58). Activated T cells from MEnT mice crossed to the Eμ-bcl-2-25 strain (59), and therefore doubly transgenic for MEnT and bcl-2, were examined for apoptosis. bcl-2 overexpression could not rescue the MEnT phenotype, either following ConA activation (Fig. 5B, left panel) or if cells were activated with anti-CD3 (data not shown). Therefore, Myb proteins must be able to influence a form of apoptosis refractory to the protective effects of bcl-2.
To directly determine whether loss of c-Myb activity could affect PKB-mediated survival in activated T cells, we obtained transgenic mice expressing activated PKB (B6/PKB) in their T cells (29). These mice exhibit enhanced survival of mature T cells. We crossed B6/PKB animals with MEnT mice to generate MEnT/PKB double transgenic offspring and then examined the survival of MEnT/PKB splenocytes compared to littermate controls following ConA activation. After 2 days in culture, the apoptotic susceptibility of activated T cells (again identified by their expression of αβTCR and CD25) was determined by staining with annexin V-FITC and 7-AAD as described above. A representative experiment is shown in Fig. 6. The numbers of dead splenocytes are increased in all cases relative to our previous results, probably due to a background-specific variation in survival in culture (K. Weston, unpublished observations). Forty-nine percent of nontransgenic activated splenocytes did not stain with either annexin V-FITC or 7-AAD and were therefore still alive. As expected, the activated B6/PKB splenocytes survived far better than did the nontransgenic control cells, with 70% still alive. In contrast, only 28% of MEnT activated splenocytes were still surviving. Crucially, the MEnT/PKB splenocyte cultures contained 32% activated live cells and therefore resembled the MEnT cultures. Therefore, Myb activity is required for PKB to act as a survival factor during T-cell activation.
FIG. 6.
Myb lies downstream of PKB. Splenocytes from 5-week-old littermates derived from a MEnT × B6/PKB cross were activated with ConA for 2 days. Activated T cells were gated for expression of αβTCR and CD25 and assessed for apoptosis by staining with annexin V-FITC and 7-AAD.
DISCUSSION
The three principal means by which the IL-2R transmits signals into the cell are the Ras/MAP kinase, the PI3K, and the JAK-STAT pathways (for a review, see reference 39). Using a combination of Ras effector mutants, small molecule inhibitors, and activated and dominant-negative proteins, we have demonstrated that the c-myb gene is not regulated by MAP kinases but only by components of the PI3K pathway. The PI3K signal to c-myb appears to require PKB, as dominant-negative PKB can almost completely block the effects of activated PI3K on c-myb promoter activity. Although we have not directly examined the JAK-STAT pathway, we do not think that it plays a significant part in control of c-myb transcription, as the c-myb promoter does not contain any STAT binding sites and the two crucial regulators of the promoter are members of the E2F and NF-κB families, which do not require the JAK-STAT pathway for their expression during T-cell activation (10, 63).
Our transient-transfection experiments strongly suggest that the principal transcription factor responsible for transmitting the activating PI3K-PKB signal to the c-myb gene is a member of the E2F family and that NF-κB is necessary but not sufficient for full promoter activity. This fits well with recent reports that both these families can indeed be regulated by PI3K and PKB (10, 11, 29, 31, 33, 43, 52). However, whilst it is likely that E2F activity is induced concomitant with IL-2-mediated progression into G1 and S phase, NF-κB family members are induced immediately upon stimulation of a resting T cell (reviewed in reference 15), and regulation of NF-κB by IL-2 is a matter of dispute (3, 28). In our system, NF-κB binding activity is present in nuclear extracts made from B6.1 cells starved of IL-2 (K. Weston, unpublished observations), suggesting that regulation by IL-2 is not essential. Taken together, these data suggest that E2F induction is the most likely means by which IL-2 stimulates c-myb transcription, with NF-κB proteins acting to boost levels of c-myb mRNA once E2F binds the promoter.
The c-myb gene is regulated in T cells by a combination of transcriptional activation and release of attenuation (51, 64). Previously, IL-2 has been shown to regulate both these processes (51), and PI3K is clearly required for at least activation, as no c-myb transcript is produced when the PI3K inhibitor LY294002 is added during IL-2 stimulation of T cells. Our transient-transfection experiments do not demonstrate directly that PI3K can affect both promoter-mediated up-regulation of initiation and elongation, although we did show that a reporter construct bearing the c-myb attenuation sequence is switched on by both activated PI3K and PKB. Equally, we have not directly addressed the question of whether E2F and NF-κB affect attenuation. Runoff experiments in primary T cells in which E2F and NF-κB have been inhibited are necessary to prove this point unambiguously. However, as E2F, through which much of the PI3K signal to the c-myb promoter is transmitted, is known to promote both initiation and elongation (9) and NF-κB factors may do the same (8), we feel that the promoter is likely to be the principal determinant dictating the degree of elongation occurring.
Although a number of proteins, including c-Myb itself (27, 41), an inducible factor termed CMAT (46), and c-Jun (40), have been suggested as regulators of the human c-myb promoter, their significance is unclear, as none of the sites mapped are conserved between species and their functional relevance has not been established. In contrast, the conserved E2F site in the c-myb promoter has been recognized for some time (36), and E2F has been shown by others to be important for c-myb promoter activity (37, 53). Recently, a detailed analysis of the E2F element showed that a number of E2F family members bind the site and that SP1 can cooperate with E2F to augment transcriptional activation (14). Our data regarding the E2F site are in good agreement with these published studies. NF-κB proteins have previously been proposed to bind to sites within c-myb intron 1, but it is unclear whether or how this affects endogenous c-myb transcription, as studies have given conflicting results, suggesting either enhancement (60) or inhibition (65) of elongation. Although we do not discount the possibility that NF-κB binds other sites, our results point to a prominent role for the conserved NF-κB site that we have identified in regulation of the c-myb promoter.
In this paper, we have extended our previous results implicating Myb proteins as survival factors, and we have also placed c-Myb downstream of PKB, which is an important survival kinase in T lymphocytes (29) and many other cell types (reviewed in reference 19). We would like to suggest that the process of activation-induced cell death (AICD) is being exacerbated by MEnT and inhibited by v-Myb. AICD occurs after repeated stimulation of the TCR complex, is enhanced by IL-2 (48), and can be triggered in vitro by treatment of activated T cells with anti-CD3 antibody or ConA (67, 68). We show here that loss of Myb function sensitizes activated T cells to apoptosis under in vitro conditions which are likely to be causing some degree of AICD. Furthermore, AICD appears to kill T cells by a bcl-2-independent mechanism (47, 58), a feature of MEnT-induced apoptosis. As AICD is mediated predominantly by the Fas pathway, and Fas killing occurs in the absence of transcription (38), we propose that Myb proteins, rather than acting after the triggering of AICD, transcriptionally up-regulate protective factors which result in cells being more resistant to AICD. Thus, immediately after T cells are activated, Myb factors would play an important part in allowing T-cell expansion rather than AICD to occur. Later on during the immune response, c-myb is down-regulated (18), and therefore cells would be more susceptible to AICD. Intriguingly, in lpr/lpr and gld/gld mice, in which Fas and its receptor, respectively, are defective (61, 70), c-myb is expressed at extremely high levels in peripheral T cells (35), suggesting a complex regulatory relationship between c-myb and the Fas pathway. To shed light on this, we are currently attempting to identify the key Myb-regulated genes involved.
ACKNOWLEDGMENTS
We thank D. Cantrell, J. Downward, S. Goodbourn, R. Hay, N. Jones, A. Klippel, X. Lu, C. Marshall, S. Mittnacht, M. Nabholz, G. Thomas, R. Treisman, and R. Watson for gifts of plasmids, reagents, and cells; P. Ohashi for kindly supplying B6/PKB mice; and Doreen Cantrell, Steve Goodbourn, Chris Marshall, and Richard Treisman for criticism and advice.
This work was supported by an MRC Studentship to A. J. L., by the Human Frontier Science Program (A.C.), and by the Cancer Research Campaign.
REFERENCES
- 1.Allen R D, III, Bender T P, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078. doi: 10.1101/gad.13.9.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos F, Thompson J, Rodriguez M S, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκB alpha negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arima N, Kuziel W A, Grdina T A, Greene W C. IL-2-induced signal transduction involves the activation of nuclear NF-kappa B expression. J Immunol. 1992;149:83–91. [PubMed] [Google Scholar]
- 4.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T cell development. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 5.Badiani P, Kioussis D, Swirsky D, Lampert I, Weston K. T-cell lymphomas in v-Myb transgenic mice. Oncogene. 1996;13:2205–2212. [PubMed] [Google Scholar]
- 6.Belbrahem A, Godden-Kent D, Mittnacht S. Regulation and activity of the retinoblastoma protein family in growth factor-deprived and TGF(beta)-treated keratinocytes. Exp Cell Res. 1996;225:286–293. doi: 10.1006/excr.1996.0178. [DOI] [PubMed] [Google Scholar]
- 7.Bender T P, Thompson C, Kuehl W M. Differential expression of c-myb mRNA in murine B lymphomas by a block to transcription elongation. Science. 1987;237:1473–1476. doi: 10.1126/science.3498214. [DOI] [PubMed] [Google Scholar]
- 8.Biragyn A, Nedospasov S A. Lipopolysaccharide-induced expression of TNF-alpha gene in the macrophage cell line ANA-1 is regulated at the level of transcription processivity. J Immunol. 1995;155:674–683. [PubMed] [Google Scholar]
- 9.Blau J, Xiao H, McCracken S, O'Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan P, Babbage J W, Burgering B M, Groner B, Reif K, Cantrell D A. Phosphatidylinositol 3-kinase couples the interleukin-2 receptor to the cell cycle regulator E2F. Immunity. 1997;7:679–689. doi: 10.1016/s1074-7613(00)80388-x. [DOI] [PubMed] [Google Scholar]
- 11.Brennan P, Babbage J W, Thomas G, Cantrell D. p70s6k integrates phosphatidylinositol 3-kinase and rapamycin-regulated signals for E2F regulation in T lymphocytes. Mol Cell Biol. 1999;19:4729–4738. doi: 10.1128/mcb.19.7.4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 13.Burgering B M, Coffer P J. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- 14.Campanero M R, Armstrong M, Flemington E. Distinct cellular factors regulate the c-myb promoter through its E2F element. Mol Cell Biol. 1999;19:8442–8450. doi: 10.1128/mcb.19.12.8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantrell D A. T cell antigen receptor signal transduction pathways. Cancer Surv. 1996;27:165–175. [PubMed] [Google Scholar]
- 16.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 17.Charnay P, Treisman R, Mellon P, Chao M, Axel R, Maniatis T. Differences in human alpha- and beta-globin gene expression in mouse erythroleukemia cells: the role of intragenic sequences. Cell. 1984;38:251–263. doi: 10.1016/0092-8674(84)90547-6. [DOI] [PubMed] [Google Scholar]
- 18.Churilla A M, Braciale T J, Braciale V L. Regulation of T lymphocyte proliferation. Interleukin 2-mediated induction of c-myb gene expression is dependent on T lymphocyte activation state. J Exp Med. 1989;170:105–121. doi: 10.1084/jem.170.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datta S R, Brunet A, Greenberg M E. Cellular survival: a play in three Akts. Genes Dev. 1999;13:2905–2927. doi: 10.1101/gad.13.22.2905. [DOI] [PubMed] [Google Scholar]
- 20.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 21.Dautry F, Weil D, Yu J, Dautry-Varsat A. Regulation of pim and myb mRNA accumulation by interleukin 2 and interleukin 3 in murine hematopoietic cell lines. J Biol Chem. 1988;263:17615–17620. [PubMed] [Google Scholar]
- 22.del Peso L, Gonzalez-Garcia M, Page C, Herrera R, Nunez G. Interleukin-3-induced phosphorylation of BAD through the protein kinase Akt. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 23.Frampton J, Ramqvist T, Graf T. v-Myb of E26 leukemia virus up-regulates bcl-2 and suppresses apoptosis in myeloid cells. Genes Dev. 1996;10:2720–2731. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]
- 24.Gewirtz A M, Anfossi G, Venturelli D, Valpreda S, Sims R, Calabretta B. G1/S transition in normal human T-lymphocytes requires the nuclear protein encoded by c-myb. Science. 1989;245:180–183. doi: 10.1126/science.2665077. [DOI] [PubMed] [Google Scholar]
- 25.Gille H, Downward J. Multiple ras effector pathways contribute to G(1) cell cycle progression. J Biol Chem. 1999;274:22033–22040. doi: 10.1074/jbc.274.31.22033. [DOI] [PubMed] [Google Scholar]
- 26.Grassilli E, Salomoni P, Perrotti D, Franceschi C, Calabretta B. Resistance to apoptosis in CTLL-2 cells overexpressing B-Myb is associated with B-Myb-dependent bcl-2 induction. Cancer Res. 1999;59:2451–2456. [PubMed] [Google Scholar]
- 27.Guerra J, Withers D A, Boxer L M. Myb binding sites mediate negative regulation of c-myb expression in T-cell lines. Blood. 1995;86:1873–1880. [PubMed] [Google Scholar]
- 28.Iacobelli M, Rohwer F, Shanahan P, Quiroz J A, McGuire K L. IL-2-mediated cell cycle progression and inhibition of apoptosis does not require NF-kappa B or activating protein-1 activation in primary human T cells. J Immunol. 1999;162:3308–3315. [PubMed] [Google Scholar]
- 29.Jones R G, Parsons M, Bonnard M, Chan V S, Yeh W C, Woodgett J R, Ohashi P S. Protein kinase B regulates T lymphocyte survival nuclear factor kappaB activation, and Bcl-X(L) levels in vivo. J Exp Med. 2000;191:1721–1734. doi: 10.1084/jem.191.10.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joneson T, White M A, Wigler M H, Bar-Sagi D. Stimulation of membrane ruffling and MAP kinase activation by distinct effectors of Ras. Science. 1996;271:810–812. doi: 10.1126/science.271.5250.810. [DOI] [PubMed] [Google Scholar]
- 31.Kane L P, Shapiro V S, Stokoe D, Weiss A. Induction of NF-kappaB by the Akt/PKB kinase. Curr Biol. 1999;9:601–604. doi: 10.1016/s0960-9822(99)80265-6. [DOI] [PubMed] [Google Scholar]
- 32.Kulik G, Weber M J. Akt-dependent and -independent survival signaling pathways utilized by insulin-like growth factor I. Mol Cell Biol. 1998;18:6711–6718. doi: 10.1128/mcb.18.11.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madrid L V, Wang C Y, Guttridge D C, Schottelius A J, Baldwin A S, Jr, Mayo M W. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-κB. Mol Cell Biol. 2000;20:1626–1638. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matthews J R, Botting C H, Panico M, Morris H R, Hay R T. Inhibition of NF-kappaB DNA binding by nitric oxide. Nucleic Acids Res. 1996;24:2236–2242. doi: 10.1093/nar/24.12.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mountz J D, Steinberg A D. Studies of c-myb gene regulation in MRL-lpr/lpr mice. Identification of a 5′ c-myb nuclear protein binding site and high levels of binding factors in nuclear extracts of lpr/lpr lymph node cells. J Immunol. 1989;142:328–335. [PubMed] [Google Scholar]
- 36.Mudryj M, Hiebert S W, Nevins J R. A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 1990;9:2179–2184. doi: 10.1002/j.1460-2075.1990.tb07387.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller H, Bracken A P, Vernell R, Moroni M C, Christians F, Grassilli E, Prosperini E, Vigo E, Oliner J D, Helin K. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15:267–285. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagata S, Golstein P. The Fas death factor. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 39.Nelson B H, Willerford D M. Biology of the interleukin-2 receptor. Ady Immunol. 1998;70:1–81. doi: 10.1016/s0065-2776(08)60386-7. [DOI] [PubMed] [Google Scholar]
- 40.Nicolaides N C, Correa I, Casadevall C, Travali S, Soprano K J, Calabretta B. The Jun family members, c-Jun and JunD, transactivate the human c-myb promoter via an Ap1-like element. J Biol Chem. 1992;267:19665–19672. [PubMed] [Google Scholar]
- 41.Nicolaides N C, Gualdi R, Casadevall C, Manzella L, Calabretta B. Positive autoregulation of c-myb expression via Myb binding sites in the 5′ flanking region of the human c-myb gene. Mol Cell Biol. 1991;11:6166–6176. doi: 10.1128/mcb.11.12.6166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh I H, Reddy E P. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 43.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 44.Pang L, Sawada T, Decker S J, Saltiel A R. Inhibition of MAP kinase kinase blocks the differentiation of PC-12 cells induced by nerve growth factor. J Biol Chem. 1995;270:13585–13588. doi: 10.1074/jbc.270.23.13585. [DOI] [PubMed] [Google Scholar]
- 45.Pauza D C. Regulation of human T-lymphocyte gene expression by interleukin 2: immediate response genes include the proto-oncogene c-myb. Mol Cell Biol. 1987;7:342–348. doi: 10.1128/mcb.7.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phan S, Feeley B, Withers D, Boxer L. Identification of an inducible regulator of c-myb expression during T-cell activation. Mol Cell Biol. 1996;16:2387–2393. doi: 10.1128/mcb.16.5.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reap E A, Felix N J, Wolthusen P A, Kotzin B L, Cohen P L, Eisenberg R A. bcl-2 transgenic Lpr mice show profound enhancement of lymphadenopathy. J Immunol. 1995;155:5455–5462. [PubMed] [Google Scholar]
- 48.Refaeli Y, Van Parijs L, London C A, Tschopp J, Abbas A K. Biochemical mechanisms of IL-2-regulated Fas-mediated T cell apoptosis. Immunity. 1998;8:615–623. doi: 10.1016/s1074-7613(00)80566-x. [DOI] [PubMed] [Google Scholar]
- 49.Reif K, Nobes C D, Thomas G, Hall A, Cantrell D A. Phosphatidylinositol 3-kinase signals activate a selective subset of Rac/Rho-dependent effector pathways. Curr Biol. 1996;6:1445–1455. doi: 10.1016/s0960-9822(96)00749-x. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez-Viciana P, Warne P H, Khwaja A, Marte B M, Pappin D, Das P, Waterfield M D, Ridley A, Downward J. Role of phosphoinositide 3-OH kinase in cell transformation and control of the actin cytoskeleton by Ras. Cell. 1997;89:457–467. doi: 10.1016/s0092-8674(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 51.Rohwer F, Todd S, McGuire K L. The effect of IL-2 treatment on transcriptional attenuation in proto-oncogenes pim-1 and c-myb in human thymic blast cells. J Immunol. 1996;157:643–649. [PubMed] [Google Scholar]
- 52.Romashkova J A, Makarov S S. NF-kappaB is a target of AKT in anti-apoptotic PDGF signalling. Nature. 1999;401:86–90. doi: 10.1038/43474. [DOI] [PubMed] [Google Scholar]
- 53.Sala A, Nicolaides N C, Engelhard A, Bellon T, Lawe D C, Arnold A, Grana X, Giordano A, Calabretta B. Correlation between E2F-1 requirement in the S phase and E2F-1 transactivation of cell cycle-related genes in human cells. Cancer Res. 1994;54:1402–1406. [PubMed] [Google Scholar]
- 54.Schreck R, Meier B, Mannel D N, Droge W, Baeuerle P A. Dithiocarbamates as potent inhibitors of nuclear factor kappa B activation in intact cells. J Exp Med. 1992;175:1181–1194. doi: 10.1084/jem.175.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sekaly R P, MacDonald H R, Zaech P, Nabholz M. Cell cycle regulation of cloned cytolytic T cells by T cell growth factor: analysis by flow microfluorometry. J Immunol. 1982;129:1407–1414. [PubMed] [Google Scholar]
- 56.Shapiro L H. Myb and Ets proteins cooperate to transactivate an early myeloid gene. J Biol Chem. 1995;270:8763–8771. doi: 10.1074/jbc.270.15.8763. [DOI] [PubMed] [Google Scholar]
- 57.Stern J B, Smith K A. Interleukin-2 induction of T-cell G1 progression and c-myb progression. Science. 1986;233:203–206. doi: 10.1126/science.3523754. [DOI] [PubMed] [Google Scholar]
- 58.Strasser A, Harris A W, Huang D C S, Krammer P H, Cory S. Bcl-2 and Fas/APO-1 regulate distinct pathways to lymphocyte apoptosis. EMBO J. 1995;14:6136–6147. doi: 10.1002/j.1460-2075.1995.tb00304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Strasser A, Harris A W, Vaux D L, Webb E, Bath M L, Adams J M, Cory S. Abnormalities of the immune system induced by dysregulated bcl-2 expression in transgenic mice. Curr Top Microbiol Immunol. 1990;166:175–181. doi: 10.1007/978-3-642-75889-8_22. [DOI] [PubMed] [Google Scholar]
- 60.Suhasini M, Pilz R B. Transcriptional elongation of c-myb is regulated by NF-kappaB (p50/RelB) Oncogene. 1999;18:7360–7369. doi: 10.1038/sj.onc.1203158. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi T, Tanaka M, Brannan C I, Jenkins N A, Copeland N G, Suda T, Nagata S. Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 62.Taylor D, Badiani P, Weston K. A dominant interfering Myb mutant causes apoptosis in T cells. Genes Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 63.Thomis D C, Aramburu J, Berg L J. The Jak family tyrosine kinase Jak3 is required for IL-2 synthesis by naive/resting CD4+ T cells. J Immunol. 1999;163:5411–5417. [PubMed] [Google Scholar]
- 64.Thompson C B, Challoner P B, Neiman P E, Groudine M. Expression of the c-myb proto-oncogene during cellular proliferation. Nature. 1986;319:374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- 65.Toth C R, Hostutler R F, Baldwin A S, Jr, Bender T P. Members of the nuclear factor kappa B family transactivate the murine c-myb gene. J Biol Chem. 1995;270:7661–7671. doi: 10.1074/jbc.270.13.7661. [DOI] [PubMed] [Google Scholar]
- 66.Urbanek P, Dvorak M, Bartunek P, Pecenka V, Paces V, Travnicek M. Nucleotide sequence of chicken myb proto-oncogene promoter region: detection of an evolutionarily conserved element. Nucleic Acids Res. 1988;16:11521–11531. doi: 10.1093/nar/16.24.11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Parijs L, Ibraghimov A, Abbas A K. The roles of costimulation and Fas in T cell apoptosis and peripheral tolerance. Immunity. 1996;4:321–328. doi: 10.1016/s1074-7613(00)80440-9. [DOI] [PubMed] [Google Scholar]
- 68.Van Parijs L, Refaeli Y, Lord J D, Nelson B H, Abbas A K, Baltimore D. Uncoupling IL-2 signals that regulate T cell proliferation, survival, and Fas-mediated activation-induced cell death. Immunity. 1999;11:281–288. doi: 10.1016/s1074-7613(00)80103-x. [DOI] [PubMed] [Google Scholar]
- 69.Veis D J, Sentman C L, Bach E A, Korsmeyer S J. Expression of the Bcl-2 protein in murine and human thymocytes and in peripheral T lymphocytes. J Immunol. 1993;151:2546–2554. [PubMed] [Google Scholar]
- 70.Watanabe-Fukunaga R, Brannan C I, Copeland N G, Jenkins N A, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- 71.Watson R J. A transcriptional arrest mechanism involved in controlling constitutive levels of mouse c-myb mRNA. Oncogene. 1988;2:267–272. [PubMed] [Google Scholar]
- 72.Watson R J, Dyson P J, McMahon J. Multiple c-myb transcript cap sites are variously utilized in cells of mouse haemopoietic origin. EMBO J. 1987;6:1643–1651. doi: 10.1002/j.1460-2075.1987.tb02413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weston K. An enhancer element in the short unique region of human cytomegalovirus regulates the production of a group of abundant immediate early transcripts. Virology. 1988;162:406–416. doi: 10.1016/0042-6822(88)90481-3. [DOI] [PubMed] [Google Scholar]
- 74.White J R, Weston K. Myb is required for self-renewal in a model system of early hematopoiesis. Oncogene. 2000;19:1196–1205. doi: 10.1038/sj.onc.1203394. [DOI] [PubMed] [Google Scholar]
- 75.White M A, Nicolette C, Minden A, Polverino A, Van Aelst L, Karin M, Wigler M H. Multiple Ras functions can contribute to mammalian cell transformation. Cell. 1995;80:533–541. doi: 10.1016/0092-8674(95)90507-3. [DOI] [PubMed] [Google Scholar]
- 76.Yankulov K, Blau J, Purton T, Roberts S, Bentley D L. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 77.Zeng Z, Yang H, Huang Z Z, Chen C, Wang J, Lu S C. The role of c-Myb in the up-regulation of methionine adenosyltransferase 2A expression in activated Jurkat cells. Biochem J. 2001;353:163–168. [PMC free article] [PubMed] [Google Scholar]
- 78.Zong W X, Edelstein L C, Chen C, Bash J, Gelinas C. The prosurvival Bcl-2 homolog Bfl-1/A1 is a direct transcriptional target of NF-kappaB that blocks TNFalpha-induced apoptosis. Genes Dev. 1999;13:382–387. doi: 10.1101/gad.13.4.382. [DOI] [PMC free article] [PubMed] [Google Scholar]