Abstract
Chromatin remodeling complexes such as SWI/SNF use the energy of ATP hydrolysis to remodel nucleosomal DNA and increase transcription of nucleosomal templates. Human heat shock factor one (hHSF1) is a tightly regulated activator that stimulates transcriptional initiation and elongation using different portions of its activation domains. Here we demonstrate that hHSF1 associates with BRG1, the ATPase subunit of human SWI/SNF (hSWI/SNF) at endogenous protein concentrations. We also show that hHSF1 activation domains recruit hSWI/SNF to a chromatin template in a purified system. Mutation of hHSF1 residues responsible for activation of transcriptional elongation has the most severe effect on recruitment of SWI/SNF and association of hHSF1 with BRG1, suggesting that recruitment of chromatin remodeling activity might play a role in stimulation of elongation.
Efficient function of transcriptional activators requires interactions with the general transcription machinery and with complexes that alter the chromatin template. Complexes that alter the chromatin template can be divided broadly into those that covalently modify the template (e.g., acetyltransferases, kinases, and methylases) and those that utilize the energy of ATP hydrolysis to “remodel” nucleosome structure and/or position (reviewed in references 22, 24, and 41). While genetic and biochemical studies have demonstrated a role for ATP-dependent chromatin remodeling complexes in transcriptional activation (2, 24, 41), the mechanisms by which these complexes are targeted and the steps in the transcription process that they activate are poorly understood. The heat stress-induced transcriptional activator, heat shock factor (HSF), is responsible for coordinating the rapid transcriptional response of many heat shock protein genes, including the well-studied hsp70 gene (reviewed in reference 13). This transcriptional activator has provided a good model system for understanding mechanisms of activation.
The heat shock response is regulated via activation of HSF (reviewed in references 30, 33, and 34). HSF1 is the primary stress-activated member of a family of vertebrate HSFs. Prior to heat shock, human heat shock factor 1 (hHSF1) is found throughout the cell as an inactive monomer. Upon heat shock, it becomes hyperphosphorylated, concentrates in the nucleus, trimerizes, and binds DNA. Transactivation is controlled by HSF's regulatory domain, located in the center of the protein, which is able to repress the two C-terminal activation domains (AD1 and AD2) and render them heat inducible (see Fig. 1A) (16). AD1 and AD2 each contain residues that are important for both transcriptional initiation and elongation. Mutations in acidic residues in both AD1 and AD2 preferentially affect the ability of HSF1 to stimulate transcriptional initiation, while mutations in phenylalanine residues in both activation domains preferentially affect stimulation of elongation (5).
FIG. 1.
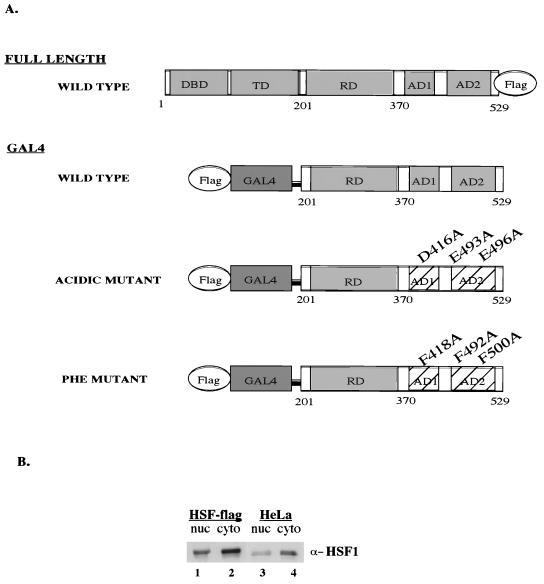
Development of stable HSF1-flag cell lines. (A) Schematic representation of HSF1 constructs. DBD, DNA binding domain; TD, trimerization domain; RD, regulatory domain; AD1, activation domain I; AD2, activation domain II. (B) Expression level of total hHSF1 (HSF1-FLAG plus endogenous HSF1), nuclear (nuc) and cytoplasmic (cyto), in the HSF1-flag cell line versus the parental HeLa cell line, obtained by Western blot analysis using the anti-HSF1 antibody (α-HSF1) (Stressgen).
In the nucleus, DNA is packaged into nucleosomes, which frequently inhibit transcription. Remodeling complexes act to move or alter the nucleosomes in a manner that allows for transcription. Two well-characterized classes of ATP-dependent chromatin remodeling complexes, named for their ATPase subunits, are the SWI/SNF family of complexes and the ISWI-based complexes (reviewed in references 52 and 54). The SWI/SNF complexes can increase DNA accessibility for activator binding (10, 28). At the same time, studies have revealed that SWI/SNF can be targeted to nucleosomal templates by template-bound activators in vitro (37, 40, 58). Experiments using chromatin immunoprecipitation analysis show that recruitment of yeast SWI/SNF (ySWI/SNF) to the HO promoter requires the initial binding of the activator SWI5 (8). The connection of activators to chromatin remodeling complexes is especially pertinent when studying the activation of the human hsp70 gene. There is a promoter-proximal pause of RNA polymerase II on the inactive Drosophila and human hsp70 genes (47), and it has been proposed that nucleosome structure in front of the elongating RNA polymerase contributes significantly to the length and regulation of the pause site (3, 21). In an in vitro transcription system, the pause is released by HSF1 more efficiently when human SWI/SNF (hSWI/SNF) is present (3).
To learn more about the role of hHSF1 in transcriptional activation, we searched for functionally interacting proteins. We utilized both a biochemical approach to identify physical interaction and an in vitro assay to study the functional consequences of the interaction. We find that BRG1, the ATPase subunit of the hSWI/SNF chromatin-remodeling complex, and chromatin remodeling activity cofractionate with HSF1. We also show, using a purified system, that hHSF1 will target hSWI/SNF to a nucleosomal template. The association of HSF1 with BRG1 and remodeling activity, as well as its recruitment of hSWI/SNF, depends on the transcriptional activation domains, primarily on the phenylalanine residues that are important for stimulation of transcriptional elongation. These studies suggest that hSWI/SNF is an important target of the HSF1 transcriptional activation domains.
MATERIALS AND METHODS
Plasmids and fusion constructs.
Plasmid pHSF1-flag for the expression of carboxy-terminally FLAG-tagged hHSF1 was generated by subcloning the human HSF1-flag construct out of pCMV-HSF1-flag and into the pBABE/puro retroviral vector (32) via the EcoRI site. pCMV-HSF1-flag was constructed in our lab by taking pCMV-huHSF-B (kindly provided by Carl Wu, National Institutes of Health, Bethesda, Md.) and inserting the FLAG epitope (DYKDDDDK) on the C-terminal end by PCR. pCMV-huHSF-B contains the cDNA of human HSF1, including the 5′ and 3′ untranslated region, cloned into the EcoRI site of the pCMV5 vector.
GAL4-HSF fusions were all created by starting with pBS108 (kindly provided by Carl Wu) which contains the full length cDNA clone of human HSF1 in Bluescript (SK+) (Stratagene). The mutations F418A, F492A, and F500A were added consecutively via site-directed mutagenesis (27) in order to create the phenylalanine mutant, pBS108-ph. The mutations E493A and E496A were added consecutively via site-directed mutagenesis, and the mutation D416A was inserted in the last step via the SacI and BstEII (partial digest) sites in order to create the acidic mutant, pBS108-ac.
The pBABE-Flag-Gal4(1-94)HSFABC plasmids (wild type, acidic mutant, and phenylalanine mutant) for the stable expression of amino-terminally FLAG-tagged GAL4-HSF-wt, -ac, and -ph, respectively, contain the FLAG epitope amino terminal to the GAL4 DNA binding domain (amino acids 1 to 94), attached to the C terminus of hHSF1, (amino acids 201 to 529). This construct was subcloned into the pBABE/puro retroviral vector via the BglII and EcoRI sites.
The pBXFlagGal(1-94)HSFABC plasmids (wild type, acidic mutant, and phenylalanine mutant) for the transient expression of amino-terminally FLAG-tagged GAL4-HSF-wt, -ac, and -ph, respectively, contain the FLAG epitope amino terminal to the GAL4 DNA binding domain (amino acids 1 to 94), attached to the C terminus of hHSF1 (amino acids 201 to 529). This construct was subcloned into the pBXG1 mammalian expression plasmid (the vector is a gift of M. Ptashne, Harvard University) via the BglII and EcoRI sites.
The pRJR-HSFABC [Gal4(1-94)] plasmids (wild type, acidic mutant, and phenylalanine mutant) for the bacterial expression of six-histidine epitope-tagged GAL4-HSF-wt, -ac, and -ph, respectively, were created by subcloning the respective Gal4-HSF inserts from the above pBXFlagGal(1-94)HSFABC plasmids into pRJR1 (46) via the XhoI and EcoRI sites.
Cell line establishment and cell culture.
The HeLa-derived, FLAG-expressing cell lines were established using a retrovirus-mediated gene transfer technique similar to that described previously (6, 14). The FLAG constructs were subcloned into the retroviral vector, pBABE/puro, as described above. The retroviral vector was transfected into Bing cells, a retroviral packaging cell line, as previously described (1). Briefly, 10 μg of plasmid DNA was used to transfect 2 × 106 Bing cells by the calcium phosphate method. Medium containing helper-free and replication-deficient retrovirus was harvested 36 to 72 h posttransfection, filtered through a 0.45-μm-pore-size Nalgene filter, and used to infect 2 × 106 freshly seeded HeLa S3 cells in the presence of 8 μg of polybrene/ml. Infected cells were grown in medium containing 1 μg of puromycin/ml, and individual colonies were isolated and expanded into cell lines. Extracts were made (12), and 10 clonal cell lines were analyzed for FLAG protein expression via Western blot analysis (ECL-Amersham Pharmacia Biotech) using the anti-FLAG antibody, D-8 polyclonal (Santa Cruz) or M5 monoclonal (Sigma).
Both HeLa and the FLAG protein-expressing cell lines were maintained on plates in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum (Sigma) at 37°C. For heat shock, medium warmed to 43°C was used to replace medium in the plates prior to a 2-h incubation at 43°C and immediate transfer to 4°C for harvesting.
Immunopurification.
In the search for copurifying proteins, many conditions were used for extract preparation and the immunopurification of HSF1 and the GAL4-HSF proteins: extraction salts including up to 340 mM KCl, up to 300 mM KOAc, up to 340 mM (NH4)2SO4, column sizes from 100 μl to 1 ml; binding detergents including 0.1% or 0.15% NP-40; binding times of 4 h, 5.5 h, and overnight (10 to 12 h); and reloading the flowthrough over the column twice. The wash salts used were 300 mM KCl, 300 mM KOAc, 300 mM NaOAc, 300 mM Na citrate, 300 mM (NH4)2SO4, 100 mM (NH4)2SO4, and 50 mM (NH4)2SO4. Detergents used in the washes were 0.1% NP-40 and 1 or 5 mM n-octylglucoside.
For the immunopurification in which BRG1 cofractionated with HSF1 and the GAL4-HSF fusions, heat-shocked, non-heat-shocked, nuclear, and cytoplasmic extracts were prepared as described previously (12). For purification, 20 mg of protein extracts was incubated overnight (10 to 12 h) at 4°C with 100 μl of anti-FLAG M2 affinity gel (Sigma) in BC-100 (100 mM KCl, 20% glycerol, 20 mM HEPES [pH 7.9], 0.2 mM EDTA, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride) containing 0.1% NP-40. After incubation, columns were poured and washed with 10 column volumes of BC-100, 80 column volumes of BC-500 (same as BC-100 except containing 500 mM KCl), and then 10 column volumes of BC-100 (all with 0.1% NP-40). Bound protein was eluted using 0.4 mg of the FLAG peptide (N-DYKDDDDK-C)/ml in BC-100 (0.1% NP-40).
Bacterial purification of GAL4-HSF fusion proteins.
GAL4-HSF1 fusion proteins (wild type and activation domain mutants) containing amino acids 1 to 94 of the GAL4 protein fused to the C terminus of human HSF1, amino acids 201 to 529 (mutations as shown in Fig. 1A), as well as the GAL4 DNA binding domain (1 to 94), were purified from Escherichia coli as His6-tagged proteins using procedures described previously (5). Protein concentrations were determined by Bradford analysis, and purity was assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). From this, the GAL4(1-94) protein was judged to be 85% pure at a dimer concentration of approximately 30 μM, GAL4-HSF1 (wild type) was 30% pure at 7.2 μM, GAL4-HSF1 (acidic mutant) was 30% pure at 6.6 μM, and GAL4-HSF1 (phenylalanine mutant) was 50% pure at 16 μM. GAL4 DNA binding activity was normalized based on gel shift analysis (data not shown).
Purification of hSWI/SNF.
Human SWI/SNF was purified as described previously (17, 50) and judged to be approximately 50% pure by silver stain analysis.
Purification of nucleosomes, polynucleosomes, and histones.
H1-depleted HeLa nucleosomes were prepared and quantitated as described previously (9, 48, 53) with the exception that the nuclear pellet that resulted following extraction by the Dignam procedure was used as the starting material (12). For the restriction enzyme accessibility assay, medium-sized polynucleosomes (average two to six nucleosomes) were used as the competitor. HeLa histones were purified via hydroxyapatite chromatography as described previously (56).
Reconstitution, purification, and assay of nucleosomal templates.
Mononucleosomes were reconstituted and purified as previously described (17) for tailed mononucleosomes. The mononucleosome disruption assay was carried out as previously described (17) for tailed mononucleosomes. Reactions contained up to 150 ng of hSWI/SNF or 15 μl of M2 column fractions from the full-length wild-type HSF-flag, GAL4-HSF-wt-flag, GAL4-HSF-ac-flag, GAL4-HSF-ph-flag, or HeLa cell lines for 30 min at 30°C in the presence or absence of 0.5 mM ATP.
Labeled circular plasmid nucleosomes were reconstituted as described previously (18) using full-length histones. Plasmid supercoiling experiments were carried out as previously described (18) with either hSWI/SNF up to 300 ng (approximately 5 nM) or 15 μl of M2 fractions from the GAL4-HSF-wt-flag, GAL4-HSF-ac-flag, GAL4-HSF-ph-flag, or HeLa cell lines, both in the presence or absence of 2.1 mM ATP. Reactions were incubated at 30°C for 30 to 45 min.
Assembly of linear 5S arrays.
The p2085S-G5E4 plasmid containing the linear array was the gift of J. Workman and K. Neely (Penn State University) (37). The 5S-G5E4 sequence was excised from the plasmid with the restriction enzymes Asp718 and ClaI (Roche Molecular Biochemicals, Inc.). DdeI (NEB, Inc.) was included in the restriction digest to facilitate band identification. The fragment was gel purified and end labeled with [32P]dCTP with a Klenow fill-in reaction at the Asp718 site. The linear DNA was assembled into a polynucleosomal template by gradient dialysis as described previously (31, 42).
Restriction enzyme accessibility assay.
Restriction enzyme accessibility experiments were carried out at 30°C in 50 μl of reaction buffer (4 mM MgCl2, ±0.5 mM ATP/MgCl2, 60 mM KCl, 7% glycerol [before addition of restriction enzyme], 16 mM HEPES [pH 7.9], 10 mM Tris [pH 7.5], 0.36 mM EDTA, 0.8 mM dithiothreitol, 0.16 mM phenylmethylsulfonyl fluoride, 20-ng/μl bovine serum albumin). Template was added to a final concentration of 0.15 ng/μl, which equaled approximately 0.08 nM linear array (12 nucleosomes/array; therefore, 1 nM nucleosomes), and reactions included 2 or 5 nM purified hSWI/SNF, ±200 or 600 nM polynucleosomes (expressed in mononucleosome units), ±2 or 5 nM GAL4-HSF proteins (bacterially purified).
Restriction enzyme, 0.3 to 0.9 U of XbaI/μl, was present throughout the reaction. The start of the reaction was addition of the template. Aliquots were removed to a deproteinating stop buffer (3% SDS, 100 mM EDTA, 50 mM Tris HCl [pH 8.0], 25% glycerol, 6-mg/ml proteinase K) at 2, 10, 40, and 80 min and allowed to incubate at 30°C for 30 min. Gel loading dye was added, and the samples were analyzed on a 1% agarose gel in 1× TBE (89 mM Tris, 89 mM boric acid, 2 mM EDTA [pH 8.0]).
Rate constants were obtained by first-order fits to the data using Kaleidagraph. Reactions did not go to completion but leveled off at endpoints of 30 to 55% uncut DNA. The nonzero endpoint was similar to that observed previously in studies measuring spontaneous restriction enzyme access (44; G. Narlikar, unpublished data).
Transient transfections and CAT assays.
Transient transfections and chloramphenicol acetyltransferase (CAT) assays were carried out as previously described (5). HeLa cells were grown, and transfections were carried out as described previously (39) for heat shock experiments with 5 μg of pBXG1 plasmids expressing GAL4-HSF-wt, GAL4-HSF-ac, and GAL4-HSF-ph, 5 μg of p540CAT reporter plasmid, and 1 μg of pXHG5 reference plasmid (30).
RESULTS
To identify proteins that associate with HSF1, we established cell lines that stably expressed HSF1 that had been tagged at the C terminus with the FLAG epitope (Fig. 1A). These cell lines were made in order to purify HSF1 and tightly associated proteins in a single step using immunoaffinity chromatography (see below). We wished to stably express HSF1 at levels similar to expression of the endogenous gene in order to maximize yield of the tagged protein while avoiding any titration of associated factors that might be caused by overexpression of HSF1. We therefore inserted the tagged HSF1 construct into a retroviral vector (pBABE) that is known to result in expression of near-endogenous levels of protein (32).
The retroviral construct was transfected into packaging cells to produce virus, HeLa cells were infected with the resultant recombinant virus, and infected cells were selected using puromycin (see Materials and Methods). Ten clonal cell lines were screened for expression of tagged HSF1 via Western analysis using an antibody specific to the FLAG epitope (Santa Cruz). One cell line that expressed levels of tagged HSF1 that were comparable to the levels of endogenous HSF1 was chosen for further study. In the cell line containing tagged protein (Fig. 1B, lanes 1 and 2), the total amount of HSF1, tagged plus endogenous, is approximately three- to fivefold higher than expression of HSF1 in the parental HeLa cell line (lanes 3 and 4). The tagged HSF1 protein is functional as shown by the ability to restore HSF1 function to mouse cell lines that lack endogenous HSF1 (L. Corey, unpublished data).
Purification of tagged HSF1.
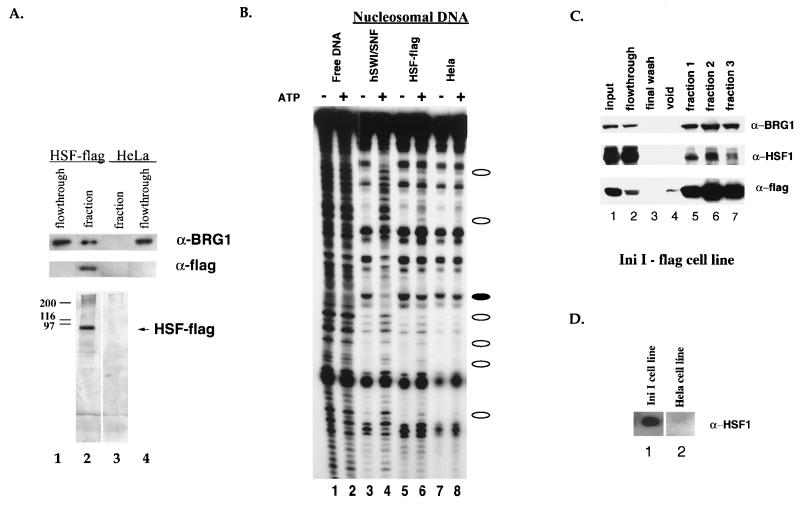
In order to purify the FLAG epitope-tagged HSF1, we made nuclear and cytoplasmic extracts from the tagged HSF1 cell line. The cell extractions and HSF1 immunopurifications were done under a wide range of salt and detergent conditions, including varying concentrations of KCl, KOAc, and (NH4)2SO4 (see Materials and Methods). All preparations resulted in substantially pure HSF1 (Fig. 2A, lane 2). There were no proteins that copurified with HSF1 in a stoichiometric manner under any buffer condition, regardless of whether HSF1 was isolated from heat-stressed cells, the nucleus, or the cytoplasm. Close inspection of silver-stained gels revealed the presence of substoichiometric bands at the approximate size of components of the human SWI/SNF family of chromatin remodeling complexes. We had previously shown that the ability of HSF1 to stimulate transcriptional elongation in vitro is enhanced by SWI/SNF (3). We used antisera specific for BRG1, a central subunit of hSWI/SNF complexes, and showed that this protein is present in HSF1 preparations but is not present after mock purification from control cells that did not express tagged HSF1 (Fig. 2A).
FIG. 2.
Association of human HSF1 with human BRG1 and chromatin remodeling activity. (A) Fractions from the M2 (anti-FLAG) purification of full-length HSF1-FLAG were resolved by SDS-PAGE and assayed by silver staining (bottom panel) or Western blot analysis using the anti-BRG1 antibody (top panel) and the anti-FLAG antibody, D-8 polyclonal (Santa Cruz) (middle panel). BRG1 flowthroughs are comparable to inputs. Heat shock fractions from cell lines expressing HSF1-FLAG (lane 2) and a HeLa cell line control (lane 3) were compared. (B) The same fractions from panel A were compared (lanes 5 to 8) to purified hSWI/SNF (lanes 3 and 4) in the DNase I mononucleosome disruption assay. Naked DNA was digested by DNase I as a control (lanes 1 and 2). Open ovals designate bands with an ATP-dependent increase in intensity, and closed ovals designate bands with an ATP-dependent decrease in intensity. Lanes 1 and 2 are at a lighter exposure in order to better see the bands in the highly digested lanes. (C) Fractions from an M2 (anti-FLAG) purification of INI1-flag, a hSWI/SNF subunit, were resolved by SDS-PAGE and assayed by Western blot analysis using the anti-BRG1 antibody (top panel), the anti-HSF1 antibody (Affinity BioReagents) (middle panel), and the anti-FLAG antibody (M5, Sigma) (bottom panel). The anti-HSF1 experiment required a longer exposure in order to visualize the HSF bands better. (D) Fractions from an M2 (anti-FLAG) purification of INI1-flag (lane 1) and a HeLa cell line control (lane 2) were resolved by SDS-PAGE and assayed by Western blot analysis using the anti-HSF1 anti-body (Stressgen).
Human SWI/SNF is an ATP-dependent remodeling complex that is able to significantly alter the structure of mononucleosomes. Therefore, we tested the purified fractions for activity in the mononucleosome disruption assay (10, 19, 28). In this assay, the ATP-dependent alteration of histone-DNA contacts is manifested by a disruption of the normal DNase I cleavage pattern of a mononucleosome. If nucleosomes are formed on DNA with rotational phasing sequences, they adopt uniform phasing which is evidenced by a 10-bp repeat when the nucleosome is cleaved with DNase I (Fig. 2B, compare free DNA in lane 1 to nucleosomal DNA in lane 3). Incubation with purified hSWI/SNF results in an ATP-dependent alteration of the DNase I pattern (lanes 3 and 4). We analyzed immunopurified HSF1 and found that it was associated with a low level of a qualitatively similar remodeling activity (lanes 5 and 6). This was despite the fact that the amount of BRG1 associated with the HSF1 fraction was estimated, via silver staining, to be approximately 10-fold less than the amount in the hSWI/SNF control reaction. We therefore performed a series of experiments designed to verify that HSF1 specifically associated with BRG1.
We asked first whether HSF1 associated with immunopurified hSWI/SNF. We purified hSWI/SNF from HeLa cells that contain the smallest SWI/SNF subunit, called INI1, with a FLAG epitope tag and tested for the presence of HSF1 by Western analysis (Fig. 2C). As reported previously, a significant portion of endogenous BRG1 copurifies with INI1 as part of hSWI/SNF (Fig. 2C, top panel) (50). HSF1 is also present in the purified hSWI/SNF, but not in a mock purification (Fig. 2C, middle panel and D). Examination of silver-stained gels reveals that HSF1 is not stoichiometric with hSWI/SNF subunits in purified hSWI/SNF (data not shown; see references 48 and 50). The observed low stoichiometry of the interaction between these two proteins suggests that they might interact transiently (see Discussion).
Association of hSWI/SNF with the transcriptional activation domains of HSF1.
We had previously shown that hSWI/SNF augments the ability of HSF1 to stimulate transcriptional elongation (3). Specific mutations in the HSF1 transcriptional activation domains inhibited the ability of HSF1 to stimulate either transcriptional initiation or elongation (5). We therefore sought to determine whether the transcriptional activation domains of HSF1 associated with hSWI/SNF. HSF1 activates transcription as a trimer, so it was not possible to test the effects of mutations in the HSF1 activation domains in the context of full-length HSF1, since the mutant protein would trimerize with the nontagged, endogenous wild-type HSF1. We therefore tested the effects of mutations in the transcriptional activation domains in the context of GAL4-HSF1 fusion proteins. In these proteins the DNA binding and trimerization domains of HSF1 are replaced by the GAL4 DNA binding domain. Cell lines were constructed (see Materials and Methods) that constitutively expressed low levels of N-terminally FLAG-tagged GAL4 fused to either the wild-type activation domains, activation domains with mutations in specific acidic amino acids, or activation domains with mutations in specific phenylalanine residues (Fig. 1A).
We first verified that the GAL4 fusion proteins functioned in the anticipated manner when transiently expressed in vivo. The G4-HSF-wt construct had previously been shown to stimulate transcription significantly and in a heat-inducible manner following transfection with a reporter construct. Acidic or hydrophobic amino acids that were critical for transcriptional activation had previously been identified using truncated portions of the activation domains (5, 39). In the experiments reported here, mutations that severely affected transcriptional activation were combined and reconstructed into the full-length transcriptional activation region of HSF1 (Fig. 1A). Wild-type and mutant fusion proteins were cotransfected with a CAT reporter construct that contained Gal4 binding sites into HeLa cells which were subsequently heat shocked. Mutation of three acidic residues decreased activation of CAT expression to 2.6% (±0.3%) of wild-type levels, while mutation of three phenylalanine residues decreased activation to 7.2% (±3.5%) of wild-type levels. Impaired activation was not caused by instability of the mutant proteins, since these proteins were expressed at higher levels than wild-type fusion proteins in HeLa cells (see below). Thus, both sets of point mutations in the HSF1 activation domains decreased overall transcriptional activation 10-fold or more. We anticipated that these different sets of mutations might differentially effect interaction with BRG1. Previously, using in vitro assays that distinguished between transcriptional initiation and transcriptional elongation, we had shown that mutations in phenylalanine residues reduced stimulation of elongation, while mutations in acidic residues reduced stimulation of initiation (5).
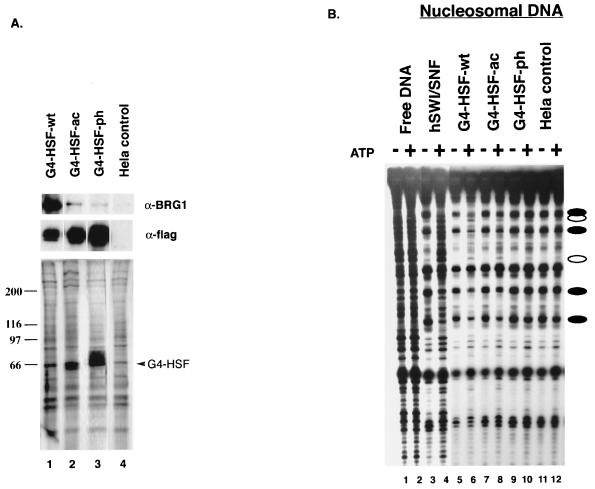
We made extracts from stable HeLa cell lines that expressed the FLAG epitope-tagged GAL4 fusion proteins and immunopurified the fusion proteins (Fig. 3A). We routinely found that G4-HSF-ph was expressed at higher levels than G4-HSF-ac and that both mutations were expressed at a higher level than G4-HSF-wt (Fig. 3A, middle panel). We estimated the amount of BRG1 that copurified with each GAL4 protein by Western analysis. More BRG1 was found in the G4-HSF-wt preparations than in G4-HSF-ac preparations, and the amount in the G4-HSF-ac preparations was always more than in G4-HSF-ph preparations (Fig. 3A, top panel, and 4B, top panel). We conclude that mutations in the transcriptional activation domains of HSF1 decrease interaction with BRG1.
FIG. 3.
Human HSF1 activation domain mutants are found associating with human BRG1 and chromatin remodeling activity in a differential manner. (A) Fractions from the M2 (anti-FLAG) purification of the GAL4-HSF-flag fusions were resolved by SDS-PAGE and assayed by silver staining (bottom panel) or Western blot analysis using the anti-BRG1 antibody (top panel) and the anti-FLAG antibody, (M5; Sigma) (middle panel). Heat shock fractions from cell lines expressing the GAL4-HSF wild type (lane 1), GAL4-HSF acidic mutant (lane 2), GAL4-HSF phenylalanine mutant (lane 3), and a HeLa cell line control (lane 4) were compared. (B) The same fractions shown in panel A were compared (lanes 5 to 12) to purified hSWI/SNF (lanes 3 and 4) in the DNase I mononucleosome disruption assay. Naked DNA was digested by DNase I (lanes 1 and 2) as a control. Open ovals designate bands with an ATP-dependent increase in intensity, and closed ovals designate bands with an ATP-dependent decrease in intensity. Lanes 1 to 4 are at a lighter exposure in order to better see the bands in the highly digested lanes.
We measured ATP-dependent nucleosome remodeling activity of the GAL4 preparations to verify that this activity mirrored the amount of BRG1 in the preparations. We observed a characteristic ATP-dependent disruption of the DNase I pattern when G4-HSF-wt preparations were used (Fig. 3B, lanes 5 and 6). The remodeling activity in the G4-HSF-wt fraction was more than the activity in G4-HSF-ac, and the activity in G4-HSF-ac was still higher than the activity in G4-HSF-ph (Fig. 3B, compare the two bands with closed ovals in the lowest positions in lanes 6, 8, 10, and 12). Thus, the amounts of remodeling activity correlated with the amount of BRG1 measured by Western analysis.
We also assessed remodeling activity using a separate topology-based protocol that is thought to be specific to the SWI/SNF family of ATP-dependent remodeling complexes (18, 28; J. Aalfs and R. E. Kingston, unpublished data). Assembly of closed circular plasmids into nucleosomes results in introduction of approximately one supercoil per nucleosome. This is most easily detected by deproteinizing the template and analyzing it by native agarose gel electrophoresis. In the examples shown (Fig. 4B, lane 1), the introduction of approximately 15 negative supercoils following removal of 15 nucleosomes caused the template to migrate rapidly; a more supercoiled template migrates faster than a less supercoiled template under these gel conditions. SWI/SNF family complexes dramatically alter topology in an ATP-dependent manner, resulting in a distribution of supercoiled species (Fig. 4B, lane 2).
FIG. 4.
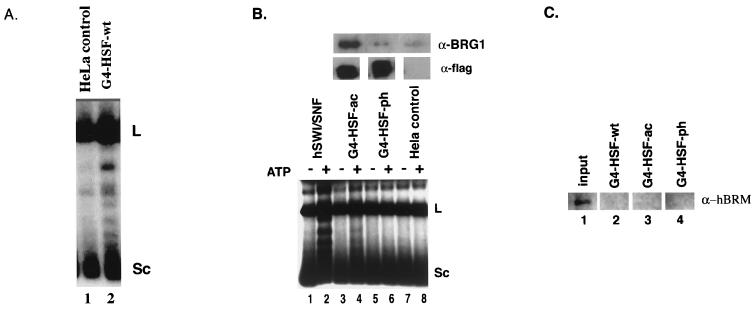
A BRG1-based SWI/SNF complex appears to be responsible for HSF1-associated remodeling activity. (A) Nuclear fractions from the M2 (anti-FLAG) purification of a HeLa cell line control (lane 1) and GAL4-HSF wild type (lane 2) were compared in the supercoiling assay, both plus ATP. L, linear DNA; Sc, completely supercoiled DNA. (B) Nuclear fractions from M2 (anti-FLAG) purification of the GAL4-HSF acidic mutant, the GAL4-HSF phenylalanine mutant, and a HeLa cell line control were resolved by SDS-PAGE and assayed by Western blot analysis using the anti-BRG1 antibody (top panel) and the anti-FLAG antibody, (M5; Sigma) (middle panel). These fractions (lanes 3 to 8) were then compared to purified hSWI/SNF (lanes 1 and 2) in the supercoiling assay. L, linear DNA; Sc, completely supercoiled DNA. (C) Nuclear fractions from M2 (anti-FLAG) purification of the GAL4-HSF wild type, GAL4-HSF acidic mutant, and the GAL4-HSF phenylalanine mutant were compared to nuclear extract input. Samples were resolved by SDS-PAGE and assayed by Western blot analysis using the anti-hBRM antibody.
When tested using this protocol, the GAL4-HSF-wt fraction altered topology when compared to a HeLa control preparation (Fig. 4A, compare lane 2 with lane 1). We tested whether the differential effects of the activation domain mutants on the physical association with BRG1 (Fig. 4B, top panel) would be mirrored in this assay by using matched amounts of different G4-HSF proteins (Fig. 4B, middle panel); the use of matched amounts of GAL4 proteins precluded the use of G4-HSF-wt in this experiment due to its low expression. We found that the G4-HSF-ac preparation had discernible ATP-dependent remodeling activity (lanes 3 and 4), while the G4-HSF-ph and the HeLa control preparations did not (lanes 5 to 8). Higher amounts of BRG1 associate with the acidic mutant than with the phenylalanine mutant (Fig. 4B, top panel); therefore, the remodeling activity correlated with the amount of BRG1 in these fractions. We thus conclude that mutations in phenylalanine residues decrease BRG1 association to a greater degree than mutations in acidic residues. These data provide further evidence that a SWI/SNF complex is responsible for the HSF1-associated remodeling activity.
SWI/SNF complexes in humans may contain BRG1 or hBRM as their central ATPase subunit (49). In order to assess whether HSF1 was also found associating with hBRM, Western blotting was done with fractions from the GAL4-HSF purifications (Fig. 4C). No association of HSF1 with hBRM was detected, although the hBRM antibody is less sensitive than the BRG1 antibody and may not detect very low levels of hBRM.
Human HSF1 transcriptional activation domains target hSWI/SNF to a chromatin template.
We next wished to assess the functional consequence of HSF1 interactions with BRG1 by determining whether HSF1 activation domains could target remodeling activity to a template. To do this we used a completely defined system that contained purified hSWI/SNF, bacterially purified GAL4-HSF1 fusion proteins, and templates assembled into nucleosomes by salt dialysis. We used a restriction enzyme access protocol previously developed to examine targeting of ySWI/SNF (58). The assembly of a restriction site into a nucleosome generally inhibits cleavage by restriction enzymes, and SWI/SNF relieves this inhibition in an ATP-dependent manner (Fig. 5B, lanes 1 and 2). We used a radiolabeled, linear template in which two sets of five 5S nucleosome positioning sequences flank DNA that can assemble into two nucleosomes; this central region also contains five Gal4 binding sites and various restriction sites (Fig. 5A) (37). If HSF1 activation domains recruit hSWI/SNF, then binding of GAL4-HSF1 fusion proteins to the template should increase local concentration of the remodeling complex, resulting in increased template accessibility to restriction enzyme cleavage.
FIG. 5.
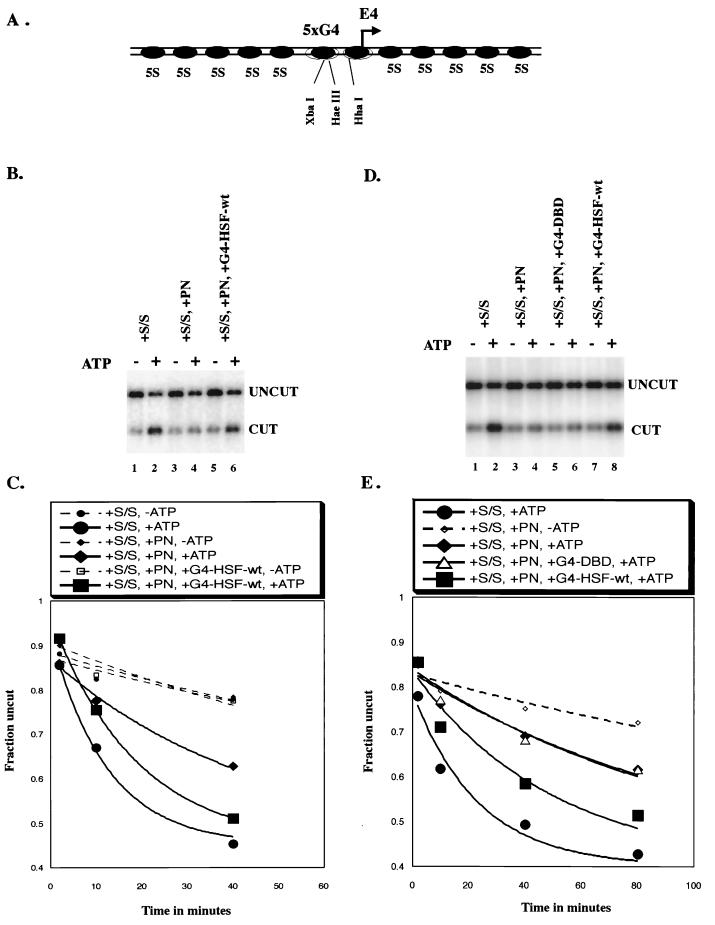
The HSF1 wild type activation domain is able to target SWI/SNF to a chromatin template. Results of a restriction enzyme accessibility assay are shown. (A) Diagram of the 5S array nucleosomal template used in the restriction enzyme accessibility assay. The two central nucleosomes are not positioned. There are five Gal4 DNA binding sites located upstream of a minimal adenovirus E4 promoter. XbaI, HaeIII, and HhaI are three restriction enzyme cleavage sites present in the nonpositioned central region. (B) Raw data of the 40-min time point for the experiment whose results are shown in panel C. (C) Graph shows the time course for XbaI cutting of the 5S array. All reactions contained 1 nM template nucleosomes and 0.3 U of XbaI/μl, which was present throughout the reaction. S/S, 2 nM human SWI/SNF complex; PN, 240 nM polynucleosomes as competitor; G4-HSF-wt, 2 nM GAL4-HSF wild-type, bacterially purified protein. Rate constants (minute−1): Average, −ATP, 7.6 × 10−3; S/S, +ATP, 81.5 × 10−3; S/S, +PN, +ATP, 22.3 × 10−3; S/S, +PN, +G4-HSF-wt, +ATP, 53.7 × 10−3. (D) Raw data of the 40-min time point for the experiment whose results are shown in panel E. (E) Graph shows the time course for XbaI cutting of the 5S array. All reactions contained 1 nM concentrations of template nucleosomes and 0.9 U of XbaI/μl, which was present throughout the entire reaction. S/S, 5 nM human SWI/SNF complex; PN, 200 nM polynucleosomes as competitor; G4-DBD, 5 nM GAL4-DNA binding domain (1 to 94). G4-HSF-wt, 5 nM GAL4-HSF wild type. All GAL4-HSF proteins were bacterially purified. Rate constants (minute−1): S/S, +ATP, 45.4 × 10−3; S/S, +PN, −ATP, 5.8 × 10−3; S/S, +PN, +ATP, 11.2 × 10−3; S/S, +PN, +G4-DBD, +ATP, 12.5 × 10−3; S/S, +PN, +G4-HSF-wt, +ATP, 24.9 × 10−3.
To best observe recruitment of SWI/SNF remodeling activity, we used a protocol in which the template containing Gal4 binding sites was present at low levels and an unlabeled competing nucleosomal template with no Gal4 binding sites (polynucleosomes) was present in excess. When added to the reaction, hSWI/SNF partitions between these templates. If GAL4-HSF protein increased the affinity of hSWI/SNF, then more SWI/SNF would partition to the GAL4-bound template and we would observe an increased rate of restriction enzyme access, reflecting an increased rate of remodeling. The remodeling rate was determined by removing aliquots from the restriction enzyme cleavage assay at various times and quenching at each time point. Data from the 40-min time point is shown for the different reactions (Fig. 5B and D and 6A). Fits to the entire time course, as shown in single representative experiments in Fig. 5C and E and 6B, give individual rate constants. The ratios between the individual rate constants were highly reproducible in repeat experiments (Fig. 6C). We thus were able to determine the effect of different mutations in HSF1 on the ability of GAL4-HSF fusion proteins to target remodeling activity to the template.
FIG. 6.
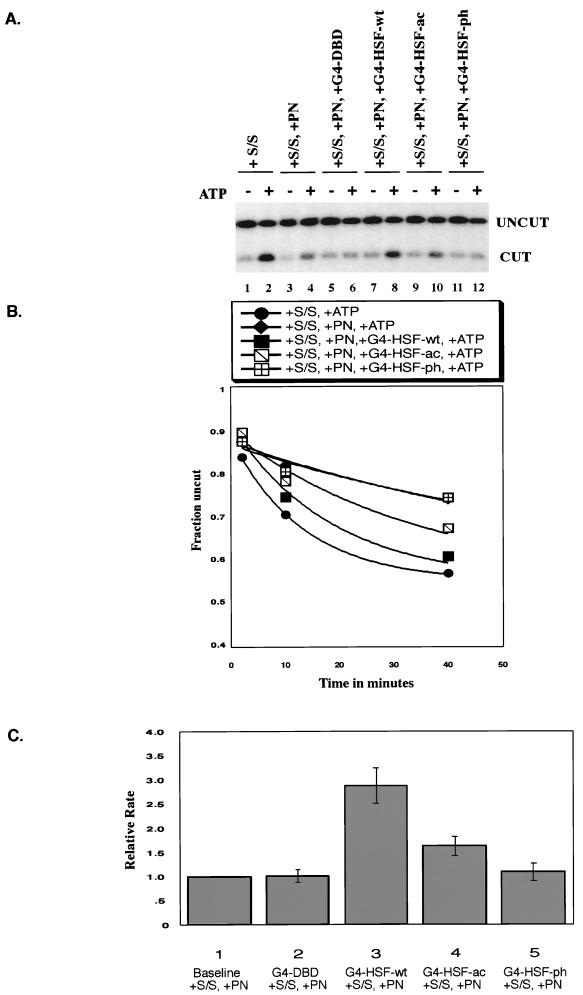
GAL4-HSF activation domain mutants target hSWI/SNF in a differential manner. Restriction enzyme accessibility assay results are shown. (A) Raw data of the 40-min time point for the experiment depicted in panel B. (B) The graph shows the time course for XbaI cutting of the 5S array. All reactions contained 1 nM concentrations of template nucleosomes and 0.6 U of XbaI/μl, which was present throughout the reaction. S/S, 5 nM human SWI/SNF complex; PN, 200 nM polynucleosomes as competitor; G4-DBD, 5 nM GAL4-DNA binding domain (1 to 94); G4-HSF-wt, 5 nM GAL4-HSF wild type. G4-HSF-ac, 5 nM GAL4-HSF acidic mutant; G4-HSF-phe, 5 nM GAL4-HSF phenylalanine mutant. All GAL4 proteins were bacterially purified. Rate constants (minute−1): S/S, +ATP, 76.1 × 10−3; S/S, +PN, +ATP, 14.0 × 10−3; S/S, +PN, +G4-HSF-wt, +ATP, 54.0 × 10−3; S/S, +PN, +G4-HSF-ac, +ATP, 28.8 × 10−3; S/S, +PN, +G4-HSF-ph, +ATP, 13.2 × 10−3. (C) Bar graph summarizing the relative rate constants for XbaI cutting in four different experiments (experiment depicted in panel B included). All reactions contained 1 nM template nucleosomes and 0.6 to 0.9 U of XbaI/μl, which was present throughout the reaction. S/S, 5 nM human SWI/SNF complex; PN, 200 to 600 nM polynucleosomes as the competitor; G4-DBD, 5 nM GAL4-DNA binding domain (1 to 94); G4-HSF-wt, 5 nM GAL4-HSF wild type; G4-HSF-ac, 5 nM GAL4-HSF-acidic mutant; G4-HSF-phe, 5 nM GAL4-HSF phenylalanine mutant. Each bar represents the standard error.
In the experiment shown in Fig. 5B and C, addition of hSWI/SNF without any cold competing template increased the rate of restriction enzyme cleavage 11-fold in an ATP-dependent manner. Addition of a 240-fold molar excess of unlabeled template (120-fold excess over SWI/SNF) decreased this stimulation to 3-fold. Including G4-HSF-wt in the reaction in the presence of competing polynucleosomes increased stimulation of cleavage to 7-fold. GAL4-HSF-wt alone did not alter restriction enzyme cleavage, as demonstrated by the comparison between reactions lacking ATP. In this and all subsequent experiments, restriction enzyme was present in excess. We conclude that G4-HSF-wt is able to target remodeling activity to a template.
To determine whether targeting by G4-HSF-wt required a transcriptional activation domain, we compared G4-HSF-wt to the GAL4 DNA binding domain alone (amino acids 1 to 94). Using equal concentrations of active GAL4 protein (normalized by DNA binding), the GAL4 DNA binding domain alone did not increase remodeling in the presence of competitor, while, as described above, G4-HSF-wt did (Fig. 5D and E). Increasing the concentration of the GAL4 DNA binding domain by threefold also did not lead to targeting of remodeling activity, verifying that the results were not altered by changes in the concentration of the GAL4 protein (data not shown).
If targeting of SWI/SNF activity correlated with the ability of proteins to cofractionate with BRG1, then we would expect phenylalanine mutations to significantly decrease targeting and acidic mutations to decrease targeting to a lesser degree. This hypothesis was confirmed by the data in Fig. 6A and B. Quantification of repeat experiments showed that when normalized to reactions with competing polynucleosomes and no activator, G4-HSF-wt increased remodeling by approximately threefold, G4-HSF-ac increased remodeling less than twofold, and G4-HSF-ph showed no significant stimulation of remodeling over baseline levels (Fig. 6C). We conclude that the HSF1 transcriptional activation domains are able to target hSWI/SNF remodeling and that this recruitment is most significantly affected by mutations in phenylalanine residues.
DISCUSSION
From the experiments above, we conclude that HSF1 associates with BRG1, the ATPase subunit of SWI/SNF, at endogenous concentrations in vivo. We demonstrate that GAL4-HSF fusion proteins are able to recruit purified hSWI/SNF to a chromatin template in vitro and that the HSF transcriptional activation domains play a major role in this recruitment. Targeting of hSWI/SNF in this protocol is decreased by mutations in residues necessary for stimulation of transcriptional initiation but is abolished by mutations in residues necessary for stimulation of transcriptional elongation. Therefore, this work suggests that targeting is involved in regulation of transcriptional elongation.
HSF1 and BRG1 associate in a substoichiometric manner when purified from extracts. In fact, despite significant effort under numerous conditions, we have been unable to identify any factor or complex that interacts stoichiometrically with HSF1 in solution. This differs from activators such as the steroid receptors and VP16 that have robust associations with mediator/coactivator complexes which allow purification of these complexes via affinity chromatography (20, 35, 45). The HSF1 transcriptional activation domains are among the most potent that have been characterized (16), yet this robust ability to stimulate transcription does not appear to correspond with a robust interaction in solution with any component of the transcription machinery.
This substoichiometric association occurs whether purification relies on immunoaffinity steps directed against HSF1 (Fig. 2A), INI1 (Fig. 2C), or GAL4-HSF fusion proteins (Fig. 3A). The association data is buttressed by two important observations. First, the ability of GAL4-HSF to recruit hSWI/SNF to a chromatin template, as assayed by restriction enzyme accessibility, demonstrates the ability of hHSF1 to functionally interact with the SWI/SNF complex. This threefold HSF1-dependent increase in remodeling is similar to the effect that the GR (glucocorticoid receptor) has on the stimulation of SWI/SNF activity (40). Second, there is a differential effect of the initiation and elongation mutants on the association of SWI/SNF components with HSF1. This difference, seen in both the purification and the recruitment studies, demonstrates the specificity of the interaction.
The substoichiometric cofractionation of HSF1 and BRG1 could be indicative of an interaction that is transient or that requires other components (DNA, proteins) to strengthen it. It is reasonable to expect that contact between an activator and a remodeling complex would be transient. For example, in the elongation step of transcription, recruitment of a remodeling complex might be required to destabilize the nucleosome structure in front of a paused RNA polymerase. However, it has been proposed that once activated, the elongation process itself remodels the template, leaving it open for subsequent elongation without further need for the remodeler (4, 29). Like most activators, HSF1 is expected to interact with multiple targets in the general transcription machinery and the chromatin modifying machinery. These interactions appear to take place using the same face of the activation domain; mutations in acidic residues of the HSF1 activation domains have different effects than do mutations in phenylalanine residues, yet the acidic and phenylalanine residues are interspersed in the activation domains (Fig. 1A) (5). Transient interactions would allow the same face of the activator to contact multiple targets in a short period of time.
In a natural context, the interaction between HSF1 and BRG1 might be stabilized by interactions with DNA or other components of the transcription machinery. The interaction that we have detected occurs in solution at endogenous levels of both proteins. For this interaction to be relevant to regulation of transcription, it is anticipated that it will occur when HSF1 is both bound to a promoter region and in the presence of general transcription factors (RNA polymerase, TFIID, etc.). For example, in an analysis of recruitment of yeast SWI/SNF by GAL4 fusion proteins, successful targeting required more than 53 bp of DNA adjacent to the Gal4 binding sites (58). This suggests that interactions in solution were not robust and that additional interactions with DNA were required to observe targeting. Both the BRG1 protein alone and the hSWI/SNF complex bind to nucleosomal DNA (10; Narlikar, unpublished), and SWI/SNF complexes have been found to cofractionate with RNA polymerase, suggesting a direct interaction between these remodelers and proteins associated with polymerase (7, 38, 55). Thus, a combination of these binding affinities might stabilize the association of hSWI/SNF with a promoter region following recruitment by HSF1. Given these considerations, the observed associations between HSF1 and BRG1 in solution reflect the potential strength of the HSF1 recruitment of hSWI/SNF.
There are likely to be several related chromatin remodeling complexes that contain BRG1. We do not know whether one specific complex associates with HSF1 in the studies reported here, or whether multiple different complexes are associated. Immunopurified HSF1 fractions contain the characteristic nucleosome remodeling activity of BRG1-containing hSWI/SNF complexes, and immunopurification of an epitope-tagged INI1 extract results in cofractionation of HSF1, suggesting that HSF1 is associated with one of the hSWI/SNF family complexes that contain BRG1 and INI1. Available antisera against other components of this family of complexes are not sufficiently sensitive to address further the nature of the associated complex (data not shown). Studies on the precise composition and function of BRG1-based complexes are ongoing and should produce more information on this issue in the future.
Different domains of activators recruit SWI/SNF.
Several labs have reported an association between SWI/SNF family chromatin remodeling complexes and activators (8, 15, 23, 25, 37, 40, 57, 58). Different domains of activators are required for physical interaction with SWI/SNF. The zinc finger DNA binding domain of the erythroid factor EKLF, as well as other zinc finger proteins, interacts with hSWI/SNF and recruits remodeling activity to assist in transcriptional activation (23). Other studies have shown, similar to the observations described above, that certain acidic transcriptional activation domains interact with yeast SWI/SNF (37, 58). Our work is consistent with these findings; the acidic activation domains of HSF demonstrate an ability to associate with BRG1 in vivo and target human SWI/SNF to a nucleosomal template in vitro. It has also been reported that hydrophobic clusters in the activation domains of GCN4 associate with SWI/SNF (36). Our results show a requirement for phenylalanine residues in recruiting hSWI/SNF. The effect of the mutated phenylalanine residues, shown to be important for HSF1 activation of elongation, in recruitment of hSWI/SNF leads us to hypothesize that this recruitment is important for stimulation of transcriptional elongation.
The hsp70 gene, as well as many other mammalian genes (reviewed in reference 51), utilizes a block in transcriptional elongation as a key regulatory step. Nucleosome formation increases the duration of the pause of RNA polymerase II on the hsp70 gene in vitro, and addition of hSWI/SNF augments the ability of HSF1 transcriptional activation domains to stimulate release of the pause (3). Phenylalanine residues in the HSF1 activation domains are required for full stimulation of transcriptional elongation in vitro (5). We propose that the transcriptional activation domains of hHSF1 directly recruit a BRG1-based chromatin remodeling complex to the hsp70 gene and that this complex alters the inhibiting nucleosome structure, thereby aiding in transcriptional elongation. Such a mechanism might play a role in the transcriptional control of other genes, especially genes that are regulated by promoter-proximal pausing, such as c-myc, c-fos, and some HIV genes (11, 26, 43).
ACKNOWLEDGMENTS
We thank M. Phelan, G. Narlikar, and N. Francis for support and advice, Cellex Biosciences for growing the INI1 cell line, and members of our lab for comments on the manuscript.
This work was supported by grants from the National Institutes of Health and Hoechst AG to R.E.K., and a grant from the NIH to S.S.
REFERENCES:
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1996. [Google Scholar]
- 2.Bjorklund S, Almouzni G, Davidson I, Nightingale K P, Weiss K. Global transcription regulators of eukaryotes. Cell. 1999;96:759–767. doi: 10.1016/s0092-8674(00)80586-3. [DOI] [PubMed] [Google Scholar]
- 3.Brown S A, Imbalzano A N, Kingston R E. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- 4.Brown S A, Kingston R E. Disruption of downstream chromatin directed by a transcriptional activator. Genes Dev. 1997;11:3116–3121. doi: 10.1101/gad.11.23.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown S A, Weirich C S, Newton E M, Kingston R E. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998;17:3146–3154. doi: 10.1093/emboj/17.11.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang C M, Ge H, Wang Z, Hoffmann A, Roeder R G. Unique TATA-binding protein containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho H, Orphanides G, Sun X, Yang X J, Ogryzko V, Lees E, Nakatani Y, Reinberg D. A human RNA polymerase II complex containing factors that modify chromatin structure. Mol Cell Biol. 1998;18:5355–5363. doi: 10.1128/mcb.18.9.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosma M P, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 9.Côté J, Utley R T, Workman J L. Basic analysis of transcription factor binding to nucleosomes. Methods Mol Gen. 1995;6:108–128. doi: 10.1016/s0076-6879(96)74024-7. [DOI] [PubMed] [Google Scholar]
- 10.Côté J, Quinn J, Workman J L, Peterson C L. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 11.Cullen B R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990;63:655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- 12.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. In: Wu R, Grossman L, Moldave K, editors. Methods in enzymology. Vol. 101. New York, N.Y: Academic Press; 1983. pp. 582–598. [DOI] [PubMed] [Google Scholar]
- 13.Farkas G, Leibovitch B A, Elgin S C. Chromatin organization and transcriptional control of gene expression in Drosophila. Gene. 2000;253:117–136. doi: 10.1016/s0378-1119(00)00240-7. [DOI] [PubMed] [Google Scholar]
- 14.Fondell J D, Ge H, Roeder R G. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fryer C J, Archer T K. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 16.Green M, Schuetz T J, Sullivan E K, Kingston R E. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol Cell Biol. 1995;15:3354–3362. doi: 10.1128/mcb.15.6.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyon J R, Narlikar G J, Sif S, Kingston R E. Stable remodeling of tailless nucleosomes by the human SWI-SNF complex. Mol Cell Biol. 1999;19:2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guyon J R, Narlikar G J, Sullivan E K, Kingston R E. Stability of a human SWI/SNF remodeled nucleosomal array. Mol Cell Biol. 2001;21:1132–1144. doi: 10.1128/MCB.21.4.1132-1144.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Imbalzano A N, Kwon H, Green M R, Kingston R E. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 20.Ito M, Yuan C X, Malik S, Gu W, Fondell J D, Yamamura S, Fu Z Y, Zhang X, Qin J, Roeder R G. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell. 1999;3:361–370. doi: 10.1016/s1097-2765(00)80463-3. [DOI] [PubMed] [Google Scholar]
- 21.Izban M G, Luse D S. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- 22.Jones K A, Kadonaga J T. Exploring the transcription-chromatin interface. Genes Dev. 2000;14:1992–1996. [PubMed] [Google Scholar]
- 23.Kadam S, McAlpine G S, Phelan M L, Kingston R E, Jones K A, Emerson B M. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kingston R E, Narlikar G J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 1999;13:2339–2352. doi: 10.1101/gad.13.18.2339. [DOI] [PubMed] [Google Scholar]
- 25.Kowenz-Leutz E, Leutz A. A C/EBP isoform recruits the SWI/SNF complex to activate myeloid genes. Mol Cell. 1999;4:735–743. doi: 10.1016/s1097-2765(00)80384-6. [DOI] [PubMed] [Google Scholar]
- 26.Krumm A, Hickey L B, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- 27.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzmol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 28.Kwon H, Imbalzano A N, Khavari P A, Kingston R E, Green M R. Nucleosome disruption and enhancement of activator binding by a human SWI/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 29.Lee M-S, Garrard W T. Transcription-induced nucleosome ‘splitting’: an underlying structure for DNase I sensitive chromatin. EMBO J. 1991;10:607–615. doi: 10.1002/j.1460-2075.1991.tb07988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lis J, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- 31.Luger K, Rechsteiner T J, Richmond T J. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 32.Morgenstern J P, Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morimoto R I. Cells in stress: transcriptional activation of heat shock genes. Science. 1993;259:1409–1410. doi: 10.1126/science.8451637. [DOI] [PubMed] [Google Scholar]
- 34.Morimoto R I. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 35.Naar A M, Beaurang P A, Zhou S, Abraham S, Solomon W, Tjian R. Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature. 1999;398:828–832. doi: 10.1038/19789. [DOI] [PubMed] [Google Scholar]
- 36.Natarajan K, Jackson B, Zhou H, Winston F, Hinnebusch A. Transcriptional activation by gcn4p involves independent interactions with the SWI/SNF complex and the SRB/mediator. Mol Cell. 1999;4:657–664. doi: 10.1016/s1097-2765(00)80217-8. [DOI] [PubMed] [Google Scholar]
- 37.Neely K, Hassan A, Wallberg A, Steger D, Cairns B, Wright A, Workman J. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol Cell. 1999;4:649–655. doi: 10.1016/s1097-2765(00)80216-6. [DOI] [PubMed] [Google Scholar]
- 38.Neish A S, Anderson S F, Schlegel B P, Wei W, Parvin J D. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res. 1998;26:847–853. doi: 10.1093/nar/26.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Newton E M, Knauf U, Green M, Kingston R E. The regulatory domain of human heat shock factor 1 is sufficient to sense heat stress. Mol Cell Biol. 1996;16:839–846. doi: 10.1128/mcb.16.3.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Östlund F A, Blomquist P, Kwon H, Wrange O. Glucocorticoid receptor-glucocorticoid response element binding stimulates nucleosome disruption by the SWI/SNF complex. Mol Cell Biol. 1997;17:895–905. doi: 10.1128/mcb.17.2.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson C L, Logie C. Recruitment of chromatin remodeling machines. J Cell Biochem. 2000;78:179–185. doi: 10.1002/(sici)1097-4644(20000801)78:2<179::aid-jcb1>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 42.Phelan M L, Sif S, Narlikar G J, Kingston R E. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 43.Pinaud S, Mirkovitch J. Regulation of c-fos expression by RNA polymerase elongation competence. J Mol Biol. 1998;280:785–798. doi: 10.1006/jmbi.1998.1905. [DOI] [PubMed] [Google Scholar]
- 44.Polach K J, Lowary P T, Widom J. Effects of core histone tail domains on the equilibrium constants for dynamic DNA site accessibility in nucleosomes. J Mol Biol. 2000;298:211–223. doi: 10.1006/jmbi.2000.3644. [DOI] [PubMed] [Google Scholar]
- 45.Rachez C, Lemon B D, Suldan Z, Bromleigh V, Gamble M, Naar A M, Erdjument-Bromage H, Tempst P, Freedman L P. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 46.Reece R J, Rickles R J, Ptashne M. Overproduction and single-step purification of GAL4 fusion proteins from Escherichia coli. Gene. 1993;126:105–107. doi: 10.1016/0378-1119(93)90596-u. [DOI] [PubMed] [Google Scholar]
- 47.Rougvie A E, Lis J T. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- 48.Schnitzler G, Sif S, Kingston R E. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 49.Sif S, Saurin A J, Imbalzano A N, Kingston R E. Purification and characterization of mSin3A-containing Brg1 and hBrm chromatin remodeling complexes. Genes Dev. 2001;15:603–618. doi: 10.1101/gad.872801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sif S, Stukenberg P T, Kirschner M W, Kingston R E. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spencer C A, Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990;5:777–785. [PubMed] [Google Scholar]
- 52.Sudarsanam P, Winston F. The Swi/Snf family nucleosome-remodeling complexes and transcriptional control. Trends Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- 53.Vettese-Dadey M, Walter P, Chen H, Juan L-J, Workman J L. Role of the histone amino termini in facilitated binding of a transcription factor, GAL4-AH, to nucleosome cores. Mol Cell Biol. 1994;14:970–981. doi: 10.1128/mcb.14.2.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vignali M, Hassan A H, Neely K E, Workman J L. ATP-dependent chromatin-remodeling complexes. Mol Cell Biol. 2000;20:1899–1910. doi: 10.1128/mcb.20.6.1899-1910.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson C J, Chao D M, Imbalzano A N, Schnitzler G R, Kingston R E, Young R A. RNA polymerase II holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- 56.Workman J L, Taylor I C, Kingston R E, Roeder R G. Control of class II gene transcription during in vitro nucleosome assembly. Methods Cell Biol. 1991;35:419–447. doi: 10.1016/s0091-679x(08)60582-8. [DOI] [PubMed] [Google Scholar]
- 57.Yoshinaga S K, Peterson C L C L, Herskowitz I I, Yamamoto K R. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]
- 58.Yudkovsky N, Logie C, Hahn S, Peterson C. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 1999;13:2369–2374. doi: 10.1101/gad.13.18.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]