Abstract
TP53 is the most commonly mutated gene in human cancer with over 100,000 literature citations in PubMed. This is a heavily studied pathway in cancer biology and oncology with a history that dates back to 1979 when p53 was discovered. The p53 pathway is a complex cellular stress response network with multiple diverse inputs and downstream outputs relevant to its role as a tumor suppressor pathway. While inroads have been made in understanding the biology and signaling in the p53 pathway, the p53 family, transcriptional readouts, and effects of an array of mutants, the pathway remains challenging in the realm of clinical translation. While the role of mutant p53 as a prognostic factor is recognized, the therapeutic modulation of its wild-type or mutant activities remain a work-in-progress. This review covers current knowledge about the biology, signaling mechanisms in the p53 pathway and summarizes advances in therapeutic development.
Keywords: p53, CDKN1A, DR5, Puma, miRNA, cell cycle, apoptosis, cancer, therapeutics
A.Tumor Suppressor TP53: “Guardian of the Genome”
TP53 is a gene that encodes for the p53 tumor suppressor protein, commonly referred to as the “Guardian of the Genome” [1]. Its main biological function appears to involve the protection of the DNA integrity of the cell. TP53 plays additional roles in development, aging and cell differentiation [2]. For example, p53-nullizygous genetic models exhibit phenotypes related to aging, pluripotency and development characterized by early aging onset, induction of cell pluripotency and inability of embryos undergo gastrulation, respectively [3–5]. The p53 protein is a transcription factor that controls the output of many biological processes according to the type of cellular stress signal input. Stress signals known to activate p53 include oncogene activation, DNA damage, and replication stress [6]. In response to these stresses, p53 undergoes post-translational modifications, promotes the transcription of genes involved in specific cell responses according to the stress type thereby controlling the cell’s fate [7]. Vastly studied biological processes where p53 has been shown to play a role are cell cycle arrest, senescence, DNA repair, and apoptosis (Figure 1). Over the last few years, researchers have elucidated many other pathways that p53 participates in such as autophagy, cell metabolism, ferroptosis, and pathways that involve the generation of reactive oxygen species. In some of these pathways, p53 does not execute signals directly as a transcription factor, but rather from its interaction with other proteins. For example, in apoptosis, activation of this pathway can occur via the interaction of p53 with anti-apoptotic proteins located in the mitochondria.
Figure 1. Overview of p53 activation, regulation, and transcriptional cellular response output.
Cellular stresses including DNA damage, oncogene activation, hypoxia, and replication/translation stress activate sensor proteins ATM, ATR, Chk1, Chk2, DNA-PK, and p14ARF. These kinases phosphorylate p53 leading to its stabilization, oligomerization, and binding to the p53RE. P53 stability is mainly regulated by MDM2, which is also a p53-target thus forming a negative feedback loop. Further protein modifiers and cofactors that bind to the p53 protein regulate the transcriptional activity of its target genes. This multi-step process of p53 activation ultimately regulates the stress input to the appropriate biological response outcome. Created with BioRender.com
The road to p53 discovery
P53 is found to be mutated in over 50% of human cancers and the remaining of the 100% involve biological inactivation of its pathway including MDM2 amplification, loss of p14ARF and mutations in activating kinases like ATM and Chk2 [8]. Loss of p53 pathway function gives cancer cells a survival advantage to bypass the resolution of oncogenic signals and DNA damage to continue abnormal proliferation. It is not taken as a surprise that when p53 was first discovered in 1979 it was thought to be an oncogene given that its mutated protein form was found in abundance in many cancer tissues. Accordingly, p53 initial discovery was found to be associated with the large T-antigen simian virus 40 (SV40) where the introduction of this virus resulted in transformed malignant cells [9, 10]. Early studies showed that transfection of available p53 cDNA clones was found to involve cooperation between p53 and the Ras proto-oncogene to transform primary rat embryonic fibroblasts [11–13]. The p53 kDa protein, as referred to back then, was uncovered by researchers a decade later (1989) to be a tumor suppressor. Baker et al. sought to study regions of the chromosome 17p commonly lost in colorectal cancer, the same region of the location of the TP53 gene. The research group cloned and sequenced the remaining allele of the 17p chromosome regions in carcinoma tumors and compared it to the previously published p53 sequences [14–17]. Baker et al. then elucidated that there was a T to C change at codon 143 in one tumor and a G to C at codon 175 in a second tumor, both resulting in an amino acid change in the p53 protein [18]. Moreover, the sequence was compared to patients’ matched samples and their normal tissue did not carry the observed point mutation indicating that the mutation was not present in the patient’s germline and rather acquired specifically in the tumor [18]. Again, the notion of mutations harbored in this region coding for p53 challenged the concept of p53 as an oncogene and recognized it as a tumor suppressor as a better fit. In 1992, it was first published the p53 knockout mice were generated by microinjection of a p53 deleted construct into mouse embryonic stem cells into mouse blastocysts in a C57BL/6 mice background. Homozygous mice lacking p53 surprisingly had no developmental defects, nonetheless, they had a noticeable higher tumor burden developing tumors as early as 10 weeks, including lymphomas and sarcomas [19]. This and many other studies that followed [20] changed the established paradigm of p53 and then, p53 became one of the most sought out tumor suppressor genes studied in cancer research history.
P53 Structure and Function:
P53 protein structure comprises five main regions: the transactivation domain, proline-rich domain, DNA binding domain, tetramerization domain, and a regulatory domain [21]. The transactivation domain (TAD) is located at the N-terminus and is subdivided into two regions: TAD1 and TAD2. These domains allow the binding of p53 to different cofactors and both are required for p53-mediated suppression of tumorigenesisin response to stress such as acute DNA damage [22]. Nonetheless, each transactivation domain provides p53 the cofactor binding specificity that in turn influences the cell’s ultimate response to a particular stress. The TAD region also allows the binding of its negative regulator MDM2 [23]. In the context of the acute DNA-damage response and ras oncogene expression, disruption of TAD1 abolishes the wild-type p53 response and instead, behaves like a p53-null cell albeit some genes are still induced [22]. On the other hand, TAD2 disruption retains similar wild-type functions capable of inducing p53 target genes p21, Bax, Noxa, and Puma as well as the ability to induce cell cycle arrest and apoptosis. Simultaneous deletion of TAD1 and TAD2 completely abolishes p53 function resulting in a p53-null response including the inability to induce senescence and predisposition to tumor lesions in vivo [22]. These findings indicate TADs provide the p53 protein a level of specificity for gene transcription, whether this holds true to in response to other stress stimuli and in vivo cancer models remains to be elucidated.
Next to the TADs, p53 protein structure contains a proline-rich domain (PRD) also known as the polyproline (PP) region of repeated PXXP sequences where P denotes proline and X is any amino acid. Like its TAD, the PRD is dispensable for the ability of p53 to bind to DNA. Although there is evidence that the PRD is required to efficiently suppress colony formation of tumor cells in vitro [24], there are more convincing data that deletion of the PRD does not interfere with the ability of p53 to suppress colony formation of transformed cells and undergo cell cycle arrest but rather cells must undergo apoptosis [25, 26]. At the transcriptional regulation level, studies have identified this region to be important for the expression of a subset of p53 target genes including GAS1 and PIG3 [27, 28]. This region is of importance for wild-type p53 stability as deletions in the PRD causes p53 nuclear export, becoming prone to ubiquitination and MDM2-mediated degradation. Particularly in the presence of MDM2, deletion of the PRD domain decreases p53 transcriptional activity suggesting that deletions/mutations in this region have deleterious consequences in wild-type p53 function [29]. PRD role was also demonstrated in vivo where its deletion compromised thymocytes to undergo apoptosis upon irradiation [30]. Interestingly, the PRD has different consequences between the interaction with MDM2 and MDM4, an MDM2-related protein and p53 negative regulator [31]. Impaired PRD generated tumors of higher weight compared to WT p53 mice, in the presence of MDM2 and Mdm4. Nonetheless, simultaneous deletion of Mdm4 and p53 PRD, rescued mouse tumor weights comparable to WT p53 mice but not in mice with MDM2+/−. This indicated that deletion of the PRD in p53 is more susceptible to Mdm4 deletion and that Mdm4 but not MDM2 renders p53 transcriptionally inactive.
The central core region residue 102 to 292 contains the DNA-binding domain (DBD) of p53. This region allows p53 to exert its function as a transcription factor in a sequence-specific manner by the recognition of the p53 responsive element (RE). This specific DNA sequence that p53 binds to consists of two copies separated by 0–13 bp of 5’-RRRCWWGYYY-3’ where R is a purine, C is cytosine, W can be adenine or thymine, G is guanine and Y is a pyrimidine [32]. For a gene to be transcriptionally regulated by p53, it must contain two copies of the 10-bp p53 RE, and this can be located at the promoter, intron, or downstream of the gene. In vitro Chromatin immunoprecipitation (ChIP) assays studies have suggested that p53 can bind to half and three quarter RE sites and that distance between the repeats inversely influences gene reporter transcription assays [33]. However, evidence of many of these new potential p53 targets expanded by the flexible definition of p53RE has not been validated in vivo or others have reported these genes to have the established p53 RE.
There are two other important regions at the C-terminus of the p53 protein: the tetramerization domain and the C-terminal regulatory domain. The tetramerization domain (TD) allows four p53 proteins to oligomerize as a tetramer allowing the appropriate protein conformation to bind to DNA for sequence recognition [34]. Deletions in this region not only affect the ability of p53 to bind to DNA but also interferes with its interaction with other proteins [35, 36]. The TD has also been shown to be necessary for p53 post-translational modifications like phosphorylation and ubiquitination. Importantly, ubiquitination by MDM2 returns p53 to basal levels once the stress has ceased and this requires p53 to be at its oligomerized state [37].
The role of the C-terminus domain has been thought to be a regulatory domain. Early in its discovery, the conception was that it negatively regulates p53 and it was not required for DNA binding [38]. It is now understood that this region inhibits the p53 DNA binding domain until the C-terminus regulatory domain is subjected to post-translational modification upon stress signals [39–41]. This results in the abrogation of the C-terminus regulatory inhibitory effect. Reported post-translational modifications at the C-terminus regulatory domain include acetylation and phosphorylation. The p53 cofactor, p300 has been shown to acetylate p53 at its C-regulatory domain to facilitate p53 binding to DNA [42]. Although, binding of p300 was suggested not to be required for p53 binding to DNA but may be needed for the transcriptional initiation [43]. Post-translational modifications are required for p53 to change from an inactive conformation to an active conformation that will allow the DBD to bind to p53RE [44]. Another controversial notion is that the regulatory domain transiently allows p53 to bind to non-specific DNA sequences [45]. Rather than binding to non-specific DNA sequences to trigger the transcription of p53 dependent targets, an acetylated C-terminal facilitates rapid binding of the DBD to specific DNA sequences (p53 RE) and stabilization of p53-DNA complexes [46]. It also has been suggested that rather than facilitating the DBD binding to p53RE, the acetylated C-terminal domain facilitates later processes required for DNA binding once p53 has been stabilized [46]. A different perspective has been proposed where hyperacetylation at the C-terminal can lead to the termination of the p53 binding to DNA [46]. Along this line, others have shown that acetylation of the C-terminal is required for subsequent p53 phosphorylation at Thr55 to cease the p21-mediated DNA-damage response [47]. The C-terminal also contains the nuclear export signal as well as the nuclear localization signal both important for p53 to exert its function as a transcription factor in the nucleus and to export p53 to the cytoplasm for its degradation. The p53 degradation process involves the ubiquitination at its C-terminal domain by MDM2 [46], which can occur at the nucleus [48] or following its translocation to the cytoplasm for proteasome mediated degradation.
P53 isoforms have differential roles in cancer
The p53 protein exists in multiple isoforms as a consequence of two promoters at the TP53 gene, post-transcriptional events such alternative mRNA splicing and internal ribosome entry site. To date, 12 isoforms have been recognized namely p53, p53 (β, γ), Δ40p53 (α β, γ), Δ133p53 (α β, γ), and Δ160p53 (α β, γ) [49]. The isoforms denote the extent of their N-terminal deletion, and β and γ have in addition deletions at the C-terminal yet carry unique sequences due to the alternative splicing of intron 9 [49]. P53 isoforms are expressed in normal and tumor tissue and can be simultaneously expressed with full length p53 [50]. In general, Δ isoforms have a dominant-negative effect toward full length p53 and consequently prevent p53-mediated apoptosis [50]. On the other hand, β isoforms can enhance p53 target gene expression [50]. Specific roles in cancer of each p53 isoform are less characterized. Nonetheless, some of these have been shown to correlate with decreased overall patient survival (Δ133p53α) and progression-free survival (p53γ) [51, 52].
P53 regulation by post-translational modifications as determinants of cell fate
TP53 is a highly regulated gene throughout the signal transduction that initiates its activation until the termination of the stress response that culminates with its degradation. Regulation of p53 activity is necessary for the cell’s appropriate response to the stress type and the resolution of the stress signal to avoid inhibition of cell growth under conditions of normal cell growth. Regulation of p53 is mediated by post-translational modifications including phosphorylation, acetylation, ubiquitination, methylation, neddylation, glycosylation, and sumoylation [53]. In general, modifications allow p53 to become stabilized, undergo an active conformational change, oligomerize as a tetramer, interact with cofactors, bind to the p53RE, and to restore its normal protein levels [53–55]. The first post-translational modification identified in p53 was phosphorylation [56–59] being Ser312 and Ser389 the first sites recognized [60] nonetheless the role of these modifications was not known at the time. Phosphorylation, specifically at the C-terminus was then proven to be necessary for p53 sequence-specific DNA binding [41]. Many studies have now elucidated multiple phosphorylated sites spanning the p53 C-terminal domain as well as sites in its N-terminal domain with very few reported to date to occur in its DBD. Phosphorylation sites at serine and threonine residues have been largely reviewed and they differ on the stress type along with the cofactor interacting with p53 [7, 53, 61]. Activation of kinases in response to DNA damage, viral infection, metabolic changes have been demonstrated to phosphorylate p53, a step required for p53 activation and transcriptional orchestration of p53 target genes. The location of the phosphate group added as well as the number of phosphate groups simultaneously added are relevant to the p53 response. Studies performed have reported that phosphorylation at the human Thr18 site elicits the most prominent response nonetheless, when all sites in the N-terminus are phosphorylated their effect synergizes [62]. For example, the phosphorylation at Thr18 increases the affinity between p53 and p300 which in turn decreases the interaction of p53 and MDM2 resulting in p53 release and activation [62, 63]. Moreover, when all sites in the N-terminal are phosphorylated, the interaction between MDM2 and p53 decreases further compared to single phosphorylation at Thr18 [62]. In addition to the location and multiplicity of the phosphorylated sites, the stress type results in distinct phosphorylation timing length and signatures, in the case of multiple phosphorylated sites, their phosphorylation can be interdependent [64]. For example, human Ser20 site is phosphorylated upon DNA damage and shown to be necessary for transcriptional activation of p21 and MDM2 but dispensable for p53 protein stabilization [65, 66]. Human Ser15 phosphorylation by ATM and ATR contributes to p53 stabilization by inhibiting the interaction with its negative regulator MDM2 [67, 68]. Additionally, phosphorylation at human Ser46 has been shown to contribute to p53 mediated apoptosis [69].
Interdependent post-translational modifications are not limited among phosphorylation sites. Phosphorylation at the N-terminus has also been shown to be required for p300/PCAF mediated p53 acetylation at the C-terminus that in turn is required for p53 sequence-specific DNA binding upon DNA damage [70]. Furthermore, in the context of conformational change leading to p53 activation, oligomerization is required for the K382 acetylation by p300 at the C-terminus [71]. This provides additional evidence of the importance of the stepwise processes of post-translational modification that lead to p53 activation. Acetylation of p53 has been reported at its C-terminus (K305, K370, K372, K373, K381, K382, and K386) and few sites in its DNA binding domain (K120 & K164). Similar to phosphorylation, acetylation of p53 occurs in response to genotoxic and non-genotoxic stress and this is mediated by p300/CBP/PCAF and Tip60/hMOF acetyltransferases. Moreover, acetylation at the C-terminus protects from ubiquitination [71] hence preventing p53 MDM2-mediated degradation and increased stabilization. In addition, a specific p53 mediated cell fate response to acetylation sites has been observed. Acetylation has been shown to be required for growth arrest and apoptosis but dispensable for MDM2 induction [72]. Tip60/hMOF acetyltransferases have been shown to acetylate K120 site in response to DNA damage and result in the transcription of genes involved in apoptosis like Bax and Puma [73, 74]. Mutation K120R compromises the expression of apoptotic genes but not those involved in cell cycle arrest like p21 [73, 74]. This suggests that acetylation at site K120 provides specificity towards the apoptotic pathway, presumably in the case of DNA damage.
Acetylation and ubiquitination appear to be mutually exclusive [75]. Once p53 is acetylated p53 it is no longer susceptible to ubiquitination [76]. While acetylation in p53 lysine residues is associated with p53’s active conformation and transcriptional activation, ubiquitination of p53 is responsible for p53 degradation and cellular localization. Monoubiquitination of p53 triggers its nuclear export while the polyubiquitination targets p53 for proteasomal degradation [77]. Ubiquitination is primarily mediated by MDM2. Nonetheless, other E3 ubiquitin ligases such as Pirh2 (p53-induced RING-H2 domain protein) have been reported. Like MDM2, Pirh2 interacts with p53 at its transactivation domain. This ubiquitin ligase has a higher preference for the tetrameric form of p53 and targets it for degradation both in vitro and in vivo [78, 79]. Other ubiquitin E3 ligases like ARF-BP1, CARP1/2, C terminus of Hsc70- interacting protein (CHIP), COP1, E1B55K/E4orf6, E4F1, E6-AP, ICP0, MSL2, MKRN1, synoviolin, Topors, TRIM2454, Ubc13, and WWP1 have been shown to ubiquitinate p53 [80].
Lysine residues are also subjected to methylation and sumoylation. Both of these post-translational modifications are associated with the inhibition of p53 activity upon cellular stress. Sumoylation is mediated by a family of small ubiquitin-related modifiers (SUMO) comprising of SUMO-1, SUMO-2, and SUMO-3. SUMO-1 was first shown to sumoylate p53 at K386 and rather than decrease p53 activity, sumoylation activated p53 the transcriptional response in response to UV exposure [81]. Both MDM2 and ARF were demonstrated to mediate p53 sumoylation and subsequent nucleolar targeting but the transcriptional consequences were not studied [82]. SUMO-2 and SUMO-3 also mediate p53 sumoylation and this leads to cell senescence upon H2O2 treatment [83]. On the other hand, SUMO-1 mediated p53 sumoylation occurs at the same K386 site in vitro and in vivo and this prevents acetylation by p300, consequently inhibiting the p53 binding to DNA [84]. Nevertheless, if acetylated, p53 can still undergo sumoylation resulting in a rescue of the inhibitory DNA binding effect of sumoylation alone [84]. In the same study, sumoylation did not interfere with p53 bound to DNA although it stimulated the recruitment of a cofactor that resulted in the repression of p53 transcriptional activity.
P53 arginine and lysine residues are also subjected to methylation, neddylation, and β-hydroxybutyrylation (at lysine). These post-translational modifications are less studied but in general, they lead to a decrease in p53 transcriptional activity, inhibiting processes such as apoptosis and cell cycle arrest [85, 86]. Except for methylation, the addition of a methyl group at K372 by SET9 increases p53 stability, transcriptional activity, and expression of p53 target genes CDKN1A and MDM2 [87]. Although methylation by Smyd2 at p53 site K370 results in an inhibitory effect of p53 activity, dimethylation at this same site promotes the interaction of 53BP1 binding cofactor resulting in enhanced transcriptional activity and protein expression of puma, MDM2, and p21 upon adriamycin treatment [88].
P53 plays a role in many biological responses
Domains of the p53 protein are subjected to post-translational modifications and this allows p53 stabilization, oligomerization, and transactivation. Sensor proteins such as ATM, ATR, Chk1, Chk2, HIPK2, DNA-PK, and p14ARF are responsible for initial signal transduction upon cellular stress resulting in p53 post-translational modifications that lead to its activation [7]. P53 has been shown to become activated and integrate many cellular stresses including DNA damage, oncogene activation, hypoxia, replication/translation stress as well as cellular metabolic changes. It is not surprising that due to the nature of all the processes that p53 participates in, its activity is tightly controlled. P53 ultimate biological response is not only influenced by the cellular stress type, but also by the protein modifiers/cofactors that bind to the p53 protein before executing the transcription of p53 target genes. These protein modifiers are responsible for p53 activation through specific post-translational modifications at key sites that provide an initial guide for the p53 response. Cofactor recruitment at the p53 RE further regulates the transcriptional outcome of p53 (Figure 1, 3). Adding to the complexity of p53 cell stress to outcome decoding mechanism, the cellular response is context-dependent. Thus, the response outcome is influenced by the presence and availability of signal transducers, modifiers, and cofactors that exist in a given cell type. Moreover, p53 not only exerts its tumor-suppressive functions through transactivational mechanisms but also by protein-protein interactions, where proteins also vary across cell types. An example of protein-protein interactions in the context of the apoptotic response is the interaction of p53 with Bak which leads to its oligomerization and consequent cytochrome c release [89]. P53 can also interact with anti-apoptotic proteins Bcl-2 and Bcl-XL and this interaction results in outer membrane permeabilization followed by cytochrome c release [90, 91] Taken together, p53 appropriate activation is essential for finetuning a stress signal to its corresponding biological response outcome to prevent cancer formation.
Figure 3. Current p53 therapeutic strategies.
Several approaches have been taken to target one of the most commonly mutated genes in tumors, p53. Unfortunately, none of the identified drugs are approved by the FDA. Thus, the current work aims to address the unmet need of small molecules that restore the p53-pathway and its functional signaling in tumor suppression.
An array of p53 mediated cellular outcomes have been characterized including cell cycle arrest, apoptosis, DNA repair, senescence, angiogenesis, cellular metabolism, reactive oxygen species (ROS), autophagy, and ferroptosis. Among them, the most understood pathways are cell cycle arrest and apoptosis, which we will focus on in greater detail. Although several p53 target genes are activated upon cell cycle arrest induction, CDKN1A is the main induced gene in p53-mediated cell cycle arrest. The CDKN1A gene codes for the p21WAF1 protein, a cyclin-dependent kinase inhibitor 1 that can directly interact and inhibit CDK complexes resulting in cell cycle arrest [92, 93]. Through the interaction of CDK’s, p21 inhibits the phosphorylation of Rb which then remains bound to E2Fs preventing the transcription necessary for cell-cycle progression. Because p21 can interact with multiple CDK complexes expressed throughout the cell cycle, this results in cell cycle arrest at different cell cycle phases, although it has a higher affinity to CDK’s involved in G1/S phase [94]. Specifically, p21 interaction with cyclin E/CDK2 and cyclin D/CDK4 promotes Rb binding to E2F resulting in G1 arrest [95, 96]. On the other hand, p21 association with cyclin B/CDK1 results in G2/M cell cycle arrest [97]. Here, p21 expression during G2/M may serve an assembly function to activate cyclin B/CDK1 at low stoichiometry [97]. Others have reported that both p21 and p27 can help cyclin D/CDK complexes assemble at low stoichiometry while being inhibitory at high stoichiometry [98, 99]. P53 can also control the cell cycle through transcriptional regulation and direct interaction with PCNA and 14-3-3σ, thereby inhibiting DNA replication and G2/M arrest, respectively [100, 101]. Notably, some of these mechanisms are less well-studied or clear. GADD45, a p53 target gene, has been shown to arrest cells in G1/S as well as in G2/M phase [102, 103]. GADD45 can lead to cell cycle arrest in G1/S through the interaction with CR6-interacting factor 1 (CRIF1) which promotes the inhibition of CDK2 and CDK1 complexes as well as direct inhibition of CDK1 activity G2/M [104].
Activation of cell cycle checkpoints, as a consequence of cellular stress results in cell cycle arrest. Checkpoint activation is triggered by checkpoint sensors and activators of p53 including ATM and ATR, which leads to p53 stabilization. Subsequently, p53 can halt the cell’s progression through the cell cycle in order to either activate DNA repair pathways, cell senescence or apoptosis. If the damage is sustainable for repair, p53 will cause cell cycle arrest to promote the transcription of proteins that will repair the damage. The proteins involved in DNA repair that are controlled by p53 include DDB2, PCNA, POLH, RRM2B, and XPC [6]. The activation of these proteins allows the recruitment of further components of the DNA repair machinery and the execution of appropriate DNA repair pathways. If the damage is extensive and cannot be repaired, p53 will induce the transcription of proteins involved in cell death resulting in apoptosis. Although p53 is mostly know as a transcriptional activator, studies have shown that p53 can indirectly repress many cell cycle genes by the induction of p21 expression. The p21 protein then binds to the DREAM repressor complex which represses genes controlled by E2Fs and CHR transcription factors [105, 106].
P53 is involved in the induction of intrinsic and extrinsic mediated apoptosis. Here, p53 transcription-dependent and independent mechanisms can prompt apoptotic cell death. For example, p53 can transcriptionally upregulate FAS, and DR5 which are receptors that trigger extrinsic apoptosis [107]. Intrinsic apoptosis does not involve extracellular receptors, rather the expression and interplay between members of the BCL-2 family members located mainly in the mitochondria are responsible for apoptosis. P53 transactivates pro-apoptotic family members Bax, Bak, Bid, Noxa, and Puma which results in the abrogation and/or degradation of anti-apoptotic Bcl-2 family members, MOMP, the release of cytochrome C from the mitochondria, and activation of caspases [108–110]. Alternatively, p53 can interact directly with Bcl-2 family members on the mitochondria. For example, p53 interaction with Bax leads to Bax homo-oligomerization and pore formation in the mitochondria outer membrane resulting in cytochrome c release [111]. Despite p53 being able to induce apoptosis in various ways, ultimately the type of apoptosis depends on the cellular context and apoptotic machinery available in the cell type.
Post-transcriptional control byp53 through microRNAs (miRNAs) induction
In addition to p53 regulating gene expression directly by binding to genes’ p53 RE, p53 modulates gene products through the induction of miRNAs that are controlled by p53 at the transcriptional level. MiRNAs induced by p53 have been shown to participate in the control of protein expression involved in cell cycle progression, senescence, apoptosis, metastasis, angiogenesis, cellular stemness and metabolic processes including glycolysis [112].
Several miRNAs have been reported to be transcriptionally controlled by p53 including the miR-34 family, miR-145, miR-107, miR-192 and miR-215 [113]. The first group of miRNAs identified to be stimulated by p53 was the miR-34 family (miR-34a-c) which downregulates the expression of proteins involved in the progression of cell cycle and stimulation of cell proliferation and survival, and immune checkpoint including Cyclin E2, CDK4, CDK6, BCL-2, and PDL-1 [114, 115]. Other miRNAs are involved in the repression of oncogenes and response to hypoxia and angiogenesis including miR-145 and miR-107 that regulate the expression of c-Myc oncogene and HIF-1β, respectively [116, 117]. The miR-107 is also involved in the regulation of the G1-S cell cycle progression by controlling the expression of CDK6 and RBL2 [118]. MiR-192 as well as miR-215 are upregulated by p53 upon genotoxic stress and they regulate gene expression products that participate in cell cycle progression at the G1 and G2-M cell cycle checkpoints including RAD51, TOP1, MCM3, RB1, PIM1, CDC7, MCM10, and MCM6 [119]. In addition to miRNAs that regulate cell cycle genes, miR-205 controls cell cycle progression through E2F1 downregulation in addition to metastatic processes by LAMC1, which is involved in cell adhesion and migration [120]. Other miRNAs regulated by p53 involved in metastasis through the EMT process are miR-34 which targets Snail1[121] and the miR-200 family found to inhibit the expression of ZEB1/2 [122]. P53 also regulates its own stability and activity by inducing miRNAs that target Mdm2 and Mdmx (Mdm4) including miR-192, miR-194, miR-215, miR-143, miR-145 and miR-34a [123–125].
TP73 and TP63 are p53 family members
TP73 (Chr.1p36.33) and TP63 (Chr.3q28) genes that encode transcription factors p73 and p63, respectively, are TP53 homologous structures. The most conserved domain between the p53 family members is the DNA binding domain, followed by the oligomerization domain and the TA is the least shared domain. These resemblances allow p73 and p63 the capability of oligomerizing, binding to canonical p53 REs, and transactivation of p53 target genes [126–128]. Thus, p73 and p63 are also involved in anti-tumor processes such as control of cell proliferation and apoptosis. On the other hand, the divergence between them allows p73 and p63 to be regulated and participate in other biological processes differently from p53. For example, the variation in the TA domain does not allow p73 to be regulated by MDM2 [129]. In addition, p73 and p63 null mice are viable but display developmental defects unlike p53 null mice where development is normal but there is a spectrum of tumor malignancies [19, 130, 131]. This suggests that in addition to the tumor-suppressive functions of p73 and p63, they might have higher functional pressure towards the control of cell differentiation at least during the early development process.
Unlike p53, p73 and p63 tumor-suppressive functions are not abrogated by mutation occurrence nor by allelic loss. Rather, the nature of having two promoters gives rise to two proteins which can be either tumor-suppressive (TA) or pro-tumorigenic associated (ΔN, lacking the TA domain). In fact, several malignancies express the ΔN isoforms [132, 133]. In addition, alternative mRNA splicing events generate different protein isoforms. TP73 has 7 TA isoforms (α, β, γ, δ, ε, ξ, η) and 7 ΔN isoforms (ΔN-α, ΔN-β, ΔN-γ, ΔN-δ, ΔN-ε, ΔN-ξ, ΔN-η) [134]. TP63 has 5 TA isoforms (α, β, γ, δ, ε) and 5 ΔN isoforms (ΔN-α, ΔN-β, ΔN-γ, ΔN-δ, ΔN-ε) [134]. TA-γ isoforms are more similar in structure to p53 and in the case of p63 it correlates with its transcriptional activity but in p73, TA-α is a more potent transcriptional activator and inducer of apoptosis [135]. A key difference in the p73 and p63 structure to that of p53 is the presence of the sterile α motif (SAM). This domain allows p73 and p63 to interact with proteins involved in developmental processes [136].
P73 and p63 are activated upon DNA damage and other cellular stresses such as hypoxia. Their function, like p53, is regulated by post-translational modifications like ubiquitination, acetylation, and phosphorylation. Following activation, p73 and p63 form homo-tetramers, and they can also form hetero-tetramers with each other. Some have concluded that neither p73 or p63 form hetero-tetramers with wild-type p53 [137], while others have shown that p73 can bind to p53 and this interaction leads to the induction of Puma and Bax [138]. This difference might be attributable to the type of stress signal and p53 phosphorylated site. Nonetheless, p73 and p63 appear to be required for apoptosis upon DNA damage in the presence of functional p53 in certain model systems [139]. P73 can transactivate p53 target genes like Puma, Noxa, RAD17, and p21 [128, 140, 141]. Similarly, p63 can upregulate p53 target genes GADD45, PIG3, p21, and Bax [142, 143]. Both p73 and p63 can regulate genes that are unique to each transcription factor activity and not shared with p53 [144]. There is limited information in regards to the distinct gene regulation of each TAp73 and TAp63 isoforms and their role in tumorigenesis. However, it is known that each isoform can be differentially expressed in tissues as well as having different activity in regulating gene expression [143]. In the case of p63, its high expression is mostly limited to the female germline [145] albeit it is also expressed in other normal and tumor tissues at lower levels [146]. Importantly, expression of ΔN isoforms is associated with inhibition of tumor-suppressive function of TA isoforms. TA isoforms despite not interacting with wild-type p53 are capable of interacting with mutant p53 and thus resulting in inhibition of TAp73 and TAp63 tumor suppressive functions [147].
TP53 mutations can result in a stable protein with different pro-tumorigenic outcomes
Somatic mutations in p53 are found across a variety of cancer types mainly in colorectal, head & neck, esophageal, female genital organs (cervical, ovarian, uterine, vaginal & vulvar), lung and pancreas [148]. Compared to other tumor suppressors, p53 is unique as mutations can influence its function into different outcomes: loss-of-function (LOF), dominant-negative (DN), and gain-of-function (GOF). In addition, mutations in p53 are rarely truncating mutations, and the TP53 gene is typically not silenced by hypermethylation of the gene locus [149]. When p53 is said to have a loss-of-function it no longer performs its regular tumor-suppressive functions including cell cycle arrest, DNA repair, and apoptosis. Given that p53 binds as a tetramer, mutant p53 can bind and inhibit wild-type p53, if the wild-type allele is not lost [150]. Moreover, multiple studies have shown that certain mutations in p53 result in gain-of-function activities where p53 no longer behaves as a tumor suppressor but rather as an oncogene. For example, some mutations in p53 help cancer cells sustain proliferation, become more aggressive allowing metastasis and resistance to chemotherapy.
TP53 GOF mutations is reflected in the clinical setting, as patients that harbor p53 mutations in their tumors have a worse prognosis than patients who do not have mutated p53 [151]. For example, Li-Fraumeni syndrome (LFS) is a rare inherited disorder (frequency of 0.005– 0.02% worldwide) characterized by mutations in the TP53 gene. In this case, a single copy of the gene predisposes patients to several malignancies even in their early lifespan. LFS patients have a 50% chance of developing tumors by the age of 40 and 90% by age 60 [152]. Compared to non-carriers, LFS patients have approximately a 24-fold higher chance of developing tumors, and this is even more penetrant in females with 93% risk by the age of 50 [152]. Common tumors observed in LFS patients are breast, osteosarcomas, brain, leukemias, and adrenocortical carcinomas. Moreover, patients carrying R248Q mutations display earlier onset tumor development and higher tumor burden than patients with G245S/+ or null/+ in their germline [153]. These variations feature the differences between LOF and GOF in addition to the diversity in the outcome of p53 mutants.
Somatic TP53 mutation frequency is higher in its DBD accounting for approximately 95% of p53 observed mutations. Although rare, mutations at the TAD and C-terminus have been reported. A total of 73% of mutations are missense, with R175, G245, R248, R249, R273, and R282 the most commonly mutated sites and also referred to as p53’s “hotspot” mutations [148]. These hotspot mutations have a selection frequency of 2–7%, a high frequency to occur by chance that reflects the pro-tumorigenic advantage found in a variety of malignancies [148].
In general, p53 mutants can be classified as class I structural mutants and class II contact mutants. Structural mutants such as R175H, R196*, Y220C, R245, R249S, and R282W have lower thermodynamic stability and hence are characterized by their unfolded structure compared to the wild-type p53 structure [154]. On the other hand, contact mutants like R248Q, R248W, R273H, R273C, R280K, and R282W retain a folded protein structure but the mutation at the core DBD prevents its binding to the DNA p53RE [154]. In regards to the GOF outcome, there is no correlation between the GOF potential and the class of mutant [155]. Nonetheless, categorization between structural and contact mutation in p53 has yielded different approaches in therapeutic targeting. For example, R175H and R273H mutants have been shown to be compromised with Zn2+ binding thus, treatment with zinc metallochaperones can rescue the wild-type p53 structure but not in R280K mutant [156]. Knockdown of endogenous mutant p53 generated isogenic cell line panels overexpressing different mutant p53 and in vivo knock-in models of various p53 mutants have provided a deeper understanding of the modus operandi spectrum of different p53 mutants [157–159]. Such studies have highlighted that not only the site of the mutation is relevant but the type of amino acid substitution can impact the outcome of mutant p53 [160]. Moreover, the cellular context provides an additional layer of complexity to mutant p53 behavioral outcomes. The vast majority of the GOF potential of mutants that have been studied are those named as hotspot mutations albeit others to a lesser extent. GOF mutants have the capacity to enable the cell to have more invasive and migratory potential. In addition, mutant p53 can alter key proteins involved in the cell cycle/DNA repair process for the cell to become highly proliferative. For example, the association of mutant p53 and NF-Y upon DNA damage results in the increased expression of G2/M phase regulating proteins (Cyclin A, B1, CDK1, Cdcd25C) resulting in enhanced proliferation [161]. Additional genes involved in cell replication that mutant p53 regulates are c-myc as well as proliferating cell nuclear antigen (PCNA) both that result in cell proliferation and avoid apoptosis [162, 163]. Through a different mechanism, mutant p53 can induce the expression of miRNAs resulting in the silencing of genes involved in the cell cycle. An example, the miR128-2 expression is induced by mutant p53 resulting in silencing of the cell cycle involved transcription factor E2F5 [164]. Inhibition of E2F5 by miR128-2 ultimately leads to chemotherapy resistance. Mutant p53 has also been shown to promote drug resistance through enhanced transcription of the MDR1 gene although not by binding directly to the MDR1 promoter, rather interacts and cooperates with the ETS-1 transcription factor to promote the expression of MDR1 and increase cell survival upon a variety of chemotherapy agent [165, 166]. MDR1 is a good example of genes that are differently regulated between wild-type and mutant p53. In the case of wild-type p53, MDR1 is repressed by p53 [167].
There is evidence of mutant p53 regulates the transcription of genes at the level of promoter binding, in this example, R175H p53 mutant was shown by chromatin immunoprecipitation analysis to bind to the EGFR1 promoter [168]. Binding to the EGFR1 promoter ultimately provides the cell a means to escape cisplatin mediated apoptosis. Along this line, mutant p53 avoids inducing apoptosis and enhances cell survival upon etoposide treatment through upregulation of NF-κB2 [169], a transcription factor that mediates expression of cell cycle and anti-apoptotic genes. NF-κB2 upregulation by mutant p53 also enhances epithelial-mesenchymal transition (EMT) potential through pro-migratory effects mediated by chemokine expression [170]. In relation to EMT, pro-angiogenesis factor VEGF-A secretion is stimulated by mutant p53 [171].
It is evident that wild-type p53 and mutant p53 GOF have different gene signatures that allow tumor suppression and tumorigenesis, respectively [158]. In addition to changes at the transcriptional level, others have reported distinct miRNA profile, proteome, and secretome expression profiles by wild-type p53 and mutant p53 [172–175]. Collectively, these studies have underscored the involvement of mutant p53 in promoting tumorigenesis at the transcriptional level to changes in the tumor microenvironment (Figure 2). Moreover, understanding the differences in cellular regulation between wild-type p53 and mutant p53, has yielded new downstream targets and pathways involved in mutant p53 GOF that have been and continue to be explored for cancer therapy.
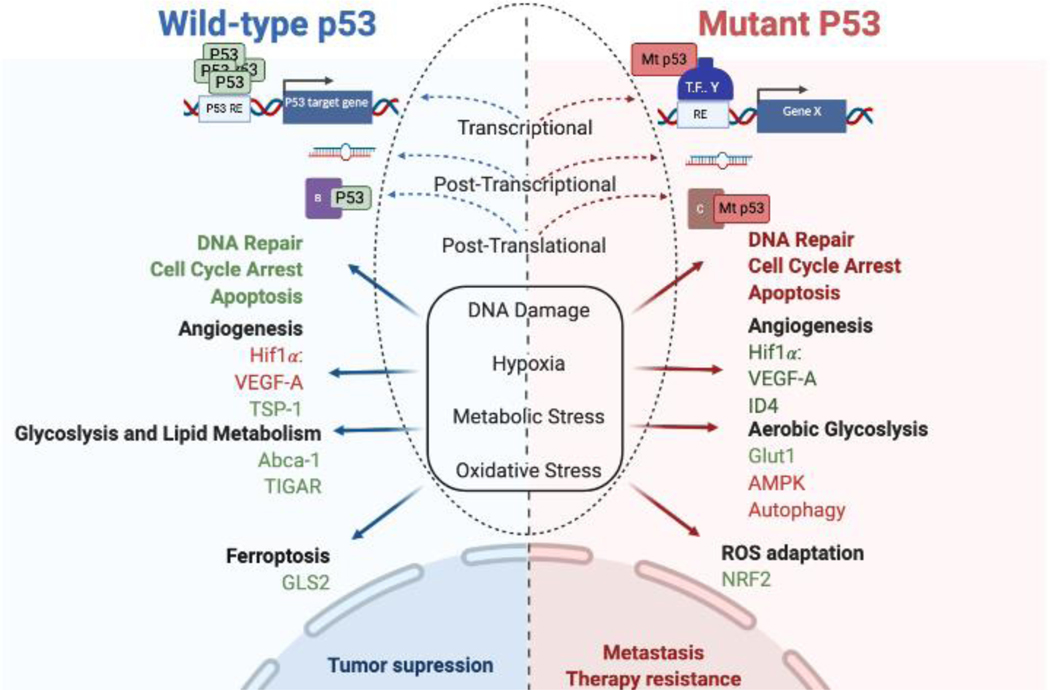
Figure 2. Opposing effects of wild-type and mutant p53 regulation in cancer biology.
Wild-type and mutant p53 respond to a variety of cellular stresses (DNA damage, hypoxia, metabolic stress, and oxidative stress). Their regulation occurs at the transcriptional, post-transcriptional, and post-translational level. At the transcriptional level, wild-type p53 binds directly at p53 response elements (RE) whereas mutant p53 binds to other transcription factors (TF), example Y to regulate a gene X, as indicated. Both wild-type and mutant p53 can regulate the expression of microRNAs and interact directly/indirectly with other proteins to induce a cellular response. Wild-type p53 biological response is tumor suppression, unlike mutant p53 where its response leads to metastasis and therapy resistance. Green color: positively regulated, red color negative regulated. Created with BioRender.com
Like wt-p53, mutant p53 undergoes post-translational modifications
Like wild-type p53, mutant p53 can be post-translationally modified albeit the biological response outcome is different from that of wild-type p53. Interestingly, sites commonly post-translationally modified in wild-type p53 are rarely mutated across a variety of cancer types. This is in part due to the common hotspot mutations in p53 that are not which are sites post-translationally modified. Nonetheless, there are sites including across the different p53 domains that are post translationally modified and found to be mutated in human tumors. Post-translational modifications in wild-type p53 allows protein activation, specificity for cofactor interactions, binding to DNA, and transactivation selectivity of p53 target genes towards tumor suppression. In the case of mutant p53, post-translational modifications do not impact the ability of mutant p53 binding to specific DNA sequences for tumorigenesis functions. In fact, to date, there is not a defined DNA sequence that mutant p53 binds to as wild-type p53 binds to its corresponding p53 RE. Instead, mutant p53 interacts with other proteins like transcription factors and interferes with their transcriptional program. Mutant p53 has been reported to be phosphorylated, ubiquitylated, acetylated, and methylated. How post-translational modifications affect mutant p53 is less defined as with wild-type p53. Studies have reported a correlation between mutant p53 hyperphosphorylation at site Ser392 and the oncogenic potential of the R175H mutant [176] although this appears to be cancer cell type-dependent as hyperphosphorylation is not seen in some breast cancer cell lines [177]. Other phosphorylation sites such as Ser15 provide mutant p53 the interaction with other cofactors and transcription factors to offer the GOF advantage [178]. Similarly, hyperacetylation of mutant p53 at Lys373 and Lys382 preferentially localizes mutant p53 in the nucleus [7, 179]. In addition, cell stress signaling such as glucose restriction can impact mutant p53 differently than wild-type p53. In this stress condition, mutant p53 is acetylated and results in cell metabolic rewiring that provides cell survival advantage [180]. By contrast to wild-type p53 were ubiquitylation targets it for degradation to control its protein levels upon stress response resolution, mutant p53 is not frequently found to be ubiquitylated. This is mainly due to the lack of MDM2 gene transactivation and an absent negative feedback loop. As a result, mutant p53 becomes highly stable in a variety of tumor types. Nonetheless, certain types of mutant p53 can interact with MDM2 and be targeted for degradation along through other ubiquitin ligases also found to regulate wild-type p53 stabilization [181].
Mutant p53 mechanisms of destabilization
Like wild-type p53, mutant p53 is targeted for degradation through the ubiquitin proteasome system (UPS). Several interacting E3 ubiquitin ligases have been reported to be involved in ubiquitinating mutant p53. MDM2 can ubiquitinate and/or direct certain types of mutant p53 for proteasomal degradation [181, 182]. Moreover, the ubiquitinated state of mutant p53 does not necessarily correlate with its degradation rate. For example, hyper-ubiquitinated mutant p53 remains stable but rather affects its localization aggregating mostly in the cytoplasm rather than in the nucleus albeit this is mutant p53 type-dependent [181]. Enhanced stability of mutant p53 mainly results from the inability of mutant p53 to transcriptionally regulate wild-type p53 target genes consequently it abrogates the negative feedback loop through Mdmd2. The fact that the interaction between mutant p53 and MDM2 is dependent on the MDM2 RING domain rather than its E3 ligase [181] suggests that other E3 ligases are can ubiquitinate mutant p53. This was shown to be the case for E3 ligases Cop1 and CHIP but not ARF-BP1 [181]. To avoid degradation due to its unfolded nature, mutant p53 can escape proper protein surveillance by binding to heat-shock proteins (Hsp). In this scenario, mutant p53 binds to Hsp90 leading protection from both MDM2 and CHIP E3 ligases [183]. Interestingly, Hsp90 is upregulated across different malignancies [184] which may be one of the reasons that facilitate the advantage for mutant p53 stabilization and GOF.
Another alternative mechanism that controls mutant p53 protein levels is autophagy. Macro-autophagy here on referred as autophagy is the process of intracellular clearance upon damaged cellular components or the need to recycle components to compensate for energy expense and reach cellular homeostasis. Targeted cellular components for autophagy-mediated degradation are enclosed by vesicles formed in an orderly concerted manner ultimately fusing with the lysosome where the cargo is degraded. Metabolic stress such as glucose restriction can result in mutant p53 degradation through the autophagy machinery and this appears to be dependent on p53 deacetylated status [180]. In the case of mutant p53, degradation can occur both via autophagy and chaperon mediated autophagy (CMA), the latter of which is independent of vesicle formation. CMA is a selective type of autophagy process mediated by the heat-shock cognate protein of 70 kDa (Hsc70). Substrate proteins bound to Hsc70 are directed to the lysosome and internalized into the lysosomal compartment by lysosome-associated membrane protein type 2a (Lamp-2A). Aggregated mutant p53 becomes polyubiquitinated at K63 by CHIP followed by interaction with Hsc70 and Lamp-2A resulting in lysosomal-dependent degradation upon metabolic stress, hypoxia, and non-active cell proliferative conditions [185, 186].
Regulation of p53 by miRNAs
Several miRNAs have been reported to silence p53 expression directly or indirectly and p53 also regulates miRNA expression. The distinction of miRNAs to target mutant vs wild-type p53 requires direct targeting of the specific mutated site of the mutant p53 mRNA. To date, the identified miRNAs have been shown to silence wild-type p53 and none that are specific to mutant p53. In the case of mutant p53, one can speculate that miRNAs that target key positive regulators of mutant p53 can result in mutant p53 decrease expression. Along this line, synthetic siRNA and shRNAs oligonucleotides have been synthesized to target specifically mutant p53 and decrease tumor growth in vivo [187, 188].
Approaches to target p53 and restore the p53-pathway are ultimately dependent on the p53 status that is wild-type p53, or mutant p53 matters as well as the type of p53 mutation. Strategies over the years have included of p53 gene therapy, stabilization of wild-type p53/degradation of mutant p53, restoration of mutant to wild-type p53 structural conformation, and small molecules that restore the p53 signaling pathway by activating downstream target of p53 even in p53-null cells (Figure 3). P53-null tumors benefit from p53 gene therapy, tumors retaining both copies of wild-type p53 strategy is to target the stabilization of wild-type p53 through the inhibition of p53 negative regulators and both strategies can be applied to tumors that have lost one and retained the other (p53+/−). The first established adenoviral gene therapy delivering wild-type p53 called Gendicine is currently approved in China for head and neck cancer but failed clinical trials in the USA in 2008 [189]. Similar to Gendicine, Advexin underwent Phase I-III clinical trials and yielded a statistically significant correlation between therapy efficiency and tumor response as well as improving patient median survival [190]. Advexin also showed a good safety profile as a monotherapy and in combination with other chemotherapy agents/radiotherapy in a variety of tumor types nonetheless, it remains to be approved by the FDA. These replication impaired adenoviral p53 therapies were proven not as effective due to their low transduction efficiency in human tumors.
Several gene therapies were developed to improve transduction efficiency involving viral active replication including ONYX 015 approved in China and H101 currently in clinical trials in China [191]. In the USA, these therapies are not currently being pursued in clinical trials. ONYX 015 does not deliver the wild-type p53 gene but rather a mutated E1B viral gene that was initially thought to incorporate and lyse cancer cells that had mutated p53 [192]. Additional studies confirmed that instead, ONYX 015 as well as similar gene therapy H101, efficiency and sensitivity of viral transduction is independent of p53 status but rather depends on the cell cycle phase stage [193].
The key negative regulator of p53 is MDM2 and studies dedicated to understanding their interaction have enabled the concept of disrupting their interaction as a therapeutic avenue which ultimately leads to wild-type p53 stabilization. High-throughput screening and de novo design approaches have identified MDM2 inhibitors such as nutlins (Nutlin-3), benzodiazepinediones (TDP521252 and TDP665759), spiro-oxindioles (MI-219) and their derivatives as well as the first MDM4/MDMX inhibitor SJ-172550 [194]. These strategies have proven to be effective in treating tumors retaining at least one copy of wild-type p53. MDM2 inhibitors bind to a hydrophobic pocket in the MDM2 protein that is responsible for interacting with p53 and have yielded positive responses in pre-clinical studies. MDM2 antagonists have proven to be safe to normal tissues, resulting in activation of wild-type p53 tumor-suppressive functions and hence this approach has been extensively pursued into clinical trial development. The first MDM2 inhibitor to become of interest was nutlin-3a because of its selectivity towards wild-type p53 [195]. Nonetheless, it was not pursued beyond the preclinical setting due to its suboptimal bioavailability. Nutlin-3 analogue, RG7112, was the first in the nutlin class of small molecules to enter clinical trials owing to its higher efficacy [196]. RG117 underwent Phase 1 clinical trials for the treatment of solid and hematological malignancies (clinicaltrials.gov, 2021). Phase I clinical trials in patients with hematological malignancies showed p53 protein stabilization followed by enhanced transcription of p53 dependent target genes [197]. Progression to Phase II trials in either type of malignancy was not followed due to toxic side effects including neutropenia and thrombocytopenia [198]. Consequently, a second nutlin related compound Idasanutlin (RG7388) was optimized [199]. Idasanutlin (RG7388) currently in clinical trials for solid tumors and hematological malignancies, showed tolerability and safety as a single agent and underwent Phase III clinical trials in acute leukemia patients in combination with cytarabine [200]. Idasanutlin clinical treatment in patients with acute myeloid leukemia showed a durable response including complete response of >12 months [201]. Clinical results for patients treated with Idasanutlin have encouraged other Phase I/II clinical trials currently active for leukemia, breast cancer and neuroblastoma, as a monotherapy or in combination with other drugs including venetoclax and chemotherapy agents (clinicaltrials.gov, 2021).
MDM4/MDMX have structural similarities to MDM2 and hence a few described MDM2 inhibitors also inhibit MDM4/MDMX albeit not all and with similar efficiency. Contrary to MDM2, MDM4/MDMX does not target wild-type p53 directly since it is not an E3 ligase rather it inhibits p53 transcriptional activity. MDM4/MDMX specific inhibitors were recently described and have yet to enter clinical trials. Given that some tumors can result in MDM2 inhibitor resistance due to MDM4/MDMX overexpression, studies combining both strategies are optimistic. KRT-232, a dual inhibitor of MDM2 and MDM4/MDMX has entered Phase I clinical trials as a single therapy as well as in combination with front line therapies in various cancer types. Another example of dual MDM2 and MDM4/MDMX inhibitor is ALRN-6924 which functions as a p53-stapled peptide [202]. ALRN-6924 is currently in Phase1/2 clinical trials for the treatment of hematological malignancies and advanced solid tumors alone or in combination with topotecan (clinicaltrials.gov, 2021). Interestingly, MDM2 has emerged as a factor in the hyperprogression phenotype observed with immune checkpoint therapy. Our group published that the MDM2 inhibitor AMG-232 promotes killing of MDM2-overexpressing tumor cells by T-cells and synergizes for T-cell killing when combined with anti-PD1 therapy [203]. In addition to AMG-232 therapy effects, similar observations were made that T-cells more efficiently killed MDM2-overexpressing tumor cells when MDM2 expression was knocked down.
Studies where the second site induced mutations to the mutant p53 restored the wild-type p53 conformation and function introduced the concept that the mutant p53 protein structure is malleable and its unfolded nature can be rescued to function as wild-type p53 [204]. Several screens of small molecules have identified compounds that stabilize the mutant p53 among these, CP-31398, PRIMA-1 (and its methylated analog APR-246), MIRA-1, PhiKan083, PK7088, and NSC319726/ZMC 1, and arsenic trioxide (ATO). CP31398, PRIMA-1, MIRA-1, and APR-246 stabilize mutant p53 protein as indicated by the enhanced recognition of the wild-type p53 specific antibody PAb1620 and loss of mutant p53 specific PAb240 [205–208]. Moreover, treatment with these small molecules resulted in transcriptional activation of p53 target genes such as p21, DR5, PUMA ultimately leading to apoptosis and cell cycle arrest in vitro and decreased tumor growth in vivo with good toxicity profiles [208, 209]. Recently, the small molecule ATO has been found to stabilize mutant p53 proteins, specifically those mutants categorized as structural mutants albeit with differences in their transcriptional function restoration [210]. Pre-clinical safety profiles of these small molecules prompted clinical trials. APR-246 is currently undergoing Phase I-III clinical trials in hematological malignancies as well as in solid tumors including ovarian, bladder, and non-small cell lung cancers (clinicaltrials.gov, 2020). Particularly, APR-246 underwent Phase I trial as a monotherapy agent and is currently in Phase III in combination with azacytidine as well as serval other chemotherapeutic agents (clinicaltrials.gov, 2020). Combination with azacytidine yielded great synergy in vitro and in vivo and thus exciting results are of hope in this trial [211]. Recently, the FDA granted “Fast Track” designation to APR-246 (eprenetapopt). Encouraging clinical trial results have been obtained with APR-246 in myelodysplastic syndromes (MDS) patients with p53 mutations, having an overall response rate (ORR) of 75%, and a 57% complete remission (CR) rate.
PhiKan083 and PK7088 specifically restore the function of the Y220C mutant p53, the 9th most frequent mutation. This specific mutation generates a cavity in the p53 protein structure due to loss of hydrophobicity at the DNA core resulting in a druggable “pocket”. Like the previously mentioned compounds, PhiKan083 and PK7088 also result in wild-type p53 protein conformation as indicated by the PAb1620 but the transcriptional upregulation of p53 target genes such as p21, NOXA, Bax is more efficiently enhanced by PK7088 [212–214]. To date, PhiKan083 and PK7088 therapeutic application has not been tested in pre-clinical models and hence their translational applicability in clinical trials has yet to be described. NSC319726/ZMC-1 belongs to a group of zinc metallochaperones (thiosemicarbazones). This group of small molecules restores specifically mutations that affect the affinity of p53 binding to zinc, which is required for wild-type p53 activity [215]. P53 mutants in this category include hotspot mutation R175H, and treatment with NSC319726/ZMC 1 increases intracellular Zn2+ allowing p53R175H harboring cells to be saturated with Zn2+ ions resulting in the rescue of wild-type p53 conformation, transcriptional activation, and decreased tumor volume in vivo [216, 217]. ZMC-1 showed toxicity in pre-clinical models and was not pursued further [218]. Nonetheless, 3-AP (Triapine) is also a zinc metallochaperones currently in Phase I-II clinical trials for advanced and metastatic solid tumors and various hematological malignancies as well as Phase III clinical trials in cervical carcinoma all of which are being studied in combination with other chemotherapy agents [219]. Additionally, COTI-2 is a thiosemicarbazone derivative was shown to be a p53 reactivator leading to apoptosis in vitro with decreased tumor volume in vivo [220]. COTI-2 is currently undergoing Phase I trials in various solid malignancies in combination with cisplatin. A study reported that unlike from the other thiosemicarbazone, it does not affect Zn2+ intracellular levels [218] indicating that COTI-2 is not mutant p53 specific. The involvement of the Akt pathway was also reported to be part of COTI-2 mechanism of action [220].
The fact that cancer cells can be “addicted” to mutant p53 and that silencing mutant p53 results in the abrogation of the GOF downstream effects, has prompted great interest in therapies targeting mutant p53 for degradation. Therapies targeting mutant p53 for degradation have mainly been focused on Hsp90 inhibitors (Geldanamycin, 17-AAG, Ganestesbip) and histone deacetylase inhibitors (Vorinostat (SAHA) and Romidepsin). These inhibitors affect the HDAC6/Hsp90 axis where HDAC positively regulates Hsp90, and p53 binds to Hsp90 to avoid degradation hence acquire protein stabilization. Thus inhibitors of HDAC’s and Hsp90 result in mutant p53 destabilization. This class of small molecules not only result in mutant p53 degradation but also p53 transcriptional activity, apoptosis, and in vivo efficacy [221–225]. Geldanamycin derivative, 17-AAG (Tanespimycin), and 17-AAG hydroquinone hydrochloride salt derivative (IPI-504) have undergone several clinical trials and completed Phase I- II for various hematological malignancies, breast cancer, non-small cell lung cancer, and several other advanced malignancies and Phase III for multiple myeloma showing anti-tumor efficacy and well-tolerated profile [226]. Deacetylase inhibitors Vorinostat (2006) and Romidepsin (2009) are FDA approved for the treatment of cutaneous T cell lymphoma with a 30% and 34% response rate, respectively [227, 228]. Other small molecules that lead to mutant p53 degradation are macroautophagy inhibitor Spautin-1 and SIRT1 activator YK-3–237. Spautin-1 blocks macroautophagy leading to CMA mediated mutant p53 degradation [185] whereas YK-3–237 decreases acetylated mutant p53 expression resulting in its decreased protein expression presumably through a proteasomal mediated mechanism [229]. Pre-clinical anti-tumor effects of both of these compounds have not been described. Despite these strategies killing mutant p53 harboring cells and not wild-type p53, affecting the stability of mutant p53 and resulting in the induction of p53 target genes, they are not specific to mutant p53 as HDACs have effect on other genes and proteins [230]. Thus, they augment toxicities in normal tissues as well.
Immunotherapy is one of the most important developments in the fields of oncology and cancer therapeutics. It has been reported that p53 is capable of modulating innate and adaptive immune responses by regulating the expression of Toll-like receptors and PD-L1, among other mechanisms [114, 231]. In addition to strategies where p53 pathway restoration leads to an immune response, production of neoantigens due to p53 mutation has been utilized to specifically promote recognition and subsequent killing of cancer cells. Recently, a research group at Johns Hopkins University developed a bispecific antibody that recognizes a p53 R175H neoantigen peptide complexed to human leukocyte antigen (HLA) [232]. Recognition of this antibody, namely H2-scDb, activated T-cells driving T-cell mediated killing of cancer cells harboring the p53R175H mutant in vitro and in vivo. Despite limitations of this bispecific antibody design towards other p53 mutations and host immune T cells, this study highlights the importance of pursuing p53 neoantigen-based immunotherapies, a tumor suppressor that is found mutated in over 50% of human cancers.
P73 and p63 are part of the p53 family members that can activate genes in common with wild-type p53. With the advantage that p73 and p63 are not commonly mutated in tumors, p53-pathway restoring compounds through the activation of p53 family members are of interest. Reactivate transcriptional activity (RETRA) mechanism of action involves the release of p73 from mutant p53 and activating p53 target genes p21 and puma in a p73 dependent manner [233]. Similarly, small molecule Prodigiosin induces p21, puma, DR5, and apoptosis in a p73 dependent manner in vitroand in vivo [234]. Prodigiosin involves the upregulation of p73 and simultaneous reduction of the ΔNp73 oncogenic isoform protein levels [235]. On the other hand, NSC59984 results in p53-pathway restoration through simultaneous upregulation of p73 and degradation of mutant p53 protein expression [236]. To date, these p53-pathway restoring small molecules have yet to reach clinical trials.
In sum, the current approaches rely on either wild-type p53, loss of p53, or presence of mutant p53 (some being specific to the missense substitution). Given the complexity of the p53 mutations and resulting LOF, DN, and GOF provides a good rationale for a general drug targeting approach looking to restore downstream transcriptional responses.
Acknowledgements
W.S.E-D. is an American Cancer Society Research Professor.
Grant Support
The work was supported, in part, by the ACS and NIH grant CA176289 to W.S.E-D.
Footnotes
Conflict of Interest Disclosure:
W.S.E-D. is a Founder of p53-Therapeutics, Inc., a biotech company focused on developing small molecule anti-cancer therapies targeting mutant p53. Dr. El-Deiry has disclosed his relationship with p53-Therapeutics and potential conflict of interest to his academic institution/employer and is fully compliant with institutional policy that is managing this potential conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane DP, Cancer. p53, guardian of the genome. Nature, 1992. 358(6381): p. 15–6. [DOI] [PubMed] [Google Scholar]
- 2.Jain AK and Barton MC, p53: emerging roles in stem cells, development and beyond. Development, 2018. 145(8). [DOI] [PubMed] [Google Scholar]
- 3.Hong H, et al. , Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature, 2009. 460(7259): p. 1132–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cordenonsi M, et al. , Integration of TGF-beta and Ras/MAPK signaling through p53 phosphorylation. Science, 2007. 315(5813): p. 840–3. [DOI] [PubMed] [Google Scholar]
- 5.Tyner SD, et al. , p53 mutant mice that display early ageing-associated phenotypes. Nature, 2002. 415(6867): p. 45–53. [DOI] [PubMed] [Google Scholar]
- 6.Kastenhuber ER and Lowe SW, Putting p53 in Context. Cell, 2017. 170(6): p. 1062–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bode AM and Dong Z, Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer, 2004. 4(10): p. 793–805. [DOI] [PubMed] [Google Scholar]
- 8.Lain S. and Lane D, Improving cancer therapy by non-genotoxic activation of p53. Eur J Cancer, 2003. 39(8): p. 1053–60. [DOI] [PubMed] [Google Scholar]
- 9.Linzer DI LA, Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell, 1979. 17(1): p. 43–52. [DOI] [PubMed] [Google Scholar]
- 10.Lane DP and Crawford LV, T antigen is bound to a host protein in SV40-transformed cells. Nature, 1979. 278(5701): p. 261–3. [DOI] [PubMed] [Google Scholar]
- 11.Parada LF, et al. , Cooperation between gene encoding p53 tumour antigen and ras in cellular transformation. Nature, 1984. 312(5995): p. 649–51. [DOI] [PubMed] [Google Scholar]
- 12.Eliyahu D, Michalovitz D, and Oren M, Overproduction of p53 antigen makes established cells highly tumorigenic. Nature, 1985. 316(6024): p. 158–60. [DOI] [PubMed] [Google Scholar]
- 13.Eliyahu D, et al. , Participation of p53 cellular tumour antigen in transformation of normal embryonic cells. Nature, 1984. 312(5995): p. 646–9. [DOI] [PubMed] [Google Scholar]
- 14.Zakut-Houri R, et al. , Human p53 cellular tumor antigen: cDNA sequence and expression in COS cells. EMBO J, 1985. 4(5): p. 1251–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lamb P. and Crawford L, Characterization of the human p53 gene. Mol Cell Biol, 1986. 6(5): p. 1379–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buchman VL, et al. , A variation in the structure of the protein-coding region of the human p53 gene. Gene, 1988. 70(2): p. 245–52. [DOI] [PubMed] [Google Scholar]
- 17.Harris N, et al. , Molecular basis for heterogeneity of the human p53 protein. Mol Cell Biol, 1986. 6(12): p. 4650–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker SJ FE, Nigro JM, Hamilton SR, Preisinger AC, Jessup JM, vanTuinen P, Ledbetter DH, Barker DF, Nakamura Y, White R, Vogelstein B, Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science (New York, N.Y.), 1989. 244(4901): p. 217–221. [DOI] [PubMed] [Google Scholar]
- 19.Donehower LA HM, Slagle BL, McArthur MJ, Montgomery CA Jr, Butel JS, Bradley A, Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature, 1992. 356(6366): p. 215–221. [DOI] [PubMed] [Google Scholar]
- 20.Lozano G. and Liu G, Mouse models dissect the role of p53 in cancer and development. Semin Cancer Biol, 1998. 8(5): p. 337–44. [DOI] [PubMed] [Google Scholar]
- 21.Harris CC, Structure and function of the p53 tumor suppressor gene: clues for rational cancer therapeutic strategies. J Natl Cancer Inst, 1996. 88(20): p. 1442–55. [DOI] [PubMed] [Google Scholar]
- 22.Brady CA, et al. , Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell, 2011. 145(4): p. 571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin J, et al. , Several hydrophobic amino acids in the p53 amino-terminal domain are required for transcriptional activation, binding to mdm-2 and the adenovirus 5 E1B 55-kD protein. Genes Dev, 1994. 8(10): p. 1235–46. [DOI] [PubMed] [Google Scholar]
- 24.Walker KK LA, Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proceedings of the National Academy of Sciences of the United States of America, 1996. 93(26): p. 15335–15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakamuro D SP, White E, Prendergast GC, The polyproline region of p53 is required to activate apoptosis but not growth arrest. Oncogene, 1997. 15(8): p. 887–898. [DOI] [PubMed] [Google Scholar]
- 26.Baptiste N, et al. , The proline-rich domain of p53 is required for cooperation with anti-neoplastic agents to promote apoptosis of tumor cells. Oncogene, 2002. 21(1): p. 9–21. [DOI] [PubMed] [Google Scholar]
- 27.Ruaro EM CL, Del Sal G, Haffner R, Oren M, Levine AJ, Schneider C, A proline-rich motif in p53 is required for transactivation independent growth arrest as induced by Gas1. Proceedings of the National Academy of Sciences of the United States of America, 1997. 94(9): p. 4675–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venot C MM, Dureuil C, Conseiller E, Bracco L, Debussche L, The requirement for the p53 proline-rich functional domain for mediation of apoptosis is correlated with specific PIG3 gene transactivation and with transcriptional repression. The EMBO journal, 1998. 17(16): p. 4668–4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berger M, et al. , A role for the polyproline domain of p53 in its regulation by Mdm2. J Biol Chem, 2001. 276(6): p. 3785–90. [DOI] [PubMed] [Google Scholar]
- 30.Lowe SW SE, Smith SW, Osborne BA, Jacks T, p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature, 1993. 362(6423): p. 847–849. [DOI] [PubMed] [Google Scholar]
- 31.Toledo F, et al. ,A mouse p53 mutant lacking the proline-rich domain rescues Mdm4 deficiency and provides insight into the Mdm2-Mdm4-p53 regulatory network. Cancer Cell, 2006. 9(4): p. 273–85. [DOI] [PubMed] [Google Scholar]
- 32.el-Deiry WS, et al. , Definition of a consensus binding site for p53. Nat Genet, 1992. 1(1): p. 45–9. [DOI] [PubMed] [Google Scholar]
- 33.Jordan JJ, et al. , Noncanonical DNA motifs as transactivation targets by wild type and mutant p53. PLoS Genet, 2008. 4(6): p. e1000104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Halazonetis TD KA, Conformational shifts propagate from the oligomerization domain of p53 to its tetrameric DNA binding domain and restore DNA binding to select p53 mutants. he EMBO journal, 1993. 12(13): p. 5057–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wagner P, et al. , Fine mapping and regulation of the association of p53 with p34cdc2. Oncogene, 1998. 16(1): p. 105–11. [DOI] [PubMed] [Google Scholar]
- 36.Gabizon R, et al. , Specific recognition of p53 tetramers by peptides derived from p53 interacting proteins. PLoS One, 2012. 7(5): p. e38060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maki CG, Oligomerization is required for p53 to be efficiently ubiquitinated by MDM2. J Biol Chem, 1999. 274(23): p. 16531–5. [DOI] [PubMed] [Google Scholar]
- 38.Anderson ME WB, Reed M, Wang P, Tegtmeyer P, Reciprocal interference between the sequence-specific core and nonspecific C-terminal DNA binding domains of p53-implications for regulation.Molecular and cellular biology 1997. 17(11): p. 6255–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y, et al. , Facilitated search for specific genomic targets by p53 C-terminal basic DNA binding domain. Cancer Biol Ther, 2004. 3(11): p. 1102–8. [DOI] [PubMed] [Google Scholar]
- 40.Barlev NA LL, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL, Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Molecular cell 2001. 8(6): p. 1243–1254. [DOI] [PubMed] [Google Scholar]
- 41.Hupp TR MD, Midgley CA, Lane DP, Regulation of the specific DNA binding function of p53. Cell, 1992. 71(5): p. 875–886. [DOI] [PubMed] [Google Scholar]
- 42.Gu W RR, Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain.pdf>. Cell, 1997. 90(4): p. 595–606. [DOI] [PubMed] [Google Scholar]
- 43.Espinosa JM EB, Transcriptional Regulation by p53 through Intrinsic DNA/Chromatin Binding and Site-Directed Cofactor Recruitment. Molecular cell, 2001. 8(1): p. 57–69. [DOI] [PubMed] [Google Scholar]
- 44.Warnock LJ, et al. , Influence of tetramerisation on site-specific post-translational modifications of p53: comparison of human and murine p53 tumor suppressor protein. Cancer Biol Ther, 2008. 7(9): p. 1481–9. [DOI] [PubMed] [Google Scholar]
- 45.Bayle JH EB, Levine AJ, The carboxyl-terminal domain of the p53 protein regulates sequence-specific DNA binding through its nonspecific nucleic acid-binding activity. Proceedings of the National Academy of Sciences of the United States of America, 1995. 92(12): p. 5729–5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laptenko O, et al. , The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell, 2015. 57(6): p. 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu Y, et al. , Phosphorylation of p53 by TAF1 inactivates p53-dependent transcription in the DNA damage response. Mol Cell, 2014. 53(1): p. 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shirangi TR, Zaika A, and Moll UM, Nuclear degradation of p53 occurs during down-regulation of the p53 response after DNA damage. FASEB J, 2002. 16(3): p. 420–2. [DOI] [PubMed] [Google Scholar]
- 49.Khoury MP and Bourdon JC, p53 Isoforms: An Intracellular Microprocessor? Genes Cancer, 2011. 2(4): p. 453–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourdon JC, et al. , p53 isoforms can regulate p53 transcriptional activity. Genes Dev, 2005. 19(18): p. 2122–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bischof K, et al. , High expression of the p53 isoform gamma is associated with reduced progression-free survival in uterine serous carcinoma. BMC Cancer, 2018. 18(1): p. 684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nutthasirikul N, et al. , Ratio disruption of the 133p53 and TAp53 isoform equilibrium correlates with poor clinical outcome in intrahepatic cholangiocarcinoma. Int J Oncol, 2013. 42(4): p. 1181–8. [DOI] [PubMed] [Google Scholar]
- 53.Sullivan KD, et al. , Mechanisms of transcriptional regulation by p53. Cell Death Differ, 2018. 25(1): p. 133–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai C. and Gu W, p53 post-translational modification: deregulated in tumorigenesis. Trends Mol Med, 2010. 16(11): p. 528–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collavin L, Lunardi A, and Del Sal G, p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ, 2010. 17(6): p. 901–11. [DOI] [PubMed] [Google Scholar]
- 56.Jenkins JR, et al. , Cloning and expression analysis of full length mouse cDNA sequences encoding the transformation associated protein p53. Nucleic Acids Res, 1984. 12(14): p. 5609–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crawford LV, et al. , Detection of a common feature in several human tumor cell lines--a 53,000-dalton protein. Proc Natl Acad Sci U S A, 1981. 78(1): p. 41–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rotter V, p53, a transformation-related cellular-encoded protein, can be used as a biochemical marker for the detection of primary mouse tumor cells. Proc Natl Acad Sci U S A, 1983. 80(9): p. 2613–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rotter V, Boss MA, and Baltimore D, Increased concentration of an apparently identical cellular protein in cells transformed by either Abelson murine leukemia virus or other transforming agents. J Virol, 1981. 38(1): p. 336–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Samad A, Anderson CW, and Carroll RB, Mapping of phosphomonoester and apparent phosphodiester bonds of the oncogene product p53 from simian virus 40-transformed 3T3 cells. Proc Natl Acad Sci U S A, 1986. 83(4): p. 897–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yogosawa S. and Yoshida K, Tumor suppressive role for kinases phosphorylating p53 in DNA damage-induced apoptosis. Cancer Sci, 2018. 109(11): p. 3376–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teufel DP, Bycroft M, and Fersht AR, Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene, 2009. 28(20): p. 2112–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng H, et al. , Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure, 2009. 17(2): p. 202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saito S, et al. , Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J Biol Chem, 2003. 278(39): p. 37536–44. [DOI] [PubMed] [Google Scholar]
- 65.Chehab NH, et al. , Phosphorylation of Ser-20 mediates stabilization of human p53 in response to DNA damage. Proc Natl Acad Sci U S A, 1999. 96(24): p. 13777–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jabbur JR, Huang P, and Zhang W, DNA damage-induced phosphorylation of p53 at serine 20 correlates with p21 and Mdm-2 induction in vivo. Oncogene, 2000. 19(54): p. 6203–8. [DOI] [PubMed] [Google Scholar]
- 67.Shieh SY, et al. , DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell, 1997. 91(3): p. 325–34. [DOI] [PubMed] [Google Scholar]
- 68.Tibbetts RS, et al. , A role for ATR in the DNA damage-induced phosphorylation of p53. Genes Dev, 1999. 13(2): p. 152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feng L, Hollstein M, and Xu Y, Ser46 phosphorylation regulates p53-dependent apoptosis and replicative senescence. Cell Cycle, 2006. 5(23): p. 2812–9. [DOI] [PubMed] [Google Scholar]
- 70.Sakaguchi K, et al. , DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev, 1998. 12(18): p. 2831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Itahana Y, Ke H, and Zhang Y, p53 Oligomerization is essential for its C-terminal lysine acetylation. J Biol Chem, 2009. 284(8): p. 5158–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tang Y, et al. , Acetylation is indispensable for p53 activation. Cell, 2008. 133(4): p. 612–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tang Y, et al. , Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol Cell, 2006. 24(6): p. 827–39. [DOI] [PubMed] [Google Scholar]
- 74.Sykes SM, et al. , Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol Cell, 2006. 24(6): p. 841–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ito A, et al. , p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J, 2001. 20(6): p. 1331–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li M, et al. , Acetylation of p53 inhibits its ubiquitination by Mdm2. J Biol Chem, 2002. 277(52): p. 50607–11. [DOI] [PubMed] [Google Scholar]
- 77.Li M, et al. , Mono- versus polyubiquitination: differential control of p53 fate by Mdm2. Science, 2003. 302(5652): p. 1972–5. [DOI] [PubMed] [Google Scholar]
- 78.Sheng Y, et al. , Molecular basis of Pirh2-mediated p53 ubiquitylation. Nat Struct Mol Biol, 2008. 15(12): p. 1334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Leng RP, et al. , Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell, 2003. 112(6): p. 779–91. [DOI] [PubMed] [Google Scholar]
- 80.Che Z, et al. , Role of post-translational modifications in regulation of tumor suppressor p53 function. Frontiers of Oral and Maxillofacial Medicine, 2020. 2. [Google Scholar]
- 81.Rodriguez MS, et al. , SUMO-1 modification activates the transcriptional response of p53. EMBO J, 1999. 18(22): p. 6455–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen L. and Chen J, MDM2-ARF complex regulates p53 sumoylation. Oncogene, 2003. 22(34): p. 5348–57. [DOI] [PubMed] [Google Scholar]
- 83.Li T, et al. , Expression of SUMO-2/3 induced senescence through p53- and pRB-mediated pathways. J Biol Chem, 2006. 281(47): p. 36221–7. [DOI] [PubMed] [Google Scholar]
- 84.Wu SY and Chiang CM, Crosstalk between sumoylation and acetylation regulates p53-dependent chromatin transcription and DNA binding. EMBO J, 2009. 28(9): p. 1246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xirodimas DP, et al. , Mdm2-mediated NEDD8 conjugation of p53 inhibits its transcriptional activity. Cell, 2004. 118(1): p. 83–97. [DOI] [PubMed] [Google Scholar]
- 86.Liu K, et al. , p53 beta-hydroxybutyrylation attenuates p53 activity. Cell Death Dis, 2019. 10(3): p. 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huang J, et al. , Repression of p53 activity by Smyd2-mediated methylation. Nature, 2006. 444(7119): p. 629–32. [DOI] [PubMed] [Google Scholar]
- 88.Huang J, et al. , p53 is regulated by the lysine demethylase LSD1. Nature, 2007. 449(7158): p. 105–8. [DOI] [PubMed] [Google Scholar]
- 89.Pietsch EC, et al. , Oligomerization of BAK by p53 utilizes conserved residues of the p53 DNA binding domain. J Biol Chem, 2008. 283(30): p. 21294–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tomita Y, et al. , WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem, 2006. 281(13): p. 8600–6. [DOI] [PubMed] [Google Scholar]
- 91.Mihara M ES, Zaika A, Petrenko O, Chittenden T, Pancoska P, Moll UM, p53 has a direct apoptogenic role at the mitochondria. Molecular cell 2003. 11(3): p. 577–90. [DOI] [PubMed] [Google Scholar]
- 92.el-Deiry WS, et al. , WAF1, a potential mediator of p53 tumor suppression. Cell, 1993. 75(4): p. 817–25. [DOI] [PubMed] [Google Scholar]
- 93.Harper JW, et al. , The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell, 1993. 75(4): p. 805–16. [DOI] [PubMed] [Google Scholar]
- 94.Harper JW, et al. , Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell, 1995. 6(4): p. 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Stewart ZA, Leach SD, and Pietenpol JA, p21(Waf1/Cip1) inhibition of cyclin E/Cdk2 activity prevents endoreduplication after mitotic spindle disruption. Mol Cell Biol, 1999. 19(1): p. 205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nakanishi M, et al. , Direct interaction of p21 cyclin-dependent kinase inhibitor with the retinoblastoma tumor suppressor protein. Biochem Biophys Res Commun, 1999. 263(1): p. 35–40. [DOI] [PubMed] [Google Scholar]
- 97.Dash BC and El-Deiry WS, Phosphorylation of p21 in G2/M promotes cyclin B-Cdc2 kinase activity. Mol Cell Biol, 2005. 25(8): p. 3364–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sherr CJ and Roberts JM, Living with or without cyclins and cyclin-dependent kinases. Genes Dev, 2004. 18(22): p. 2699–711. [DOI] [PubMed] [Google Scholar]
- 99.Sherr CJ and Roberts JM, CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev, 1999. 13(12): p. 1501–12. [DOI] [PubMed] [Google Scholar]
- 100.Hermeking H, et al. , 14–3-3sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell, 1997. 1(1): p. 3–11. [DOI] [PubMed] [Google Scholar]
- 101.Xu J. and Morris GF, p53-mediated regulation of proliferating cell nuclear antigen expression in cells exposed to ionizing radiation. Mol Cell Biol, 1999. 19(1): p. 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kastan MB, et al. , A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell, 1992. 71(4): p. 587–97. [DOI] [PubMed] [Google Scholar]
- 103.Wang XW, et al. , GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci U S A, 1999. 96(7): p. 3706–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jin S, et al. , GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene, 2002. 21(57): p. 8696–704. [DOI] [PubMed] [Google Scholar]
- 105.Fischer M, et al. , Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F target gene analyses identifies cell cycle gene regulatory networks. Nucleic Acids Res, 2016. 44(13): p. 6070–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Engeland K, Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ, 2018. 25(1): p. 114–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sheikh MS and Fornace AJ Jr., Death and decoy receptors and p53-mediated apoptosis. Leukemia, 2000. 14(8): p. 1509–13. [DOI] [PubMed] [Google Scholar]
- 108.Michalak EM, et al. , In several cell types tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ, 2008. 15(6): p. 1019–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sax JK, et al. , BID regulation by p53 contributes to chemosensitivity. Nat Cell Biol, 2002. 4(11): p. 842–9. [DOI] [PubMed] [Google Scholar]
- 110.Miyashita T, et al. , Tumor suppressor p53 is a regulator of bcl-2 and bax gene expression in vitro and in vivo. Oncogene, 1994. 9(6): p. 1799–805. [PubMed] [Google Scholar]
- 111.Chipuk JE, et al. , Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science, 2004. 303(5660): p. 1010–4. [DOI] [PubMed] [Google Scholar]
- 112.Liao JM, et al. , New insights into p53 functions through its target microRNAs. J Mol Cell Biol, 2014. 6(3): p. 206–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hermeking H, MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer, 2012. 12(9): p. 613–26. [DOI] [PubMed] [Google Scholar]
- 114.Cortez MA, et al. , PDL1 Regulation by p53 via miR-34. J Natl Cancer Inst, 2016. 108(1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bommer GT, et al. , p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol, 2007. 17(15): p. 1298–307. [DOI] [PubMed] [Google Scholar]
- 116.Sachdeva M, et al. , p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci U S A, 2009. 106(9): p. 3207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yamakuchi M, et al. , P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A, 2010. 107(14): p. 6334–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bohlig L, Friedrich M, and Engeland K, p53 activates the PANK1/miRNA-107 gene leading to downregulation of CDK6 and p130 cell cycle proteins. Nucleic Acids Res, 2011. 39(2): p. 440–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Georges SA, et al. , Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res, 2008. 68(24): p. 10105–12. [DOI] [PubMed] [Google Scholar]
- 120.Piovan C, et al. , Oncosuppressive role of p53-induced miR-205 in triple negative breast cancer. Mol Oncol, 2012. 6(4): p. 458–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim NH, et al. , A p53/miRNA-34 axis regulates Snail1-dependent cancer cell epithelial-mesenchymal transition. J Cell Biol, 2011. 195(3): p. 417–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kim T, et al. , p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med, 2011. 208(5): p. 875–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mandke P, et al. , MicroRNA-34a modulates MDM4 expression via a target site in the open reading frame. PLoS One, 2012. 7(8): p. e42034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J, et al. , Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene, 2013. 32(1): p. 61–9. [DOI] [PubMed] [Google Scholar]
- 125.Pichiorri F, et al. , Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Cancer Cell, 2010. 18(4): p. 367–81. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 126.Jost CA, Marin MC, and Kaelin WG Jr., p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature, 1997. 389(6647): p. 191–4. [DOI] [PubMed] [Google Scholar]
- 127.Yang A, et al. , p63, a p53 homolog at 3q27–29, encodes multiple products with transactivating, death-inducing, and dominant-negative activities. Mol Cell, 1998. 2(3): p. 305–16. [DOI] [PubMed] [Google Scholar]
- 128.Melino G, et al. , p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J Biol Chem, 2004. 279(9): p. 8076–83. [DOI] [PubMed] [Google Scholar]
- 129.Gu J, et al. , Identification of a sequence element from p53 that signals for Mdm2-targeted degradation. Mol Cell Biol, 2000. 20(4): p. 1243–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yang A, et al. , p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature, 2000. 404(6773): p. 99–103. [DOI] [PubMed] [Google Scholar]
- 131.Yang A, et al. , p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature, 1999. 398(6729): p. 714–8. [DOI] [PubMed] [Google Scholar]
- 132.Graziano V. and De Laurenzi V, Role of p63 in cancer development. Biochim Biophys Acta, 2011. 1816(1): p. 57–66. [DOI] [PubMed] [Google Scholar]
- 133.Deyoung MP and Ellisen LW, p63 and p73 in human cancer: defining the network. Oncogene, 2007. 26(36): p. 5169–83. [DOI] [PubMed] [Google Scholar]
- 134.Ferraiuolo M, et al. , Oncogenic Intra-p53 Family Member Interactions in Human Cancers. Front Oncol, 2016. 6: p. 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Moll UM and Slade N, p63 and p73: roles in development and tumor formation. Mol Cancer Res, 2004. 2(7): p. 371–86. [PubMed] [Google Scholar]
- 136.Thanos CD and Bowie JU, p53 Family members p63 and p73 are SAM domain-containing proteins. Protein Sci, 1999. 8(8): p. 1708–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Davison TS, et al. , p73 and p63 are homotetramers capable of weak heterotypic interactions with each other but not with p53. J Biol Chem, 1999. 274(26): p. 18709–14. [DOI] [PubMed] [Google Scholar]
- 138.Wolf ER, et al. , Mutant and wild-type p53 form complexes with p73 upon phosphorylation by the kinase JNK.Sci Signal, 2018. 11(524). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Flores ER, et al. , p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature, 2002. 416(6880): p. 560–4. [DOI] [PubMed] [Google Scholar]
- 140.Fontemaggi G, et al. , Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J Biol Chem, 2002. 277(45): p. 43359–68. [DOI] [PubMed] [Google Scholar]
- 141.Al-Bahlani S, et al. , P73 regulates cisplatin-induced apoptosis in ovarian cancer cells via a calcium/calpain-dependent mechanism. Oncogene, 2011. 30(41): p. 4219–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bernassola F, et al. , The promyelocytic leukaemia protein tumour suppressor functions as a transcriptional regulator of p63. Oncogene, 2005. 24(46): p. 6982–6. [DOI] [PubMed] [Google Scholar]
- 143.Helton ES, Zhu J, and Chen X, The unique NH2-terminally deleted (DeltaN) residues, the PXXP motif, and the PPXY motif are required for the transcriptional activity of the DeltaN variant of p63. J Biol Chem, 2006. 281(5): p. 2533–42. [DOI] [PubMed] [Google Scholar]
- 144.Lin YL, et al. , p63 and p73 transcriptionally regulate genes involved in DNA repair. PLoS Genet, 2009. 5(10): p. e1000680. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 145.Suh EK, et al. , p63 protects the female germ line during meiotic arrest. Nature, 2006. 444(7119): p. 624–8. [DOI] [PubMed] [Google Scholar]
- 146.Di Como CJ, et al. , p63 expression profiles in human normal and tumor tissues. Clin Cancer Res, 2002. 8(2): p. 494–501. [PubMed] [Google Scholar]
- 147.Stindt MH, et al. , Functional interplay between MDM2, p63/p73 and mutant p53. Oncogene, 2015. 34(33): p. 4300–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Bouaoun L SD, Ardin M, Hollstein M, Byrnes G, Zavadil J, Olivier M, TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data., in Hum Mutat. 2016. p. 865–76. [DOI] [PubMed] [Google Scholar]
- 149.Kazanets A, et al. , Epigenetic silencing of tumor suppressor genes: Paradigms, puzzles, and potential. Biochim Biophys Acta, 2016. 1865(2): p. 275–88. [DOI] [PubMed] [Google Scholar]
- 150.Willis A, et al. , Mutant p53 exerts a dominant negative effect by preventing wild-type p53 from binding to the promoter of its target genes. Oncogene, 2004. 23(13): p. 2330–8. [DOI] [PubMed] [Google Scholar]
- 151.Robles AI and Harris CC, Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol, 2010. 2(3): p. a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Malkin D, Li-fraumeni syndrome. Genes Cancer, 2011. 2(4): p. 475–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hanel W, et al. , Two hot spot mutant p53 mouse models display differential gain of function in tumorigenesis. Cell Death Differ, 2013. 20(7): p. 898–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Hainaut P. and Pfeifer GP, Somatic TP53 Mutations in the Era of Genome Sequencing. Cold Spring Harb Perspect Med, 2016. 6(11). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Muller PA and Vousden KH, Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell, 2014. 25(3): p. 304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Garufi A, et al. , A fluorescent curcumin-based Zn(II)-complex reactivates mutant (R175H and R273H) p53 in cancer cells. J Exp Clin Cancer Res, 2013. 32: p. 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Song H, Hollstein M, and Xu Y, p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat Cell Biol, 2007. 9(5): p. 573–80. [DOI] [PubMed] [Google Scholar]
- 158.Walerych D, Lisek K, and Del Sal G, Mutant p53: One, No One, and One Hundred Thousand. Front Oncol, 2015. 5: p. 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Freed-Pastor WA and Prives C, Mutant p53: one name, many proteins. Genes Dev, 2012. 26(12): p. 1268–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Yoshikawa K, et al. , Mutant p53 R248Q but not R248W enhances in vitro invasiveness of human lung cancer NCI-H1299 cells. Biomed Res, 2010. 31(6): p. 401–11. [DOI] [PubMed] [Google Scholar]
- 161.Di Agostino S, et al. , Gain of function of mutant p53: the mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell, 2006. 10(3): p. 191–202. [DOI] [PubMed] [Google Scholar]
- 162.Frazier MW, et al. , Activation of c-myc gene expression by tumor-derived p53 mutants requires a discrete C-terminal domain. Mol Cell Biol, 1998. 18(7): p. 3735–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lanyi A, et al. , ‘Gain of function’ phenotype of tumor-derived mutant p53 requires the oligomerization/nonsequence-specific nucleic acid-binding domain. Oncogene, 1998. 16(24): p. 3169–76. [DOI] [PubMed] [Google Scholar]
- 164.Donzelli S, et al. , MicroRNA-128–2 targets the transcriptional repressor E2F5 enhancing mutant p53 gain of function. Cell Death Differ, 2012. 19(6): p. 1038–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Sampath J, et al. , Mutant p53 cooperates with ETS and selectively up-regulates human MDR1 not MRP1. J Biol Chem, 2001. 276(42): p. 39359–67. [DOI] [PubMed] [Google Scholar]
- 166.Chin KV, et al. , Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science, 1992. 255(5043): p. 459–62. [DOI] [PubMed] [Google Scholar]
- 167.Thottassery JV, et al. , p53-dependent regulation of MDR1 gene expression causes selective resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A, 1997. 94(20): p. 11037–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Weisz L, et al. , Transactivation of the EGR1 gene contributes to mutant p53 gain of function. Cancer Res, 2004. 64(22): p. 8318–27. [DOI] [PubMed] [Google Scholar]
- 169.Scian MJ, et al. , Tumor-derived p53 mutants induce NF-kappaB2 gene expression. Mol Cell Biol, 2005. 25(22): p. 10097–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Yeudall WA, et al. , Gain-of-function mutant p53 upregulates CXC chemokines and enhances cell migration. Carcinogenesis, 2012. 33(2): p. 442–51. [DOI] [PubMed] [Google Scholar]
- 171.Khromova NV, et al. , p53 hot-spot mutants increase tumor vascularization via ROS-mediated activation of the HIF1/VEGF-A pathway. Cancer Lett, 2009. 276(2): p. 143–51. [DOI] [PubMed] [Google Scholar]
- 172.Grespi F, et al. , Differential regulated microRNA by wild type and mutant p53 in induced pluripotent stem cells. Cell Death Dis, 2016. 7(12): p. e2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jones M. and Lal A, MicroRNAs, wild-type and mutant p53: more questions than answers. RNA Biol, 2012. 9(6): p. 781–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Polotskaia A, et al. , Proteome-wide analysis of mutant p53 targets in breast cancer identifies new levels of gain-of-function that influence PARP, PCNA, and MCM4. Proc Natl Acad Sci U S A, 2015. 112(11): p. E1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Pavlakis E. and Stiewe T, p53’s Extended Reach: The Mutant p53 Secretome. Biomolecules, 2020. 10(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Matsumoto M, et al. , Prognostic significance of serine 392 phosphorylation in overexpressed p53 protein in human esophageal squamous cell carcinoma. Oncology, 2004. 67(2): p. 143–50. [DOI] [PubMed] [Google Scholar]
- 177.Yap DB, et al. , Ser392 phosphorylation regulates the oncogenic function of mutant p53. Cancer Res, 2004. 64(14): p. 4749–54. [DOI] [PubMed] [Google Scholar]
- 178.Zerbini LF, et al. , Blockage of NF-kappaB induces serine 15 phosphorylation of mutant p53 by JNK kinase in prostate cancer cells. Cell Cycle, 2005. 4(9): p. 1247–53. [DOI] [PubMed] [Google Scholar]
- 179.Minamoto T, et al. , Distinct pattern of p53 phosphorylation in human tumors. Oncogene, 2001. 20(26): p. 3341–7. [DOI] [PubMed] [Google Scholar]
- 180.Rodriguez OC, et al. , Dietary downregulation of mutant p53 levels via glucose restriction: mechanisms and implications for tumor therapy. Cell Cycle, 2012. 11(23): p. 4436–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Lukashchuk N. and Vousden KH, Ubiquitination and degradation of mutant p53. Mol Cell Biol, 2007. 27(23): p. 8284–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Haupt Y, et al. , Mdm2 promotes the rapid degradation of p53. Nature, 1997. 387(6630): p. 296–9. [DOI] [PubMed] [Google Scholar]
- 183.Peng Y, et al. , Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J Biol Chem, 2001. 276(44): p. 40583–90. [DOI] [PubMed] [Google Scholar]
- 184.Jolly C. and Morimoto RI, Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst, 2000. 92(19): p. 1564–72. [DOI] [PubMed] [Google Scholar]
- 185.Vakifahmetoglu-Norberg H, et al. , Chaperone-mediated autophagy degrades mutant p53. Genes Dev, 2013. 27(15): p. 1718–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Maan M. and Pati U, CHIP promotes autophagy-mediated degradation of aggregating mutant p53 in hypoxic conditions. FEBS J, 2018. 285(17): p. 3197–3214. [DOI] [PubMed] [Google Scholar]
- 187.Martinez LA, et al. , Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc Natl Acad Sci U S A, 2002. 99(23): p. 14849–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ubby I, et al. , Cancer therapeutic targeting using mutant-p53-specific siRNAs. Oncogene, 2019. 38(18): p. 3415–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ma G, et al. , Gene medicine for cancer treatment: commercially available medicine and accumulated clinical data in China. Drug Des Devel Ther, 2009. 2: p. 115–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Chen GX, et al. , Clinical utility of recombinant adenoviral human p53 gene therapy: current perspectives. Onco Targets Ther, 2014. 7: p. 1901–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Garber K, China approves world’s first oncolytic virus therapy for cancer treatment. J Natl Cancer Inst, 2006. 98(5): p. 298–300. [DOI] [PubMed] [Google Scholar]
- 192.Bischoff JR, et al. , An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science, 1996. 274(5286): p. 373–6. [DOI] [PubMed] [Google Scholar]
- 193.Goodrum FD and Ornelles DA, p53 status does not determine outcome of E1B 55kilodalton mutant adenovirus lytic infection. J Virol, 1998. 72(12): p. 9479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Reed D, et al. , Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem, 2010. 285(14): p. 10786–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Vassilev LT, et al. , In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science, 2004. 303(5659): p. 844–8. [DOI] [PubMed] [Google Scholar]
- 196.Tovar C, et al. , MDM2 small-molecule antagonist RG7112 activates p53 signaling and regresses human tumors in preclinical cancer models. Cancer Res, 2013. 73(8): p. 258797. [DOI] [PubMed] [Google Scholar]
- 197.Olivier M, Hollstein M, and Hainaut P, TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol, 2010. 2(1): p. a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ray-Coquard I, et al. , Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol, 2012. 13(11): p. 1133–40. [DOI] [PubMed] [Google Scholar]
- 199.Ding Q, et al. , Discovery of RG7388, a potent and selective p53-MDM2 inhibitor in clinical development. J Med Chem, 2013. 56(14): p. 5979–83. [DOI] [PubMed] [Google Scholar]
- 200.Montesinos P, et al. , MIRROS: a randomized, placebo-controlled, Phase III trial of cytarabine +/− idasanutlin in relapsed or refractory acute myeloid leukemia. Future Oncol, 2020. 16(13): p. 807–815. [DOI] [PubMed] [Google Scholar]
- 201.Yee K MG, Vey N, et al. , Phase 1/1b study of RG7388, a potent MDM2 antagonist, in acute myelogenous leukemia (AML) patients. . Blood, 2014. 124(21): p. 116. [Google Scholar]
- 202.Carvajal LA, et al. , Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci Transl Med, 2018. 10(436). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Sahin I, et al. , AMG-232 sensitizes high MDM2-expressing tumor cells to T-cellmediated killing. Cell Death Discov, 2020. 6: p. 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Wieczorek AM, et al. , Structure-based rescue of common tumor-derived p53 mutants. Nat Med, 1996. 2(10): p. 1143–6. [DOI] [PubMed] [Google Scholar]
- 205.Foster BA, et al. , Pharmacological rescue of mutant p53 conformation and function. Science, 1999. 286(5449): p. 2507–10. [DOI] [PubMed] [Google Scholar]
- 206.Zhang Q, et al. , APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis, 2018. 9(5): p. 439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Bykov VJ, et al. , Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J Biol Chem, 2005. 280(34): p. 30384–91. [DOI] [PubMed] [Google Scholar]
- 208.Bykov VJ, et al. , Restoration of the tumor suppressor function to mutant p53 by a lowmolecular-weight compound. Nat Med, 2002. 8(3): p. 282–8. [DOI] [PubMed] [Google Scholar]
- 209.Takimoto R, et al. , The mutant p53-conformation modifying drug, CP-31398, can induce apoptosis of human cancer cells and can stabilize wild-type p53 protein. Cancer Biol Ther, 2002. 1(1): p. 47–55. [DOI] [PubMed] [Google Scholar]
- 210.Chen S, et al. , Arsenic Trioxide Rescues Structural p53 Mutations through a Cryptic Allosteric Site.Cancer Cell, 2020. [DOI] [PubMed] [Google Scholar]
- 211.Sallman DA, To target the untargetable: elucidation of synergy of APR-246 and azacitidine in TP53 mutant myelodysplastic syndromes and acute myeloid leukemia. Haematologica, 2020. 105(6): p. 1470–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Paulmurugan R, et al. , A protein folding molecular imaging biosensor monitors the effects of drugs that restore mutant p53 structure and its downstream function in glioblastoma cells. Oncotarget, 2018. 9(30): p. 21495–21511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Boeckler FM, et al. , Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci U S A, 2008. 105(30): p. 10360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Liu X, et al. , Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res, 2013. 41(12): p. 6034–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Butler JS and Loh SN, Structure, function, and aggregation of the zinc-free form of the p53 DNA binding domain. Biochemistry, 2003. 42(8): p. 2396–403. [DOI] [PubMed] [Google Scholar]
- 216.Yu X, et al. , Allele-specific p53 mutant reactivation. Cancer Cell, 2012. 21(5): p. 61425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Blanden AR, et al. , Synthetic metallochaperone ZMC1 rescues mutant p53 conformation by transporting zinc into cells as an ionophore. Mol Pharmacol, 2015. 87(5): p. 825–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lindemann A, et al. , COTI-2, A Novel Thiosemicarbazone Derivative, Exhibits Antitumor Activity in HNSCC through p53-dependent and -independent Mechanisms. Clin Cancer Res, 2019. 25(18): p. 5650–5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yu X, et al. , Thiosemicarbazones Functioning as Zinc Metallochaperones to Reactivate Mutant p53. Mol Pharmacol, 2017. 91(6): p. 567–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Salim KY, et al. , COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget, 2016. 7(27): p. 41363–41379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Li D, Marchenko ND, and Moll UM, SAHA shows preferential cytotoxicity in mutant p53 cancer cells by destabilizing mutant p53 through inhibition of the HDAC6-Hsp90 chaperone axis. Cell Death Differ, 2011. 18(12): p. 1904–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Li D, et al. , Functional inactivation of endogenous MDM2 and CHIP by HSP90 causes aberrant stabilization of mutant p53 in human cancer cells. Mol Cancer Res, 2011. 9(5): p. 577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Wang C. and Chen J, Phosphorylation and hsp90 binding mediate heat shock stabilization of p53. J Biol Chem, 2003. 278(3): p. 2066–71. [DOI] [PubMed] [Google Scholar]
- 224.Whitesell L, et al. ,Geldanamycin-stimulated destabilization of mutated p53 is mediated by the proteasome in vivo. Oncogene, 1997. 14(23): p. 2809–16. [DOI] [PubMed] [Google Scholar]
- 225.Alexandrova EM, et al. , Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature, 2015. 523(7560): p. 352–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Wagner AJ, et al. , A phase I study of the HSP90 inhibitor retaspimycin hydrochloride (IPI-504) in patients with gastrointestinal stromal tumors or soft-tissue sarcomas. Clin Cancer Res, 2013. 19(21): p. 6020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Grant C, et al. , Romidepsin: a new therapy for cutaneous T-cell lymphoma and a potential therapy for solid tumors. Expert Rev Anticancer Ther, 2010. 10(7): p. 9971008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Mann BS, et al. , FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist, 2007. 12(10): p. 1247–52. [DOI] [PubMed] [Google Scholar]
- 229.Yi YW, et al. , Targeting mutant p53 by a SIRT1 activator YK-3–237 inhibits the proliferation of triple-negative breast cancer cells. Oncotarget, 2013. 4(7): p. 984–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Patterson J, et al. , IPI-504, a novel and soluble HSP-90 inhibitor, blocks the unfolded protein response in multiple myeloma cells. Cancer Chemother Pharmacol, 2008. 61(6): p. 923–32. [DOI] [PubMed] [Google Scholar]
- 231.Shatz M, Menendez D, and Resnick MA, The human TLR innate immune gene family is differentially influenced by DNA stress and p53 status in cancer cells. Cancer Res, 2012. 72(16): p. 3948–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Hsiue EH, et al. , Targeting a neoantigen derived from a common TP53 mutation. Science, 2021. 371(6533). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Kravchenko JE, et al. , Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc Natl Acad Sci U S A, 2008. 105(17): p. 6302–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Hong B, et al. , Prodigiosin rescues deficient p53 signaling and antitumor effects via upregulating p73 and disrupting its interaction with mutant p53. Cancer Res, 2014. 74(4): p. 1153–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Prabhu VV, et al. , Small-Molecule Prodigiosin Restores p53 Tumor Suppressor Activity in Chemoresistant Colorectal Cancer Stem Cells via c-Jun-Mediated DeltaNp73 Inhibition and p73 Activation. Cancer Res, 2016. 76(7): p. 1989–99. [DOI] [PubMed] [Google Scholar]
- 236.Zhang S, et al. , Small-Molecule NSC59984 Restores p53 Pathway Signaling and Antitumor Effects against Colorectal Cancer via p73 Activation and Degradation of Mutant p53. Cancer Res, 2015. 75(18): p. 3842–52. [DOI] [PMC free article] [PubMed] [Google Scholar]