Abstract
The “pocket” proteins pRb, p107, and p130 are a family of negative growth regulators. Previous studies have demonstrated that overexpression of pRb can repress transcription by RNA polymerase (Pol) I. To assess whether pRb performs this role under physiological conditions, we have examined pre-rRNA levels in cells from mice lacking either pRb alone or combinations of the three pocket proteins. Pol I transcription was unaffected in pRb-knockout fibroblasts, but specific disruption of the entire pRb family deregulated rRNA synthesis. Further analysis showed that p130 shares with pRb the ability to repress Pol I transcription, whereas p107 is ineffective in this system. Production of rRNA is abnormally elevated in Rb−/− p130−/− fibroblasts. Furthermore, overexpression of p130 can inhibit an rRNA promoter both in vitro and in vivo. This reflects an ability of p130 to bind and inactivate the upstream binding factor, UBF. The data imply that rRNA synthesis in living cells is subject to redundant control by endogenous pRb and p130.
The related “pocket” proteins pRb, p107, and p130 are potent suppressors of growth and proliferation that act by modulating transcription (11, 28). They are abundant proteins that bind and regulate many cellular targets, which together may account for their pleiotropic biological effects (reviewed in references 9, 16, 28, and 44). Some of the most important targets are the E2F transcription factors, which play key roles in promoting cell cycle progression (9, 28). Binding of pocket proteins to E2F can block expression of genes that are required for DNA replication (9, 16, 28, 44). Id2 is another protein that is bound by pRb (18, 24, 33), and recent genetic evidence shows that it is a critical target for pocket proteins in vivo (25). As well as these and other factors used by RNA polymerase (Pol) II, pRb has been shown to be able to repress transcription by Pols I and III (4, 51). The rRNA and tRNA produced by these polymerases account for ∼95% of all cellular RNA and therefore constitute a very substantial investment in biosynthetic machinery (48). Repression of Pols I and III could potentially provide a powerful mechanism to help pRb restrain the accumulation of mass that constitutes cell growth (30, 50).
There is considerable evidence that pRb and its relatives control Pol III transcription in vivo, including transient transfection assays and genetic experiments (5, 41–43, 51). The most compelling findings are nuclear run-on data, which show that synthesis of tRNA and 5S rRNA is significantly elevated in Rb−/− fibroblasts when compared to that in matched wild-type cells (41, 51). In the case of Pol I, on the other hand, the in vivo relevance of pRb-mediated transcriptional repression is less clearly established. Immunofluorescence analyses provided the initial indication that pRb may interact with the Pol I machinery in cells; these studies revealed that pRb accumulates in the nucleolus as U937 cells differentiate and down-regulate Pol I transcription (4, 39). pRb also accumulates in the nucleoli of confluent fibroblasts, which again correlates with a decrease in rRNA synthesis (14). Nucleolar localization, however, does not necessarily mean that pRb is involved in controlling Pol I transcription, since this organelle is sometimes used to sequester proteins with no known role in ribosome biogenesis, such as Mdm2, Cdc14, and p19Arf (3, 10, 49). Colocalization data therefore cannot be taken as proof that pRb regulates Pol I in vivo. More direct evidence has come recently from transient transfections, which have shown that a pRb expression vector can repress a cotransfected ribosomal DNA (rDNA) promoter (14, 36). However, such experiments rely on overexpression, which can sometimes force interactions that may not occur at physiological concentrations. Given the importance of the pRb family in controlling cell growth, we were keen to determine the role of pRb and its relatives in regulating Pol I within the cell. We have therefore adopted a genetic approach to test the effect of endogenous pRb on pre-rRNA synthesis in vivo. We find no evidence of overexpression in fibroblasts derived from Rb−/− mice. However, rRNA levels are abnormally elevated in cells lacking the entire pRb family or a combination of pRb and p130, which suggests redundancy in the control mechanism. Consistent with this, we find that the pocket protein p130 shares with pRb an ability to bind and repress the Pol I-specific factor UBF (upstream binding factor). The results indicate that p130 and pRb display functional overlap in down-regulating rRNA synthesis in cells.
MATERIALS AND METHODS
Cell culture, transfection, and RNA analysis.
Rb+/+ and Rb−/− fibroblasts were cultured as described previously (41). Primary cultures of wild-type, double knockout, and triple knockout embryonic fibroblasts were grown in BHK-21 medium supplemented with 1 mM sodium pyruvate, 1× nonessential amino acids, 0.1 mM 2-mercaptoethanol, 100 U of penicillin per ml, 100 μg of streptomycin per ml, and 10% fetal calf serum. RNA extraction and primer extension analyses were performed as previously described (1). Northern blotting and reverse transcription (RT)-PCR were carried out as described previously (42). Culture and transfection of NIH 3T3 cells and RNA analysis were carried out as described previously (46). The pMr1930-BH reporter is an artificial ribosomal minigene construct containing a murine rDNA promoter fragment (from −1930 to +292) fused to a fragment that contains two terminator elements (46).
Protein purification and transcription in vitro.
Expression and purification of glutathione S-transferase (GST) fusion proteins, partial purification of the Pol I transcription machinery, and in vitro transcription assays were all conducted as described previously (47). The pMrWT template contains a 324-bp fragment of murine rDNA, which includes sequence from −170 to +155; this plasmid was truncated with NdeI to yield a 371-nucleotide runoff transcript (46).
Protein binding assays.
Expression of UBF, construction of UBF mutants, and pull-down assays have been described previously (47). Coimmunoprecipitation was performed as previously described (43).
RESULTS
Pol I activity is not deregulated in pRb-knockout fibroblasts.
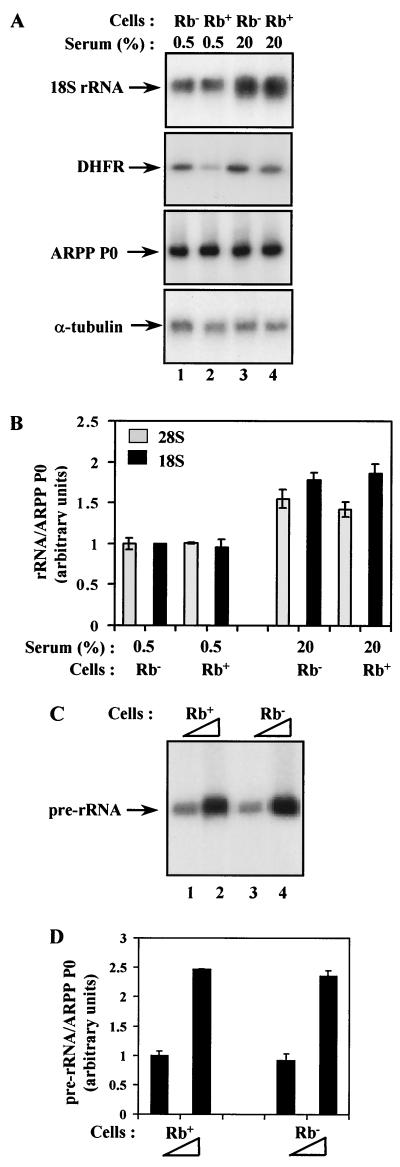
Since previous studies had shown that pRb has a potent effect on Pol I transcription when overexpressed either in vitro or in vivo, we expected to find that rRNA synthesis is deregulated in pRb-knockout mice. Unexpectedly, however, there was very little difference in the amount of 18S rRNA when Northern blotting was used to compare matched pairs of Rb+/+ and Rb−/− fibroblasts (Fig. 1A, upper panel). Since Pol I activity is known to correlate with growth rate (12, 34, 38), we examined both rapidly cycling cells cultured under high-serum conditions and quiescent cells arrested under low-serum conditions, but in neither case was any consistent change observed as a result of knocking out pRb. As a positive control, we looked at expression of the gene for dihydrofolate reductase (DHFR), which has been shown to respond to pRb in vivo (27, 42); the level of DHFR mRNA was elevated in our pRb-knockout cells, as expected (Fig. 1A, middle panel). As loading controls, we used an mRNA encoding α-tubulin and an mRNA for the acidic ribosomal phosphoprotein P0 (ARPP P0), which has been shown not to be regulated by the pocket proteins (17) (Fig. 1A, lower two panels). The 18S rRNA level was quantitated and normalized against the ARPP P0; this showed that there is no significant difference in the abundance of 18S rRNA between embryonic fibroblasts of wild-type and pRb-knockout mice (Fig. 1B). Identical results were obtained for the 28S rRNA (Fig. 1B).
FIG. 1.
Pol I activity is not elevated in fibroblasts from Rb−/− mice. (A) Northern blot analysis of total RNA (10 μg) from Rb−/− (lanes 1 and 3) or Rb+/+ (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). The upper panel shows the blot probed with an 18S rRNA gene fragment, and the bottom two panels show the same blot, which has been stripped and reprobed with ARPP P0 and α-tubulin gene fragments. In the second panel, the same RNA samples were analyzed by RT-PCR for expression of DHFR mRNA. (B) The 18S rRNA signals were normalized against the ARPP P0 signals, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath. Data obtained in the same way for 28S rRNA are also presented. (C) Primer extension analysis of 2 μg (lanes 1 and 3) or 5 μg (lanes 2 and 4) of total RNA from Rb+/+ (lanes 1 and 2) or Rb−/− (lanes 3 and 4) fibroblasts that were actively growing in 20% serum. (D) The pre-rRNA signals were normalized against ARPP P0, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath.
Since the steady-state level of mature rRNA does not necessarily reflect Pol I activity, we monitored the synthesis of nascent pre-rRNA molecules by primer extension assays. Because the primer used hybridizes to sequences at the 5′ end of the primary rRNA transcript, which are processed rapidly during transcription, the assay accurately reflects the rate of Pol I transcription initiation (21). Again, there was no detectable increase in rRNA synthetic activity in pRb-deficient cells (Fig. 1C and D). Thus, although overexpression of pRb strongly represses rDNA transcription (4, 14, 36, 47), loss of endogenous pRb does not affect Pol I transcription in mouse embryonic fibroblasts.
Pol I activity is elevated in fibroblasts lacking both pRb and p130.
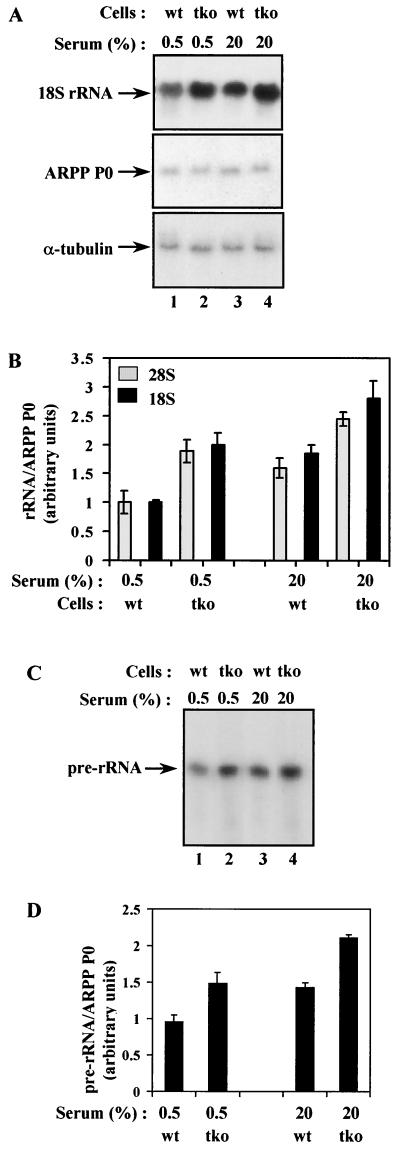
The fact that Rb−/− cells display normal levels of rRNA raised the possibility that the repression reported in vitro and in transient transfections might be an artifact due to overexpression of pRb. An alternative explanation would be that the pocket proteins provide redundant control of this system. Because p107 and p130 are 30 to 35% identical to pRb, they might compensate for its loss and prevent deregulation in Rb−/− cells (28). There are a number of precedents for such a scenario, such as E2F-4, which in T lymphocytes can be bound and regulated by all three pocket proteins (29). To assess whether the Pol I system is subject to redundant control, we analyzed rRNA levels in fibroblasts derived from Rb−/−/p107−/−/p130−/− triple knockout mice (8, 40). As shown in Fig. 2A, there was a consistent increase in the abundance of 18S rRNA when triple knockout cells were compared with wild-type cells. This effect is specific, since knocking out the pocket proteins has no significant effect on expression of the ARPP P0 and α-tubulin mRNAs. After normalization against the ARPP P0 control, the abundance of 18S rRNA was ∼50% higher in proliferating knockout cells than in the wild-type cells (Fig. 2B). The same was true of 28S rRNA (Fig. 2B). These increases were also observed if rRNA expression was normalized against cell number instead of a control mRNA (P. H. Scott and R. J. White, unpublished observations). In addition, monitoring of pre-rRNA synthesis by primer extension revealed an ∼50% increase in the primary Pol I transcript (Fig. 2C and D). The elevated rRNA levels and rate of Pol I transcription initiation in the triple knockout, but not the Rb−/−, fibroblasts, implies that there is functional redundancy in the ability of pocket proteins to inhibit cellular rRNA synthesis.
FIG. 2.
Pol I activity is abnormally elevated in fibroblasts from triple knockout mice. (A) Northern blot analysis of total RNA (10 μg) from wild-type (wt; lanes 1 and 3) or Rb−/− p107−/− p130−/− triple knockout (tko; lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). The upper panel shows the blot probed with an 18S rRNA gene fragment, and the panels underneath show the same blot that has been stripped and reprobed with ARPP P0 and α-tubulin gene fragments. (B) The 18S rRNA signals were normalized against the ARPP P0 signals, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath. Data obtained in the same way for 28S rRNA are also presented. (C) Primer extension analysis of 5 μg of total RNA from wild-type (lanes 1 and 3) or triple knockout (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or cultured for 24 h in 0.5% serum (lanes 1 and 2). (D) The pre-rRNA signals were normalized against ARPP P0, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath.
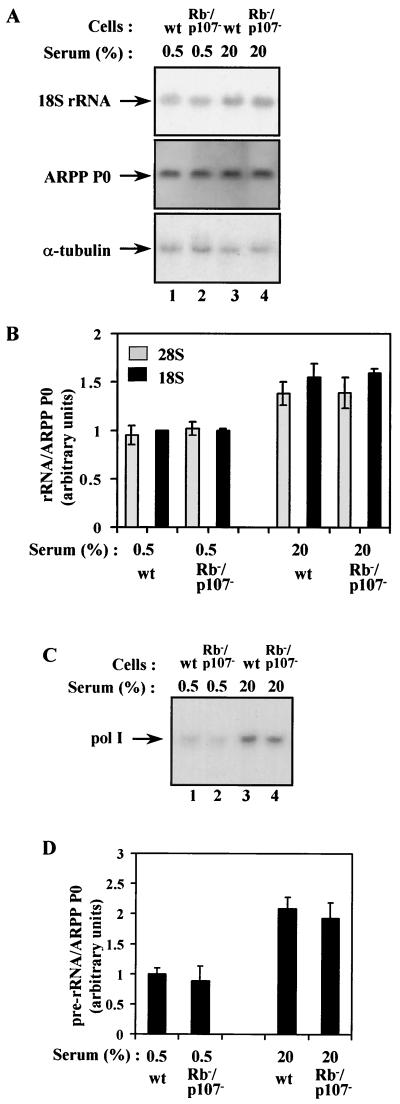
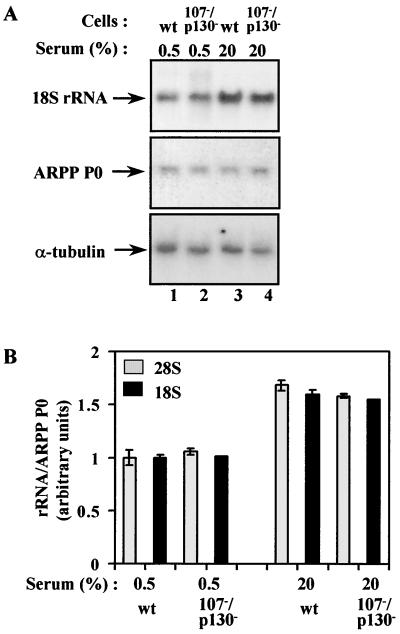
Double knockout fibroblasts were examined to determine which members of the pocket protein family are involved in controlling Pol I activity in vivo. When cultured in either 20% serum or 0.5% serum, there was no significant difference in the levels of 18S and 28S rRNA or the unprocessed primary transcript between wild-type and Rb−/− p107−/− double knockout cells (Fig. 3). The abundance of rRNA also appeared normal in p107−/− p130−/− double knockout cells (Fig. 4). In contrast, the 18S and 28S rRNAs are overexpressed by ∼50% in serum-starved Rb−/− p130−/− double knockout cells (Fig. 5A and B). Similarly, combined loss of pRb and p130 causes an ∼40% increase in expression of the primary Pol I transcript (Fig. 5C and D). Interpretation of these experiments is complicated by differences in proliferation rate between the various genetic backgrounds. For example, the triple knockout fibroblasts proliferate most rapidly (8, 40), and we cannot exclude that this contributes to their elevated Pol I activity. However, elevated proliferation is also shown by the Rb−/− p107−/− double knockout cells, but their rRNA levels are normal. Conversely, the Rb−/− p130−/− fibroblasts proliferate at the same rate as wild-type cells, and yet show a clear increase in rRNA production. The differences in Pol I activity seen in double knockout cells therefore cannot be explained by variations in growth or proliferation. These observations suggest that p130 shares with pRb the ability to suppress Pol I transcription in vivo. They also indicate that p107 lacks this capacity in mouse embryonic fibroblasts, although it might behave differently in other cell types.
FIG. 3.
Pol I activity is not elevated in fibroblasts from Rb−/− p107−/− double knockout mice. (A) Northern blot analysis of total RNA (10 μg) from wild-type (wt; lanes 1 and 3) or Rb−/− p107−/− double knockout (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). The upper panel shows the blot probed with an 18S rRNA gene fragment, and the panels underneath show the same blot that has been stripped and reprobed with ARPP P0 and α-tubulin gene fragments. (B) The 18S rRNA signals were normalized against the ARPP P0 signals, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath. Data obtained in the same way for 28S rRNA are also presented. (C) Primer extension analysis of 5 μg of total RNA from wt (lanes 1 and 3) or Rb−/− p107−/− double knockout (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). (D) The pre-rRNA signals were normalized against ARPP P0, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath.
FIG. 4.
Pol I activity is not elevated in fibroblasts from p130−/− p107−/− double knockout mice. (A) Northern blot analysis of total RNA (10 μg) from wild-type (wt; lanes 1 and 3) or p130−/− p107−/− double knockout (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). The upper panel shows the blot probed with an 18S rRNA gene fragment, and the panels underneath show the same blot that has been stripped and reprobed with ARPP P0 and α-tubulin gene fragments. (B) The 18S rRNA signals were normalized against the ARPP P0 signals, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath. Data obtained in the same way for 28S rRNA are also presented.
FIG. 5.
Pol I activity is abnormally elevated in fibroblasts from Rb−/− p130−/− double knockout mice. (A) Northern blot analysis of total RNA (10 μg) from wild-type (wt; lanes 1 and 3) or Rb−/− p130−/− double knockout (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). The upper panel shows the blot probed with an 18S rRNA gene fragment, and the panels underneath show the same blot that has been stripped and reprobed with ARPP P0 and α-tubulin gene fragments. (B) The 18S rRNA signals were normalized against the ARPP P0 signals, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath. Data obtained in the same way for 28S rRNA are also presented. (C) Primer extension analysis of 5 μg of total RNA from wild-type (lanes 1 and 3) or Rb−/− p130−/− double knockout (lanes 2 and 4) fibroblasts that were actively growing in 20% serum (lanes 3 and 4) or made quiescent by culture for 24 h in 0.5% serum (lanes 1 and 2). (D) The pre-rRNA signals were normalized against ARPP P0, as determined by PhosphorImager, and the mean values from three independent experiments are represented graphically underneath.
Repression of Pol I transcription by p130 in vivo and in vitro.
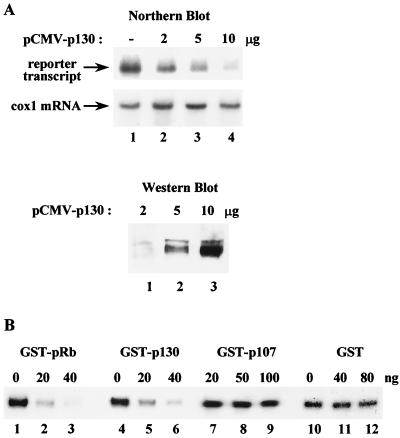
To test further the ability of p130 to regulate Pol I transcription in vivo, increasing amounts of pCMV-p130 were transfected into murine fibroblasts together with an rDNA reporter (pMr1930-BH). Pol I transcripts were detected by hybridization to a labeled reporter-specific RNA probe. Overexpression of p130 significantly reduced transcription of the reporter plasmid in a dose-dependent and specific manner (Fig. 6A). This is unlikely to be an indirect response to cell cycle arrest, since overexpression of p21 under the same conditions blocks proliferation without affecting rRNA synthesis (2).
FIG. 6.
Overexpression of p130 represses Pol I transcription in vitro and in vivo. (A) NIH 3T3 cells (4 × 105) were cotransfected with 10 μg of an rDNA minigene (pMr1930-BH), together with 2 μg (lane 2), 5 μg (lane 3), or 10 μg (lane 4) of pCMV-p130 expression plasmid. RNA was isolated after 44 h, and transcripts from the reporter construct were detected on a Northern blot with a 32P-labeled riboprobe complementary to pUC-derived sequences present in pMr1930-BH. Cytochrome oxidase 1 (cox1) mRNA was used as an internal control. (Bottom panel) To monitor p130 expression in the transfected cells, equal amounts of cell extracts (10 μg) were subjected to Western blot analysis with anti-p130 antibodies. (B) p130 represses Pol I transcription in vitro. Increasing amounts of GST-pRb (lanes 1 to 3), GST-p130 (lanes 4 to 6), GST-p107 (lanes 7 to 9), or GST (lanes 10 to 12) were added to a reconstituted transcription system containing 15 ng of linearized template pMrWT, partially purified TIF-IA/IC, TIF-IB, Pol I, and 15 ng of UBF.
Evidence for a direct effect of p130 comes from the use of a reconstituted system containing partially purified transcription factors and Pol I. Titration of increasing amounts of GST-p130 produced a dose-dependent inhibition of rDNA transcription (Fig. 6B, lanes 4 to 6). The degree of p130-mediated repression was close to that obtained with GST-pRb (lanes 1 to 3), although a little less potent. This effect was specific, since even greater amounts of GST alone had only a marginal effect on transcription (lanes 10 to 12). Whereas 20 ng of GST-Rb or GST-p130 was sufficient to give clear repression, 100 ng of GST-p107 failed to inhibit Pol I transcription in this assay (lanes 7 to 9), which is consistent with the inability of p107 to compensate in vivo for the loss of pRb and p130 in double knockout fibroblasts (Fig. 6B).
Preassembly of a Pol I transcription complex confers protection against p130.
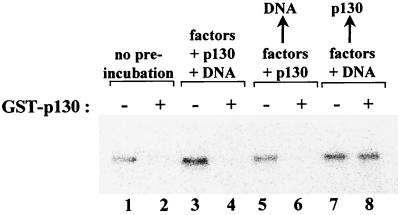
We investigated whether the response to p130 is influenced by prior assembly of a stable preinitiation complex on an rRNA promoter (Fig. 7). GST-p130 was preincubated with transcription factors and Pol I in the presence or absence of template DNA (lanes 4 and 6, respectively), or alternatively was added after preinitiation complex assembly (lane 8). Whereas p130 caused efficient repression if present during formation of the preinitiation complex (lanes 2, 4, and 6), no repression occurred when p130 was added after assembly was complete (lane 8). Thus, the Pol I machinery is much more susceptible to p130 when free in solution and receives substantial protection once it is assembled on a promoter. The fact that the preformed Pol I transcription complex is relatively immune to p130-directed repression suggests that p130 acts at some stage during complex formation.
FIG. 7.
p130 acts during transcription complex assembly to inhibit Pol I transcription. Pol I and transcription factors were preincubated for 20 min at 30°C with buffer (lanes 3 and 5) or with 30 ng of GST-p130 (lanes 4 and 6); template DNA was added at the start (lanes 3 and 4) or end (lanes 5 and 6) of the preincubation. In lanes 7 and 8, the factors and Pol I were preincubated for 20 min at 30°C, together with template DNA, before GST-p130 (30 ng) was added (lane 8). Following preincubation, transcription was initiated by the addition of nucleotides. Lanes 1 and 2 show the transcription reactions without preincubation in the absence (lane 1) or presence (lane 2) of GST-p130 (30 ng).
The Pol I-specific factor UBF is bound and repressed by p130.
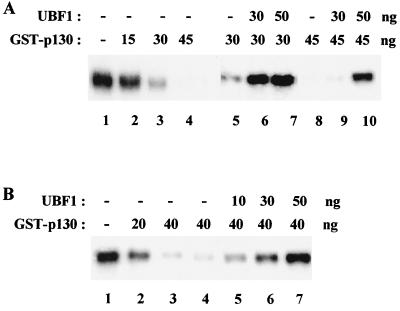
When the reconstituted system was supplemented with more of the Pol I-specific factor UBF1, larger amounts of GST-p130 were required to decrease transcription (Fig. 8A). This suggests that p130 represses rRNA synthesis by targeting UBF, as was found previously for pRb (4, 47). In support of this, equimolar amounts of recombinant UBF1 were sufficient to fully overcome p130-mediated repression (Fig. 8B). We conclude that p130, like pRb, targets UBF and interferes with transcription complex formation.
FIG. 8.
UBF is sufficient to relieve repression by p130. (A) The extent of p130-mediated repression is sensitive to the amount of UBF. Increasing amounts of recombinant GST-p130 (∼110 kDa), as indicated, were added to a reconstituted transcription system, which was supplemented with 30 ng (lanes 6 and 9) or 50 ng (lanes 7 and 10) of UBF1 (∼97 kDa). (B) UBF is sufficient to overcome repression by p130. In vitro transcription activity in the absence (lane 1) and presence of 20 ng (lane 2) and 40 ng (lanes 3 to 7) of GST-p130. Following preincubation of transcription factors and Pol I with GST-p130, transcription reactions were supplemented with 10 ng (lane 5), 30 ng (lane 6), or 50 ng (lane 7) of recombinant UBF1 as indicated and assayed for transcriptional activity in the presence of template DNA.
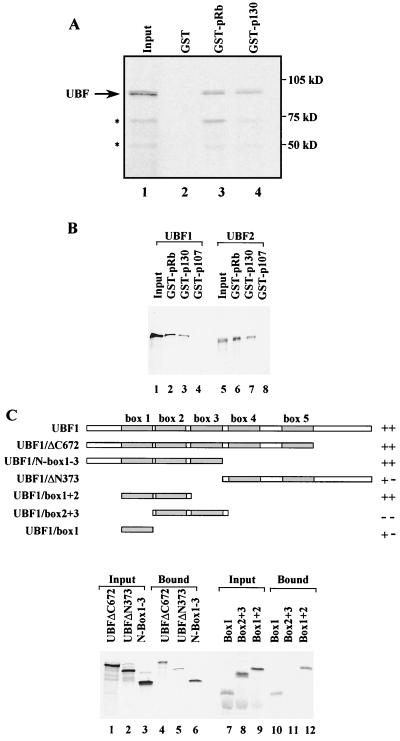
Having established that UBF is a functional target of p130, pull-down assays were used to test for a physical interaction. GST-p130 was immobilized on glutathione beads and tested for its ability to associate with 35S-labeled UBF. Like GST-pRb, GST-p130 was found to bind to UBF (Fig. 9A). This interaction is specific, since it was not observed with GST. Although p130 clearly binds to UBF, the efficiency of association is somewhat lower (about 60%) when compared to that of pRb. In contrast to these robust interactions, little or no binding was obtained with GST-p107 (Fig. 9B). This is consistent with the failure of p107 to repress Pol I transcription in vitro (Fig. 6B) or to compensate in vivo for the loss of pRb and p130 (Fig. 5). The alternative splice forms UBF1 and UBF2 behave similarly in these assays: both bind to GST-pRb and GST-p130, but not to GST alone or GST-p107 (Fig. 9B). A series of UBF1 mutants were used to map which regions contribute to the interaction with p130 (Fig. 9C). Deletion of the C-terminal part of UBF (ΔC672 and N boxes 1 to 3) did not impair the interaction with GST-p130 significantly (Fig. 9C lanes 1, 2, 4, and 6). HMG boxes 1 and 2 together are bound efficiently, whereas box 1 alone interacts only weakly, and boxes 2 and 3 together do not bind at all (Fig. 9C, lanes 7 to 12). Thus, a region encompassing HMG boxes 1 and 2 allows efficient interaction with p130. In addition, the C-terminal acidic tail of UBF1 binds with low affinity to p130 (ΔN373, lanes 2 and 5).
FIG. 9.
Recombinant p130 interacts with UBF. (A) [35S]methionine-labeled UBF1 was synthesized by in vitro translation and incubated with immobilized GST (lane 2), GST-pRb(379–928) (lane 3), or GST-p130(372–1139) (lane 4). Lane 1 shows 10% of the input that was used for the pull-down experiments. Proteins retained after extensive washing were resolved on an 8% polyacrylamide gel and then visualized by autoradiography. The asterisks mark partial forms of UBF resulting from degradation or incomplete translation. (B) [35S]methionine-labeled UBF1 (lanes 1 to 4) and UBF2 (lanes 5 to 8) were synthesized by in vitro translation and incubated with immobilized GST-pRb(379–928) (lanes 2 and 6), GST-p130(372–1139) (lanes 3 and 7), or GST-p107(385–1068) (lanes 4 and 8). Lanes 1 and 5 show 10% of the input that was used for the pull-down experiments. Proteins retained after extensive washing were resolved on an 8% polyacrylamide gel and then visualized by autoradiography. (C) Scheme showing the structural organization of UBF1 and the mutants used for the interaction studies; the individual HMG boxes are indicated. [35S]methionine-labeled UBF1 mutants, as indicated, were synthesized by in vitro translation and incubated with immobilized GST-p130(372–1139). The input (lanes 1 to 3 and 7 to 9) represents 10% of the protein that was used for pull-down experiments. Bound proteins were separated on 10% (lanes 1 to 6) or 15% (lanes 7 to 12) sodium dodecyl sulfate-polyacrylamide gels and visualized by autoradiography.
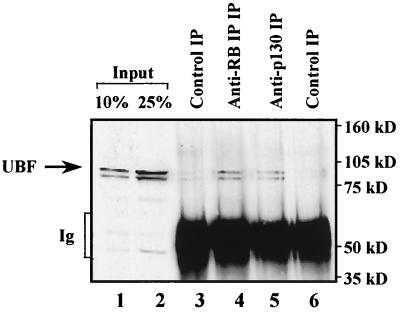
Since pull-down assays rely on the use of overexpressed recombinant proteins, we also carried out coimmunoprecipitations to look for an interaction between endogenous factors present at physiological ratios. Antisera against both pRb and p130 were found to coimmunoprecipitate UBF from cell extracts, whereas only low background levels of UBF were detected in controls that used protein A alone or an irrelevant antiserum against Oct-1 (Fig. 10). These data provide evidence for a physical association between endogenous p130 and endogenous UBF.
FIG. 10.
Endogenous cellular p130 associates with endogenous UBF. Whole-cell extract (150 μg) of BALB/c 3T3 fibroblasts was immunoprecipitated with antibody against Oct-1 (lane 3), pRb (lane 4), or p130 (lane 5) or was mock treated with protein A-Sepharose beads in the absence of antibody (lane 6). Precipitated material was resolved on a sodium dodecyl sulfate–7.8% polyacrylamide gel and then analyzed by Western blotting with anti-UBF antibody H-300. Lanes 1 and 2 show 10 and 25%, respectively, of the input extract.
DISCUSSION
This article presents several complementary lines of evidence to show that p130 shares with pRb the ability to regulate Pol I transcription. First, we demonstrate that the level of cellular rRNA is increased in primary fibroblasts, which lack pRb and p130, but not in cells which are bereft of only pRb. Although Pol I activity generally correlates with growth rate (12, 34, 38), its elevation in these double knockout mutants cannot be explained in this way, since the Rb−/− p130−/− fibroblasts show wild-type rates of growth and proliferation. Second, rDNA transcription could be suppressed by overexpression of p130 in transient transfection experiments. Again, this is unlikely to be a secondary response to cell cycle arrest, since Pol I transcription is not decreased if the cyclin-dependent kinase inhibitor p21 is overexpressed under the same conditions (2). Third, these effects are likely to be direct, because recombinant p130 inhibits rRNA synthesis that has been reconstituted in vitro with purified transcription factors. Fourth, both splice forms of UBF bind specifically to GST-p130, and endogenous UBF can be coimmunoprecipitated with cellular p130. Furthermore, the ability of UBF to support transcription in vitro is inhibited by recombinant p130. These data, taken together, suggest that p130 regulates Pol I transcription in vivo by associating with UBF, although additional mechanisms cannot be excluded. During the course of this work, another group has also shown that p130 will repress the rRNA promoter when overexpressed in transfected cells and will coimmunoprecipitate with UBF (13). We have added substantially to their findings by showing that recombinant p130 can bind and repress UBF, as well as by mapping the site of interaction. Most importantly, we have provided the crucial genetic evidence that has previously been lacking, which shows that the Pol I transcription system is a bona fide target for pRb and p130 when present at naturally occurring concentrations within the cell. Although the increase in rRNA synthesis is only ∼40 to 50% following loss of these pocket proteins, this is likely to make a very substantial difference to nuclear activity, given the quantitative dominance of Pol I compared with the other RNA polymerases.
There is significant functional overlap among the pRb family proteins (11, 28). For example, each member can inhibit cell growth and proliferation when overexpressed in tumor cells (7, 37, 52). Several protein targets can interact with pRb, p107, and p130, including E2F, Id2, and the oncoprotein products of various DNA tumor viruses (9, 16, 28, 44). The ability to bind and repress TFIIIB is also shared by all three members of the pRb family (43). However, the fact that Rb−/− mice die at midgestation (6, 19, 26) clearly shows that at least some functions of pRb cannot be performed by its relatives. Further evidence that this is the case is provided by the fact that the Rb gene is often mutated in tumors, whereas mutations are extremely rare in the genes encoding p107 and p130 (15, 28). It was shown previously that p107 lacks the ability of pRb to repress Pol I transcription in vitro (47). This can be explained by its apparent inability to bind to UBF, an observation we share with Hannan et al. (13). These data are supported strongly by our genetic analysis, which reveals that p107 is unable to substitute for the other pocket proteins in suppressing rRNA synthesis in mouse embryonic fibroblasts. In general, there is considerable functional overlap between p107 and p130, which are ∼50% identical (11, 28). Nevertheless, we provide evidence here that p130 differs from p107 in being able to bind and repress UBF. We are aware of one other example of a factor that binds pRb and p130, but not p107; this is the transcriptional repressor HBP1, which is produced during muscle cell differentiation (45). It is interesting to note that HBP1 also resembles UBF in containing HMG domains.
pRb and p130 prevent growth and proliferation under inappropriate conditions (11, 28). Their pleiotropic effects on cellular behavior are likely to reflect an ability to regulate multiple targets involved in several key physiological pathways. The capacity to bind and repress E2F is believed to play a principal role in allowing pRb and its relatives to block cell cycle progression (11, 28). However, arresting the cell cycle alone is generally insufficient to prevent accumulation of mass and can result in unbalanced growth and abnormally large cells (20, 22, 23, 30–32, 50). The coordinated response elicited by pRb may therefore require direct effects on the biosynthetic machinery. A key example of such effects may be provided by its ability to suppress the synthesis of rRNA and tRNA by Pols I and III, which together can contribute up to 80% of all nuclear transcription and ∼95% of cellular RNA (35, 48). Restraint of these polymerases is very likely to have a major impact on the accumulation of mass that constitutes cell growth.
ACKNOWLEDGMENTS
Sonia Ciarmatori and Pamela Scott contributed equally to this work.
This work was funded in part by grant SP2314/0101 to R.J.W. from the Cancer Research Campaign, grants to J.G. and R.V. from the Deutsche Forschungsgemeinschaft, and the Fonds der Chemischen Industrie. J.E.S. was supported by a postgraduate studentship from the Biotechnology and Biological Sciences Research Council. P.H.S. is a Wellcome Trust Research Fellow, and R.J.W. is a Jenner Research Fellow of the Lister Institute of Preventive Medicine.
REFERENCES
- 1.Alzuherri H M, White R J. Regulation of RNA polymerase I transcription in response to F9 embryonal carcinoma stem cell differentiation. J Biol Chem. 1999;274:4328–4334. doi: 10.1074/jbc.274.7.4328. [DOI] [PubMed] [Google Scholar]
- 2.Budde A, Grummt I. p53 represses ribosomal gene transcription. Oncogene. 1999;18:1119–1124. doi: 10.1038/sj.onc.1202402. [DOI] [PubMed] [Google Scholar]
- 3.Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. 2000;2:E107–E112. doi: 10.1038/35014078. [DOI] [PubMed] [Google Scholar]
- 4.Cavanaugh A H, Hempel W M, Taylor L J, Rogalsky V, Todorov G, Rothblum L I. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 5.Chu W-M, Wang Z, Roeder R G, Schmid C W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J Biol Chem. 1997;272:14755–14761. doi: 10.1074/jbc.272.23.14755. [DOI] [PubMed] [Google Scholar]
- 6.Clarke A R, Maandag E R, van Roon M, van der Lugt N M T, van der Valk M, Hooper M L, Berns A, te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 7.Claudio P P, Howard C M, Baldi A, De Luca A, Fu Y, Condorelli G, Sun Y, Colburn N, Calabretta B, Giordano A. p130/Rb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res. 1994;54:5556–5560. [PubMed] [Google Scholar]
- 8.Dannenberg J-H, Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G1 control causing immortalisation and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–3064. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12:2245–2262. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- 10.Garcia S N, Pillus L. Net results of nucleolar dynamics. Cell. 1999;97:825–828. doi: 10.1016/s0092-8674(00)80794-1. [DOI] [PubMed] [Google Scholar]
- 11.Grana X, Garriga J, Mayol X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene. 1998;17:3365–3383. doi: 10.1038/sj.onc.1202575. [DOI] [PubMed] [Google Scholar]
- 12.Grummt I. Regulation of mammalian ribosomal gene transcription by RNA polymerase I. Prog Nucleic Acid Res Mol Biol. 1999;62:109–154. doi: 10.1016/s0079-6603(08)60506-1. [DOI] [PubMed] [Google Scholar]
- 13.Hannan K M, Hannan R D, Smith S D, Jefferson L S, Lun M, Rothblum L I. Rb and p130 regulate RNA polymerase I transcription: Rb disrupts the interaction between UBF and SL-1. Oncogene. 2000;19:4988–4999. doi: 10.1038/sj.onc.1203875. [DOI] [PubMed] [Google Scholar]
- 14.Hannan K M, Kennedy B K, Cavanaugh A H, Hannan R D, Hirschler-Laszkiewicz I, Jefferson L S, Rothblum L I. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene. 2000;19:3487–3497. doi: 10.1038/sj.onc.1203690. [DOI] [PubMed] [Google Scholar]
- 15.Helin K, Holm K, Niebuhr A, Eiberg H, Tommerup N, Hougaard S, Poulsen H S, Spang-Thomsen M, Norgaard P. Loss of the retinoblastoma protein-related p130 protein in small cell lung carcinoma. Proc Natl Acad Sci USA. 1997;94:6933–6938. doi: 10.1073/pnas.94.13.6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herwig S, Strauss M. The retinoblastoma protein: a master regulator of cell cycle, differentiation and apoptosis. Eur J Biochem. 1997;246:581–601. doi: 10.1111/j.1432-1033.1997.t01-2-00581.x. [DOI] [PubMed] [Google Scholar]
- 17.Hurford R K, Cobrinik D, Lee M-H, Dyson N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997;11:1447–1463. doi: 10.1101/gad.11.11.1447. [DOI] [PubMed] [Google Scholar]
- 18.Iavarone A, Garg P, Lasorella A, Hsu J, Israel M A. The helix-loop-helix protein Id-2 enhances cell proliferation and binds to the retinoblastoma protein. Genes Dev. 1994;8:1270–1284. doi: 10.1101/gad.8.11.1270. [DOI] [PubMed] [Google Scholar]
- 19.Jacks T, Fazeli A, Schmitt E M, Bronson R T, Goodell M A, Weinberg R A. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 20.Johnston G C, Pringle J R, Hartwell L H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977;105:79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- 21.Kass S, Craig N, Sollner-Webb B. Primary processing of mammalian rRNA involves two adjacent cleavages and is not species specific. Mol Cell Biol. 1987;7:2891–2898. doi: 10.1128/mcb.7.8.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Killander D, Zetterberg A. A quantitative cytochemical investigation of the relationship between cell mass and initiation of DNA synthesis in mouse fibroblasts in vitro. Exp Cell Res. 1965;40:12–20. doi: 10.1016/0014-4827(65)90285-5. [DOI] [PubMed] [Google Scholar]
- 23.Kung A L, Sherwood S W, Schimke R T. Differences in the regulation of protein synthesis, cyclin B accumulation, and cellular growth in response to the inhibition of DNA synthesis in Chinese hamster ovary and HeLa S3 cells. J Biol Chem. 1993;268:23072–23080. [PubMed] [Google Scholar]
- 24.Lasorella A, Iavarone A, Israel M A. Id2 specifically alters regulation of the cell cycle by tumor suppressor proteins. Mol Cell Biol. 1996;16:2570–2578. doi: 10.1128/mcb.16.6.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasorella A, Noseda M, Beyna M, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–598. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 26.Lee E Y-H P, Chang C-Y, Hu N, Wang Y-C J, Lai C-C, Herrup K, Lee W-H, Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 27.Luo R X, Postigo A A, Dean D C. Rb interacts with histone deacetylase to repress transcription. Cell. 1998;92:463–473. doi: 10.1016/s0092-8674(00)80940-x. [DOI] [PubMed] [Google Scholar]
- 28.Mulligan G, Jacks T. The retinoblastoma gene family: cousins with overlapping interests. Trends Genet. 1998;14:223–229. doi: 10.1016/s0168-9525(98)01470-x. [DOI] [PubMed] [Google Scholar]
- 29.Mulligan G J, Wong J, Jacks T. p130 is dispensable in peripheral T lymphocytes: evidence for functional compensation by p107 and pRB. Mol Cell Biol. 1998;18:206–220. doi: 10.1128/mcb.18.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasmyth K. Another role rolls in. Nature. 1996;382:28–29. doi: 10.1038/382028a0. [DOI] [PubMed] [Google Scholar]
- 31.Neufeld T P, de la Cruz A F, Johnston L A, Edgar B A. Coordination of growth and cell division in the Drosophila wing. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 32.Neufeld T P, Edgar B A. Connections between growth and the cell cycle. Curr Opin Cell Biol. 1998;10:784–790. doi: 10.1016/s0955-0674(98)80122-1. [DOI] [PubMed] [Google Scholar]
- 33.Norton J D, Deed R W, Craggs G, Sablitzky F. Id helix-loop-helix proteins in cell growth and differentiation. Trends Cell Biol. 1998;8:58–65. [PubMed] [Google Scholar]
- 34.Paule M R. RNA polymerase I transcription. Berlin, Germany: Springer-Verlag; 1998. [Google Scholar]
- 35.Paule M R, White R J. Transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283–1298. doi: 10.1093/nar/28.6.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pelletier G, Stefanovsky V Y, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum L I, Cote J, Moss T. Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell. 2000;6:1059–1066. doi: 10.1016/s1097-2765(00)00104-0. [DOI] [PubMed] [Google Scholar]
- 37.Qin X, Chittenden T, Livingston D M, Kaelin W G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992;6:953–964. doi: 10.1101/gad.6.6.953. [DOI] [PubMed] [Google Scholar]
- 38.Reeder R H. Regulation of RNA polymerase I transcription in yeast and vertebrates. Prog Nucleic Acid Res Mol Biol. 1999;62:293–327. doi: 10.1016/s0079-6603(08)60511-5. [DOI] [PubMed] [Google Scholar]
- 39.Rogalsky V, Todorov G, Moran D. Translocation of retinoblastoma protein associated with tumour cell growth inhibition. Biochem Biophys Res Commun. 1993;192:1139–1146. doi: 10.1006/bbrc.1993.1535. [DOI] [PubMed] [Google Scholar]
- 40.Sage J, Mulligan G J, Attardi L D, Miller A, Chen S, Williams B, Theodorou E, Jacks T. Targeted disruption of the three Rb-related genes leads to loss of G1 control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scott P H, Cairns C A, Sutcliffe J E, Alzuherri H M, McLees A, Winter A G, White R J. Regulation of RNA polymerase III transcription during cell cycle entry. J Biol Chem. 2001;276:1005–1014. doi: 10.1074/jbc.M005417200. [DOI] [PubMed] [Google Scholar]
- 42.Sutcliffe J E, Brown T R P, Allison S J, Scott P H, White R J. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol Cell Biol. 2000;20:9192–9202. doi: 10.1128/mcb.20.24.9192-9202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sutcliffe J E, Cairns C A, McLees A, Allison S J, Tosh K, White R J. RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130. Mol Cell Biol. 1999;19:4255–4261. doi: 10.1128/mcb.19.6.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taya Y. RB kinases and RB-binding proteins: new points of view. Trends Biochem Sci. 1997;22:14–17. doi: 10.1016/s0968-0004(96)10070-0. [DOI] [PubMed] [Google Scholar]
- 45.Tevosian S G, Shih H H, Mendelson K G, Sheppard K-A, Paulson K E, Yee A S. HBP1: a HMG box transcriptional repressor that is targeted by the retinoblastoma family. Genes Dev. 1997;11:383–396. doi: 10.1101/gad.11.3.383. [DOI] [PubMed] [Google Scholar]
- 46.Voit R, Hoffmann M, Grummt I. Phosphorylation by G1-specific cdk-cyclin complexes activates the nucleolar transcription factor UBF. EMBO J. 1999;18:1891–1899. doi: 10.1093/emboj/18.7.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voit R, Schafer K, Grummt I. Mechanism of repression of RNA polymerase I transcription by the retinoblastoma protein. Mol Cell Biol. 1997;17:4230–4237. doi: 10.1128/mcb.17.8.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner J R. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 49.Weber J D, Taylor L J, Roussel M F, Sherr C J, Bar-Sagi D. Nucleolar Arf sequesters Mdm2 and activates p53. Nat Cell Biol. 1999;1:20–26. doi: 10.1038/8991. [DOI] [PubMed] [Google Scholar]
- 50.White R J. Regulation of RNA polymerases I and III by the retinoblastoma protein: a mechanism for growth control? Trends Biochem Sci. 1997;22:77–80. doi: 10.1016/s0968-0004(96)10067-0. [DOI] [PubMed] [Google Scholar]
- 51.White R J, Trouche D, Martin K, Jackson S P, Kouzarides T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature. 1996;382:88–90. doi: 10.1038/382088a0. [DOI] [PubMed] [Google Scholar]
- 52.Zhu L, van den Heuvel S, Helin K, Fattaey A, Ewen M, Livingston D, Dyson N, Harlow E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993;7:1111–1125. doi: 10.1101/gad.7.7a.1111. [DOI] [PubMed] [Google Scholar]